Introduction
Hepatocellular carcinoma (HCC) is a digestive tumor
with a high morbidity and mortality worldwide. HCC is the commonest
cancer of the liver and the sixth commonest cancer in worldwide
(1). The overall survival of HCC
patients is poor due to its unresponsiveness to early diagnosis and
its drug resistance (2). Over the
past few decades, efforts have been made to characterize the
molecular and genetic mechanisms of HCC (3). However, further research to uncover
new therapeutic targets for HCC is urgently needed.
Accumulated evidence has demonstrated that a type of
non-coding RNA, circular RNA (circRNA), is commonly expressed in
human tissues and participates in multiple biological processes
(4). Abnormal expression of
circRNAs is associated with cancer pathogenesis, including HCC
development and progression (5,6).
circRNAs have been identified to function as miRNA sponges
(7–9) and circRNAs might indirectly modulate
target gene expression by sponging miRNAs at the
post-transcriptional level. The present study performed circRNA
sequencing to reveal the dysregulated circRNAs in HCC tissues
compared with nontumorous samples. circ-CCT3 originates from
chr1:156303337-156304709 in the host gene CCT3. The spliced
length of circ-CCT3 is 211 nt. circ-CCT3 has not been studied
previously, to the best of the authors' knowledge. The present
study identified that upregulation of circ-CCT3 facilitated HCC
cell progression by sponging miR-1287-5p to regulate TEA domain
transcription factor 1 (TEAD1) expression. TEAD1 transcriptional
activity is widely believed to be modulated by the presence or
absence of nuclear Yes-associated protein (YAP)/transcriptional
activator with PDZ-binding domain (TAZ) (10). The present study found that TEAD1
could directly activate patched 1 (PTCH1) and lysyl oxidase (LOX)
transcription. In brief a novel circRNA, circ-CCT3 was identified,
which may be a potential therapeutic target for HCC.
Materials and methods
Study participants
A total of 68 HCC tissues and neighboring
nontumorous specimens (≥2 cm from the edge of the tumor) were
collected from HCC patients who underwent partial hepatectomy at
the Second Affiliated Hospital of Qiqihar Medical University
between January 2013 and June 2015 and written informed consent was
obtained from all patients. There were 57 males and 11 females in
the cohort. The average age was 59.2 years (range, 35–78 years).
All patients were followed up after surgery until mortality or
survival of >5 years. All tissues were snap-frozen and then
transferred into a −80°C freezer. The present study was authorized
by the Institutional Review Board of the Second Affiliated Hospital
of Qiqihar Medical University.
circRNA sequencing
Total RNA was extracted from four pairs of HCC
tissue samples with TRIzol® (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. RNA quantity and
quality were then determined using an ND-1000 spectrophotometer
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.). RNA
integrity number (RIN) analysis was performed using an Agilent 2100
Bioanalyser and RNA 6000 LabChip kit with Agilent 2100 Expert
software (Agilent Technologies). The isolated RNA from tissue
samples with RIN ≥7 was considered usable in the study. A cDNA
library was established using an RNA Sample Prep kit and
circRNA-seq was conducted on an Illumina HiSeq 2500 platform
(Illumina, Inc.). The original sequencing data were preprocessed
through cutadapt v3.2 (cutadapt.readthedocs.io/en/stable/) and
FastQC v0.11.9 (bioinformatics.babraham.ac.uk/projects/fastqc/) and
clean readings were subsequently recorded in hg38 and circBase
through TopHat 2.1.1 (ccb.jhu.edu/software/tophat/index.shtml) and MapSplice
2.2.1 (netlab.uky.edu/p/bioinfo/MapSplice2). The circRNAs
with different expression levels were determined through the R
package EdgeR v3.14.0 (R-project.org/) (11)
with a fold alternation >2 and P<0.05.
HCC cells and transfection
Normal human liver cells (Chang) and liver cancer
cells (HepG2, Huh7, HCCLM3 and SK-Hep-1) were purchased from the
Chinese Academy of Sciences. Cells were cultivated at 37°C, 5%
CO2 in a mixture composed of 10% fetal bovine serum
(FBS) and 90% DMEM (HyClone; Cytiva). Cells were harvested at
70–80% confluence to perform the subsequent experiments.
Short hairpin RNA (shRNA) against circ-CCT3
(sh-CCT3-1/-2), TEAD1 (sh-TEAD1), sh negative control (NC),
miR-1287-5p mimics, inhibitor, mimics-NC and inhibitor-NC were
synthesized by Shanghai GenePharma Co., Ltd. TEAD1 vector and its
NC vector were purchased from Shanghai GeneChem Co., Ltd. Transient
transfection was performed using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. The targeted sequences of shRNA-circ-CCT3
were: sh-circ-CCT3-1, 5′-AGTTTTATTAGAGACAAAGCA-3′ and
sh-circ-CCT3-2, 5′-TAATTATTCTTGACTATTGCA-3′.
Reverse transcription-quantitative
(RT-q) PCR and western blotting
TRIzol® (Thermo Fisher Scientific, Inc.)
was used to isolate total RNA from tissue samples and cells
according to the manufacturer's instructions. RNA isolation was
performed at 48 h after transfection (the cells were grown to ~80%
density). cDNA was synthesized by Capital-Bio with Oligo (dT) in
accordance with the manufacturer's protocol. Primers were designed
by Shanghai Sangong Pharmaceutical Co., Ltd. RT-qPCR experiments
were conducted on a real-time system using SYBR Green Master Mix
(Roche GmbH) following the manufacturer's instructions. The
reaction volume was 10 µl. Thermocycling conditions were as
follows: 90°C for 5 min, then 90°C for 15 sec, 60°C for 30 sec for
45 cycles. For circRNA and mRNA quantification, GAPDH was used as
the internal reference. For miRNA quantification, U6 was used as
the internal control. The primers used were as follows: circ-CCT3
forward, 5′-AATTAGCCGGACCCAGGATG-3′ and reverse,
5′-ACAATGCCTCCCATTGGGTC-3′; CCT3 forward,
5′-AAGTCCATGATCGAAATTAGCCG-3′ and reverse,
5′-TGCTCAGCTACAGACAGCATT-3′; TEAD1 forward,
5′-ATGGAAAGGATGAGTGACTCTGC-3′ and reverse,
5′-TCCCACATGGTGGATAGATAGC-3′; PTCH1 forward,
5′-CCAGAAAGTATATGCACTGGCA-3′ and reverse,
5′-GTGCTCGTACATTTGCTTGGG-3′; LOX forward, 5′-CGGCGGAGGAAAACTGTCT-3′
and reverse, 5′-TCGGCTGGGTAAGAAATCTGA-3′; GAPDH forward,
5′-GGGAGCCAAAAGGGTCAT-3′ and reverse, 5′-GAGTCCTTCCACGATACCAA-3′
and U6 forward, 5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′. Each reaction was performed in
triplicate. The 2−ΔΔCq method was employed to analyze
gene expression (12).
Immunoblotting was carried out as per our previous
study (13). In brief, the cells
were lysed with RIPA lysis buffer (Beyotime Institute of
Biotechnology). BCA method was used to detect the concentration of
proteins. Protein (30 µg/lane) was fractionated by SDS-PAGE
vertical electrophoresis (10% gel), followed by transfer onto a
0.45 µm PVDF membrane. The membrane was blocked with 5% skimmed
milk (BD Biosciences) diluted in Tris-buffered saline containing
0.05% Tween-20 for 1 h at room temperature and then probed with
primary antibodies to TEAD1 (1:1,000; cat. no. ab133533; Abcam) and
GAPDH (1:10,000; cat. no. ab181602; Abcam) at 4°C overnight. After
washing and incubating with horseradish peroxidase-conjugated goat
anti-rabbit IgG secondary antibody (1:5,000; cat. no. ab6721;
Abcam) for 2 h at room temperature, BeyoECL Plus kit (Beyotime
Institute of Biotechnology) was used to visualize the blots. ImageJ
1.50i software (National Institutes of Health) was used to analyze
the blots.
Dual-luciferase reporter gene
assay
To explore the interaction between the
circ-CCT3/TEAD1 3′-UTR and miR-1287-5p, the circ-CCT3/TEAD1 3′-UTR
vector was constructed using the pmirGLO Luciferase Reporter Vector
(Promega Corporation) according to the manufacturer's protocol.
Transfections were performed in accordance with the instructions of
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) to co-transfect circ-CCT3/TEAD1 3′-UTR vector
with NC miRNA or miR-1287-5p mimics into cells. The Dual-Luciferase
Reporter Assay System (Promega Corporation) was used to evaluate
the relative luciferase signal at 36 h post-transfection. The
specific target activity was expressed as the relative activity
ratio of firefly luciferase to Renilla luciferase.
miR-1287-5p mimics sequence was 5′-UGCUGGAUCAGUGGUUCGAGUC-3′.
Mimics-NC sequence was 5′-UUCUCCGAACGUGUCACGU-3′ (Shanghai
GenePharma Co., Ltd.).
Chromatin immunoprecipitation
(ChIP)
DNA and proteins were crosslinked in cells using
formaldehyde (Sinopharm Chemical Reagent Co., Ltd.). Cell lysates
were sonicated to generate chromatin fragments of 200–300 bp (20
kHz; 4 pulses of 12 sec each, followed by 30 sec rest on ice
between each pulse). Centrifugation was performed at 14,000 × g for
10 min at 4°C before protease inhibitor mixture II buffer (EMD
Millipore) at a ratio of 9:1 and 60 µl protein G agarose was added
and then cultured at 4°C for 1 h. After removing the agarose by
centrifugation at 4,000 × g for 1 min at 4°C, antibody against
TEAD1 (1:80; cat. no. ab133533; Abcam) or IgG (1:50; cat. no.
ab6721; Abcam) was added into the supernatant. The precipitated DNA
was purified using the ChIP DNA Clean & Concentrator kit
(A&D Technology) according to the manufacturer's protocol. The
precipitated chromatin DNA was recovered and assessed by qPCR.
Cell counting kit-8 (CCK-8) and
colony-forming assays
CCK-8 was used to measure the viability of treated
cells. Cells were placed in 96-well plates and then 10 µl of CCK-8
solution (Dojindo) was added to each well according to the
manufacturer's instructions. The absorbance was estimated using a
spectrophotometer at 450 nm. Transfected cells were plated in 2.5
cm dishes for 10 days. The visible colonies were then fixed and
stained for observation.
Apoptosis assays
HCCLM3 and Huh7 cells (10,000 cells/well)
transfected accordingly were harvested by trypsin (HyClone; Cytiva)
digestion. A total of 5 µl Annexin V-FITC and 10 µl PI were added
to each sample for 20 min in the dark at room temperature.
Afterwards, cell apoptosis was assessed by flow cytometry (FCM;
FACScan; BD Biosciences). FlowJo v10 software (Tree Star, Inc.) was
used for apoptosis analysis. The percentage of early + late
apoptotic cells was deemed as apoptotic rate. For the acridine
orange/ethidium bromide (AO/EB) double fluorescence staining assay,
transfected cells were cultured in an incubator, followed by
staining with prepared AO/EB mixing solution (1:1) at room
temperature for 5 min (Beijing Solarbio Science & Technology
Co., Ltd.).
Wound healing assay
For wound-healing assay, 2×105
transfected HCCLM3 and Huh7 cells were seeded into a 2.5 cm dish
and cultured with complete medium until 90% confluency. Wounds were
made using a 200 µl sterile pipette tip. The detached cells were
removed by washing twice with PBS and then, the cells were cultured
with serum-free medium for 24 h. A light microscope was used for
visualizing and capturing images (magnification, ×100). ImageJ
version 1.50i software (National Institutes of Health) was used to
analyze cell migration.
Transwell experiments
A Transwell chamber (Corning Inc.) was used for the
migration assay (with 8-µm polycarbonate nucleopore filters). For
the invasion assay, the Transwell compartment was coated with
Matrigel (precooled at 4°C overnight) and placed in an incubator at
37°C for 4 h to form a reconstructed basement membrane. The cells
(~5×104 cells) resuspended in serum-free medium were
seeded into the top compartment of the Transwell chamber, while the
bottom compartment was filled with medium containing 10% FBS. After
incubation at 37°C for 24 h, the cells on the upper side of
membrane were removed. The migrated/invaded cells were fixed with
paraformaldehyde for 20 min at room temperature, stained with 0.5%
crystal violet for 20 min and measured with a light microscope
(Olympus Corporation).
Bioinformatics and statistical
analyses
Target prediction was performed by starBase v2.0
software (starbase.sysu.edu.cn), which is based
on seed region matching of miRNAs (14). The circRNA-miRNA interaction network
was drawn by Circular RNA Interactome software v2020-01-30
(15). The Cancer Genome Atlas
(TCGA) database (cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga)
was analyzed by starBase v2.0 software. Kaplan-Meier estimate was
used to measure the overall survival rate of patients. The
differences between two groups and multiple groups were analyzed by
using unpaired t-tests and one-way analysis of variance with
Tukey's test, respectively. Pearson's correlation coefficient
analysis was performed to analyze the expression correlation
between circ-CCT3, miR-1287-5p and TEAD1. P<0.05 was considered
to indicate a statistically significant difference.
Results
circ-CCT3 is elevated in HCC tissues
and indicates poor prognosis
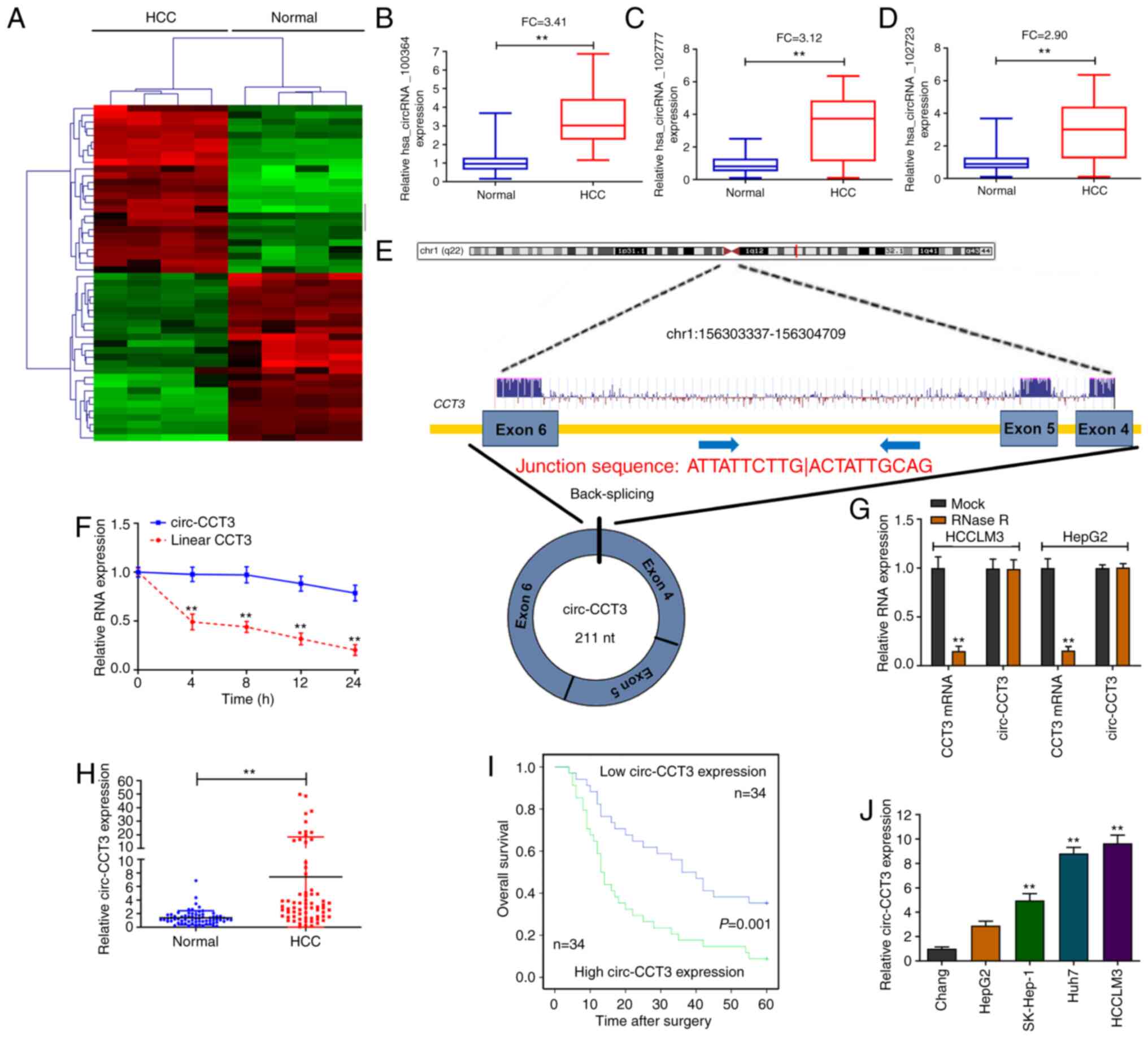
A circRNA microarray was carried out to identify the
dysregulated circRNAs in HCC tissue specimens. The heatmap for the
top 20 up/downregulated circRNAs is shown in Fig. 1A. Then, RT-qPCR was performed to
explore the three most elevated circRNAs (hsa_circRNA_100364,
hsa_circRNA_102777 and hsa_circRNA_102723) in 20 paired
HCC/nontumorous specimens. As Fig.
1B-D shows, all three circRNAs were upregulated in HCC
specimens. Hsa_circRNA_100364 was the most overexpressed circRNA
and was selected for subsequent study. Hsa_circRNA_100364 was
spliced from exons 4–6 of the CCT3 gene. The full sequence length
of hsa_circRNA_100364 (circ-CCT3) was 211 nt (Fig. 1E). Additionally, it was found that
the half-life of circ-CCT3 was longer than that of its linear
isoform (CCT3 mRNA; Fig. 1F).
Moreover, circ-CCT3 was more stable than CCT3 mRNA after treatment
with RNase R (Fig. 1G). As shown in
Fig. 1H, circ-CCT3 expression was
significantly higher in HCC tissue samples than in nontumorous
tissues. The 68 HCC patients were divided into two groups based on
the median cutoff to analyze the clinical significance of circ-CCT3
expression in tissues. It was found that high circ-CCT3 expression
was linked to a worse overall survival rate (P=0.001) for patients
after surgical resection (Fig. 1I).
circ-CCT3 expression was higher in Huh7, HCCLM3 and SK-Hep-1 cells
compared with Chang cells (Fig.
1J), suggesting that circ-CCT3 might promote HCC progression.
Therefore, HCCLM3 and Huh7 cells were considered suitable for use
in knockdown experiments.
circ-CCT3 regulates HCC cell
progression
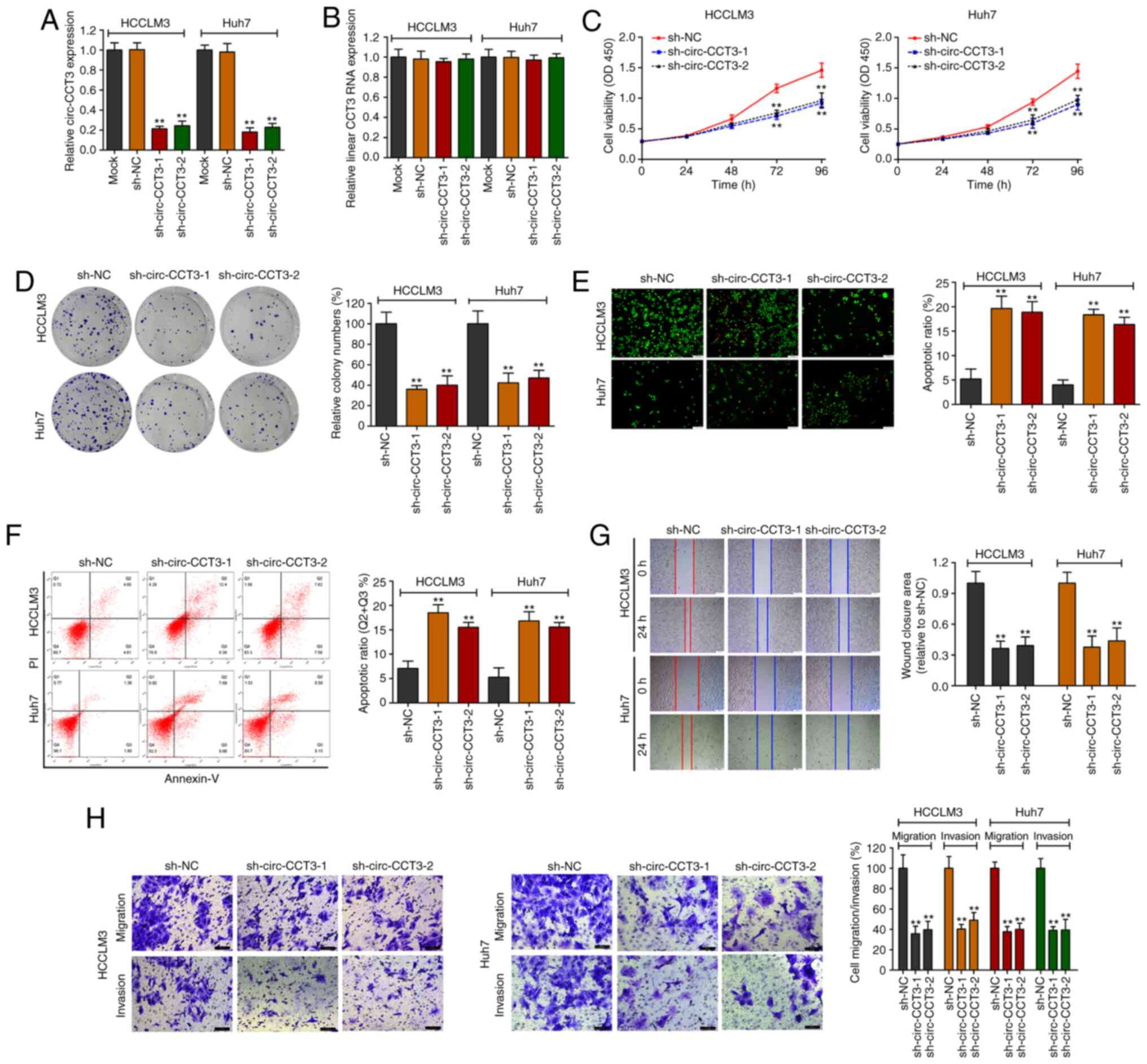
As expected, two shRNAs led to downregulation of
circ-CCT3 expression in HCCLM3 and Huh7 cell lines, as confirmed by
RT-qPCR (Fig. 2A). In contrast,
CCT3 mRNA expression was unaffected following transfection
(Fig. 2B). The functional
experiments were then conducted. Reduced circ-CCT3 markedly
suppressed cell proliferation, as analyzed by CCK-8 and
colony-forming assays (Fig. 2C and
D). Consistently, AO/EB staining and flow cytometry assays
showed that downregulation of circ-CCT3 led to increased apoptosis
in HCCLM3 and Huh7 cells (Fig. 2E and
F). Notably, HCCLM3 and Huh7 cells had decreased migratory
potential following knockdown of circ-CCT3 as demonstrated by
Transwell and scratched wound assays (Fig. 2G and H). The Transwell assay also
identified decreased invasion of cells in the circ-CCT3-KD group
compared with the control group.
circ-CCT3 upregulates TEAD1 expression
via sponging miR-1287-5p in HCC
circ-CCT3 regulates the growth and aggressiveness of
HCC cells, but the underlying mechanism remains to be elucidated.
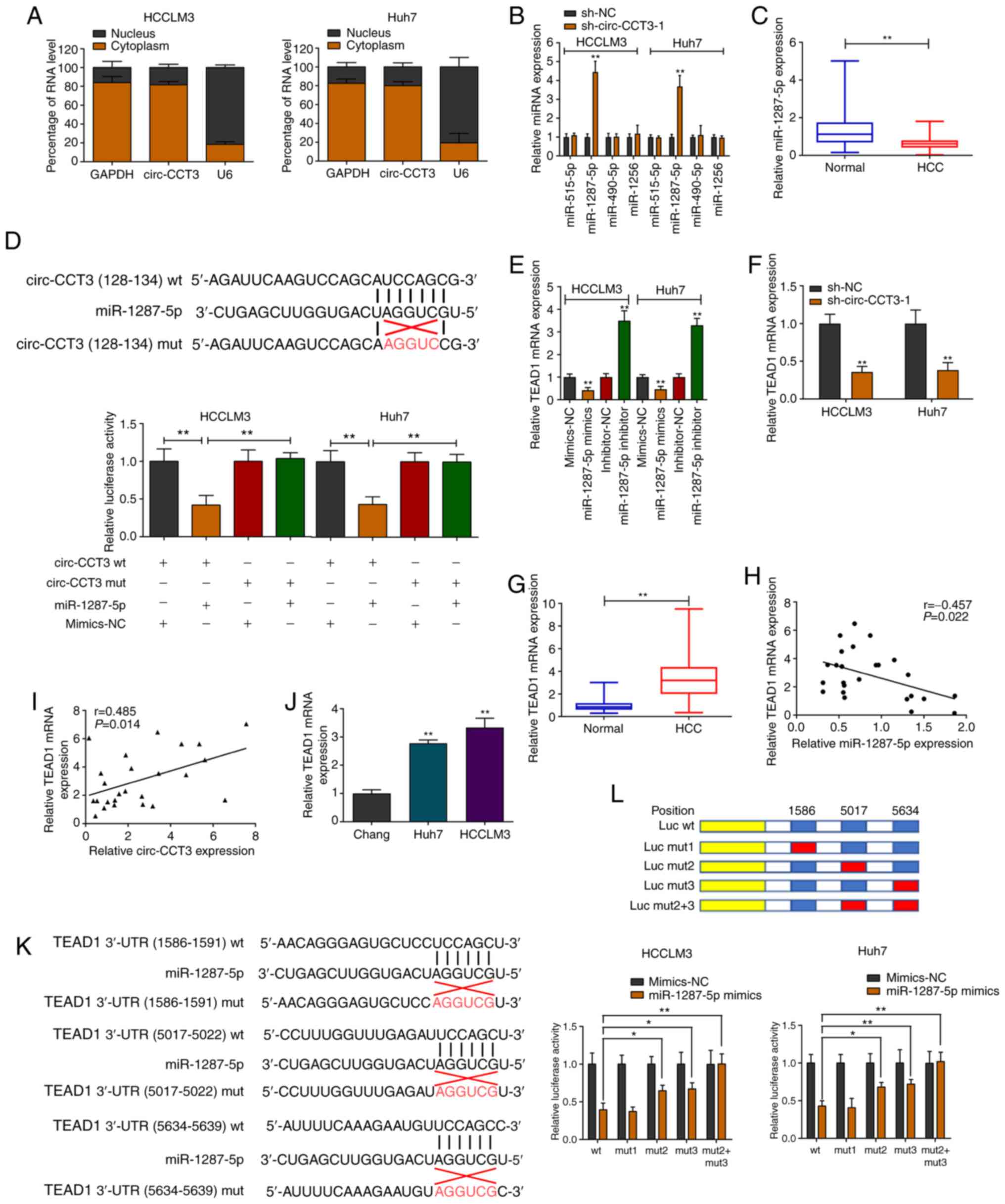
As shown in Fig. 3A, circ-CCT3 was
primarily localized in the cytoplasm of cells. An online database
predicted the miRNAs that may be bound to circ-CCT3. The expression
of predicted miRNAs was evaluated after knockdown of circ-CCT3 in
HCCLM3 and Huh7 cell lines. The data indicated that only
miR-1287-5p expression levels were enhanced in circ-CCT3-depleted
cells (Fig. 3B). Additionally,
miR-1287-5p expression was decreased in HCC tissue samples
(Fig. 3C). Luciferase reporter
vectors were constructed a for wild-type (wt) and mutant (mut)
circ-CCT3. The vectors were cotransfected with miR-1287-5p mimics
or mimics-NC in HCCLM3 and Huh7 cells. After 36 h, miR-1287-5p
mimics notably inhibited luciferase activity compared with the
negative control (Fig. 3D). The
potential target gene of miR-1287-5p was next investigated using
the starBase v2.0 database. TEAD1 expression was negatively
regulated by miR-1287-5p (Figs. 3E
and S1A). Additionally, silencing
circ-CCT3 attenuated TEAD1 expression in both HCC cell lines
(Fig. 3F). TEAD1 mRNA was elevated
in HCC samples relative to nontumorous tissues (Fig. 3G). Pearson's correlation analysis
indicated a negative correlation between miR-1287-5p and TEAD1 mRNA
expression (Fig. 3H). A positive
correlation of circ-CCT3 and TEAD1 mRNA expression was identified
in 25 pairs of HCC tissues (Fig.
3I). Notably, TEAD1 was overexpressed in HCC cells compared
with normal cells (Fig. 3J). There
are three potential binding sites for miR-1287-5p within the 3′-UTR
of TEAD1 (Fig. 3K). To validate the
binding between the TEAD1 3′-UTR and miR-1287-5p, constructs for
wt-TEAD1 3′-UTR (Luc wt) and mut-TEAD1 3′-UTR (Luc mut; Fig. 3K-L) were generated. miR-1287-5p
overexpression reduced luciferase activity in cells with wt-TEAD1
3′-UTR compared to cells with mutated binding sites for miR-1287-5p
(Luc mut2/mut3). Furthermore, simultaneously mutating sites 2 and 3
did not affect luciferase activity. The above results indicated
that miR-1287-5p could interact with the second (5017–5022) and
third (5634–5639) predicted sites of the 3′-UTR of TEAD1 (Fig. 3L).
circ-CCT3 executes oncogenic
properties by targeting the miR-1287-5p/TEAD1 axis
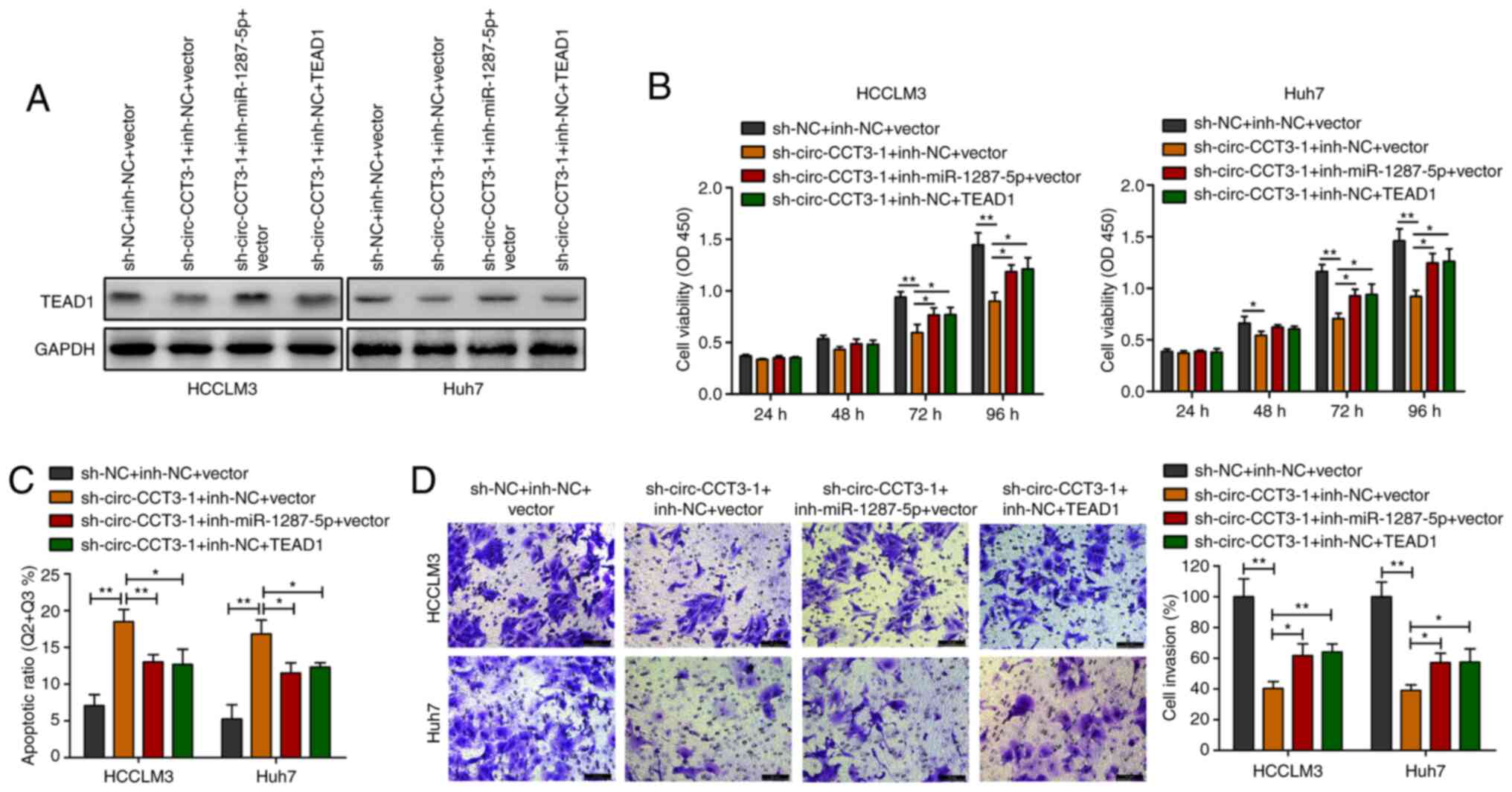
sh-TEAD1 and TEAD1 vectors were used to
silence/overexpress TEAD1 expression in HCC cells (Fig. S1B). HCC cells were co-transfected
with sh-circ-CCT3-1 and miR-1287-5p inhibitor or TEAD1 vector,
followed by western blotting. circ-CCT3 inhibition downregulated
TEAD1 expression, whereas cotransfection with miR-1287-5p inhibitor
or TEAD1 vector increased TEAD1 expression levels (Fig. 4A). CCK-8 and Transwell experiments
showed that decreasing miR-1287-5p or elevating TEAD1 reversed the
inhibition of HCC cell viability and invasion caused by
sh-circ-CCT3-1 (Fig. 4B and D).
Flow cytometric analysis demonstrated that knockdown of circ-CCT3
triggered HCC cell apoptosis. However, this effect was partially
reversed by silencing miR-1287-5p or upregulating TEAD1 (Fig. 4C).
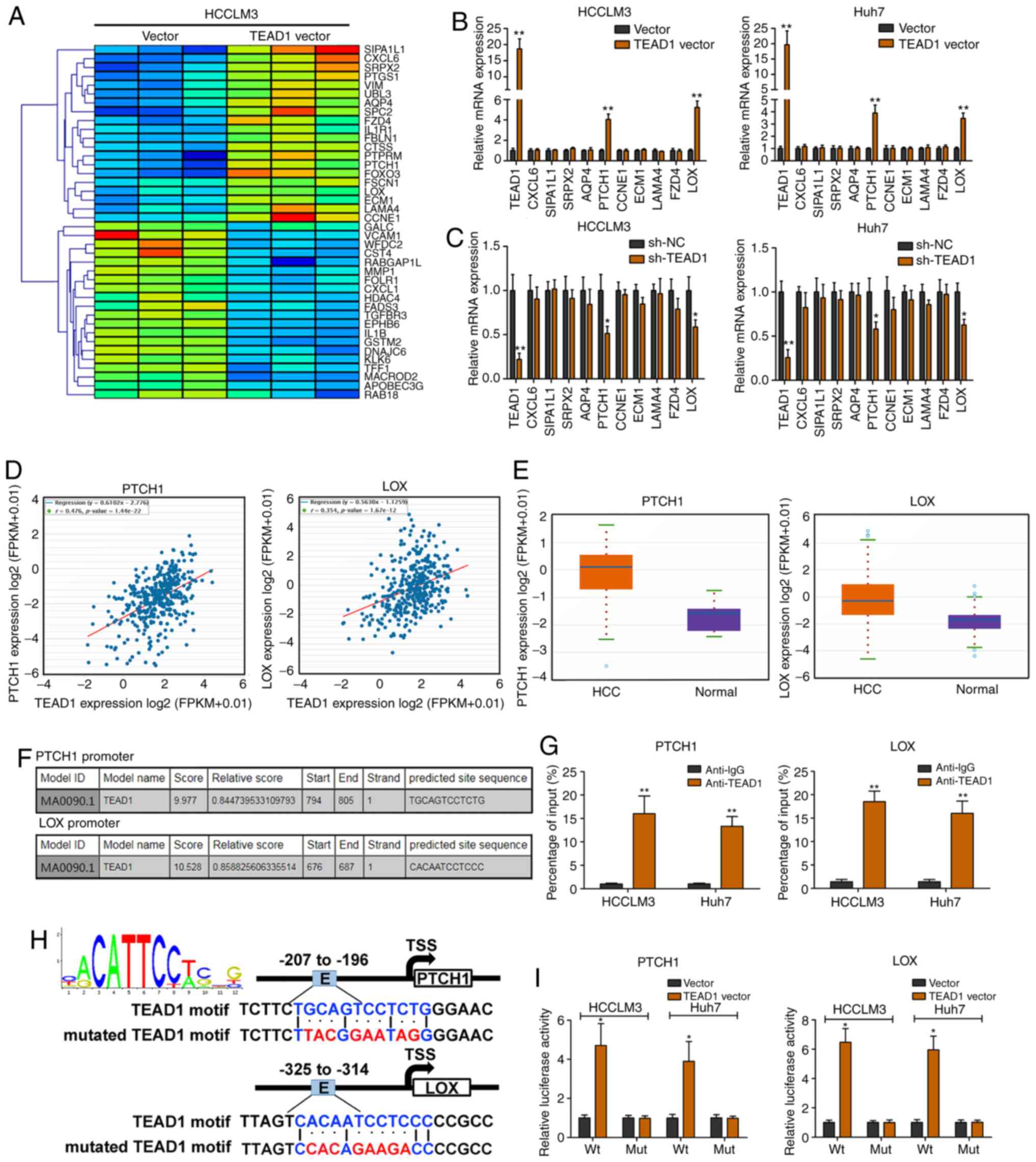
TEAD1 activates PTCH1 and LOX
transcription in HCC
RNA-seq was performed to explore the potential
target gene of TEAD1 in HCCLM3 cells (Fig. 5A). TEAD1 was ectopically expressed
by transfection with the TEAD1 vector (Fig. 5B). PTCH1 and LOX expression levels
were significantly elevated upon TEAD1 overexpression (Fig. 5B). Silencing of TEAD1 led to PTCH1
and LOX depletion (Fig. 5C). In
addition, TEAD1 expression was positively correlated with PTCH1 and
LOX expression levels analyzed with the TCGA dataset (Fig. 5D). Moreover, PTCH1 and LOX
expression levels were markedly elevated in HCC tissues compared
with nontumorous tissues analyzed with the TCGA dataset (Fig. 5E). As shown in Fig. 5F, TEAD1 was predicted to bind to the
promoter regions of PTCH1 and LOX. ChIP revealed significant
TEAD1-binding activity on the endogenous PTCH1/LOX promoter region
(Fig. 5G). A luciferase reporter
assay demonstrated that TEAD1 could bind to the predicted binding
sites of the PTCH1 and LOX promoters (Fig. 5H and I).
Discussion
In the present study, a circRNA microarray analysis
was conducted to explore the dysregulated circRNAs in HCC tissues.
circ-CCT3 was identified as the most upregulated circRNA by
RT-qPCR. Therefore, the present study focused on a novel identified
circRNA, circ-CCT3, originating from chr1 (156303337-156304709) of
the host gene CCT3. circRNAs possess the modulatory potency of
target genes (16) and act as
promising biomarkers; therefore, a number of circRNAs have been
explored in the onset and progression of diverse types of cancer
(17,18). Several circRNAs may be used as
potential diagnostic or prognostic biomarkers in human cancers
(18). The present study also
identified circ-CCT3 as an unfavorable prognostic marker for
patients with HCC.
The functions of circ-CCT3 in HCC cell progression
were assessed. A knockdown study indicated that depleted circ-CCT3
inhibited cell proliferation, migration and invasion and triggered
cell apoptosis. Previous studies revealed that miR-1287-5p serves
an important role in breast (19),
colorectal (20) and cervical
(21) cancers. A previous study
indicated that miR-1287-5p inhibits HCC progression by targeting
PIK3R3 (22). The present study
found that knockdown of circ-CCT3 enhanced miR-1287-5p expression
and that downregulation of miR-1287-5p resulted in a reversal
effect of sh-circ-CCT3-1 in HCC cells.
It appeared that the 3′-UTR of TEAD1 contained the
binding sites for miR-1287-5p, which was verified by a
dual-luciferase reporter gene assay. In most cases, miRNAs regulate
mRNA expression at the post-transcriptional level. circRNAs can act
as sponges of miRNAs, thus indirectly regulating downstream mRNAs
(23). TEAD1 transcriptional
activity is widely believed to be modulated by the presence or
absence of nuclear YAP/TAZ (24).
Nevertheless, several studies have shown that TEAD itself is
regulated through other mechanisms (25). It has been firmly established that
TEAD1 acts as a pleiotropic transcription factor to control cell
proliferation, apoptosis, metabolism, adhesion, DNA replication,
differentiation and angiogenesis (26,27).
The present study observed that circ-CCT3 elevated TEAD1 expression
by sponging miR-1287-5p in HCC. Functional experiments showed that
the inhibitory effect of sh-circ-CCT3-1 on cell proliferation and
invasion was attenuated by co-transfection with the TEAD1 vector,
indicating the circ-CCT3/miR-1287-5p/TEAD1 regulatory axis in HCC.
The TEAD1 oncogene is broadly overexpressed in a number of
late-stage cancers and is often associated with tumorigenesis by
causing inappropriate gene expression (28). The results of the present study
suggested that up/downregulation of TEAD1 resulted in a significant
increase/decrease in PTCH1 and LOX expression. PTCH1 is involved in
cancer progression and chemoresistance (29). PTCH1 is the Hedgehog (HH) receptor
at the cell surface or in primary cilia, which binds to HH to
initiate ligand-dependent signaling (30). Several studies have indicated its
important role in the generation and transduction of HH signaling
(31,32). LOX is one of five members of the LOX
family. It serves a primary, catalytic activity-related, role in
the assembly of the extracellular matrix, a dynamic structural and
regulatory framework that is essential for cell fate,
differentiation and communication (33,34).
ChIP and luciferase reporter assays suggested that TEAD1 could
directly bind to the promoter regions of PTCH1 and LOX, thereby
elevating their transcription levels.
In summary, circ-CCT3 acted as a sponge for
miR-1287-5p to enhance TEAD1 expression, which subsequently
contributed to the activation of PTCH1 and LOX and consequently
promotes tumorigenesis and progression. The present study
identified a novel circRNA, circ-CCT3, which may be used as a
potential therapeutic target for HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Research
Foundation of Qiqihar Academy of Medical Sciences (grant no.
QMSI2019M-27).
Availability of data and materials
The datasets used in the present study are available
from the corresponding author.
Authors' contributions
WL, TZ, GD, LH, BZ, JY, YP, FG, LZ and MZ performed
the experiments. WL, MZ, QY, HL, YW and CZ analyzed and interpreted
the data. WL, YW and CZ wrote the manuscript. WL and TZ confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was authorized by the
Institutional Review Board of The Second Affiliated Hospital of
Qiqihar Medical University and written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kalogeridi MA, Zygogianni A, Kyrgias G,
Kouvaris J, Chatziioannou S, Kelekis N and Kouloulias V: Role of
radiotherapy in the management of hepatocellular carcinoma: A
systematic review. World J Hepatol. 7:101–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khemlina G, Ikeda S and Kurzrock R: The
biology of Hepatocellular carcinoma: Implications for genomic and
immune therapies. Mol Cancer. 16:1492017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartha I, di Iulio J, Venter JC and
Telenti A: Human gene essentiality. Nat Rev Genet. 19:51–62. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiong DD, Dang YW, Lin P, Wen DY, He RQ,
Luo DZ, Feng ZB and Chen G: A circRNA-miRNA-mRNA network
identification for exploring underlying pathogenesis and therapy
strategy of hepatocellular carcinoma. J Transl Med. 16:2202018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang B, Liu Z, Cao K, Shan W, Liu J, Wen
Q and Wang R: circ-SPECC1 modulates TGFβ2 and autophagy under
oxidative stress by sponging miR-33a to promote hepatocellular
carcinoma tumorigenesis. Cancer Med. 9:5999–6008. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Afify AY, Ibrahim SA, Aldamsisi MH,
Zaghloul MS, El-Ekiaby N and Abdelaziz AI: Competing endogenous
RNAs in hepatocellular carcinoma-the pinnacle of rivalry. Semin
Liver Dis. 39:463–475. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilczynska A and Bushell M: The complexity
of miRNA-mediated repression. Cell Death Differ. 22:22–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bitarte N, Bandres E, Boni V, Zarate R,
Rodriguez J, Gonzalez-Huarriz M, Lopez I, Javier Sola J, Alonso MM,
Fortes P, et al: MicroRNA-451 is involved in the self-renewal,
tumorigenicity, and chemoresistance of colorectal cancer stem
cells. Stem Cells. 29:1661–1671. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heng BC, Zhang X, Aubel D, Bai Y, Li X,
Wei Y, Fussenegger M and Deng X: An overview of signaling pathways
regulating YAP/TAZ activity. Cell Mol Life Sci. 78:497–512. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
R Core Team: (v3.14.0; 2016), . R: A
language and environment for statistical computing. R Foundation
for Statistical Computing; Vienna, Austria: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computingFebruary
10–2015
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu J, Zhang B, Zhang H, Qi Y and Wang Y,
Wang W and Wang Y and Wang Y: E2F1-induced upregulation of long
non-coding RNA LMCD1-AS1 facilitates cholangiocarcinoma cell
progression by regulating miR-345-5p/COL6A3 pathway. Biochem
Biophys Res Commun. 512:150–155. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42D:D92–D97. 2014. View Article : Google Scholar
|
|
15
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA hsa_circ_0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 496:455–461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schwarzenbacher D, Klec C, Pasculli B,
Cerk S, Rinner B, Karbiener M, Ivan C, Barbano R, Ling H,
Wulf-Goldenberg A, et al: miR-1287-5p inhibits triple negative
breast cancer growth by interaction with phosphoinositide 3-kinase
CB, thereby sensitizing cells for PI3Kinase inhibitors. Breast
Cancer Res. 21:202019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui G, Zhao H and Li L: Long noncoding RNA
PRKCQ-AS1 promotes CRC cell proliferation and migration via
modulating miR-1287-5p/YBX1 axis. J Cell Biochem. 121:4166–4175.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X
and Ding Y: Circular RNA circSLC26A4 Accelerates Cervical Cancer
Progression via miR-1287-5p/HOXA7 Axis. Mol Ther Nucleic Acids.
19:413–420. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu J, Tang L, Xu Y, Ge K, Huang J, Gu M,
Zhong J and Huang Q: mir-1287 suppresses the proliferation,
invasion, and migration in hepatocellular carcinoma by targeting
PIK3R3. J Cell Biochem. 119:9229–9238. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han TS, Hur K, Cho HS and Ban HS:
Epigenetic Associations between lncRNA/circRNA and miRNA in
Hepatocellular Carcinoma. Cancers (Basel). 12:26222020. View Article : Google Scholar
|
|
24
|
Mammoto A, Muyleart M, Kadlec A, Gutterman
D and Mammoto T: YAP1-TEAD1 signaling controls angiogenesis and
mitochondrial biogenesis through PGC1α. Microvasc Res. 119:73–83.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Landin-Malt A, Benhaddou A, Zider A and
Flagiello D: An evolutionary, structural and functional overview of
the mammalian TEAD1 and TEAD2 transcription factors. Gene.
591:292–303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Huang T, Zhang J, Wong CC, Zhang
B, Dong Y, Wu F, Tong JHM, Wu WKK, Cheng ASL, et al: TEAD1/4 exerts
oncogenic role and is negatively regulated by miR-4269 in gastric
tumorigenesis. Oncogene. 36:6518–6530. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tome-Garcia J, Erfani P, Nudelman G,
Tsankov AM, Katsyv I, Tejero R, Bin Zhang, Walsh M, Friedel RH,
Zaslavsky E, et al: Analysis of chromatin accessibility uncovers
TEAD1 as a regulator of migration in human glioblastoma. Nat
Commun. 9:40202018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan Y, Gao Y, Rao J, Wang K, Zhang F and
Zhang C: YAP-1 promotes tregs differentiation in hepatocellular
carcinoma by enhancing TGFBR2 transcription. Cell Physiol Biochem.
41:1189–1198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Olesen UH, Bojesen S, Gehl J and
Haedersdal M: Anticancer drugs and the regulation of Hedgehog genes
GLI1 and PTCH1, a comparative study in nonmelanoma skin cancer cell
lines. Anticancer Drugs. 28:1106–1117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qi C, Di Minin G, Vercellino I, Wutz A and
Korkhov VM: Structural basis of sterol recognition by human
hedgehog receptor PTCH1. Sci Adv. 5:eaaw64902019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Skoda AM, Simovic D, Karin V, Kardum V,
Vranic S and Serman L: The role of the Hedgehog signaling pathway
in cancer: A comprehensive review. Bosn J Basic Med Sci. 18:8–20.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qi X and Li X: Mechanistic insights into
the generation and transduction of hedgehog signaling. Trends
Biochem Sci. 45:397–410. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Laczko R and Csiszar K: Lysyl Oxidase
(LOX): Functional contributions to signaling pathways.
Biomolecules. 10:10932020. View Article : Google Scholar
|
|
34
|
Saatci O, Kaymak A, Raza U, Ersan PG,
Akbulut O, Banister CE, Sikirzhytski V, Tokat UM, Aykut G, Ansari
SA, et al: Targeting lysyl oxidase (LOX) overcomes chemotherapy
resistance in triple negative breast cancer. Nat Commun.
11:24162020. View Article : Google Scholar : PubMed/NCBI
|