Introduction
Currently, the occurrence of liver cancer,
particularly hepatocellular carcinoma (HCC), is increasing
worldwide (1), and liver cancer is
the third most common cause of cancer-related mortality (2). Compared with that of the general
population, chronic hepatitis B virus (HBV) carriers have a higher
risk of contracting HCC, and HBV is the most direct cause of HCC
(3). The HBV DNA molecule, which is
a circular double-stranded DNA containing part of a single-stranded
region, transforms into covalently closed circular (ccc)DNA during
its life cycle (4). As a marker of
HBV infection, hepatitis B e-antigen (HBeAg) positivity is
associated with high viral loads, which further leads to high
expression levels of hepatitis B surface antigen (HBsAg) (5). Therefore, it is urgent to investigate
novel biomarkers for the development and metastasis of HBV-related
liver cancer from the influence of antigen activity and DNA
replication.
MicroRNAs (miRNAs or miRs), a type of small,
non-coding RNA of 18~25 nucleotides in length, mainly regulate mRNA
expression by modulating their translation and degradation via
combining with the 3′ untranslated region (3′ UTR) of the target
mRNA (6). miRNAs partly involved in
the progress of HBV-related HCC have attracted much attention. For
example, miR-384 expression is reduced in HBV-induced HCC, and its
dysregulation has acute effects on proliferation, metastasis and
lipogenesis (7). miR-204 and
miR-1236 are downregulated in HBV-producing cells, and their
overexpression can inhibit HBV DNA replication (8). miR-106b, which was observed to be
upregulated in patients with HCC by miRNA array analysis, is
associated with poor prognosis of patients with HBV-associated HCC
(9). These data indicate that
different miRNAs have variable regulatory efficacy in HBV-HCC. The
role of miR-1271-5p is partly reported in HCC (10). However, its role in HBV-associated
liver cancer remains to be elucidated.
Aquaporins (AQPs) are transport channels that
promote water permeation through cell membranes (11). Water channels serve vital roles in
numerous biological processes, particularly the maintenance of
water homeostasis (12). AQP5, a
member of the AQP family, is a primary water-selective channel.
Emerging data suggest that AQP5 expression may be involved in the
growth and development of a number of systemic malignancies. For
instance, AQP5 is overexpressed in squamous cell carcinoma
(12), non-small cell lung
(13), lung (14) and breast cancer (15). Thus, AQP5 may function as an
oncogene in numerous types of cancer. However, studies on AQP5
expression and function in HBV-associated liver cancer remain
limited.
In the present study, the abundance of miR-1271-5p
and AQP5 was measured in HBV-related liver cancer tissues and
cells. The role of miR-1271-5p was analyzed according to the levels
of HBeAg and HBsAg, HBV DNA replication, cell viability, apoptosis,
migration and invasion in vitro, as well as tumor growth
in vivo. The interaction between miR-1271-5p and AQP5 was
verified, thus providing a potential regulatory mechanism of
miR-1271-5p.
Materials and methods
Specimens
The present study was approved by the Ethics
Committee of The Second Affiliated Hospital of Shandong First
Medical University (Tai'an, China). A total of 32 HBV-induced HCC
tissues and paired adjacent healthy tissues were collected from The
Second Affiliated Hospital of Shandong First Medical University
from October 2015 to February 2018. All samples were placed
immediately in liquid nitrogen and then stored at −80°C until use.
All patients (including 20 males and 12 females, aged 36–62 years)
had signed the informed consent form before tissue resection.
Cell lines and culture
The hepatoblastoma cell line HepG2.2.15 and the HCC
cell line Huh7 were purchased from the BeNa Culture Collection. The
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS), 100 µg/ml
streptomycin and 100 U/ml penicillin (all from Gibco; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2.
Cell transfection
To mimic HBV infection, a plasmid with 1.3 mer HBV
genomic DNA (pcDNA3.1-1.3 mer) was transfected into Huh7 cells
using FuGENE® HD Transfection reagent (Promega
Corporation), thus generating the HBV-HCC cell line Huh7-1.3.
miR-1271-5p mimic (miR-1271-5p; 40 nM;
5′-CUUGGCACCUAGCAAGCACUCA-3′) or miR-1271-5p inhibition
(anti-miR-1271-5p; 60 nM; 5′-UGAGUGCUUGCUAGGUGCCAAG-3′) and their
corresponding control miR-negative control (NC) or anti-miR-NC were
purchased from Guangzhou RiboBio Co., Ltd. The small interference
(si)RNA against AQP5 (si-AQP5; 2.5 µg;
5′-GACAGACUGGUUCAUUGAAUG-3′), the scrambled siRNA (Scramble; 2.5
µg), pcDNA3.1-AQP5 (AQP5; 1 µg) and pcDNA3.1 empty vector (vector;
1 µg) were all obtained from Shanghai GenePharma Co., Ltd. These
transfection assays were conducted using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Blank transfection served as the control.
The experiments described below were performed at 48 h after
transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from tissues or cells
(5×106 cells) using TRIzol® reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
cDNA was synthesized using miRNA cDNA Synthesis kit (Applied
Biological Materials Inc.) for miR-1271-5p or PrimeScript RT
Reagent kit (Takara Bio, Inc.) for AQP5 according to the
manufacturer's protocol using the following reaction conditions:
42°C for 15 min and 85°C for 5 min. qPCR was performed using TB
Green Premix Ex Taq II (Takara Bio, Inc.) on an ABI 7900 (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The thermocycling conditions were as
follows: Denaturation at 95°C for 30 sec, 95°C for 5 sec and 60°C
for 1 min for a total of 40 cycles. GAPDH or small nuclear RNA U6
acted as an internal reference for AQP5 or miR-1271-5p,
respectively. The relative expression was calculated using the
2−ΔΔCq method (16). The
primers used were: miR-1271-5p, forward,
5′-CTTGGCACCTAGCAAGCACTCA-3′ and reverse,
5′-GCGAGCACAGAATTAATACGAC-3′; AQP5, forward,
5′-GCCATCCTCTATTTCTACCTGC-3′ and reverse,
5′-GCTTCAAACTCTTCGTCTTCC-3′; GAPDH, forward,
5′-CATCACCATCTTCCAGGAGCG-3′ and reverse,
5′-TGACCTTGCCCACAGCCTTG-3′; and U6, forward,
5′-AGAGCCTGTGGTGTCCG-3′ and reverse,
5′-CATCTTCAAAGCACTTCCCT-3′.
ELISA
ELISA was employed to analyze the levels of HBsAg
and HBeAg using ELISA kits (cat. nos. INS1030201 and INS1030203;
Huangshi Irons Biotechnology Co., Ltd.) according to the
manufacturer's protocols. Antibodies against HBsAg (1:2,000; cat.
no. ab9193; Abcam) and HBeAg (1:2,000; cat. no. ab91273; Abcam)
were used for ELISA. The inhibitory rate was analyzed according to
the formula: Inhibitory rate
(%)=(Ccontrol-Ctested)/Ccontrol
×100%, where C is the concentration.
Detection of HBV DNA level
Viral DNA was isolated from transfected HepG2.2.15
and Huh7-1.3 cells using QIAamp DNA Mini kits (Qiagen GmbH)
according to the manufacturer's protocols. An HBV PCR Fluorescence
Quantitative Detection kit (Hangzhou Bioer Co., Ltd.) was used for
quantitative detection of HBV DNA by quantitative PCR in accordance
with the manufacturer's protocols. The average Cq values were used
to ascertain the level of HBV DNA. The calculation of the
inhibitory rate was performed as aforementioned.
Cell Counting Kit-8 (CCK-8) assay
To evaluate cell viability, a CCK-8 reagent
(Beyotime Institute of Biotechnology) was used in accordance with
the manufacturer's protocols. In brief, transfected Huh7-1.3 and
HepG2.2.15 cells were seeded in a 96-well plate (Corning Inc.
2×103 cells/well) for 24, 48 and 72 h. Cells at
different stages were collected and treated with the CCK-8 solution
for another 2 h at room temperature. The absorbance at 450 nm was
measured using a microplate reader (Thermo Fisher Scientific, Inc.)
at room temperature.
Flow cytometry assay
For apoptosis, an Annexin V-FITC/PI kit (BD
Pharmingen; BD Biosciences) was used to perform the flow cytometry
assay according to the manufacturer's protocol. Transfected cells
(5×104) were digested with 0.25% trypsin, washed with
PBS and fixed in 70% cold ethanol at 4°C for 12 h. Upon washing
with PBS, the cells were stained with FITC-Annexin V and PI for 30
min in the dark. Finally, fluorescence was recorded with a CytoFLEX
flow cytometer (Beckman Coulter, Inc.), and the apoptotic cells
were analyzed using Kaluza software (version 2.1; Beckman Coulter,
Inc.). The apoptotic cells was calculated as the percentage of late
(Annexin V-FITC)+/PI+ and early apoptotic
cells (Annexin V-FITC)+/PI−.
Western blot analysis
RIPA lysis solution (Beyotime Institute of
Biotechnology) was used to extract total proteins from cells or
tumor tissue. After quantifying the concentration of the lysates
using BCA Protein Assay kit (Beyotime Institute of Biotechnology),
the proteins (40 µg) were separated by 12% SDS-PAGE and transferred
onto PVDF membranes (Bio-Rad Laboratories, Inc.) on ice.
Subsequently, the membranes were blocked with 5% skimmed milk at
room temperature for 1 h and then incubated with primary antibodies
against Bax (1:1,000; cat. no. ab182734), Bcl-2 (1:1,000; cat. no.
ab194583), cleaved-caspase-3 (1:500; cat. no. ab49822), AQP5
(1:2,000; cat. no. ab92320), proliferating cell nuclear antigen
(PCNA; 1:2,000; cat. no. ab29) and GAPDH (1:10,000; cat. no.
ab8245; all from Abcam) overnight at 4°C. On the following day, the
membranes were incubated with a secondary antibody conjugated to
horseradish peroxidase (1:5,000; cat. nos. ab205718 and ab205719;
Abcam) at room temperature for 2 h after washing with TBS. The
blots were visualized by an enhanced chemiluminescence kit
(Beyotime Institute of Biotechnology), and images were captured for
densitometry using ImageJ software (version 1.46; National
Institutes of Health).
Transwell assay
A part of the upper surface of Transwell chambers (8
µm) was treated with Matrigel (Corning Inc.) at 4°C overnight
before the experiment. Transfected HepG2.2.15 and Huh7-1.3 cells
were collected by centrifugation (326 × g; 5 min; 4°C) and
resuspended in serum-free DMEM. The cells (5×104) were
transferred to the top of Transwell chambers without Matrigel for
migration assays or to the top of Transwell chambers containing
Matrigel for invasion assays. The bottom of the Transwell chambers
was filled with DMEM containing 10% FBS. After incubating for 24 h,
the migrated or invaded cells through the membrane were fixed with
95% ethanol for 15 min and stained with 0.1% crystal violet for 20
min at room temperature. Cells passing through the basement
membranes were counted using a Leica DC 300F light microscope
(magnification, ×100; Leica Microsystems GmbH).
Prediction of target genes
The online software TargetScan Human 7.2 (targetscan.org/vert_72/) was used to predict the
possible target genes of miR-1271-5p and to analyze the binding
sites between them.
Dual-luciferase reporter assay
The luciferase reporter vector PGL3-basic (Promega
Corporation) was employed to verify the association between
miR-1271-5p and AQP5. Specifically, PGL3-AQP5 wild-type (wt) fusion
vector containing AQP5 3′ UTR wt sequence and PGL-AQP5 mutant (mut)
fusion vector containing AQP5 3′ UTR mut sequence were assembled
and transfected into HepG2.2.15 and Huh7-1.3 cells together with
miR-1271-5p, miR-NC, anti-miR-1271-5p or anti-miR-NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the luciferase activity was
estimated using the Dual-Luciferase Reporter Assay System (Promega
Corporation), according to the manufacturer's protocol. Relative
luciferase activity was assessed by comparison with Renilla
luciferase activity.
RNA binding protein
immunoprecipitation (RIP) assay
HepG2.2.15 and Huh7-1.3 cells (1×105)
transfected with miR-1271-5p or miR-NC were collected and lysed
with RIP lysis buffer containing protease inhibitor and RNase
inhibitor from an EZ-Magna RIP kit (EMD Millipore). The above
reactant was incubated with magnetic beads combined with an
antibody against Ago2 or with an lgG control for 6 h at 4°C.
Finally, the reaction products were purified and analyzed by
RT-qPCR to monitor the enrichment of AQP5.
In vivo experiments
The animal experiments were approved by the
Institutional Animal Care and Use Committee of The Second
Affiliated Hospital of Shandong First Medical University. In brief,
12 BALB/c nude mice (male, 6-week old, mean weight 14.6 g)
purchased from Shanghai SLAC Laboratory Animal Co., Ltd. were
divided into 2 groups (n=6) and housed in pathogen-free conditions
(temperature, 23°C; humidity, 55%; light/dark cycle, 12/12h). The
right flank of the back was subcutaneously injected with HepG2.2.15
cells (5×106) transfected with lentiviral vectors with
miR-1271-5p or miR-NC. Tumor volumes (length × width2
×0.5) were recorded every 7 days using a vernier caliper. Tumor
weight was measured for 5 weeks until the mice were sacrificed via
cervical dislocation. The mice were sacrificed when >20% body
weight was lost. The excisional tumor tissues were used for further
analyses, including RT-qPCR and western blotting as
aforementioned.
Statistical analysis
All tests were independently conducted ≥ 3 times.
Experimental data are presented as the mean ± standard deviation,
and were analyzed by SPSS 17.0 (SPSS, Inc.). Student's t-test
(paired or unpaired) was used to estimate the statistical
differences between two groups, and one-way or two-way analysis of
variance and Tukey's post hoc test were used for comparison of
multiple groups. Spearman's correlation analysis was used to assess
the correlation between miR-1271-5p and AQP5 expression. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-1271-5p is downregulated, while
AQP5 is upregulated, in HBV-HCC tissues
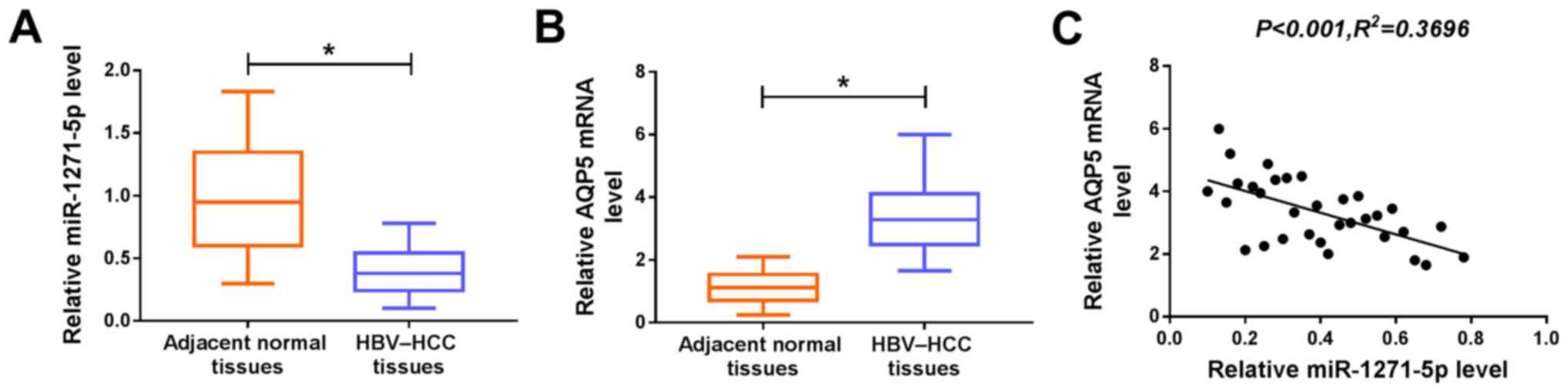
To determine the abundance of miR-1271-5p and AQP5
in HBV-HCC tissues, RT-qPCR analysis was performed to analyze their
expression. The expression of miR-1271-5p was significantly lower
in HBV-HCC tissues compared with that in adjacent normal tissues
(Fig. 1A), while the expression of
AQP5 mRNA was enhanced in HBV-HCC tissues (Fig. 1B). Correlation analysis revealed a
negative correlation between AQP5 and miR-1271-5p levels (Fig. 1C). The data suggested that
miR-1271-5p and AQP5 may serve important roles, at least in part,
in HBV-associated HCC.
Overexpression of miR-1271-5p inhibits
the infection and replication of HBV in HBV-related liver cancer
cells
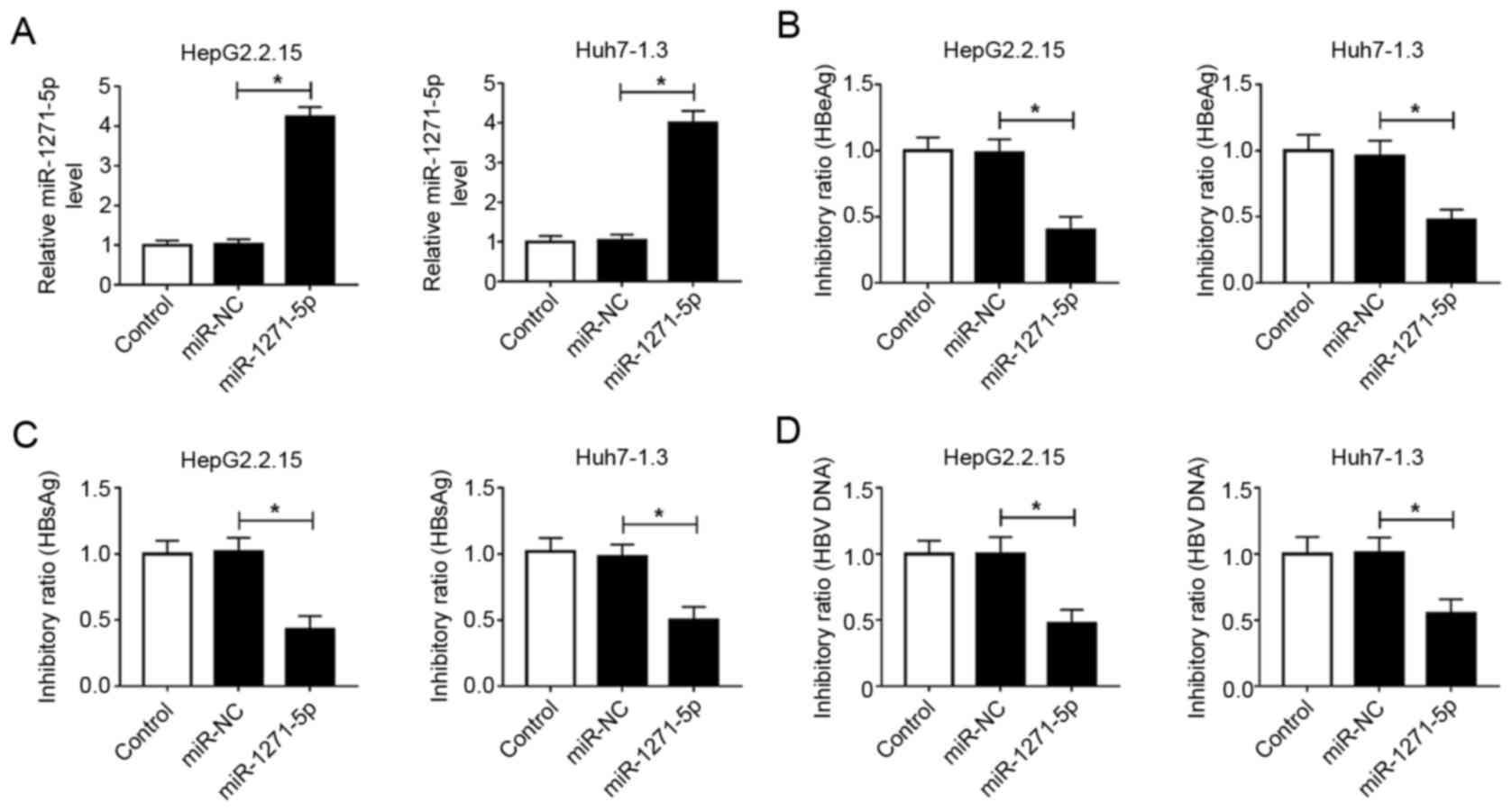
To examine the influence of miR-1271-5p on HBV
infection, analyses of antigen secretions and HBV replication were
performed. First, the efficiency of miR-1271-5p overexpression, as
detected by RT-qPCR, demonstrated that the abundance of miR-1271-5p
was increased in both HepG2.2.15 and Huh7-1.3 cells with
miR-1271-5p mimic transfection (Fig.
2A). ELISA analysis revealed that the levels of HBeAg and HBsAg
declined with the increase in miR-1271-5p compared with those in
the miR-NC group (Fig. 2B and C).
In addition, the level of HBV DNA was also notably reduced in
HepG2.2.15 and Huh7-1.3 cells transfected with miR-1271-5p mimic
compared with that of miR-NC-transfected cells (Fig. 2D). These data indicated that
miR-1271-5p enrichment contributed to alleviating HBV infection by
inhibiting the secretion of HBeAg and HBsAg and the replication of
HBV.
Overexpression of miR-1271-5p blocks
viability, migration and invasion, but induces apoptosis, in
HBV-related liver cancer cells
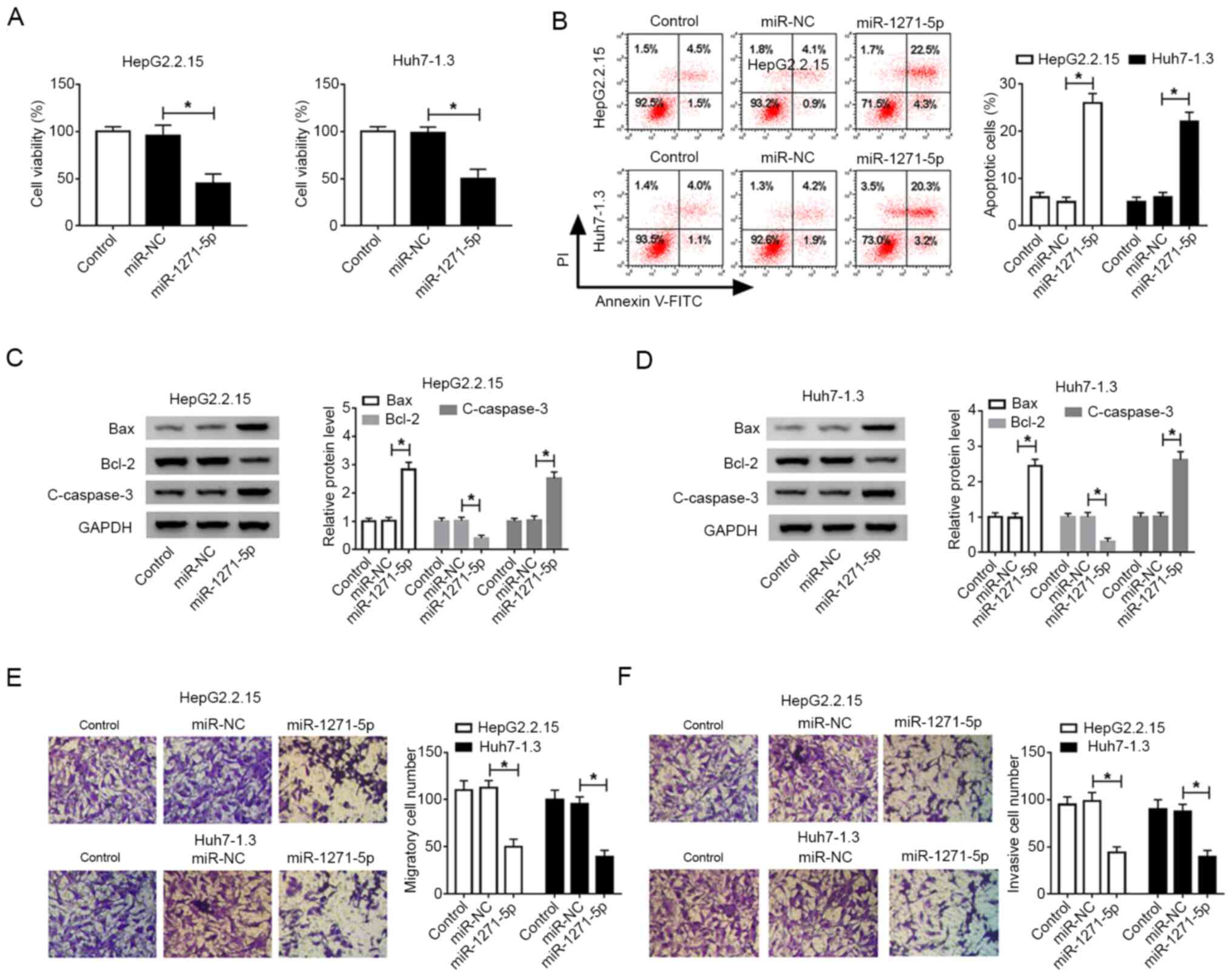
To explore the role of miR-1271-5p on the progress
of HBV-associated liver cancer, CCK-8, flow cytometry and Transwell
assays were performed on cancer cells. Cell viability was markedly
reduced by miR-1271-5p transfection both in HepG2.2.15 and Huh7-1.3
cells compared with that of miR-NC-transfected cells (Fig. 3A). By contrast, the number of
apoptotic cells was clearly increased with the accumulation of
miR-1271-5p (Fig. 3B). Several
apoptosis-related proteins were analyzed by western blotting, which
demonstrated that the levels of Bax and cleaved-caspase-3 were
enhanced, while the Bcl-2 level decreased in HepG2.2.15 and
Huh7-1.3 cells transfected with miR-1271-5p compared with that in
cells transfected with miR-NC (Fig. 3C
and D). The number of migrated and invaded cells was less in
HepG2.2.15 and Huh7-1.3 cells transfected with miR-1271-5p compared
with that of miR-NC-transfected cells (Fig. 3E and F). These data revealed that
overexpression of miR-1271-5p positively reduced the malignant
behaviors of HBV-associated liver cancer in vitro.
AQP5 knockdown reduces the levels of
HBeAg, HBsAg and HBV DNA, inhibits viability, migration and
invasion, and promotes apoptosis in cancer cells
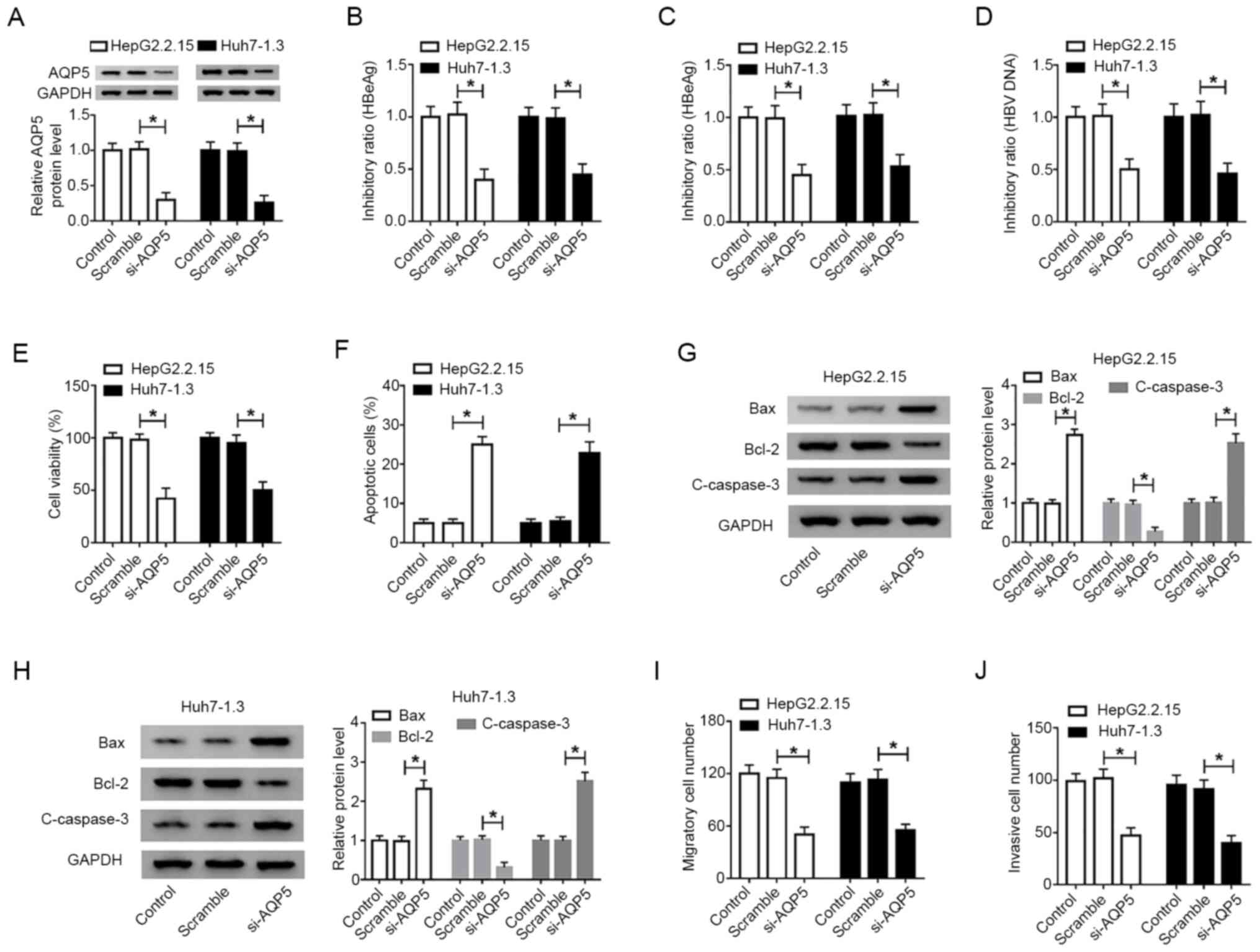
To investigate the role of AQP5 in the progress of
HBV-associated liver cancer, the expression level of AQP5 was
knocked down in cancer cells. The AQP5 protein level was decreased
in HepG2.2.15 and Huh7-1.3 cells transfected with si-AQP5 compared
that in the scramble group (Fig.
4A). ELISA suggested that the levels of HBeAg and HBsAg were
impaired with the lack of AQP5 in both HepG2.2.15 and Huh7-1.3
cells (Fig. 4B and C). Similarly,
the level of HBV DNA was reduced in cells transfected with si-AQP5
(Fig. 4D). CCK-8 assay revealed
that si-AQP5 efficiently impeded the viability of HepG2.2.15 and
Huh7-1.3 cells (Fig. 4E). Flow
cytometry revealed that si-AQP5 increased the number of apoptotic
cells (Fig. 4F). The protein levels
of Bax and cleaved-caspase-3 were enhanced, while the Bcl-2 level
was depleted, with the decrease in AQP5 in both HepG2.2.15 and
Huh7-1.3 cells (Fig. 4G and H).
Transwell assay demonstrated that the number of migrated and
invaded cells was decreased in HepG2.2.15 and Huh7-1.3 cells
transfected with si-AQP5 compared with that in the scramble group
(Fig. 4I and J). The above data
demonstrated that AQP5 knockdown attenuated malignant behaviors of
HBV-associated liver cancer in vitro.
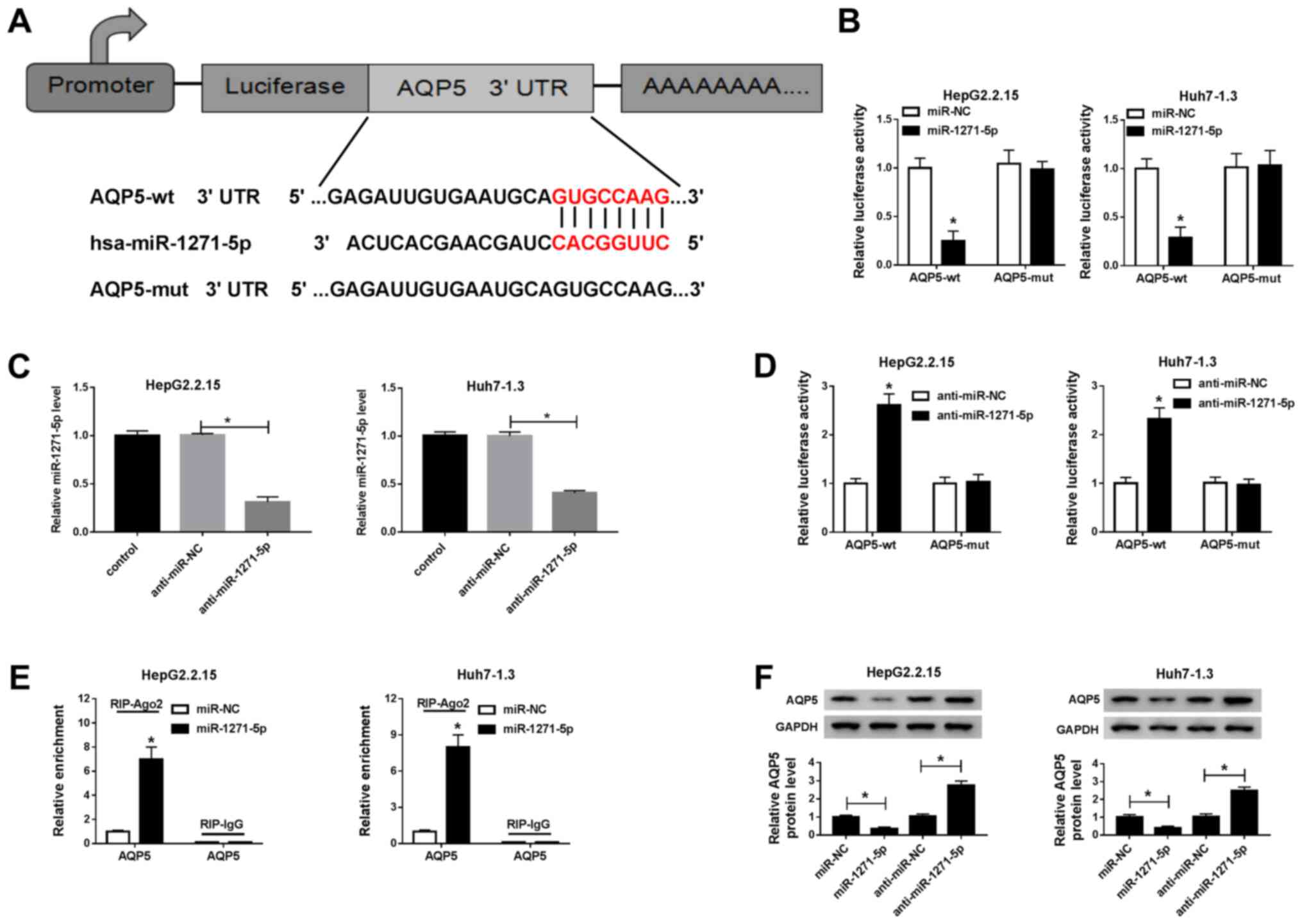
AQP5 is a target of miR-1271-5p
To understand the interaction of AQP5 with
miR-1271-5p, the putative binding sites between them were predicted
using the bioinformatics tool TargetScan. The sequences of AQP5-wt
3′ UTR and AQP5-mut 3′ UTR containing miR-1271-5p binding site or
mutated binding site are shown in Fig.
5A. Dual-luciferase reporter assay demonstrated that the
luciferase activity was considerably reduced in HepG2.2.15 and
Huh7-1.3 cells co-transfected with AQP5-wt and miR-1271-5p, whereas
the no significant difference in luciferase activity was observed
in the AQP5-mut group compared with that of the miR-NC group
(Fig. 5B). The expression levels of
miR-1271-5p were significantly decreased in HepG2.2.15 and Huh7-1.3
cells transfected with anti-miR-1271-5p compared with anti-miR-NC
(Fig. 5C). By contrast, the
inhibition of miR-1271-5p had the opposite effects to miR-1271-5p
mimic on luciferase activity in HepG2.2.15 and Huh7-1.3 cells
(Fig. 5D). Additionally, RIP assay
indicated that miR-1271-5p clearly reinforced the enrichment of
AQP5 in the RIP-Ago2 group compared with that in the RIP-lgG group
in HepG2.2.15 and Huh7-1.3 cells (Fig.
5E). The protein level of AQP5 was depleted by miR-1271-5p but
stimulated by anti-miR-1271-5p compared with their corresponding
controls in HepG2.2.15 and Huh7-1.3 cells (Fig. 5F). These data implied that AQP5 was
a direct target of miR-1271-5p and that its expression level was
regulated by miR-1271-5p.
Overexpression of AQP5 reverses the
effects of miR-1271-5p mimic in HBV-associated liver cancer
cells
To investigate the mechanism of miR-1271-5p in the
progress of HBV-mediated liver cancer, miR-1271-5p and miR-1271-5p
+ AQP5 together with their corresponding controls miR-NC and
miR-1271-5p + vector were transfected into HepG2.2.15 and Huh7-1.3
cells. The detection of AQP5 overexpression efficiency demonstrated
that AQP5 was overexpressed, since the expression of AQP5 was
significantly increased in HepG2.2.15 and Huh7-1.3 cells
transfected with AQP5 compared with that of cells transfected with
vector (Fig. S1). The data from
western blotting demonstrated that miR-1271-5p + AQP5 transfection
recovered the protein level of AQP5 that was weakened by
miR-1271-5p transfection (Fig. 6A),
suggesting that the transfection had been effective. ELISA
established that the levels of HBeAg and HBsAg suppressed by
miR-1271-5p mimic were significantly restored with the increase in
AQP5 expression (Fig. 6B and C).
Similarly, the level of HBV DNA was decreased by miR-1271-5p mimic
but increased by AQP5 overexpression (Fig. 6D). In addition, miR-1271-5p + AQP5
transfection reversed the inhibitory effect of miR-1271-5p on cell
viability (Fig. 6E). By contrast,
the apoptosis induced by miR-1271-5p was alleviated with AQP5
overexpression (Fig. 6F). Western
blot analysis demonstrated that the levels of Bax and
cleaved-caspase-3 were enhanced by miR-1271-5p but reduced by AQP5
upregulation, while the level of Bcl-2 was impaired by miR-1271-5p
but improved by AQP5 overexpression in HepG2.2.15 and Huh7-1.3
cells (Fig. 6G and H). Transwell
assay demonstrated that miR-1271-5p + AQP5 transfection reinforced
the number of migratory or invasive cells reduced by miR-1271-5p
transfection (Fig. 6I and J).
Overall, these data demonstrated that miR-1271-5p blocked the
malignant behaviors of HBV-associated liver cancer by suppressing
the expression of AQP5.
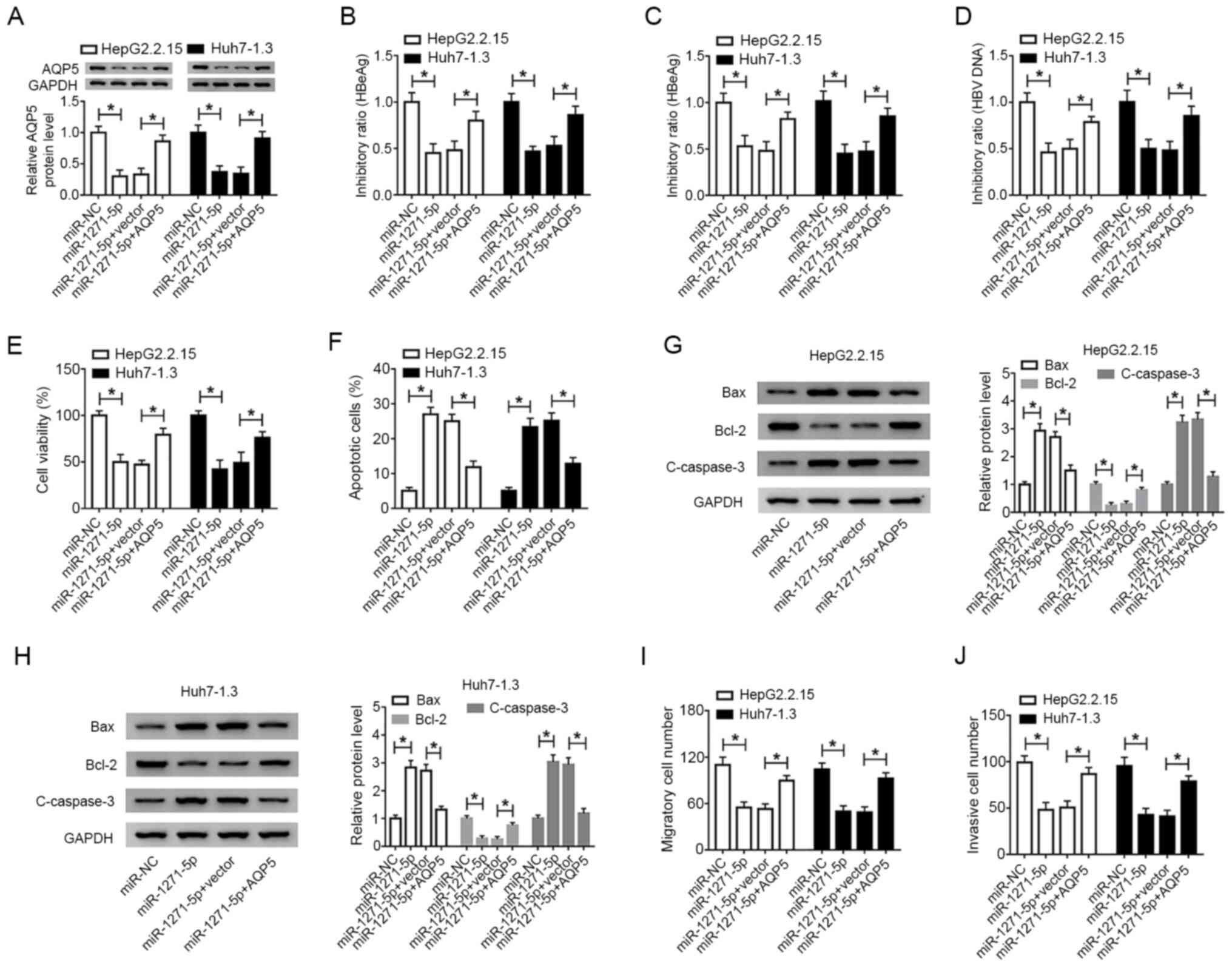 | Figure 6.miR-1271-5p regulates the levels of
HBeAg, HBsAg and HBV DNA, as well as cell viability, apoptosis,
migration and invasion, by binding to AQP5. HepG2.2.15 and Huh7-1.3
cells were transfected with miR-1271-5p, miR-NC, miR-1271-5p + AQP5
or miR-1271-5p + vector. (A) The protein level of AQP5 was detected
by western blotting. (B) HBeAg, (C) HBsAg and (D) HBV DNA levels
were detected by ELISA or quantitative PCR. (E) Cell viability was
assessed by Cell Counting Kit-8 assay. (F) Apoptosis was examined
by flow cytometry. Bax, Bcl-2 and cleaved-caspase-3 were quantified
by western blotting in (G) HepG2.2.15 and (H) Huh7-1.3 cells. (I)
Migration and (J) invasion were determined by Transwell assay.
*P<0.05. miR, microRNA; HBV, hepatitis B virus; AQP5, aquaporin
5; NC, negative control; C-caspase-3, cleaved-caspase-3. |
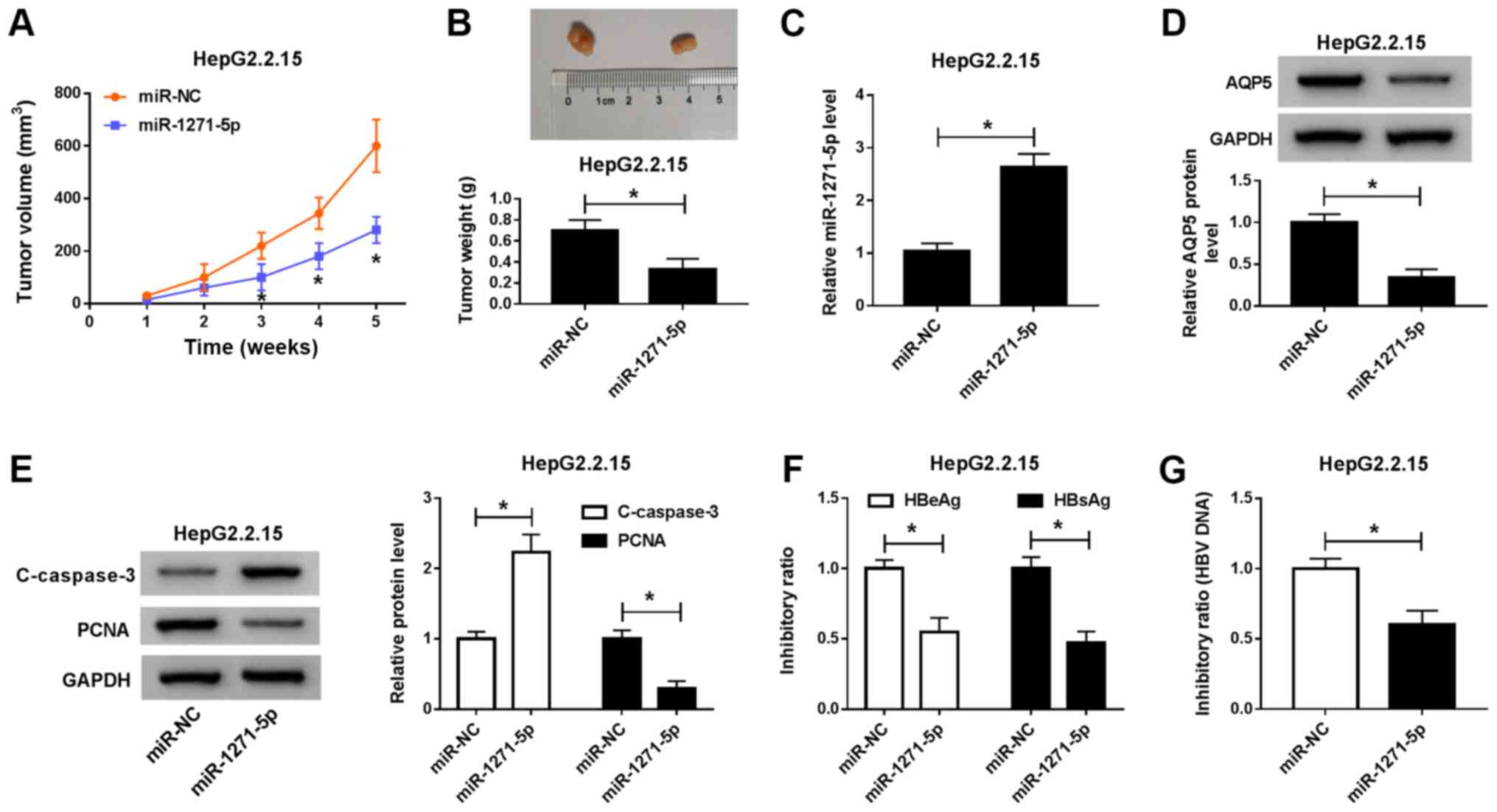
Overexpression of miR-1271-5p
obstructs tumor growth in vivo
To further validate the function of miR-1271-5p on
tumor growth in vivo, a nude mouse tumorigenicity assay was
performed. HepG2.2.15 cells transfected with miR-1271-5p or miR-NC
were subcutaneously injected into the nude mice. The results
demonstrated that tumor volume was lower in the miR-1271-5p group
compared with that in the miR-NC group from week 1 until the mice
were sacrificed (Fig. 7A). Tumor
weight was also lower in the miR-1271-5p group compared with that
in the miR-NC group, and the largest tumor diameters observed were
7.4 and 13.5 mm in the miR-1271-5p and miR-NC groups, respectively
(Fig. 7B). Expression analysis
revealed that the level of miR-1271-5p was enhanced in tumor
tissues from the miR-1271-5p group, while the protein level of AQP5
decreased in the miR-1271-5p group compared with that of the miR-NC
group (Fig. 7C and D). miR-1271-5p
overexpression also increased the level of an apoptosis-related
protein, cleaved-caspase-3, while decreasing the level of a cell
cycle-related protein, PCNA, compared with the findings in the
miR-NC group (Fig. 7E). The data
from ELISA demonstrated that the levels of HBeAg and HBsAg were
reduced in the miR-1271-5p group compared with those in the miR-NC
group (Fig. 7F). Additionally, the
level of HBV DNA was also significantly impaired in the miR-1271-5p
group compared with that in the miR-NC group (Fig. 7G). Taken together, these data
demonstrated that miR-1271-5p overexpression could inhibit tumor
growth in vivo.
Discussion
HBV is a primary causative agent of chronic liver
diseases, which is difficult to cure because integrated virus
templates-induced proteins easily initiate and accelerate chronic
liver diseases (17). Identifying
potential gene targets in the pathogenesis of HBV infection may
provide opportunities for the development of specific therapies to
combat liver cancer. In the present study, low expression of
miR-1271-5p and high expression of AQP5 were detected in
HBV-related liver cancer tissues and cells. Functional analyses
disclosed that miR-1271-5p overexpression inhibited the levels of
HBeAg, HBsAg and HBV DNA, as well the cell viability, migration and
invasion, but promoted cell apoptosis. AQP5 knockdown exerted a
similar function with miR-1271-5p overexpression. In addition,
miR-1271-5p regulated HBV infection and liver cancer development
via competitively targeting AQP5. Nude mouse tumorigenicity assay
strengthened the hypothesis that miR-1271-5p impeded tumor growth
in vivo.
Various studies have suggested that miR-1271-5p can
suppress cell proliferation and metastasis in different types of
cancer, such as endometrial cancer (18), glioblastoma (19), gastric cancer (20) and HCC (21–23).
Notably, a previous study observed that miR-1271-5p exhibits
relatively low expression in HBV-associated HCC tissues, and its
upregulation impairs HBV-DNA replication, proliferation and
metastasis by promoting the adenosine monophosphate-activated
protein kinase (AMPK) signaling pathway (24). Another study stated that miR-1271-5p
can reduce cell growth and migration by binding to forkhead box K2
in HCC (10). Consistent with these
data, the present study observed lower expression of miR-1271-5p in
HBV-associated liver cancer tissues and cells, and its
reintroduction effectively reduced HBV-DNA replication, HBeAg and
HBsAg levels, cell viability and metastasis. Thus, the function of
miR-1271-5p in HBV-associated liver cancer might be related to
tumor inhibition.
In the present study, AQP5 was confirmed as a target
of miR-1271-5p. AQP5, which is 21–24 kDa in molecular weight, is
regarded as the main structural caveolae protein from cell
membranes (25). Satisfactory
progress has been made in the research on AQP5 in HCC. He et
al (26) concluded that AQP5 is
highly expressed in HCC cells, and its inhibition blocks HCC
metastasis and EMT by regulating the NF-κB signaling pathway. Zhang
et al (25) noted that the
expression of AQP5 is accumulated in HBV-HCC tissues and cells, and
that AQP5 contributes to cell proliferation but reduces apoptosis.
In agreement with these previous studies, the present study
observed that a high level of AQP5 was present in HBV-associated
liver cancer tissues and cells. AQP5 interference helped to
ameliorate the extent of HBV infection and malignant activities of
tumor cells, suggesting that AQP5 possessed carcinogenesis effects,
at least in liver cancer.
HBsAg is a sensitive and specific biomarker of HBV
infection. The detection of HBsAg is the primary diagnostic tool
for the diagnosis, prevention and treatment of HBV (27). HBeAg is an indicator of HBV
replication, and its high abundance indicates that HBV replication
is active and that the viral load is high (28). Generally, the level of HBV DNA is an
indicator to measure the replication of HBV (29). Therefore, the levels of HBsAg, HBeAg
and HBV DNA have been used in previous studies to measure HBV
infection (30–32). For example, miR-125a-5p
overexpression can inhibit the secretion of HBsAg and HBeAg, but
does not change the replication of HBV DNA (30). Long non-coding RNA highly
upregulated in liver cancer enhances the levels of HBsAg, HBeAg and
cccDNA, and activates HBV replication in HBV-infected cells
(31). miR-302c-3p protects against
HBV replication by reducing the concentrations of HBV DNA and HBsAg
in HBV transgenic mice (32).
Similarly, in the present study, miR-1271-5p mimic or AQP5
knockdown could reduce the levels of HBsAg, HBeAg and HBV DNA,
indicating that miR-1271-5p functioned on blocking HBV replication,
while AQP5 exhibited the opposite effect.
Collectively, the data from the present study
concluded that miR-1271-5p inhibited cell viability, migration and
invasion, induced apoptosis, attenuated HBV infection in
vitro, and impeded tumor growth in vivo by targeting
AQP5. Thus, miR-1271-5p may be a promising biomarker for therapy of
HBV-associated liver cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL conceived and designed the experiments, which
were performed by LM and LD. ZL and XL analyzed the data and wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Second Affiliated Hospital of Shandong First
Medical University. Each patient signed the informed consent form.
The animal experiments were approved by the Institutional Animal
Care and Use Committee of The Second Affiliated Hospital of
Shandong First Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
De Giorgio M and Fagiuoli S: Management of
hepatocellular carcinoma. Dig Dis. 25:279–281. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amir F, Siddiqui ZI, Farooqui SR, Anwer A,
Khan S, Azmi MI, Mehmankhah M, Dohare R, Khan LA and Kazim SN:
Impact of length of replication competent genome of hepatitis B
virus over the differential antigenic secretion. J Cell Biochem.
120:17858–17871. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Downs LO, Smith DA, Lumley SF, Patel M,
Mcnaughton AL, Mokaya J, Ansari MA, Salih H, Varnai KA, Freeman O,
et al: Electronic health informatics data to describe clearance
dynamics of hepatitis B surface antigen (HBsAg) and e antigen
(HBeAg) in chronic hepatitis B virus infection. MBio. 10:e00699–19.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gu S, Jin L, Zhang F, Sarnow P and Kay MA:
Biological basis for restriction of microRNA targets to the 3
untranslated region in mammalian mRNAs. Nat Struct Mol Biol.
16:144–150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bai PS, Xia N, Sun H and Kong Y:
Pleiotrophin, a target of miR-384, promotes proliferation,
metastasis and lipogenesis in HBV-related hepatocellular carcinoma.
J Cell Mol Med. 21:3023–3043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang JY, Chen HL and Shih C: MicroRNA
miR-204 and miR-1236 inhibit hepatitis B virus replication via two
different mechanisms. Sci Rep. 6:347402016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yen CS, Su ZR, Lee YP, Liu IT and Yen CJ:
miR-106b promotes cancer progression in hepatitis B
virus-associated hepatocellular carcinoma. World J Gastroenterol.
22:5183–5192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin MF, Yang YF, Peng ZP, Zhang MF, Liang
JY, Chen W, Liu XH and Zheng YL: FOXK2, regulted by miR-1271-5p,
promotes cell growth and indicates unfavorable prognosis in
hepatocellular carcinoma. Int J Biochem Cell Biol. 88:155–161.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Direito I, Madeira A, Brito MA and Soveral
G: Aquaporin-5: From structure to function and dysfunction in
cancer. Cell Mol Life Sci. 73:1623–1640. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sekine S, Shimada Y, Nagata T, Moriyama M,
Omura T, Watanabe T, Hori R, Yoshioka I, Okumura T, Sawada S, et
al: Prognostic significance of aquaporins in human biliary tract
carcinoma. Oncol Rep. 27:1741–1747. 2012.PubMed/NCBI
|
|
13
|
Song T, Yang H, Ho JC, Tang SC, Sze SC,
Lao L, Wang Y and Zhang KY: Expression of aquaporin 5 in primary
carcinoma and lymph node metastatic carcinoma of non-small cell
lung cancer. Oncol Lett. 9:2799–2804. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Lu J, Zhou H, Du Z and Zhang G:
Silencing of aquaporin 5 inhibits the growth of A549 lung cancer
cells in vitro and in vivo. Int J Oncol. 52:1643–1650.
2018.PubMed/NCBI
|
|
15
|
Jung HJ, Park JY, Jeon HS and Kwon TH:
Aquaporin-5: A marker protein for proliferation and migration of
human breast cancer cells. PLoS One. 6:e284922011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arzumanyan A, Reis HM and Feitelson MA:
Pathogenic mechanisms in HBV- and HCV-associated hepatocellular
carcinoma. Nat Rev Cancer. 13:123–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tian Y, Chen YY and Han AL: MiR-1271
inhibits cell proliferation and metastasis by targeting LDHA in
endometrial cancer. Eur Rev Med Pharmacol Sci. 23:5648–5656.
2019.PubMed/NCBI
|
|
19
|
Yang L, Wang Y, Li YJ and Zeng CC:
Chemo-resistance of A172 glioblastoma cells is controlled by
miR-1271-regulated Bcl-2. Biomed Pharmacother. 108:734–740. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lim B, Kim HJ, Heo H, Huh N, Baek SJ, Kim
JH, Bae DH, Seo EH, Lee SI, Song KS, et al: Epigenetic silencing of
miR-1271 enhances MEK1 and TEAD4 expression in gastric cancer.
Cancer Med. 7:3411–3424. 2018.(Epub ahead of print). View Article : Google Scholar
|
|
21
|
Li C, Jiang Y, Miao R, Qu K, Zhang J and
Liu C: MicroRNA-1271 functions as a metastasis and
epithelial-mesenchymal transition inhibitor in human HCC by
targeting the PTP4A1/c-Src axis. Int J Oncol. 52:536–546.
2018.PubMed/NCBI
|
|
22
|
Qin A, Zhu J, Liu X, Zeng D, Gu M and Lv
C: MicroRNA-1271 inhibits cellular proliferation of hepatocellular
carcinoma. Oncol Lett. 14:6783–6788. 2017.PubMed/NCBI
|
|
23
|
Maurel M, Jalvy S, Ladeiro Y, Combe C,
Vachet L, Sagliocco F, Bioulac-Sage P, Pitard V, Jacquemin-Sablon
H, Zucman-Rossi J, et al: A functional screening identifies five
microRNAs controlling glypican-3: Role of miR-1271 down-regulation
in hepatocellular carcinoma. Hepatology. 57:195–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Y, Zhao ZX, Huang F, Yuan XW, Deng L
and Tang D: MicroRNA-1271 functions as a potential tumor suppressor
in hepatitis B virus-associated hepatocellular carcinoma through
the AMPK signaling pathway by binding to CCNA1. J Cell Physiol.
234:3555–3569. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z, Han Y, Sun G, Liu X, Jia X and Yu
X: MicroRNA-325-3p inhibits cell proliferation and induces
apoptosis in hepatitis B virus-related hepatocellular carcinoma by
down-regulation of aquaporin 5. Cell Mol Biol Lett. 24:132019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He Z, Dong W, Hu J and Ren X: AQP5
promotes hepatocellular carcinoma metastasis via NF-κB-regulated
epithelial-mesenchymal transition. Biochem Biophys Res Commun.
490:343–348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hadziyannis E and Laras A: Viral
biomarkers in chronic HBeAg negative HBV infection. Genes (Basel).
9:4692018. View Article : Google Scholar
|
|
28
|
Kam W, Rall LB, Smuckler EA, Schmid R and
Rutter WJ: Hepatitis B viral DNA in liver and serum of asymptomatic
carriers. Proc Natl Acad Sci USA. 79:7522–7526. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hc Y and Jh K: Persistence of hepatitis B
virus covalently closed circular DNA in hepatocytes: Molecular
mechanisms and clinical significance. Emerg Microbes Infect.
3:e642014.PubMed/NCBI
|
|
30
|
Li G, Zhang W, Gong L and Huang X:
MicroRNA 125a-5p inhibits cell proliferation and induces apoptosis
in hepatitis B virus-related hepatocellular carcinoma by
downregulation of ErbB3. Oncol Res. 27:449–458. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang
Y, Bu Y, Zhao M, Zhang S and Zhang X: Long non-coding RNA HULC
activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in
HBV-related hepatocellular carcinoma. Cancer Lett. 454:158–170.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hamada-Tsutsumi S, Naito Y, Sato S,
Takaoka A, Kawashima K, Isogawa M, Ochiya T and Tanaka Y: The
antiviral effects of human microRNA miR-302c-3p against hepatitis B
virus infection. Aliment Pharmacol Ther. 49:1060–1070. 2019.
View Article : Google Scholar : PubMed/NCBI
|