Introduction
Atherosclerosis (AS) is the main cause of
cardiovascular diseases and is a chronic progressive disease
initiated by vascular inflammation (1,2). The
endothelium is the innermost layer of blood vessels and changes in
blood environment serve an important role in vascular homeostasis.
Several factors, such as hyperlipidemia, tobacco use,
hypercysteinemia, diabetes and hypertension (3,4), may
induce vascular injury by activating endothelial cells, which then
release increased amounts of proinflammatory cytokines, chemokines
and adhesion molecules (5,6). Subsequently, circulating monocytes are
recruited to the lesion site, further triggering a series of
pathogenic processes, including excessive lipid accumulation,
tunica smooth muscle cell migration and plaque formation (7). Therefore, inhibition of endothelial
inflammatory processes may constitute a key step toward preventing
AS.
Bakkenolides are a relatively rare class of natural
products, which can be isolated from plants of the genus
Petasites, including Petasites japonicus (Sieb. et
Zucc.) F. Schmidt and Petasites tricholobus Franch (8), which have been used in Chinese folk
medicine to treat palsy, hypertension, cough and snakebite for
hundreds of years (9). Compared
with three other bakkenolides, bakkenolide-IIIa (Bak-IIIa), which
is an important active component of bakkenolides, has been reported
to display the best neuroprotective effect against oxygen-glucose
deprivation-induced neuronal injuries (9). Another study involving animal cerebral
damage models indicated that Bak-IIIa could exert neuroprotective
effects by inhibiting nuclear factor (NF)-κB activation (10). Inflammation is a fundamental, yet
complex, biological response initiated by the body to counter
injuries, as well as infections, and to eliminate the initial
causes of cell injury (11).
Although the inflammatory levels of injured neurons were not
evaluated in the two aforementioned studies, it is reasonable to
surmise that inflammation may have been decreased, considering the
significant neuroprotective effect of Bak-IIIa.
Long noncoding RNAs (lncRNAs) belong to an important
class of ncRNAs that are involved in the regulation of various
physiological and pathological processes, including AS (12). Recently, some lncRNAs have been
identified as novel regulators of vascular inflammation, which
constitutes an initial step toward AS (12,13).
For example, the lncRNA HOXA-AS3 has been revealed to be a critical
activator that promotes NF-κB-mediated endothelial inflammation
(14). In addition, the lncRNA H19
was upregulated in the serum of patients with AS and in oxidized
low-density lipoprotein (ox-LDL)-treated human umbilical vein
endothelial cells (HUVECs), whereas knockdown of its expression
significantly ameliorated inflammation, apoptosis and oxidative
stress in ox-LDL-injured HUVECs (15). In addition, the lncRNAs HOXA-AS2 and
MALAT1 have been shown to participate in the regulation of
endothelial inflammation (16,17).
The present study investigated the effect of
Bak-IIIa on lipopolysaccharide (LPS)-induced inflammatory injury in
HUVECs and identified potential lncRNAs involved in this process.
The present study may provide a candidate molecule for the
treatment of AS.
Materials and methods
Cells and cell culture
HUVECs were purchased from American Type Culture
Collection, and were cultured in DMEM supplemented with 10% fetal
bovine serum and 1% penicillin-streptomycin (all from Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in a humidified atmosphere
containing 5% CO2.
Cell viability assay
HUVECs were seeded in a 96-well plate at 8,000
cells/well. The next day, the cells were exposed to different
concentrations (0, 10, 20, 50, 100 and 200 µM) of Bak-IIIa
(CSNpharm, Inc.) for 72 h at 37°C. The supernatant was then
removed, and 100 µl medium containing 50 µg MTT was added to the
wells. After incubation at 37°C for 3 h, accumulated formazan was
dissolved in 100 µl DMSO (Sangon Biotech Co., Ltd.), and absorbance
was determined at 490 nm using a microplate reader (ELx800; BioTek
Instruments, Inc.).
To observe the effect of Bak-IIIa on LPS-induced
cellular injury, HUVECs were first exposed to 10 ng/ml LPS
(Sigma-Aldrich; Merck KGaA) for 4 h at 37°C, and then treated with
0, 10, 20 and 50 µM Bak-IIIa for 24 h at 37°C. Cell survival was
assessed using the aforementioned MTT assay. Cells treated without
LPS and Bak-IIIa were used as the control.
Enzyme-linked immunosorbent assay
(ELISA)
HUVECs were seeded in a 12-well plate at
3×104 cells/well, and were treated with LPS for 4 h, and
with 0, 10, 20 and 50 µM Bak-IIIa for 24 h, respectively.
Thereafter, cell culture supernatants were collected and filtered
through a 0.45-µm cell Nalgene syringe filter (Thermo Fisher
Scientific, Inc.). TNF-α (cat. no. PT518), interleukin (IL)-1β
(cat. no. PI305), IL-8 (cat. no. PI640) and IL-6 (cat. no. PI330)
levels in the supernatants were analyzed using corresponding ELISA
kits (Beyotime Institute of Biotechnology), according to the
manufacturer's instructions.
Microarray analysis
After LPS-treated HUVECs were treated with 0 or 50
µM Bak-IIIa for 24 h (named as LPS or Bak-IIIa group respectively),
total RNA from these two groups was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Subsequent microarray hybridization and data analysis were
performed by Bhbio Ltd. Co (http://www.shbio.com/). Briefly, purified RNA from
each sample was amplified and labeled using a Low Input Quick Amp
Labeling kit (Agilent Technologies, Inc.). Thereafter, labeled cRNA
was hybridized in a hybridization oven (Agilent Technologies,
Inc.), and the hybridized slides were washed and scanned using a
Microarray Scanner (Agilent Technologies, Inc.). Raw data were
extracted using Feature Extraction software (v10.7.1.1; Agilent
Technologies, Inc.) and normalized via a quantile algorithm.
Differentially expressed lncRNAs (DELs) were filtered through fold
change absolute values (FC abs ≥2) and the significance of P-values
(P<0.5).
Prediction of lncRNA targets
For predicting cis-regulation, genes
transcribed within 10 kb upstream or downstream of DELs were
considered as cis-targets. DELs and potential
cis-targets were paired using the UCSC genome browser
(18). For trans-target
prediction, complementary or similar sequences were first selected
via BLAST software (https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/).
Thereafter, complementary energy between the two sequences was
calculated using RNAplex software (v2.1.8) (19) and energy ≤-30 was selected as the
trans-target of DELs.
Bioinformatics analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) analyses (https://david.ncifcrf.gov/) were conducted to
determine enrichment of the predicted targets of DELs. Both
cis- and trans-targets were first classified into
corresponding GO terms or KEGG pathways according to each database,
and the top 30 GO terms or KEGG pathways showing a higher
enrichment factor and a significant P-value (<0.5) were
selected.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was isolated from cells in the LPS and
Bak-IIIa groups using TRIzol® reagent (Thermo Fisher
Scientific, Inc.) and cDNA was synthesized using PrimeScript™ One
Step RT-PCR kit (Takara Biotechnology Co., Ltd.), according to the
manufacturer's protocol. RT-qPCR was performed using SYBR Green PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.),
following the manufacturer's instructions. Thermocycling was: 94°C
for 5 min, then 40 cycles at 94°C for 5 sec and 60°C for 1 min.
Expression levels were calculated using the 2−ΔΔCq
method (20), and relative levels
of lncRNA were normalized to GAPDH. Primer sequences for lncRNAs
and GAPDH are listed in Table
I.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| LncRNA/Gene | Forward
(5′→3′) | Reverse
(5′→3′) |
|---|
| NONHSAT089168 |
TGTCATTCAGCCCAAACTCA |
CGCCAGGTTAGTTCAACACA |
| NR_015451 |
TCTCGATCTCCTGACCTCGT |
ACAGAGCAGCCTCCACCTTA |
| NR_038275 |
CCCTCCTATGTGCTCGTTTC |
GAGAAGTCCTCGTCGGTCAG |
| lnc-MUC20-9:1 |
GCCTGTCTCTGCCTCTGAAC |
CCAGGCCAGACCAATCTAAA |
| NR_034072 |
TCCCAAAGTGCTGGGATTAC |
CAGTTGGGAACTTGGGAAAA |
|
lnc-HSP90B1-3:1 |
ACTTGAGGCCAGGAGTTTGA |
AACCTCTCAGGCTCAAGCAA |
| GAPDH |
CGACCACTTTGTCAAGCTCA |
AGGGGAGATTCAGTGTGGTG |
Transfection assay
HUVECs were seeded in 6-well plates
(2×105 cells/well) and transfected with 2 µg
pCDH-CMV-MCS-EF1-GFP-T2A-Puro plasmid encoding full length
LINC00294 (Hanbio Biotechnology Co., Ltd.) or the empty vector,
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), following the manufacturer's instructions. After
transfection at 37°C for 48 h, cells were collected for RT-qPCR
validation. HUVECs transfected with the plasmid or empty vector
were categorized as LINC00294 or negative control (NC) groups,
respectively and were then used for LPS treatment, followed by cell
viability and ELISA assay.
Statistical analysis
SPSS v16.0 (SPSS, Inc.) was used for data
processing. Data are presented as the mean ± standard deviation and
each experiment was performed three times. Differences between two
groups were determined using Student's t-test, whereas one-way
analysis of variance and Tukey's post hoc test were used to compare
three or more groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Bak-IIIa ameliorates the LPS-induced
inflammatory damage in HUVECs
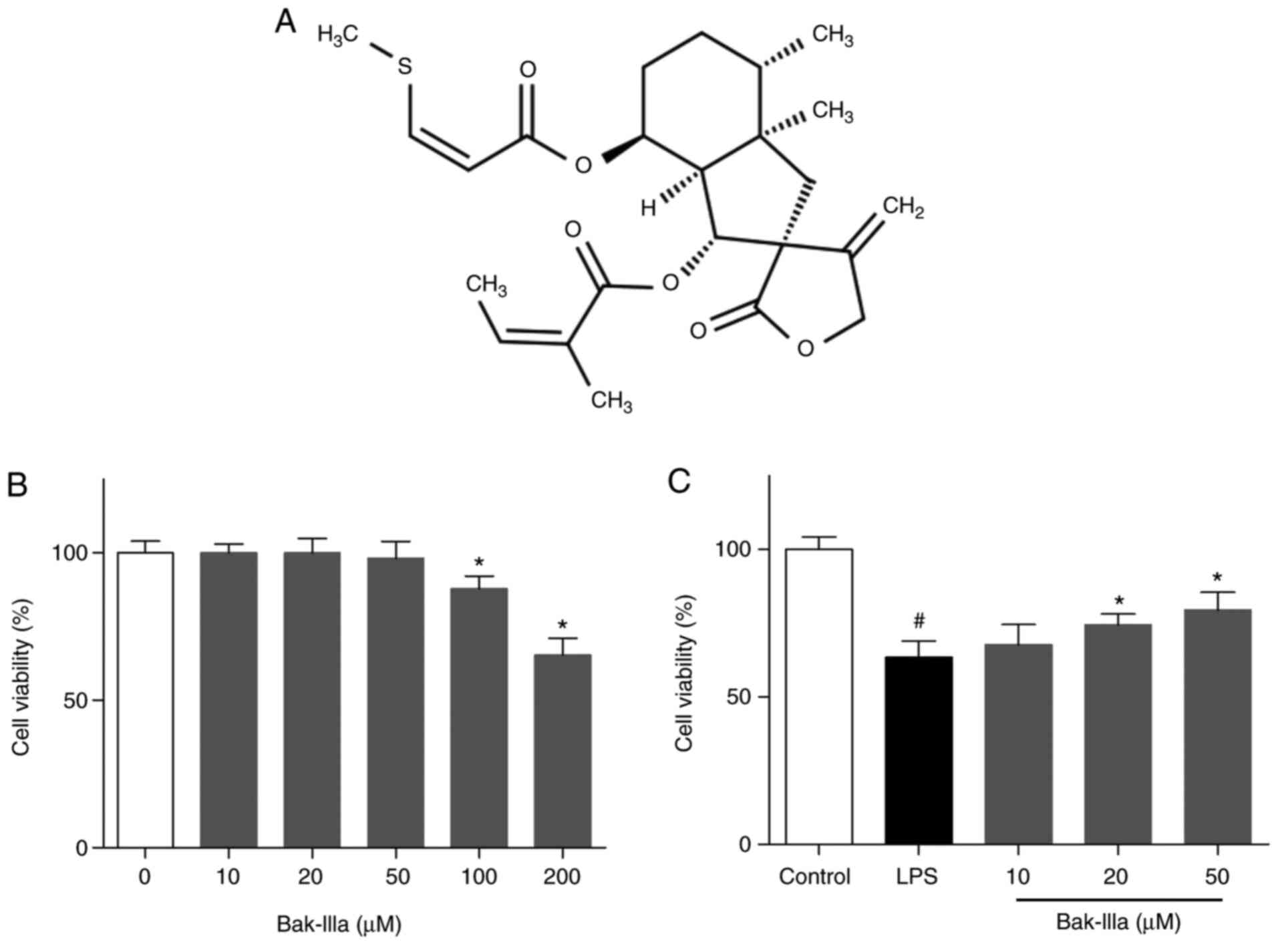
Initially, the cytotoxic effects exerted by Bak-IIIa
on HUVECs were assessed (Fig. 1A).
MTT assays indicated that the viability of HUVECs was not
significantly changed by treatment with lower concentrations (10–50
µM) of Bak-IIIa, whereas viability was reduced by 12–35% by high
doses (100 and 200 µM) (Fig. 1B).
Hence, low doses of Bak-IIIa (10–50 µM) were selected for use in
subsequent studies. HUVECs were then exposed to LPS, and the
results of the MTT assay indicated that the survival of HUVECs was
decreased to ~63% compared with the control group (Fig. 1C). Following further treatment with
20 and 50 µM Bak-IIIa, the viability of damaged HUVECs was
increased by ~11 and ~16%, respectively (Fig. 1C).
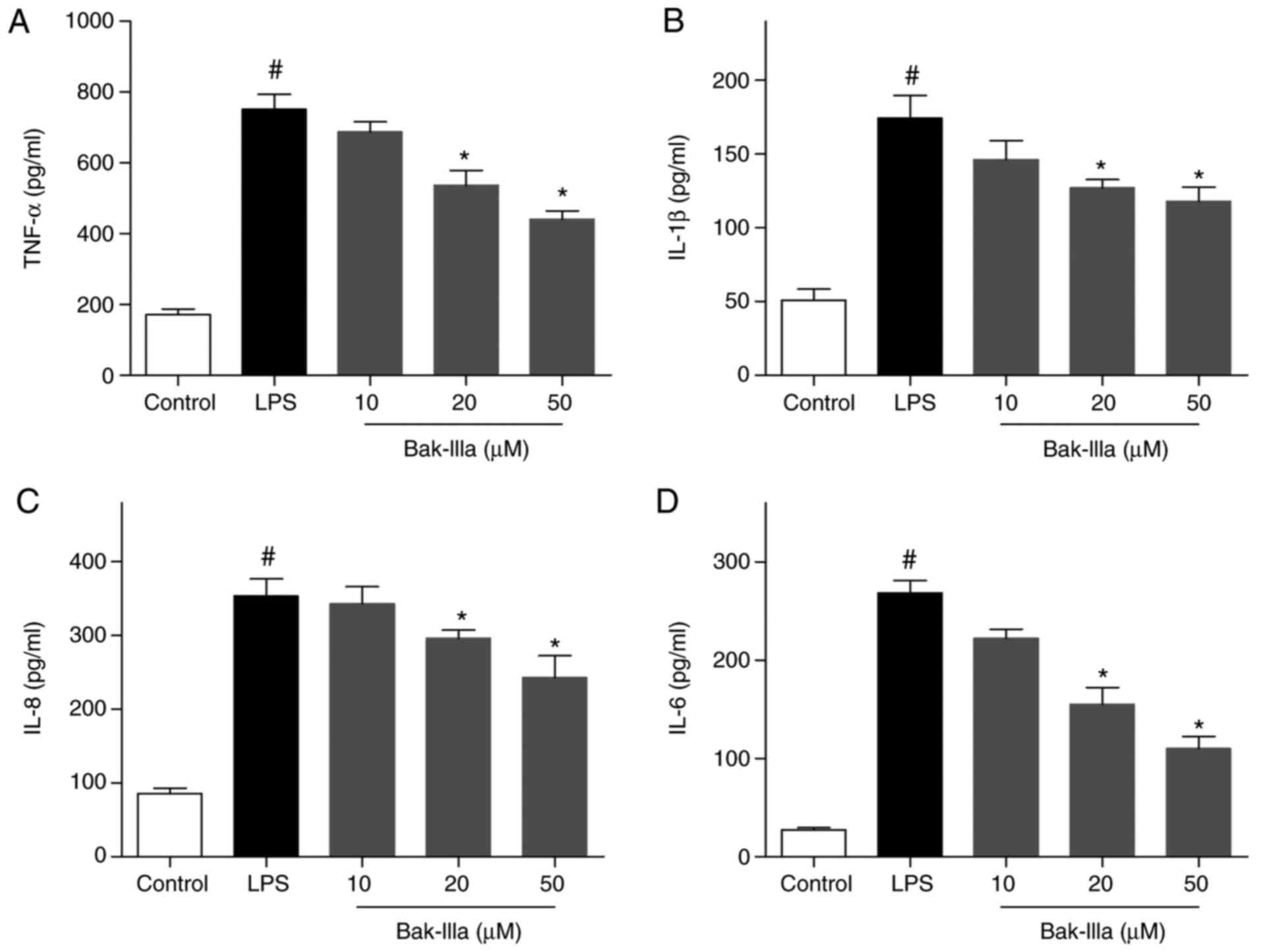
ELISA revealed that the levels of TNF-α, IL-1β, IL-8
and IL-6 were significantly increased following LPS-induced
stimulation compared with in the control group, indicating that
inflammatory damage was inflicted on the HUVECs (Fig. 2A-D). The LPS-injured HUVECs were
then treated with 10–50 µM Bak-IIIa, and the results indicated that
the levels of the four proinflammatory factors were gradually
decreased in response to increasing Bak-IIIa concentrations
(Fig. 2A-D). Of the three dose
groups, the 20 and 50 µM groups induced a significant reduction in
TNF-α, IL-1β, IL-8 and IL-6 levels compared with in the LPS group
(Fig. 2A-D), thus suggesting that
Bak-IIIa treatment was effective in ameliorating LPS-induced
inflammatory damage in HUVECs.
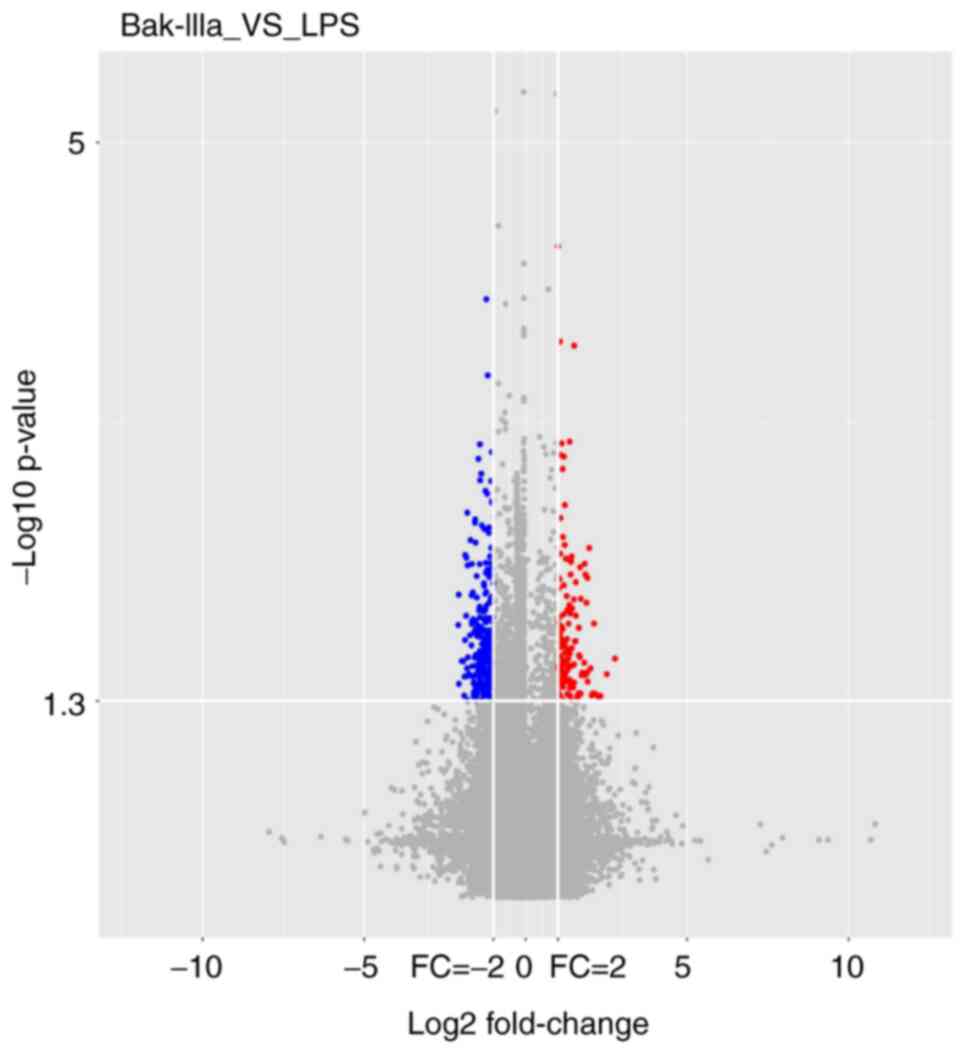
Microarray analysis
In order to investigate the mechanisms underlying
the effect of Bak-IIIa on HUVECs damaged by inflammation, cells
from the LPS and Bak-IIIa groups were subjected to lncRNA
microarray analyses, which identified 70 DELs in these two groups.
A volcano plot showed the presence of 29 significantly upregulated
(red dots) and 41 significantly downregulated (blue dots) DELs in
HUVECs that had undergone LPS-induced inflammatory damage and
subsequent Bak-IIIa treatment (Fig.
3). These 70 DELs were then ranked according to FC abs values
in descending order; the top 10 are listed in Table II. The FC abs of most DELs was ~4,
whereas the lncRNA ENST00000441971 had the highest FC abs of
~7.
 | Table II.Top 10 differentially expressed
lncRNAs after lipopolysaccharide-damaged human umbilical vein
endothelial cells were treated with bakkenolide-IIIa. |
Table II.
Top 10 differentially expressed
lncRNAs after lipopolysaccharide-damaged human umbilical vein
endothelial cells were treated with bakkenolide-IIIa.
| Number | lncRNA | P-value | FC | FC (abs) | Regulation | Source |
|---|
| 1 |
ENST00000441971 | 0.026 | 6.807 | 6.807 | Up |
ENSEMBL_GENCODE |
| 2 | lnc-NR2F1-8:1 | 0.048 | 4.810 | 4.810 | Up | lncipedia |
| 3 |
ENST00000609511 | 0.015 | 4.343 | 4.343 | Up |
ENSEMBL_GENCODE |
| 4 | NONHSAT089168 | 0.045 | 4.250 | 4.250 | Up | NONCODE |
| 5 |
ENST00000451766 | 0.039 | 0.239 | 4.190 | Down |
ENSEMBL_GENCODE |
| 6 | NONHSAT113888 | 0.005 | 3.920 | 3.920 | Up | NONCODE |
| 7 |
ENST00000618397 | 0.037 | 3.788 | 3.788 | Up |
ENSEMBL_GENCODE |
| 8 | lnc-COMTD1-1:1 | 0.006 | 0.281 | 3.560 | Down | lncipedia |
| 9 | lnc-MOK-1:2 | 0.030 | 0.284 | 3.523 | Down | lncipedia |
| 10 | NR_015451 | 0.034 | 3.320 | 3.320 | Up | RefSeq |
lncRNA target prediction and
bioinformatics analyses
To obtain a better understanding of the DELs, the
cis- and trans-targets were predicted. As a result,
52 cis-regulated targets were obtained from 44 DELs
(Table SI), whereas 386
trans-regulated targets (111 overlaps among all 497 rows)
were predicted from only 12 DELs (Table SII); nine of the 12 DELs, listed in
Table III, covered a total of 382
targets (some overlapped). In addition, the trans-target
number for NONHSAT089168, NR_015451 and lnc-MUC20-9:1 reached 100
(Table III).
 | Table III.Nine differentially expressed lncRNAs
covering 382 trans-targets. |
Table III.
Nine differentially expressed lncRNAs
covering 382 trans-targets.
| Number | lncRNA | Trans-target
number | FC | FC (abs) | Regulation | Sources |
|---|
| 1 | NONHSAT089168 | 113 | 4.250 | 4.250 | Up | NONCODE |
| 2 | NR_015451 | 154 | 3.320 | 3.320 | Up | RefSeq |
| 3 | NR_027411 | 10 | 2.650 | 2.650 | Up | RefSeq |
| 4 | NR_033350 | 10 | 2.578 | 2.578 | Up | RefSeq |
| 5 | NR_033353 | 10 | 2.435 | 2.435 | Up | RefSeq |
| 6 | NR_038275 | 13 | 2.395 | 2.395 | Up | RefSeq |
| 7 | lnc-MUC20-9:1 | 100 | 2.366 | 2.366 | Up | lncipedia |
| 8 | NR_034072 | 29 | 2.188 | 2.188 | Up | RefSeq |
| 9 |
lnc-HSP90B1-3:1 | 54 | 2.037 | 2.037 | Up | lncipedia |
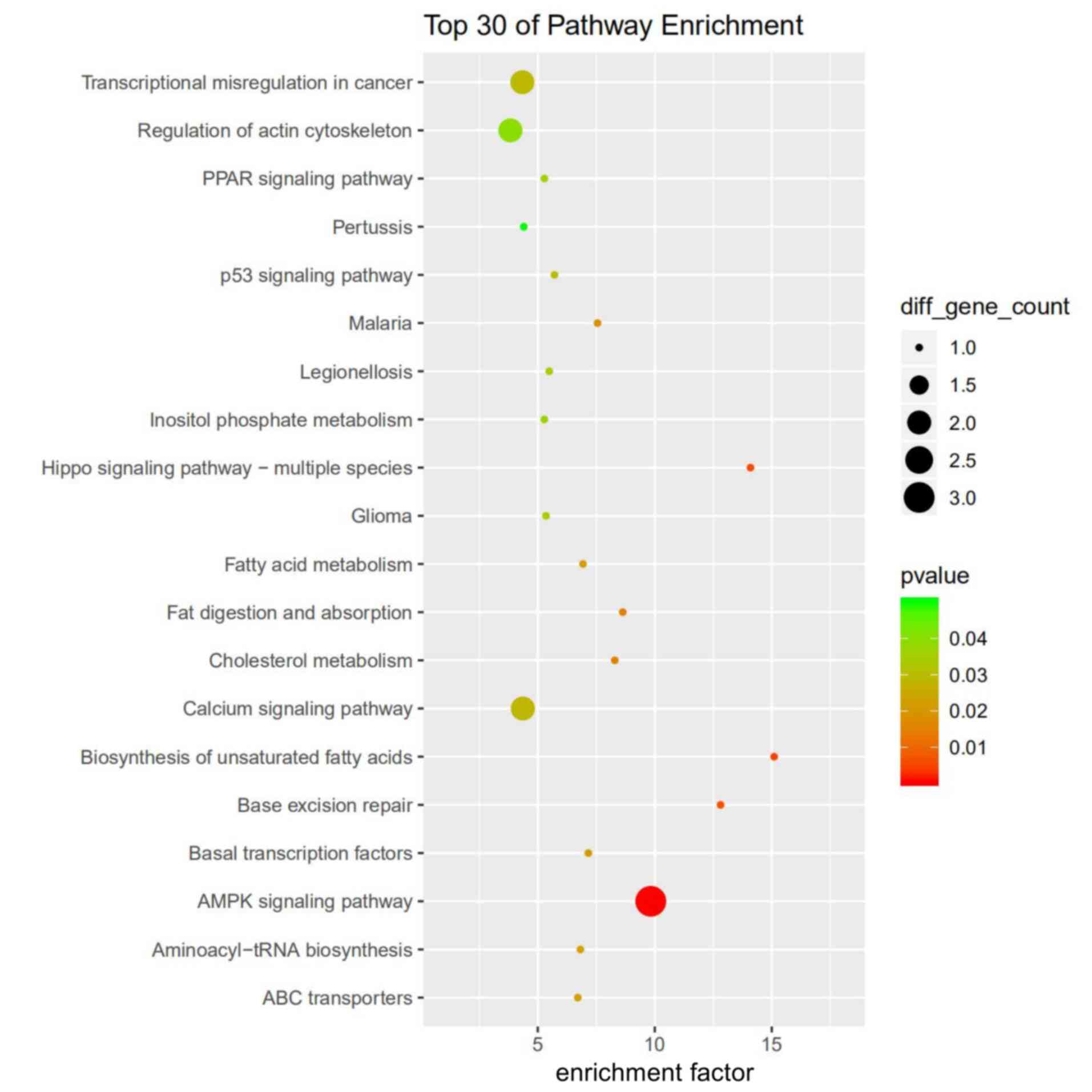
Among the top 30 targets found to be enriched
according to GO analysis, some cis-targets were involved in
ion homeostasis and ‘autophagy’ (Fig.
S1), whereas trans-targets were enriched in
‘cis-Golgi network’ and ‘IL-12 production’ (Fig. S2). KEGG pathway analysis showed
that cis-targets were mainly enriched in the ‘AMPK signaling
pathway’, whereas several targets were involved in the ‘p53
signaling pathway’ (Fig. 4).
trans-targets were enriched in ‘purine metabolism’ and
‘glycosylphosphatidylinositol-anchor biosynthesis’ (Fig. S3).
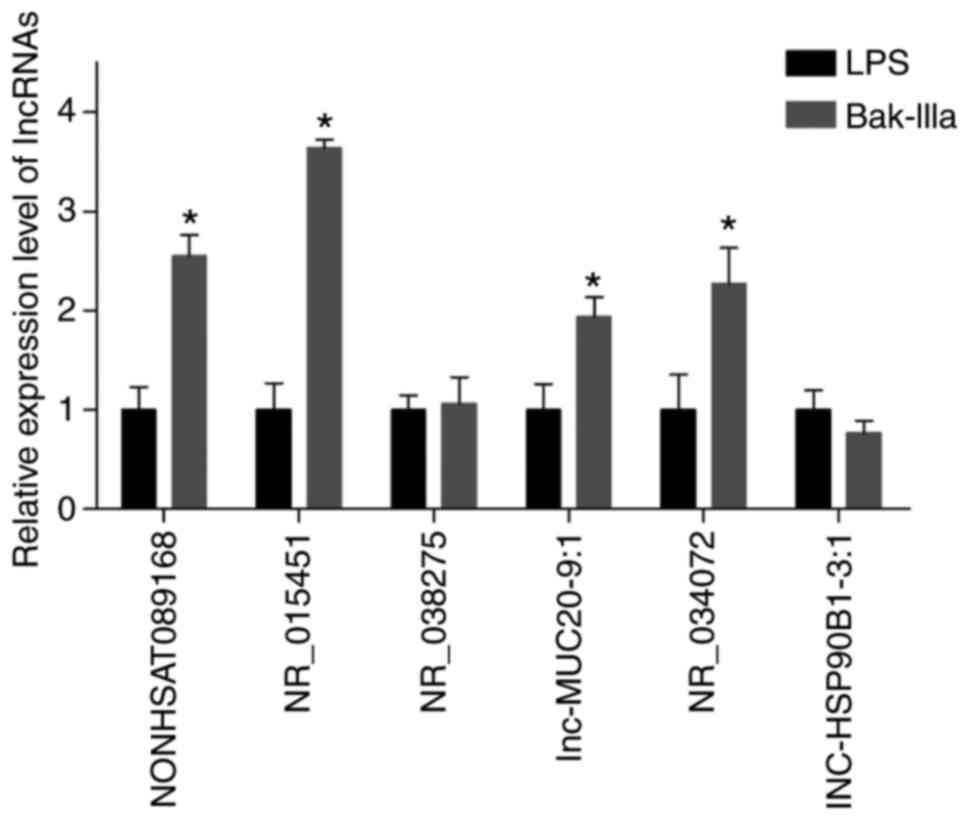
RT-qPCR validation of the DELs
corresponding to inflammation-related trans-targets
The key words ‘inflammation’ and ‘inflammatory’ were
searched during GO and KEGG analyses of trans-targets
(Tables SIII and SIV), and three inflammation-related GO
terms, ‘inflammatory response’, ‘positive regulation of
inflammatory response’, and ‘regulation of inflammatory response’,
involving a total of 17 targets (some overlapped) were screened
(Table IV). The corresponding DELs
are listed in Table V, and nine
(XIAP, CCL5, ACER3, OPRM1, IDO1, LYZ, HMGB1, LDLR and F2R) of the
17 targets were those of LINC00294. RT-qPCR indicated that the
expression levels of most of the six candidate DELs were consistent
with those revealed by the results of microarray analyses.
Subsequently, NR_015451 (also termed LINC00294), which displayed
the highest verified expression level, was selected for further
evaluation (Fig. 5).
 | Table IV.Inflammation-related GO terms and
trans-targets. |
Table IV.
Inflammation-related GO terms and
trans-targets.
| GO ID | Description | Count |
Trans-targets |
|---|
| GO:0006954 | Inflammatory
response | 17 | XIAP, CCR6, CCL5,
MEFV, ACER3, OPRM1, IDO1, AXL, TLR10, REL, LYZ, CCL22, NLRP12,
HMGB1, LDLR, CAMK1D, F2R |
| GO:0050729 | Positive regulation
of inflammatory response | 4 | IDO1, NLRP12,
TLR10, LDLR |
| GO:0050727 | Regulation of
inflammatory response | 7 | NLRP12, CCL5, MEFV,
LDLR, XIAP, IDO1, TLR10 |
 | Table V.The 17 trans-targets and
corresponding lncRNAs. |
Table V.
The 17 trans-targets and
corresponding lncRNAs.
| Trans-targets | lncRNAs |
|---|
| XIAP | NONHSAT089168,
NR_015451 |
| CCR6 | lnc-MUC20-9:1 |
| CCL5 | lnc-MUC20-9:1,
NR_015451, NR_034072 |
| MEFV |
lnc-HSP90B1-3:1 |
| ACER3 | lnc-MUC20-9:1,
NR_015451 |
| OPRM1 | NR_015451 |
| IDO1 | NONHSAT089168,
NR_015451 |
| AXL | NR_034072 |
| TLR10 | NONHSAT089168 |
| REL | NR_038275 |
| LYZ | NR_015451 |
| CCL22 |
lnc-HSP90B1-3:1 |
| NLRP12 | lnc-MUC20-9:1,
NONHSAT089168 |
| HMGB1 | NONHSAT089168,
NR_015451 |
| LDLR | NR_015451 |
| CAMK1D | lnc-MUC20-9:1 |
| F2R | NR_015451 |
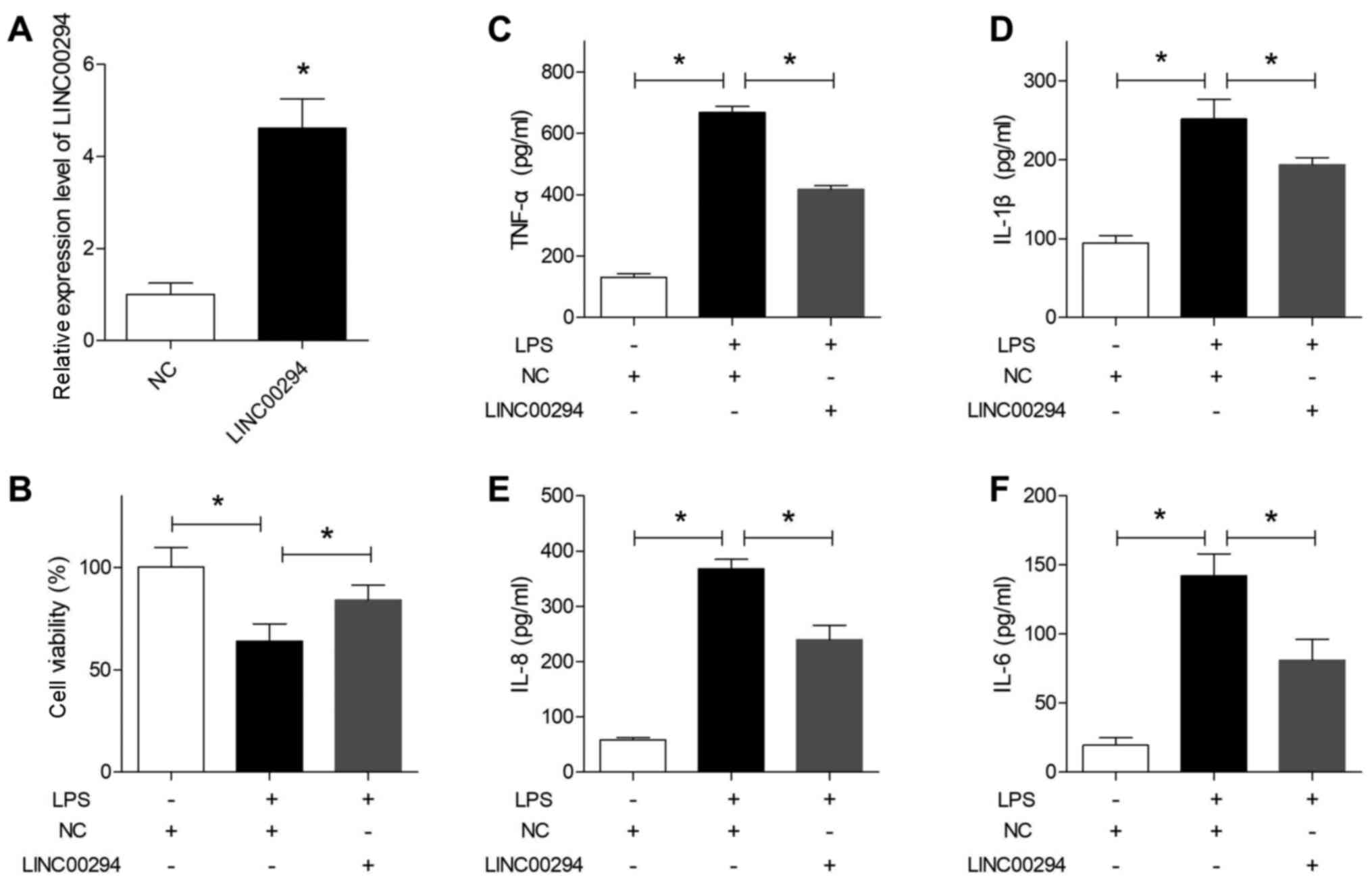
Overexpression of LINC00294 alleviates
the LPS-induced inflammatory damage in HUVECs
The RT-qPCR results indicated that LINC00294 was
successfully overexpressed in HUVECs post-transfection with the
LINC00294 plasmid (Fig. 6A).
Subsequently, cells were treated with LPS and subjected to an MTT
assay, which indicated that overexpression of LINC00294
significantly reversed the survival inhibition induced by LPS,
compared with cells in the NC group (Fig. 6B). Furthermore, ELISA indicated that
the levels of the four proinflammatory factors TNF-α, IL-1β, IL-8
and IL-6 were all significantly decreased in HUVECs overexpressing
LINC00294 (Fig. 6C-F), suggesting
that upregulation of LINC00294 significantly alleviated the
inflammatory damage in HUVECs induced by LPS.
Discussion
To the best of our knowledge, the present study was
the first to assess the effects of Bak-IIIa treatment on
LPS-induced inflammatory damage in HUVECs and the lncRNAs involved.
The present results indicated that Bak-IIIa ameliorated
inflammatory damage in HUVECs.
Vascular inflammation is usually caused by
endothelial injury, and thus, prevention of endothelial injury may
be considered a key step toward limiting the development of
vascular inflammatory processes. LPS acts as a common exogenous
factor that stimulates endothelial injury (21). Firstly, LPS treatment generally
increases cell permeability and impairs the endothelial barrier,
which constitutes an essential step toward the initiation of
inflammation (22,23). Secondly, exposure of endothelial
cells to LPS leads to inflammation via the production of
proinflammatory cytokines/chemokines, cell adhesion molecules and
nitric oxide (NO) (24). Hence,
LPS-induced inflammatory damage in HUVECs has been widely used as a
model for developing potentially effective compounds, as well as
for exploring the underlying mechanisms (25). In the present study, HUVECs were
exposed to LPS, following which the levels of TNF-α, IL-1β, IL-8
and IL-6 were all significantly enhanced, thereby substantiating
the results of previous studies (26,27).
TNF-α and IL-1β are two predominant proinflammatory factors
involved in the pathogenesis of vascular inflammation. Firstly,
increased levels of TNF-α may promote the expression of the
inducible form of NO synthase, which in turn can lead to excessive
synthesis of NO, resulting in endothelial dysfunction, apoptosis
and vascular damage (28).
Secondly, IL-1β can stimulate the synthesis of monocyte
chemoattractant protein 1, resulting in the adhesion of monocytes
and the subsequent induction of proliferative, proinflammatory
vascular smooth muscle cell phenotypes, thereby initiating the
pathological progression of vascular inflammation (29). In the present study, the levels of
TNF-α and IL-1β, as well as two other proinflammatory factors, IL-8
and IL-6, were all significantly decreased following the treatment
of HUVECs with Bak-IIIa, which effectively inhibited vascular
inflammation.
In order to clarify the potential mechanism
underlying the effect of Bak-IIIa on damaged HUVECs, the DELs
present in the two groups were analyzed and 70 were identified.
lncRNAs may regulate gene expression in a cis- or
trans-acting manner (30).
The difference between these two mechanisms is that
cis-acting lncRNAs generally influence the expression of
nearby genes, whereas trans-acting lncRNAs usually leave the
transcription site to execute an array of cellular functions by
interacting with promoters, enhancers or other proteins (30). Therefore, as trans-targets of
lncRNAs are always abundant, potential ways of interacting with
these are diverse. The lncRNA target predictions in the present
study indicated that 12 DELs covered a total of 386
trans-targets, suggesting a predominant role for these 12
DELs in the amelioration of inflammatory damage in HUVECs by
Bak-IIIa. Furthermore, a bioinformatics-based analysis of
trans-targets was performed to determine the DELs that may
be potentially involved. However, the GO and KEGG pathway
enrichment analysis results only indicated those that were
significant (P<0.5); thus, it was difficult to establish certain
associations between these and inflammation. Therefore,
‘inflammation’ and ‘inflammatory’ were searched in the
bioinformatics analyses of trans-targets; this resulted in
the screening out of 17 involved targets and six candidate
DELs.
Further validation via RT-qPCR and ELISA indicated
that upregulation of LINC00294 in the Bak-IIIa group was an
important mechanism underlying the ameliorative effect of Bak-IIIa
on LPS-induced survival inhibition and inflammatory damage in
HUVECs. Among the 17 inflammation-related targets, nine were
targets of LINC00294. C-C motif chemokine ligand 5 (CCL5) is a key
proinflammatory chemokine expressed in endothelial cells, smooth
muscle cells and macrophages, which has been reported to activate
various inflammation-related pathological processes, including
those of vascular inflammation (31,32).
CCL5 may be involved in emodin-alleviated cytotoxicity and high
glucose-induced inflammatory events in HUVECs (33), indicating that CCL5 may potentially
regulate viability and inflammatory damage in HUVECs. Furthermore,
high mobility group box 1 (HMGB1), a ubiquitously expressed nuclear
protein, has been reported to serve a predominantly proinflammatory
role in the progression of inflammation (34). Several studies confirmed that HMGB1
may have an important regulatory role in vascular endothelial
inflammation (35,36). Therefore, it may reasonably be
surmised that CCL5 and HMGB1 serve a role in Bak-IIIa-alleviated
survival inhibition and inflammatory damage in HUVECs; however,
this requires further study.
Although this study focused on the
trans-targets of DELs, KEGG pathway-based enrichment
analyses of cis-targets provided further leads regarding
potential mechanisms that may be involved. AMPK acts as a pivotal
cellular energy sensor in the regulation of bioenergy metabolism
(37), and exerts anti-inflammatory
and antioxidative effects (38).
AMPK is widely expressed in the endothelium, and modulates vascular
functions via both endothelium-dependent and -independent
mechanisms (39). Previous reports
have indicated that activation of the AMPK signaling pathway could
serve an important role in the amelioration of endothelial
inflammation and barrier dysfunction (40,41).
Hence, the AMPK signaling pathway and the cis-targets
adrenoceptor alpha 1A, tuberous sclerosis 2 and stearoyl-CoA
desaturase (SCD) that are enriched in it, as well as the
corresponding DELs, lnc-PNMA2-5:1, ENST00000570072 and lnc-SCD-1:8,
may serve a role in the alleviation of inflammatory damage in
HUVECs via Bak-IIIa.
To date, studies focusing on Bak-IIIa are very
limited, and one previous study revealed that the protective effect
of Bak-IIIa against cerebral damage were induced through the
inactivation of NF-κB signaling (10), which is not only involved in the
pathogenesis of several diseases but also has an important role in
transcriptional regulation. Increasing evidence has uncovered that
NF-κB could be the downstream effector of lncRNAs, but in some
instances, NF-κB may also be able to regulate lncRNA expression
(42). For example, cardiac and
apoptosis-related lncRNA was revealed to be translocated to the
cytoplasm after NF-κB activation in macrophages, and its expression
levels were increased in an NF-κB dependent manner (43). NF-κB has also been shown to bind to
the HOX antisense intergenic RNA promoter and induce its expression
by 16-fold in ovarian cancer cells leading to the maintenance of
DNA damage response (44).
Moreover, other transcription factors, including p53 and FOXC1,
have been reported to regulate lncRNA expression in a direct or
indirect manner (45,46). Hence, the increased level of
LINC00294 in injured-HUVECs treated with Bak-IIIa may be attributed
to the potential regulatory role of NF-κB, p53 or other
transcription factors.
There are some limitations in the present study.
First, apoptosis often occurs alongside inflammation in HUVECs
stimulated by LPS (47); however,
the role of Bak-IIIa in apoptosis of injured HUVECs was not
investigated. Secondly, the underlying mechanisms by which Bak-IIIa
induced the expression of LINC00294 and how LINC00294 protected
HUVECs remain unknown; therefore, further studies are required.
In conclusion, the present findings indicated that
Bak-IIIa ameliorated LPS-induced inflammatory damage in HUVECs by
upregulating LINC00294, and that Bak-IIIa may have potential as a
therapeutic agent against vascular inflammation.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Project from Shanghai
Natural Science Fund Project (grant no. 19ZR1449000), the Training
Program of Shanghai Tongji Hospital (grant no. GJPY1812), the
National Natural Science Foundation of China (grant no. 81702872)
and the Program of Outstanding Young Scientists of Tongji Hospital
of Tongji University (grant no. HBRC1808).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CF and SY worked on the conception and design of the
study. JX and HF performed the experiments. JX, LM and HT collected
and analyzed data. CF, SY and JX confirmed the authenticity of all
the raw data. All authors contributed to the preparation of the
manuscript, and read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Roth GA, Johnson C, Abajobir A, Abd-Allah
F, Abera SF, Abyu G, Ahmed M, Aksut B, Alam T, Alam K, et al:
Global, regional, and national burden of cardiovascular diseases
for 10 causes, 1990 to 2015. J Am Coll Cardiol. 70:1–25. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wolf MP and Hunziker P: Atherosclerosis:
Insights into vascular pathobiology and outlook to novel
treatments. J Cardiovasc Transl Res. 13:744–757. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tabas I, Garcia-Cardeña G and Owens GK:
Recent insights into the cellular biology of atherosclerosis. J
Cell Biol. 209:13–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Libby P, Buring JE, Badimon L, Hansson GK,
Deanfield J, Bittencourt MS, Tokgözoğlu L and Lewis EF:
Atherosclerosis. Nat Rev Dis Primers. 5:562019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng G, Liu Y, Lou C and Yang H: Emodin
suppresses lipopolysaccharide-induced pro-inflammatory responses
and NF-κB activation by disrupting lipid rafts in CD14-negative
endothelial cells. Br J Pharmacol. 161:1628–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeng XK, Guan YF, Remick DG and Wang X:
Signal pathways underlying homocysteine-induced production of MCP-1
and IL-8 in cultured human whole blood. Acta Pharmacol Sin.
26:85–91. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moroni F, Ammirati E, Norata GD, Magnoni M
and Camici PG: The role of monocytes and macrophages in human
atherosclerosis, plaque neoangiogenesis, and atherothrombosis.
Mediators Inflamm. 2019:74343762019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pei L, Dai D, Bao Y, Chen F, Zheng J, Li
J, Liu S and Chen X: Determination of bakkenolide A in rat plasma
using liquid chromatography/tandem mass spectrometry and its
application to a pharmacokinetic study. J Mass Spectrom.
47:962–968. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang YL, Li RP, Guo ML, Zhang G, Zhang N
and Ma YL: Bakkenolides from Petasites tricholobus and their
neuroprotective effects related to antioxidant activities. Planta
Med. 75:230–235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang Q, Li RP, Tang Y, Wang YQ, Liu C and
Guo ML: Bakkenolide-IIIa protects against cerebral damage via
inhibiting NF-κB activation. CNS Neurosci Ther. 21:943–952. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Skaper SD, Facci L, Zusso M and Giusti P:
An inflammation-centric view of neurological disease: Beyond the
neuron. Front Cell Neurosci. 12:722018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aryal B and Suárez Y: Non-coding RNA
regulation of endothelial and macrophage functions during
atherosclerosis. Vascul Pharmacol. 114:64–75. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang HN, Xu QQ, Thakur A, Alfred MO,
Chakraborty M, Ghosh A and Yu XB: Endothelial dysfunction in
diabetes and hypertension: Role of microRNAs and long non-coding
RNAs. Life Sci. 213:258–268. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu X, Chen D, Liu Y, Yu J, Qiao L, Lin S,
Chen D, Zhong G, Lu X, Wang Y, et al: Long noncoding RNA HOXA-AS3
integrates NF-κB signaling to regulate endothelium inflammation.
Mol Cell Biol. 39:e00139–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao L, Zhang Z, Li Y, Zhao P and Chen Y:
LncRNA H19/miR-let-7 axis participates in the regulation of
ox-LDL-induced endothelial cell injury via targeting periostin. Int
Immunopharmacol. 72:496–503. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu X, Liu Y, Yu J, Du J, Guo R, Feng Y,
Zhong G, Jiang Y and Lin J: LncRNA HOXA-AS2 represses endothelium
inflammation by regulating the activity of NF-κB signaling.
Atherosclerosis. 281:38–46. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li S, Sun Y, Zhong L, Xiao Z, Yang M, Chen
M, Wang C, Xie X and Chen X: The suppression of ox-LDL-induced
inflammatory cytokine release and apoptosis of HCAECs by long
non-coding RNA-MALAT1 via regulating microRNA-155/SOCS1 pathway.
Nutr Metab Cardiovasc Dis. 28:1175–1187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Casper J, Zweig AS, Villarreal C, Tyner C,
Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et
al: The UCSC genome browser database: 2018 update. Nucleic Acids
Res. 46D:D762–D769. 2018. View Article : Google Scholar
|
|
19
|
Tafer H and Hofacker IL: RNAplex: A fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song L, Kang C, Sun Y, Huang W, Liu W and
Qian Z: Crocetin inhibits lipopolysaccharide-induced inflammatory
response in human umbilical vein endothelial cells. Cell Physiol
Biochem. 40:443–452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin X, Liang Z, Yun Y and Pei L:
Intravenous transplantation of BMP2-transduced endothelial
progenitor cells attenuates lipopolysaccharide-induced acute lung
injury in rats. Cell Physiol Biochem. 35:2149–2158. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Uhlig S, Yang Y, Waade J, Wittenberg C,
Babendreyer A and Kuebler WM: Differential regulation of lung
endothelial permeability in vitro and in situ. Cell Physiol
Biochem. 34:1–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim JH, Jeong JH, Jeon ST, Kim H, Ock J,
Suk K, Kim SI, Song KS and Lee WH: Decursin inhibits induction of
inflammatory mediators by blocking nuclear factor-kappaB activation
in macrophages. Mol Pharmacol. 69:1783–1790. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang M, Pan H, Xu Y, Wang X, Qiu Z and
Jiang L: Allicin decreases lipopolysaccharide-induced oxidative
stress and inflammation in human umbilical vein endothelial cells
through suppression of mitochondrial dysfunction and activation of
Nrf2. Cell Physiol Biochem. 41:2255–2267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hou X, Yang S and Yin J: Blocking the
REDD1/TXNIP axis ameliorates LPS-induced vascular endothelial cell
injury through repressing oxidative stress and apoptosis. Am J
Physiol Cell Physiol. 316:C104–C110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Makó V, Czúcz J, Weiszhár Z, Herczenik E,
Matkó J, Prohászka Z and Cervenak L: Proinflammatory activation
pattern of human umbilical vein endothelial cells induced by IL-1β,
TNF-α, and LPS. Cytometry A. 77:962–970. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mangoni AA, Zinellu A, Sotgia S, Carru C,
Piga M and Erre GL: Protective effects of methotrexate against
proatherosclerotic cytokines: A review of the evidence. Mediators
Inflamm. 2017:96328462017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khan R, Rheaume E and Tardif JC: Examining
the role of and treatment directed at IL-1β in atherosclerosis.
Curr Atheroscler Rep. 20:532018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marques RE, Guabiraba R, Russo RC and
Teixeira MM: Targeting CCL5 in inflammation. Expert Opin Ther
Targets. 17:1439–1460. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nosalski R and Guzik TJ: Perivascular
adipose tissue inflammation in vascular disease. Br J Pharmacol.
174:3496–3513. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao Y, Zhang J, Li G, Xu H, Yi Y, Wu Q,
Song M, Bee YM, Huang L, Tan M, et al: Protection of vascular
endothelial cells from high glucose-induced cytotoxicity by emodin.
Biochem Pharmacol. 94:39–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sachdev U and Lotze MT: Perpetual change:
Autophagy, the endothelium, and response to vascular injury. J
Leukoc Biol. 102:221–235. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cai W, Duan XM, Liu Y, Yu J, Tang YL, Liu
ZL, Jiang S, Zhang CP, Liu JY and Xu JX: Uric acid induces
endothelial dysfunction by activating the HMGB1/RAGE signaling
pathway. Biomed Res Int. 2017:43919202017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang ST, Wang F, Shao M, Wang Y and Zhu
HQ: MicroRNA-126 suppresses inflammation in endothelial cells under
hyperglycemic condition by targeting HMGB1. Vascul Pharmacol.
88:48–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim WH, Lee JW, Suh YH, Lee HJ, Lee SH, Oh
YK, Gao B and Jung MH: AICAR potentiates ROS production induced by
chronic high glucose: Roles of AMPK in pancreatic beta-cell
apoptosis. Cell Signal. 19:791–805. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bijland S, Mancini SJ and Salt IP: Role of
AMP-activated protein kinase in adipose tissue metabolism and
inflammation. Clin Sci (Lond). 124:491–507. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ewart MA and Kennedy S: AMPK and
vasculoprotection. Pharmacol Ther. 131:242–253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu R, Wang MQ, Ni SH, Wang M, Liu LY, You
HY, Wu XH, Wang YJ, Lu L and Wei LB: Salidroside ameliorates
endothelial inflammation and oxidative stress by regulating the
AMPK/NF-κB/NLRP3 signaling pathway in AGEs-induced HUVECs. Eur J
Pharmacol. 867:1727972020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dennhardt S, Finke KR, Huwiler A and
Coldewey SM: Sphingosine-1-phosphate promotes barrier-stabilizing
effects in human microvascular endothelial cells via AMPK-dependent
mechanisms. Biochim Biophys Acta Mol Basis Dis. 1865:774–781. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta SC, Awasthee N, Rai V, Chava S,
Gunda V and Challagundla KB: Long non-coding RNAs and nuclear
factor-κB crosstalk in cancer and other human diseases. Biochim
Biophys Acta Rev Cancer. 1873:1883162020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Castellanos-Rubio A, Kratchmarov R,
Sebastian M, Garcia-Etxebarria K, Garcia L, Irastorza I and Ghosh
S: Cytoplasmic form of carlr lncRNA facilitates inflammatory gene
expression upon NF-κB activation. J Immunol. 199:581–588. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Özeş AR, Miller DF, Özeş ON, Fang F, Liu
Y, Matei D, Huang T and Nephew KP: NF-κB-HOTAIR axis links DNA
damage response, chemoresistance and cellular senescence in ovarian
cancer. Oncogene. 35:5350–5361. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen S, Thorne RF, Zhang XD, Wu M and Liu
L: Non-coding RNAs, guardians of the p53 galaxy. Semin Cancer Biol.
Sep 11–2020.(Epub ahead of print). doi:
10.1016/j.semcancer.2020.09.002. View Article : Google Scholar
|
|
46
|
Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo
Y, Zhang H, Wang J, Zhang Y, Huang SX, et al: FOXC1-mediated
LINC00301 facilitates tumor progression and triggers an
immune-suppressing microenvironment in non-small cell lung cancer
by regulating the HIF1α pathway. Genome Med. 12:772020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jung TW, Park HS, Choi GH, Kim D, Ahn SH,
Kim DS, Lee T and Jeong JH: Maresin 1 attenuates pro-inflammatory
reactions and ER stress in HUVECs via PPARα-mediated pathway. Mol
Cell Biochem. 448:335–347. 2018. View Article : Google Scholar : PubMed/NCBI
|