Introduction
Ovarian cancer (OC) is the seventh most common
cancer among females with an estimated 239,000 new cases being
diagnosed worldwide annually; the incidence rate (11.4 per 100,000)
is the highest in Central and Eastern Europe (1). Although the diagnosis and treatment
for OC have been increasingly successful, the 5-year survival rate
of the advanced patient with OC is only ~30% (1,2).
Therefore, it is imperative to search more molecular targets for
promoting the diagnostic and therapeutic effects on OC patients.
Studies have reported that long non-coding RNAs (lncRNAs) can
regulate biological behaviors in OC cells (3–5).
Aerobic glycolysis, known as the ‘Warburg effect’,
is characterized by oxidating glucose and producing lactate under
the catalysis of diverse enzymes and normoxic conditions (6). Glycolysis can provide the energy for
tumor cell growth and the promotion of glycolytic metabolism
indicates the malignant progression of types of cancer (7). A number of lncRNAs have been shown to
regulate the metabolic process of glycolysis in types of cancer,
including OC (8–10). It may be pertinent to seek the
useful targets in OC progression through the glycolytic metabolism.
LncRNAs are common class of regulatory ncRNAs with length >200
nucleotides and lack of protein-encoding capacity (11).
LncRNAs can participate in the initiation and
progression of various types of human tumors as oncogenes or tumor
inhibitors (12,13). O-phthalaldehyde-interacting protein
5 antisense transcript 1 (OIP5-AS1) has been found as the regulator
by acting as the sponges of microRNAs (miRNAs) in various types of
cancers. For instance, Wang et al (14) revealed that OIP5-AS1 improves cell
proliferation and results in poor prognosis through sponging
miR-378a-3p in lung cancer. In addition, OIP5-AS1 knockdown
restrains CCAAT/enhancer-binding protein α and TNF
receptor-associated factor 4 levels to retard glioma cell growth
via binding to miR-367-3p (15).
OIP5-AS1 is reported to be overexpressed in OC and facilitates the
malignant tumor growth and metastasis of OC by the miR-137/ZNF217
or miR-324-3p/NFIB axis (16,17).
However, the regulation in glycolytic process of OC and other
regulatory mechanisms underpinning OIP5-AS1 remain to be
elucidated.
Non-coding miRNAs can cause mRNA degradation and
translation inhibition by combining with the 3′-untranslated
regions (3′-UTRs) of target mRNAs, leading to a significant
involvement in biological processes (18,19).
Studies have found an inhibitory effect of miR-128-3p on the
development of a number of carcinomas (20–22).
The aberrant downregulation of miR-128 is found in OC (23,24).
As a submit of miR-128, the present study hypothesized that
miR-128-3p might also be associated with OC regulation.
Cyclin G1 (CCNG1), a member of cell cyclin protein,
is dysregulated in leiomyoma and colorectal cancer (25,26).
CCNG1 is associated with the regulation of miR-122a in
hepatocellular carcinoma as a target of miR-122a (27). Studies also suggest that CCNG1 acts
as an oncogene to enhance OC cell growth (28,29),
but it is unknown if CCNG1 is a target of miR-128-3p in OC.
The present study was mainly devoted to
investigating the role of OIP5-AS1 in OC and its functional
mechanism with miR-128-3p and CCNG1, it also attempted to provide a
novel pathogenesis for OC progression.
Materials and methods
Tissues acquisition and cell
culture
A total of 41 patients with OC were recruited to
this study between March 2015 and September 2018. A total of 41
paired tissues of OC and adjacent normal tissues (>3 cm from
tumor sites) were acquired from these patients with OC (age range
18–60 years old) subjected to surgical excision in the Shengli
Oilfield Central Hospital, after obtaining informed consents from
the patients. These tissues were divided into two groups (I+II:
n=23; III+IV: n=18) according to the tumor stage. All specimens
were promptly stored in liquid nitrogen for subsequent RNA or
protein extraction. The present study was approved and supported by
the Shengli Oilfield Central Hospital.
Human ovarian surface epithelial cells (IOSE-80) and
OC cell line OVCAR-3 were bought from American Type Culture
Collection and another OC cell line, SKOV3, was obtained from the
cell bank of the Chinese Academy of Sciences (Shanghai, China).
These cells were cultured in basic medium RPMI 1640 (Invitrogen;
Thermo Fisher Scientific, Inc.) complemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a humidified
incubator with a constant temperature of 37°C and CO2
concentration at 5%.
Cell transfection
Small interfering (si) RNA against OIP5-AS1
(si-OIP5-AS1, 5′-GGCTTTGTGTTCCTTATCACAGG-3′), siRNA against CCNG1
(si-CCNG1, 5′-TTGAAGTAAAAGATCTTCTTAGT-3′), siRNA negative control
(si-control, 5′-TTCTCCGAACGTGTCACGTTT-3′), short hairpin (sh) RNA
vector against OIP5-AS1 (sh-OIP5-AS1,
5′-CCGGGCTCCTAGGATTCCAGTTATCCTCGAGGCAGAAGGCTGAGTTTCATTTTTTTTG-3′),
shRNA vector negative control (sh-control,
5′-CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG-3′),
miR-128-3p mimic (miR-128-3p, 5′-UCACAGUGAACCGGUCUCUUU-3′), mimic
negative control (miR-control, 5′-UUGUACUACACAAAAGUACUG-3′),
miR-128-3p inhibitor (anti-miR-128-3p,
5′-AAAGAGACCGGUUCACUGUGA-3′), inhibitor negative control
(anti-control, 5′-CAGUACUUUUGUGUAGUACAA-3′) and the overexpression
vector pCE-RB-Mam-OIP5-AS1 (OIP5-AS1) vector or the empty vector
pCE-RB-Mam were all purchased from Guangzhou RiboBio Co., Ltd. Cell
transfection was performed using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were seeded
into the 24-well plates and cultured overnight to 50% coverage,
then transfected with different concentrations of RNAs or vectors
(40 nM siRNA, 40 nM shRNA vector, 40 nM mimic, 20 nM inhibitor or 2
µg pCE-RB-Mam/OIP5-AS1 vector). Further experimentation was
performed after cells were incubated at 37°C for 48 h.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
The extraction of total RNA was performed using
TRIzol® (Thermo Fisher Scientific, Inc.) from OC tissues
and cells (2×105) in accordance with the manufacturer's
protocol, followed by the quantification of RNA concentration. Then
1 µg RNA was reversely transcribed to synthesis the complementary
DNA (cDNA) using Prime Script RT regent kit (Takara Bio, Inc.)
according to the manufacturer's protocols. PCR reaction system was
prepared in 384 well-plates using a TB Green Premix EX Taq II kit
(Takara Bio, Inc.) following the manufacturer's protocol and
amplified via ABI Step One Real-time PCR System (Thermo Fisher
Scientific, Inc.). The reaction system was prepared on ice with a
20-µl volume: 10 µl TB Green Premix EX Taq II (2×), 0.8 µl forward
primer, 0.8 µl reverse primer, 0.4 µl ROX Reference Dye, 2 µl cDNA,
6 µl sterile water. Three technical replications were set for each
sample. The reaction protocols were as follows: Predenaturation at
95°C for 30 sec, 40 cycles of denaturation at 95°C for 5 sec and
annealing at 60°C for 30 sec. GAPDH and U6 were used as respective
internal control for lncRNA or mRNA and miRNA. Primers were
synthesized by Sangon Biotech Co., Ltd. and the sequences were:
OIP5-AS1 (forward: 5′-TGCGAAGATGGCGGAGTAAG-3′, reverse:
5′-TAGTTCCTCTCCTCTGGCCG-3′); miR-128-3p (forward:
5′-GCCGAGTCACAGTGAACCGGT-3′, reverse: 5′-CAGTGCAGGGTCCGAGGTAT′);
CCNG1 (forward: 5′-GTTACCGCTGAGGAGCTGCAGTC-3′, reverse:
5′-ATAGCCATCATGGATAGACTCAG-3′); GAPDH (forward:
5′-GTCTCCTCTGACTTCAACAGCG-3′, reverse: 5′-
ACCACCCTGTTGCTGTAGCCAA-3′); U6 (forward:
5′-ATTGGAACGATACAGAGAAGATT-3′, reverse: 5′-GGAACGCTTCACGAATTTG-3′).
The collected data from three independent experiments were analyzed
using the 2−∆∆Cq method (30).
3-(4, 5)-Dimethylthiazole-2-y1)-2,
5-biphenyl tetrazolium bromide (MTT) assay
Cell viability was examined by MTT assay. At 0, 24,
48 and 72 h post-transfection, 15 µl MTT solution (5 mg/ml, pH=7.4;
Thermo Fisher Scientific, Inc.) was added to cells in the 96-well
plates and incubated for another 4 h. Then the supernatants were
slowly removed and 150 µl dimethylsulfoxide (DMSO; Thermo Fisher
Scientific, Inc.) was added to each well for 10 min to resolve the
generated formazan. The optical density (OD) value of each well at
the wavelength of 490 nm was determined using a microplate reader
(Thermo Fisher Scientific, Inc.).
Flow cytometry
An FITC Annexin V Apoptosis Detection kit I (BD
Biosciences) was used to distinguish the apoptotic cells. After 48
h post-transfection, OVCAR-3 and SKOV3 cells were collected
following digestion by 0.25% trypsin (GIBCO), then cell pellets
were washed by pro-cooled phosphate buffered saline (PBS; GIBCO)
and resuspended in 1X binding buffer. Afterwards, cells were
stained by 5 µl FITC Annexin V and 5 µl PI for 15 min in the dark
at room temperature according to the manufacturer's protocol. The
apoptotic cells were examined using a BD Accuri C6 flow cytometer
(BD Biosciences) and analyzed using the BD Accuri C6 system
(32-bit) software (BD Biosciences). The apoptosis rate was
calculated as the percentage of early apoptotic cells
(Annexin+/PI−) and late apoptotic cells
(Annexin+/PI+).
Transwell migration and invasion
assays
A Transwell 24-well chamber (Corning Life Sciences)
was used to detect the abilities of cell migration and invasion.
Cell suspension in serum-free medium was pipetted into the upper
chamber at room temperature. Simultaneously, 600 µl RPMI 1640
medium with 10% FBS was added to the lower chamber at room
temperature. The chamber was incubated at 37°C for 24 h, and cells
that passed through the lower membranes were fixed with methyl
alcohol (Sangon Biotech Co., Ltd.) for 10 min and stained with
crystal violet (Sangon Biotech Co., Ltd.) for 10 min at room
temperature. Finally, the cells were counted under an inverted
light microscope (Olympus Corporation) and cell images were
acquired at ×100 magnification. For invasion assay, the lower
surface of the upper chamber was coated with Matrigel (Corning Life
Sciences) at 37°C overnight before seeding the cells.
Detection of glucose consumption and
lactate production
According to the manufacturer's protocol, the
consumption of glucose and the production of lactate were assessed
using glucose detection kit (cat. no. K188-200) and lactate
detection kit (cat. no. K209-100) purchased from Beijing Zhonghao
Biotechnology Co., Ltd.).
Western blotting assay
Total proteins from tissues and cells were extracted
by RIPA Lysis Buffer (Sangon Biotech Co., Ltd.). The obtained
proteins were quantified using BCA Protein Assay kit (Sangon
Biotech Co., Ltd.) and 30 µg proteins were separated by sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on
10% gels for 2 h. Next, the separated proteins were transferred
onto the polyvinylidene fluoride (PVDF) membranes (Thermo Fisher
Scientific, Inc.). These membranes were then immersed in 5% skimmed
milk (Sangon Biotech Co., Ltd.) diluted with phosphate buffered
saline and 0.1% Tween 20 (PBST) at room temperature for 3 h to
prevent the non-specific protein binding, followed by incubation
with primary antibodies overnight at 4°C. The primary antibodies
were: anti-hexokinase 2 (anti-HK2; Abcam; cat. no. ab209847;
1:1,000), anti-CCNG1 (Abcam; cat. no. ab170389; 1:1,000) and
anti-β-actin (Abcam; cat. no. ab8227; 1:3,000). Finally, the
chromogenic reaction was performed by an HRP/DAB (ABC) detection
kit (Abcam) after the incubation of anti-rabbit secondary antibody
(Abcam; cat. no. ab205718; 1:5,000) for 1 h at room temperature.
The gray level of each band was obtained by the statistical
analysis of ImageJ software (National Institutes of Health) using
β-actin as the endogenous reference.
Dual-luciferase reporter assay
The online software applications StarBase
(http://starbase.sysu.edu.cn) and
TargetScan (http://www.targetscan.org) were used
to seek the miRNA target of OIP5-AS1 and target gene for
miR-128-3p, respectively. To construct the recombinant luciferase
plasmids, the wild-type (WT) OIP5-AS1 and 3′UTR of CCNG1 sequences
were respectively cloned into the pGL-3 luciferase basic vector
(Promega Corporation) to acquire WT-OIP5-AS1 and WT-CCNG1, then
their mutant-types (MUT) with the mutant sites for miR-128-3p were
also constructed as MUT-OIP5-AS1 and MUT-CCNG1. Each constructed
luciferase reporter plasmid was transfected into OVCAR-3 and SKOV3
cells with miR-128-3p or miR-control by Lipofectamine 3000. After
transfection for 48 h, cells were lysed and the luciferase
intensity was determined by the dual-luciferase reporter system
(Promega Corporation) following the manufacturer's protocol. The
luciferase activity ratio of firefly and Renilla was
considered to be the relative luciferase activity.
RNA immunoprecipitation (RIP)
assay
RIP assay was performed using Magna RNA
immunoprecipitation kit (EMD Millipore). OC cells were centrifuged
at 4°C and 16,000 × g for 10 min and 2×107 cells were
lysed using RIPA lysis buffer (EMD Millipore). The cells were then
incubated with magnetic beads pro-covered with antibodies against
Argonaute2 (Anti-Ago2; Abcam; cat. no. ab32381) using
anti-Immunoglobulin G (Anti-IgG; Abcam; cat. no. ab205718) as the
negative reference. RNA was extracted using TRIzol and the RNA
enrichment was detected via RT-qPCR. Finally, the levels of
OIP5-AS1 and miR-128-3p in Anti-IgG and Anti-Ago2 groups were
compared.
RNA pull-down assay
Pierce™ Biotinylated Protein Interaction Pull-Down
kit (Thermo Fisher Scientific, Inc.) was used to perform the
pull-down assay. OVCAR-3 and SKOV3 cells were transfected with
biotin-labeled miR-128-3p (Bio-miR-128-3p) or Bio-NC (Guangzhou
RiboBio Co., Ltd) using Lipofectamine 3000. Following transfection
at 37°C for 48 h, cells were harvested by centrifugation at 4°C and
16,000 g for 10 min. The cell lysates were then incubated with
streptavidin-coupled agarose beads at 4°C overnight. Following the
isolation of RNA from the washed beads by TRIzol, the enrichment of
OIP5 was measured by RT-qPCR analysis.
Xenograft tumor assay
A total of 12 BALB/c nude mice (six-week-old, 20–25
g) were purchased from Beijing Vital River Laboratory Animal
Technology Co., Ltd. and maintained in a specific-pathogen-free
environment under temperature of 25°C, humidity of 70% and a 12 h
light/dark cycle. These mice received food and water ad
libitum. Mice were arbitrarily divided into two groups with 6
mice/group, comprising sh-control and sh-OIP5-AS1 groups. The
xenograft tumor model was established through the subcutaneous
injection of 2×106 OVCAR-3 cells stably expressed
sh-OIP5-AS1 or sh-control into the left flank of mouse's back. The
health and behavior of mice were monitored every 2 days and tumor
volume (length × width2 × 0.5) was measured using a
digital caliper every week. After cell injection for 4 weeks, tumor
volume reached 900 mm3 (<1,000 mm3, which
was set as the humane endpoint) and the mice were sacrificed. The
30% air of environment (per min) was displaced using the flow rate
of CO2 according to the current guidelines of the
American Veterinary Medical Association (31), then mice were verified to have
succumbed by monitoring the breathing. No mice died during the
4-week experimental period. The tumor tissues were excised from
mice and weighed on an electronic scale. The examination of
OIP5-AS1, miR-128-3p and CCNG1 was performed via RT-qPCR or western
blotting. This experiment was ratified by the Animal Ethics
Committee of the Shengli Oilfield Central Hospital and all
procedures followed the guidelines provided by the National
Institutes of Health for the Care and Use of Laboratory Animals,
8th edition (32).
Statistical analysis
All data were given as the mean ± standard deviation
from three independent experiments. SPSS 19.0 (IBM Corp.) was used
to conduct statistical analysis and figure plotting was performed
using GraphPad Prism 7 (GraphPad Software, Inc.). The differences
of two groups and more than two groups were analyzed through
Student's t-test and a one-way analysis of variance followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
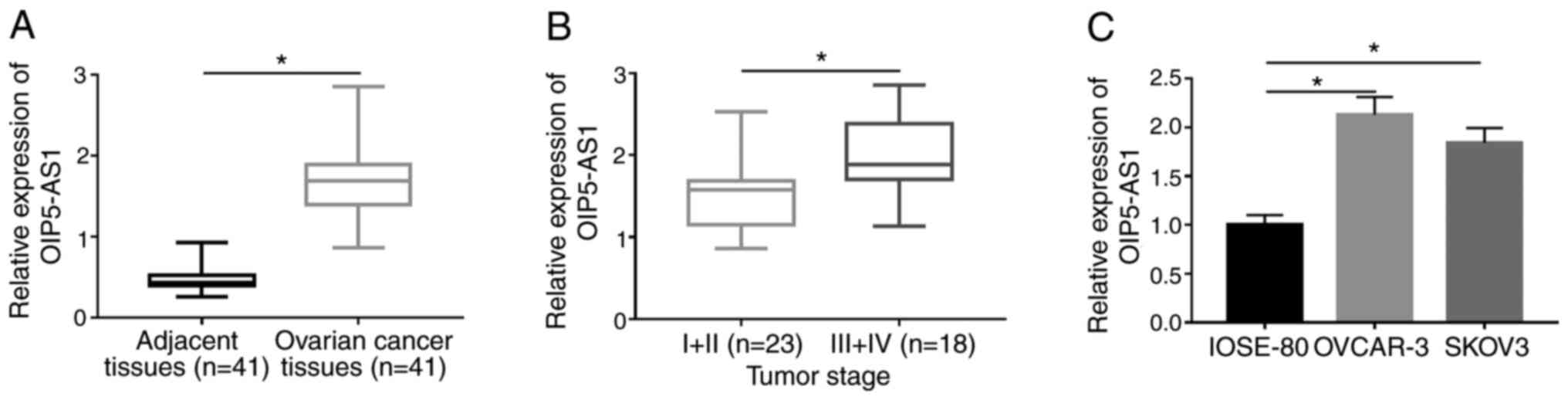
OIP5-AS1 is upregulated in OC tissues
and cells
RT-qPCR analysis was firstly conducted to examine
the expression of OIP5-AS1. OIP5-AS1 expression was markedly
increased in OC tissues compared with the adjacent normal tissues
(Fig. 1A). Among 41 OC tissues,
OIP5-AS1 was upregulated in tissues at III+IV stage compared with
those tissues at I+II stage (Fig.
1B). OIP5-AS1 expression was higher in both OVCAR-3 and SKOV3
cells compared with normal human ovarian IOSE-80 cells (Fig. 1C). This dysregulation of OIP5-AS1
implied its vital role in OC.
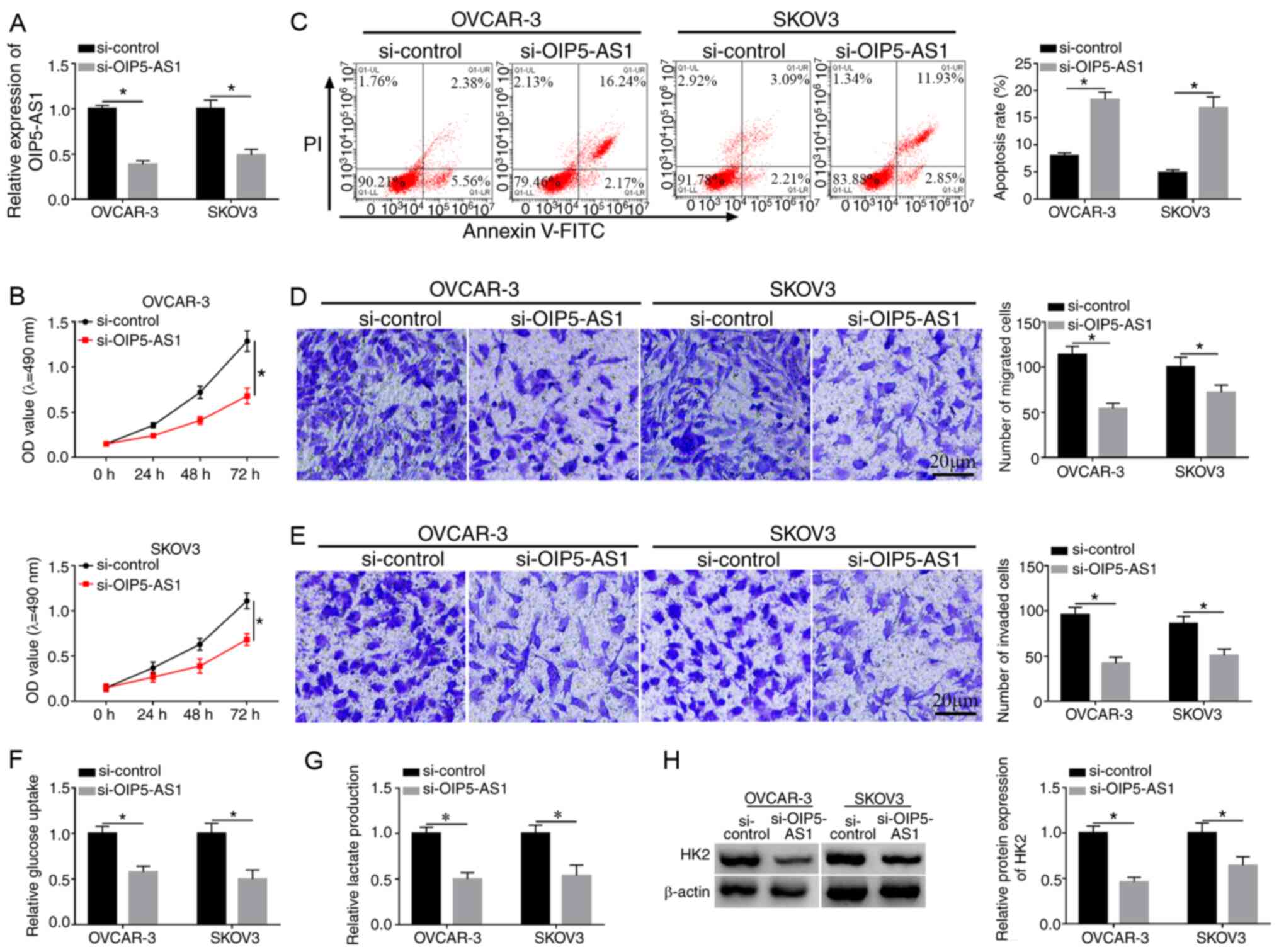
Knockdown of OIP5-AS1 represses cell
viability, migration, invasion and glycolysis while promoting
apoptosis in OC cells
OVCAR-3 and SKOV3 cells were transfected with
si-OIP5-AS1 or si-control to investigate the function of OIP5-AS1
in OC. As shown in Fig. 2A, the
decreased OIP5-AS1 expression in si-OIP5-AS1 group compared with
the si-control group demonstrated that OIP5-AS1 was successfully
knocked down in the two cell lines. MTT assay demonstrated that
cell viabilities of OVCAR-3 and SKOV3 cells were distinctly reduced
following the introduction of si-OIP5-AS1 compared with si-control
group (Fig. 2B), while the
apoptosis rate was shown to be increased (Fig. 2C). Transwell assay suggested that
the migrated and invaded cells were decreased after downregulation
of OIP5-AS1 (Fig. 2D and E).
Analysis of glycolysis was performed by the detection of glucose
consumption and lactate production, as well as the protein level of
HK2 (an important enzyme of glycolysis). The data revealed that the
transfection of si-OIP5-AS1 suppressed the consumption of glucose
(Fig. 2F), lactate production
(Fig. 2G) and HK2 protein
expression (Fig. 2H) in both
OVCAR-3 and SKOV3 cells, indicating that OIP5-AS1 knockdown caused
the inhibition of glycolysis. These results suggested that
knockdown of OIP5-AS1 repressed the progression of OC.
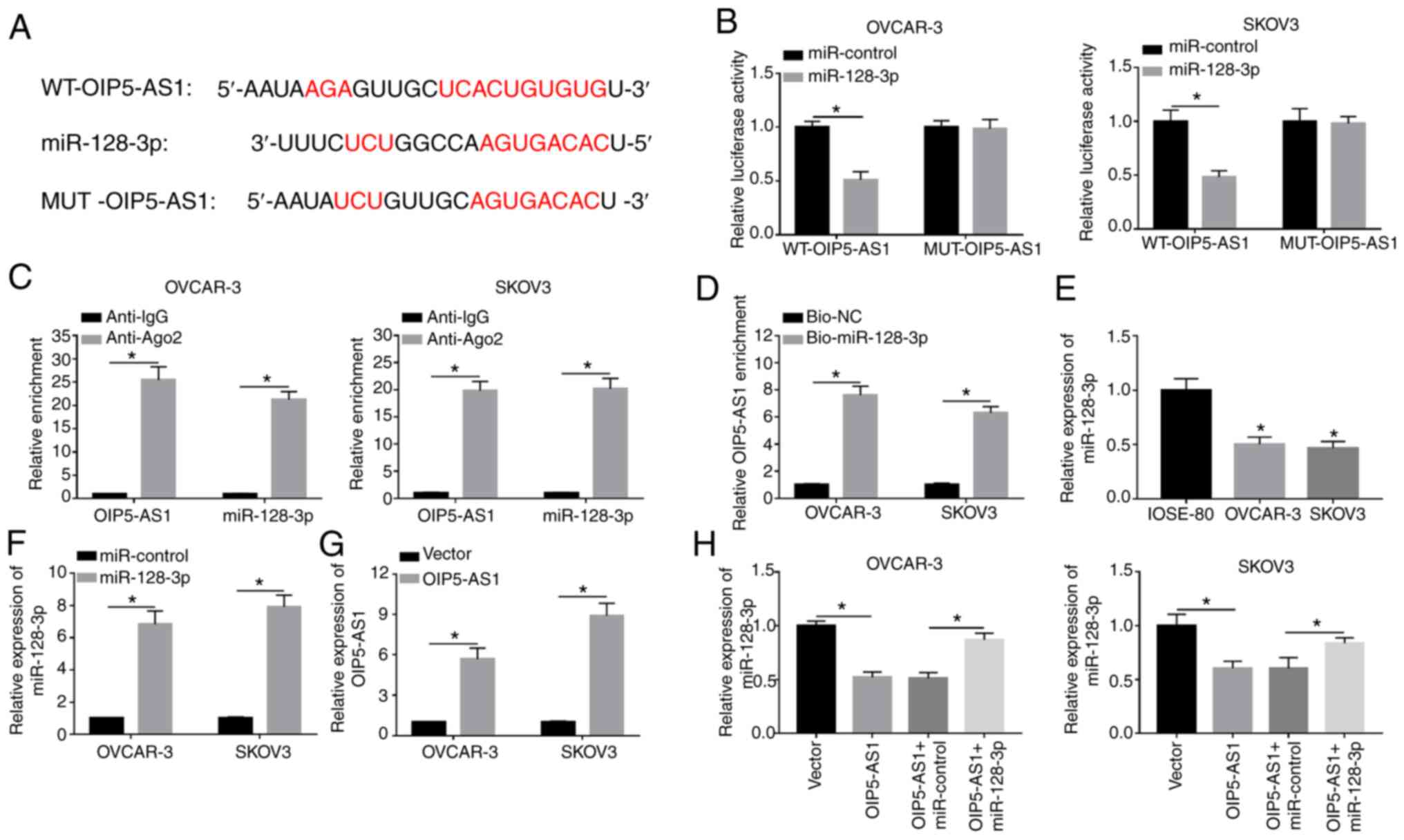
OIP5-AS1 interacts with
miR-128-3p
LncRNAs exert their regulatory effects through
sponging miRNAs generally (33). As
demonstrated in Fig. 3A, StarBase
predicted that OIP5-AS1 contained the binding sequences (UCACUGUG)
for miR-128-3p and these sites were mutated into AGUGACAC in mutant
OIP5-AS1. After performing the dual-luciferase reporter assay, it
was found that the luciferase activity of WT-OIP5-AS1 plasmid was
definitely inhibited by miR-128-3p mimic (compared with the
miR-control group) while there was no obvious change in
MUT-OIP5-AS1 group of OVCAR-3 and SKOV3 cells (Fig. 3B). RIP assay demonstrated that
OIP5-AS1 and miR-128-3p were enriched in Ago-2 pellet compared with
the IgG group (Fig. 3C). Moreover,
the enrichment of OIP5-AS1 was much higher in Bio-miR-128-3p group
compared with the Bio-NC group, suggesting that OIP5-AS1 was pulled
down by miR-128-3p (Fig. 3D).
Subsequently, the miR-128-3p expression in OC cells was examined
and the results indicated that miR-128-3p level was downregulated
in OVCAR-3 and SKOV3 cells compared with normal IOSE-80 cells
(Fig. 3E). After verifying the
successful overexpression effects on miR-128-3p and OIP5-AS1 by
miR-128-3p mimic and OIP5-AS1 vector (Fig. 3F and G), it was noticed that
miR-128-3p overexpression could recover the OIP5-AS1-induced
miR-128-3p suppression (Fig. 3H).
The results demonstrated that OIP5-AS1 could directly sponge
miR-128-3p.
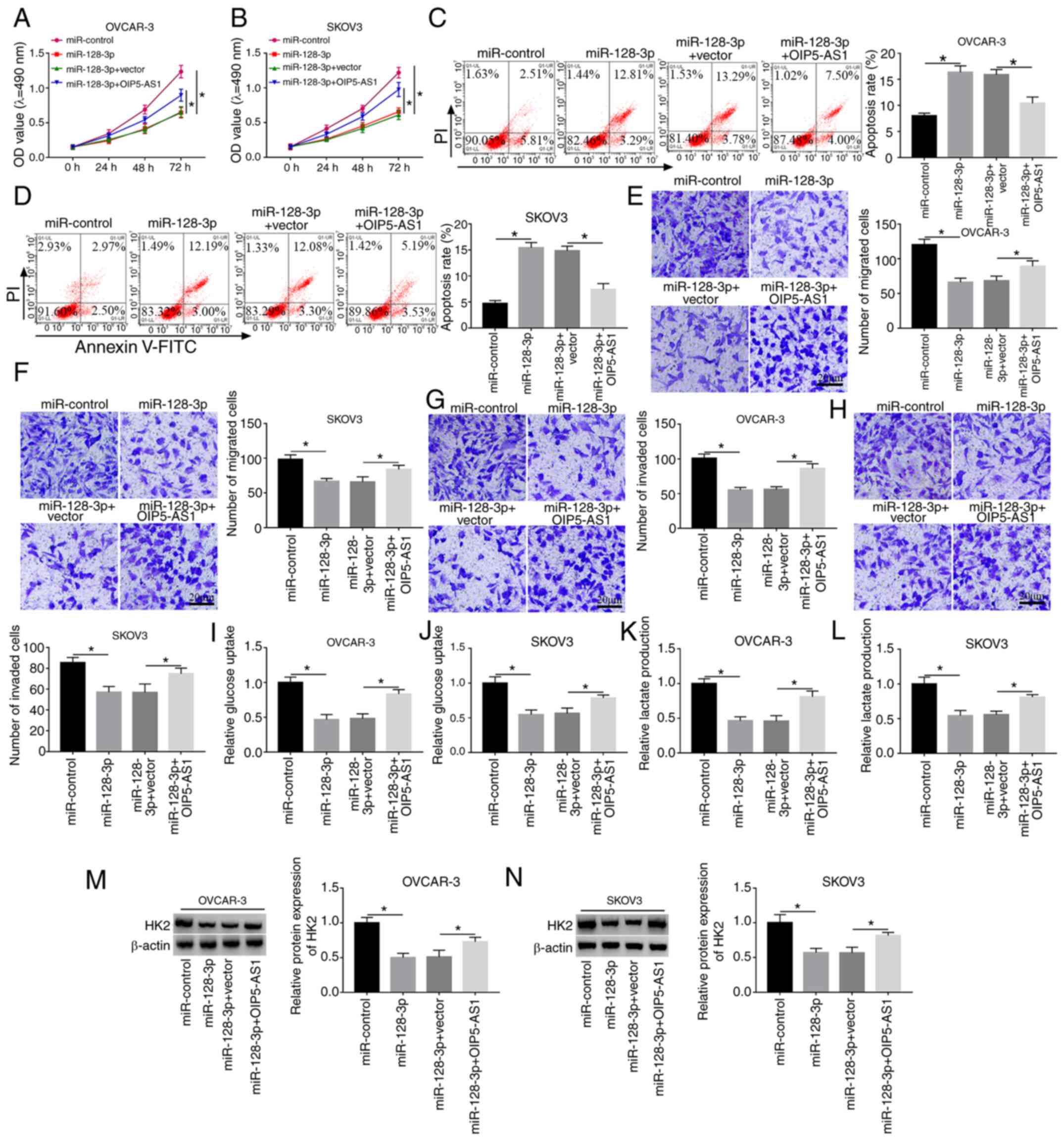
Overexpression of OIP5-AS1 ameliorates
the suppressive effects of miR-128-3p on the progression of OC
cells
To explore the regulatory mechanism of OIP5-AS1 and
miR-128-3p, OVCAR-3 and SKOV3 cells were transfected with
miR-128-3p, miR-128-3p+OIP5-AS1 or their matched controls. MTT and
flow cytometry demonstrated that miR-128-3p had a suppressive
effect on cell viability (Fig. 4A and
B) and a promoting effect on apoptosis (Fig. 4C and D) in OVCAR-3 and SKOV3 cells,
whereas the overexpression of OIP5-AS1 reversed these effects. Cell
migration (Fig. 4E and F) and
invasion (Fig. 4G and H) were
inhibited by miR-128-3p transfection, which was abrogated following
the upregulation of OIP5-AS1. OIP5-AS1 transfection counteracted
the inhibition of glucose consumption (Fig. 4I and J) and lactate production
(Fig. 4K and L) caused by
miR-128-3p. The miR-128-3p-induced downregulation of HK2 protein
level was clearly alleviated by the promotion of OIP5-AS1
expression (Fig. 4M and N). Hence,
the inhibitory effect of miR-128-3p on OC progression was abated by
OIP5-AS1 overexpression.
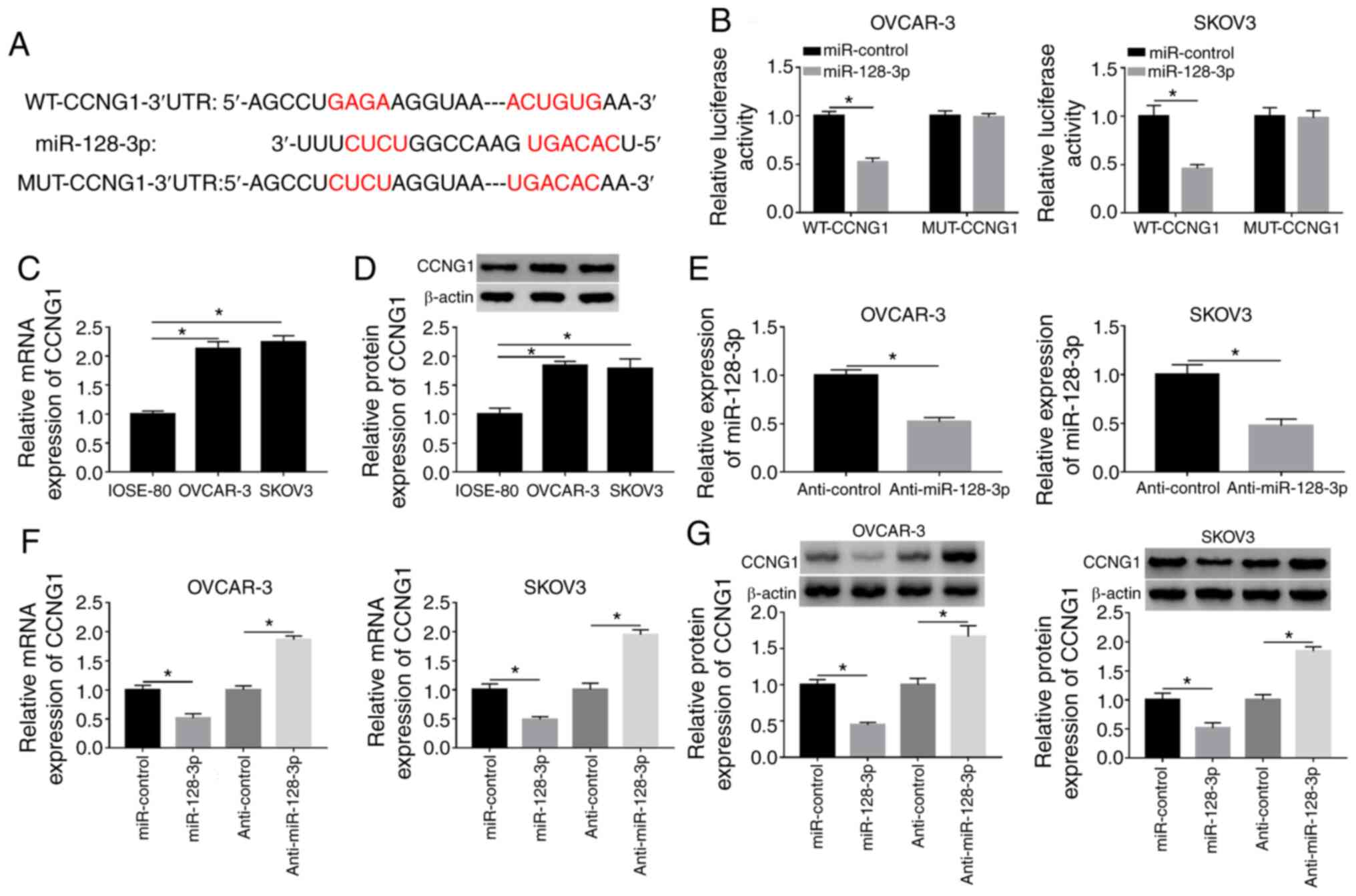
CCNG1 is a target of miR-128-3p
TargetScan software predicted the binding domain of
miR-128-3p in the 3′-UTR sequence of CCNG1 (Fig. 5A), indicating that CCNG1 might be a
potential target of miR-128-3p. Dual-luciferase reporter assay
proved the combination of miR-128-3p and CCNG1 because the
overexpression of miR-128-3p markedly repressed the luciferase
activity of WT-CCNG1 group but failed to decrease that of MUT-CCNG1
group (Fig. 5B). Then RT-qPCR and
western blotting revealed that the mRNA and protein expression
levels of CCNG1 were higher in OVCAR-3 and SKOV3 cells than those
in normal IOSE-80 cells (Fig. 5C and
D). The RT-qPCR analysis showed that the repressive impact of
anti-miR-128-3p on the expression of miR-128-3p was significant
(Fig. 5E). Subsequently, the
introduction of miR-128-3p was presented to suppress the CCNG1 mRNA
and protein levels, while the opposite effects were observed
following anti-miR-128-3p transfection (Fig. 5F and G). Collectively, miR-128-3p
directly targeted CCNG1.
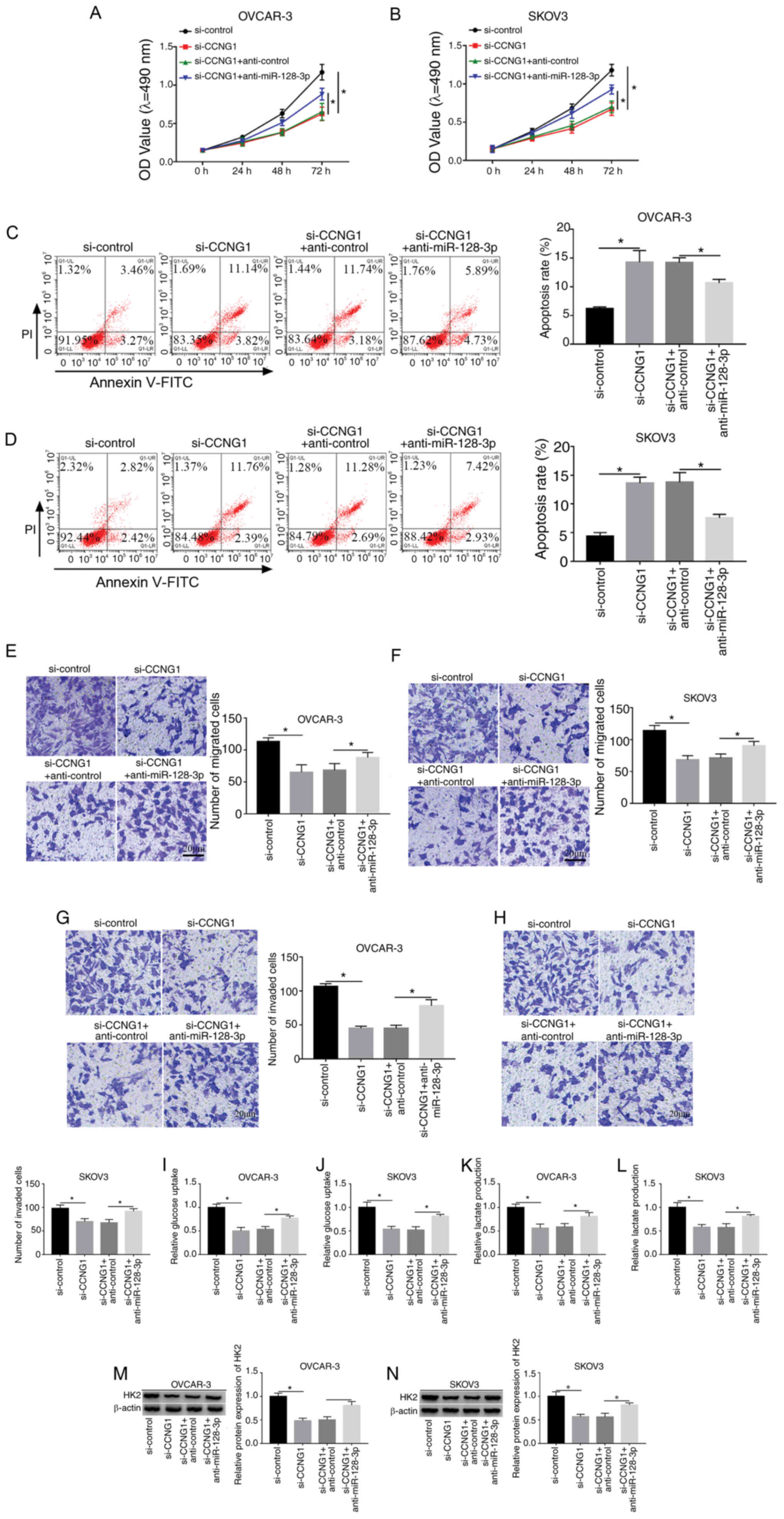
Downregulation of miR-128-3p restored
the si-CCNG1-induced effects on OC cells
To explore whether CCNG1 was associated with the
influence of miR-128-3p on OC development, transfection of
si-CCNG1, si-CCNG1 + anti-miR-128-3p or the relative controls was
conducted in OVCAR-3 and SKOV3 cells. Western blotting indicated
that si-CCNG1 transfection markedly decreased the protein level of
CCNG1, while miR-128 inhibitor relieved this expression
downregulation in OVCAR-3 and SKOV3 cells (Fig. S1). Subsequent experiments
demonstrated that anti-miR-128-3p partly abolished the
si-CCNG1-induced cell viability repression (Fig. 6A and B), apoptosis enhancement
(Fig. 6C and D) and migration
(Fig. 6E and F) or invasion
(Fig. 6G and H) inhibition.
Additionally, the decline of glucose consumption (Fig. 6I and J), lactate production
(Fig. 6K and L) and HK2 protein
level (Fig. 6M and N) caused by
CCNG1 knockdown was also weakened following the downregulation of
miR-128-3p. Above data clarified that miR-128-3p inhibition
lightened the effects of CCNG1 knockdown on OC cells, hinting that
CCNG1 inhibition was responsible for the anti-cancer effect of
miR-128-3p on OC.
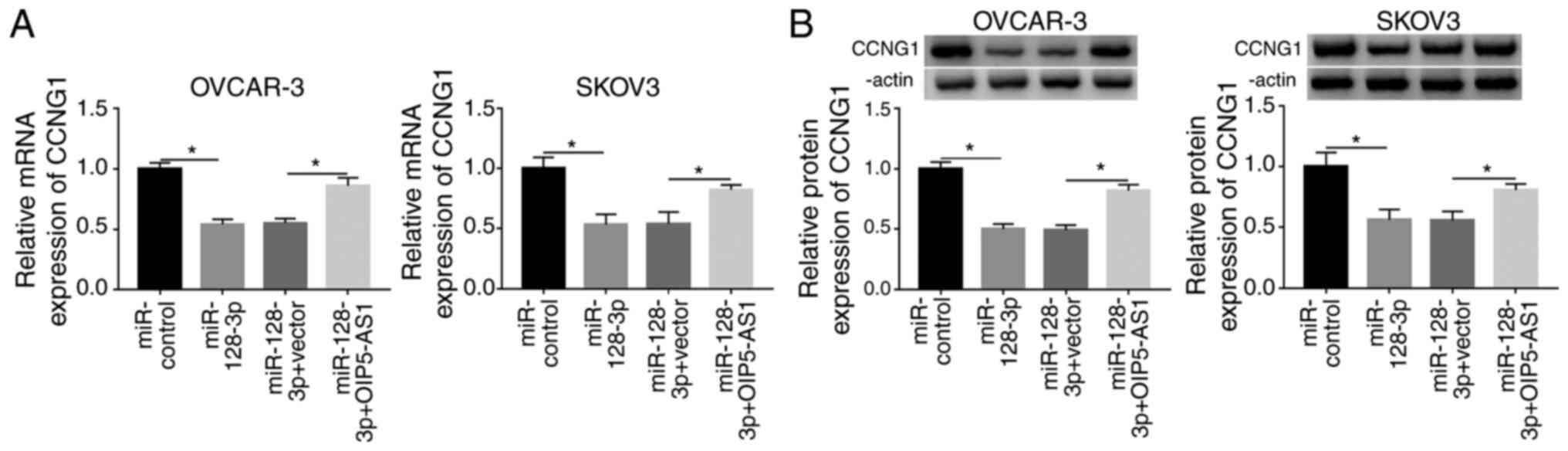
OIP5-AS1 upregulated CCNG1 expression
via sponging miR-128-3p in OC cells
To ascertain whether CCNG1 could be regulated by
OIP5-AS1, OVCAR-3 and SKOV3 cells were respectively transfected
with miR-control, miR-128-3p, miR-128-3p+vector or
miR-128-3p+OIP5-AS1, followed by the analysis of RT-qPCR and
western blotting. As shown in Fig. 7A
and B, miR-128-3p transfection resulted in reducing the CCNG1
mRNA and protein levels, which was ameliorated by ectopic high
expression of OIP5-AS1. In sum, OIP5-AS1 could upregulate the CCNG1
level by targeting miR-128-3p.
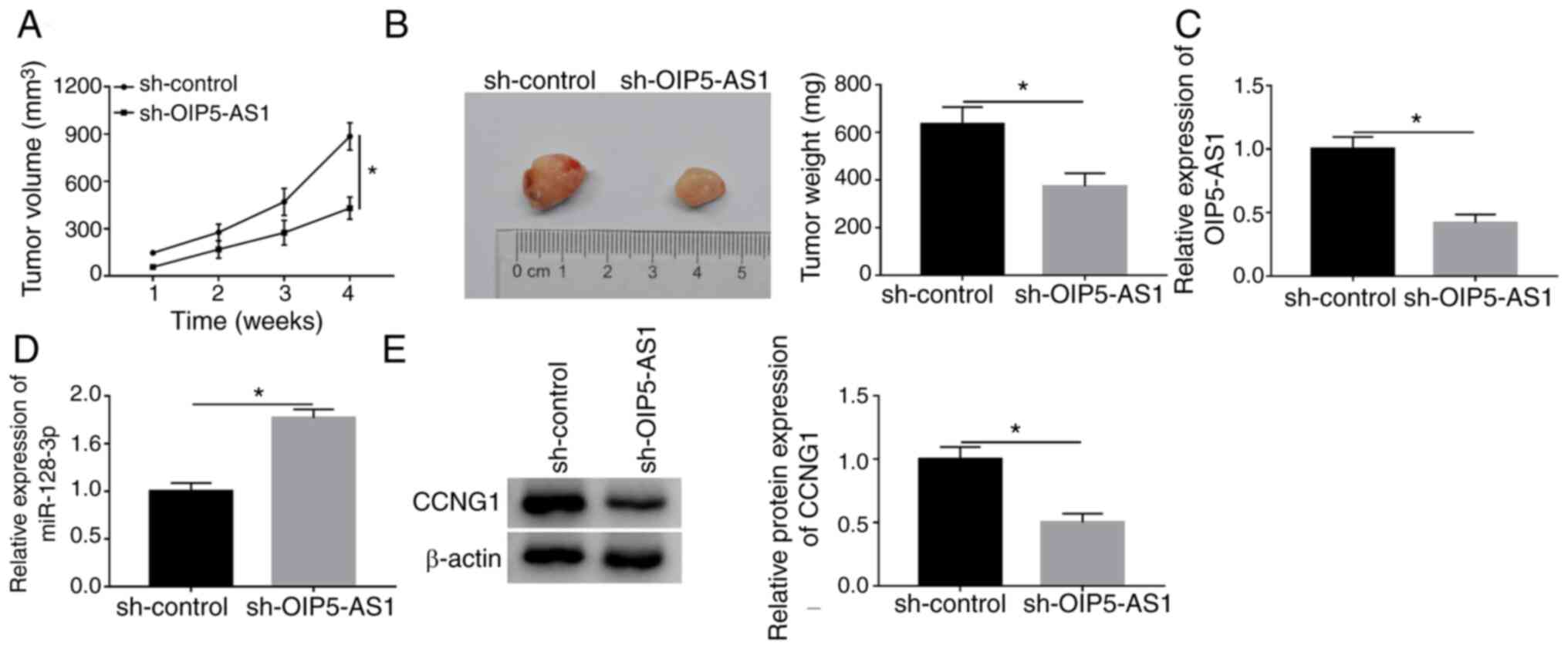
OIP5-AS1 depression inhibited tumor
growth of OC by miR-128-3p/CCNG1 axis in vivo
The xenograft tumor model was constructed to explore
the effect of OIP5-AS1 on OC in vivo. As Fig. 8A and B demonstrate, tumor volume and
weight were significantly lower in sh-OIP5-AS1 group compared with
the sh-control group, implying that OIP5-AS1 knockdown restrained
the OC carcinogenesis in vivo. In contrast to the sh-control
group, the expression of OIP5-AS1 was downregulated (Fig. 8C) but miR-128-3p was upregulated
(Fig. 8D) in sh-OIP5-AS1 group
according to the results of RT-qPCR. CCNG1 protein level was
decreased with the downregulation of OIP5-AS1 in western blot assay
(Fig. 8E). Therefore, OIP5-AS1
knockdown impeded the oncogenesis of OC by the miR-128-3p/CCNG1
axis in vivo.
Discussion
OC is one of the most common and familiar diseases
of the reproductive system with an estimated 239,000 new cases
being diagnosed worldwide annually (1), which can lead to the severe injury and
mortality. Present clinical therapies leave much to be desired due
to the dissatisfactory outcomes for OC patients. In recent years,
lncRNAs have been reported to participate in regulating various
types of cancers, including OC (34,35).
In the present study, OIP5-AS1 was considered as a diagnostic and
therapeutic biomarker in OC and the OIP5-AS1/miR-128-3p/CCNG1
network was revealed.
A previous study, using the analysis of The Cancer
Genome Consortium, suggested that OIP5-AS1 was overexpressed in
human epithelial origin types of cancer, including lung, cervical
and head and neck tumors (36).
Consistent with this finding, the present study also found that the
expression of OIP5-AS1 in OC tissues and cells was conspicuously
upregulated. It has been reported that the proliferation and
migration of glioma cells are inhibited after OIP5-AS1 knockdown
(37). In addition, a study of
cervical cancer revealed that silencing OIP5-AS1 reduced cell
proliferation of HeLa cells (38).
Bai and Li (39) stated that cell
proliferation and migration were impeded, while more apoptotic
cells were induced, following the silence of OIP5-AS1 in gastric
cancer. Knockdown of OIP5-AS1 restrained cell viability, migration,
invasion and induced apoptosis in OC cells during the current
study, which agrees with the results of OIP5-AS1 in other OC
research (16,17). The link between OIP5-AS1 and
glycolysis has yet to be reported. The present study revealed the
inhibitory effects of low OIP5-AS1 expression on glucose
consumption, lactate production and HK2 level. The acceleration of
glycolysis by OIP5-AS1 also reflected its oncogenic function in OC
evolution.
Previous studies have reported that lncRNAs can
inhibit miRNA activity as miRNA ‘sponges’ (40–42).
OIP5-AS1 is reported to interact with miRNAs in different types of
cancer, such as lung cancer and glioma (14,15).
In the present study, miR-128-3p was identified as a target of
OIP5-AS1 and OIP5-AS1 was involved in the cellular processes of OC
by sponging miR-128-3p. Sun et al (43) found that OIP5-AS1 regulates Wnt-7b
expression via targeting miR-410 in glioma cells. Nevertheless, the
target of miR-128-3p in OC cells remains to be elucidated.
Glycolytic metabolism has been found to be associated with G1/S
transition in cell cycle progression of types of cancer (44,45).
In 1996, Horne et al (46)
first discovered the high expression of CCNG1 in skeletal muscle,
kidney and ovary. Knockdown of CCNG1 can reduce the incidence of
hepatic tumor (47). The present
study found that downregulating CCNG1 repressed cell growth,
migration, invasion and glycolysis, demonstrating the pro-cancer
effect of CCNG1 on OC. Subsequently, CCNG1 was reported to promote
OC progression as the target genes of miR-1271 and miR-23b
(28,29). The data of the present study proved
that CCNG1 functioned as a downstream target of miR-128-3p and the
role of miR-128-3p in OC was attributed to the negative regulation
of CCNG1.
Notably, OIP5-AS1 has already been proved to
modulate glioma progression through the miR-410/Wnt-7b axis
(43). The results of the present
study clearly indicated that OIP5-AS1 indirectly regulated the
CCNG1 level by sponging miR-128-3p. The oncogenic role of OIP5-AS1
in OC was achieved by the regulatory network of miR-128-3p/CCNG1.
Furthermore, the xenograft tumor assay also demonstrated that the
carcinogenic effect of OIP5-AS1 in OC was dependent on the
miR-128-3p/CCNG1 axis in vivo.
There are certain limitations to the current study.
First, the effects of OIP5-AS1/miR-128-3p/CCNG1 on several
signaling pathways remain to be elucidated. Second, other targets
of miR-128-3p associated with glycolysis need to be discovered.
Given the involvement of OIP5-AS1 and miR-128-3p in glycolytic
metabolism, it would be worth exploring the potentials of
glycolytic genes including phosphoglycerate dehydrogenase and
glucose 6-phosphatase as the targets of miR-128-3p.
In conclusion, the present study revealed that
OIP5-AS1 worked as a tumorigenic factor in OC development via the
miR-128-3p/CCNG1 axis and revealed the OIP5-AS1/miR-128/CCNG1
modulatory network in OC. The present study might lay the
foundation for OC progression at the lncRNA level.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XF and XW were responsible for the conceptualization
and methodology; YaL, YuL and XS performed formal analysis and data
curation; YuL and YaL were responsible for validation and
investigation; and YuL, XF and XW prepared the original draft of
the manuscript, which they wrote, reviewed and edited. YuL and XF
confirm the authenticity of all raw data. All authors reviewed and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethical review
committee of the Shengli Oilfield Central Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu E, Liu Z and Zhou Y:
Carboplatin-docetaxel-induced activity against ovarian cancer is
dependent on up-regulated lncRNA PVT1. Int J Clin Exp Pathol.
8:3803–3810. 2015.PubMed/NCBI
|
|
4
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: LncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chai Y, Liu J, Zhang Z and Liu L:
HuR-regulated lncRNA NEAT1 stability in tumorigenesis and
progression of ovarian cancer. Cancer Med. 5:1588–1598. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ganapathy-Kanniappan S: Molecular
intricacies of aerobic glycolysis in cancer: Current insights into
the classic metabolic phenotype. Crit Rev Biochem Mol Biol.
53:667–682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vaupel P, Schmidberger H and Mayer A: The
warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang B, Zhang L, Cao Y, Chen S, Cao J, Wu
D, Chen J, Xiong H, Pan Z, Qiu F, et al: Overexpression of lncRNA
IGFBP4-1 reprograms energy metabolism to promote lung cancer
progression. Mol Cancer. 16:1542017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu Y, Sun H, Hu J and Zhang X: LncRNA
DLX6-AS1 promotes the progression of neuroblastoma by activating
STAT2 via targeting miR-506-3p. Cancer Manag Res. 12:7451–7463.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li N and Zhan X and Zhan X: The lncRNA
SNHG3 regulates energy metabolism of ovarian cancer by an analysis
of mitochondrial proteomes. Gynecol Oncol. 150:343–354. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang M, Sun X, Yang Y and Jiao W: Long
non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells
and leads to poor prognosis by targeting miR-378a-3p. Thorac
Cancer. 9:939–949. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Zheng J, Xue Y, Yu H, Gong W, Wang
P, Li Z and Liu Y: PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop
regulates the biological behavior of glioma cells. Theranostics.
8:1084–1105. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo L, Chen J, Liu D and Liu L:
OIP5-AS1/miR-137/ZNF217 axis promotes malignant behaviors in
epithelial ovarian cancer. Cancer Manag Res. 12:6707–6717. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu QY, Jiang XX, Tian HN, Guo HL, Guo H
and Guo Y: Long non-coding RNA OIP5-AS1 plays an oncogenic role in
ovarian cancer through targeting miR-324-3p/NFIB axis. Eur Rev Med
Pharmacol Sci. 24:7266–7275. 2020.PubMed/NCBI
|
|
18
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen J, Zhao D and Meng Q: Knockdown of
HCP5 exerts tumor-suppressive functions by up-regulating tumor
suppressor miR-128-3p in anaplastic thyroid cancer. Biomed
Pharmacother. 116:1089662019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao J, Li D and Fang L: MiR-128-3p
suppresses breast cancer cellular progression via targeting LIMK1.
Biomed Pharmacother. 115:1089472019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Xie L, Liang H and Cui Y: LncRNA
MIAT facilitates osteosarcoma progression by regulating
mir-128-3p/VEGFC axis. IUBMB Life. 71:845–853. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu M, Zhou K, Wu Y, Wang L and Lu S:
Linc0a0161 regulated the drug resistance of ovarian cancer by
sponging microRNA-128 and modulating MAPK1. Mol Carcinog.
58:577–587. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li R, Gong L, Li P, Wang J and Bi L:
MicroRNA-128/homeobox B8 axis regulates ovarian cancer cell
progression. Basic Clin Pharmacol Toxicol. 125:499–507. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baek WK, Kim D, Jung N, Yi YW, Kim JM, Cha
SD, Bae I and Cho CH: Increased expression of cyclin G1 in
leiomyoma compared with normal myometrium. Am J Obstet Gynecol.
188:634–639. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Perez R, Wu N, Klipfel AA and Beart RW Jr:
A better cell cycle target for gene therapy of colorectal cancer:
Cyclin G. Gastroenterology. 7:884–889. 2003.
|
|
27
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Ma L, Rao Q, Mao Y, Xin Y, Xu H, Li
C and Wang X: MiR-1271 inhibits ovarian cancer growth by targeting
cyclin G1. Med Sci Monit. 21:3152–3158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan J, Jiang JY, Meng XN, Xiu YL and Zong
ZH: MiR-23b targets cyclin G1 and suppresses ovarian cancer
tumorigenesis and progression. J Exp Clin Cancer Res. 35:312016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Albright JL: Dairy animal welfare: Current
and needed research. J Dairy Sci. 70:2711–2731. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Swearengen JR: Common challenges in
safety: A review and analysis of AAALAC findings. ILAR J.
59:127–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lou W, Ding B and Fu P: Pseudogene-derived
lncRNAs and their miRNA sponging mechanism in human cancer. Front
Cell Dev Biol. 8:852020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu DD, Chen X, Sun KX, Wang LL, Chen S and
Zhao Y: Role of the lncRNA ABHD11-AS1 in the tumorigenesis and
progression of epithelial ovarian cancer through targeted
regulation of RhoC. Mol Cancer. 16:1382017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu FF, Zheng FY, Wang HO, Zheng JJ and
Zhang Q: Downregulation of lncRNA TUBA4B is associated with poor
prognosis for epithelial ovarian cancer. Pathol Oncol Res.
24:419–425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Arunkumar G, Anand S, Raksha P,
Dhamodharan S, Prasanna Srinivasa Rao H, Subbiah S, Murugan AK and
Munirajan AK: LncRNA OIP5-AS1 is overexpressed in undifferentiated
oral tumors and integrated analysis identifies as a downstream
effector of stemness-associated transcription factors. Sci Rep.
8:70182018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu GW, Wu L, Kuang W, Chen Y, Zhu XG, Guo
H and Lang HL: Knockdown of linc-OIP5 inhibits proliferation and
migration of glioma cells through down-regulation of YAP-NOTCH
signaling pathway. Gene. 610:24–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Naemura M, Kuroki M, Tsunoda T, Arikawa N,
Sawata Y, Shirasawa S and Kotake Y: The long noncoding RNA OIP5-AS1
is involved in the regulation of cell proliferation. Anticancer
Res. 38:77–81. 2018.PubMed/NCBI
|
|
39
|
Bai Y and Li S: Long noncoding RNA
OIP5-AS1 aggravates cell proliferation, migration in gastric cancer
by epigenetically silencing NLRP6 expression via binding EZH2. J
Cell Biochem. 121:353–362. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sen R, Ghosal S, Das S, Balti S and
Chakrabarti J: Competing endogenous RNA: The key to
posttranscriptional regulation. Sci World J. 2014:8962062014.
View Article : Google Scholar
|
|
41
|
Tan JY, Sirey T, Honti F, Graham B,
Piovesan A, Merkenschlager M, Webber C, Ponting CP and Marques AC:
Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in
mouse embryonic stem cells. Genome Res. 25:655–666. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao
WZ, Peng H, Hong WW, Yin LH, Pu YP and Liang GY: Integrated
analysis of long non-coding RNA-associated ceRNA network reveals
potential lncRNA biomarkers in human lung adenocarcinoma. Int J
Oncol. 49:2023–2036. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun WL, Kang T, Wang YY, Sun JP, Li C, Liu
HJ, Yang Y and Jiao BH: Long noncoding RNA OIP5-AS1 targets Wnt-7b
to affect glioma progression via modulation of miR-410. Biosci Rep.
39:BSR201803952019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Escoté X and Fajas L: Metabolic adaptation
to cancer growth: From the cell to the organism. Cancer Lett.
356:171–175. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Icard P, Fournel L, Wu Z, Alifano M and
Lincet H: Interconnection between metabolism and cell cycle in
cancer. Trends Biochem Sci. 44:490–501. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Horne MC, Goolsby GL, Donaldson KL, Tran
D, Neubauer M and Wahl AF: Cyclin G1 and cyclin G2 comprise a new
family of cyclins with contrasting tissue-specific and cell
cycle-regulated expression. J Biol Chem. 271:6050–6061. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jensen MR, Factor VM, Fantozzi A, Helin K,
Huh CG and Thorgeirsson SS: Reduced hepatic tumor incidence in
cyclin G1-deficient mice. Hepatology. 37:862–870. 2003. View Article : Google Scholar : PubMed/NCBI
|