Introduction
Oral cancer (OC) refers to a group of tumors found
in the lining of the mouth, lips, throat, tongue or cheek (1). At present, >540,000 individuals are
diagnosed with OC annually, and the 5-year survival rate of
patients with OC is <50% (1).
Oral squamous cell carcinoma (OSCC) is the most common type of OC,
ranking eighth in terms of cancer incidence worldwide (2). This type of mouth cancer has gradually
become a public health problem due to its high incidence and low
cure rate (3). Numerous methods,
including surgery, radiotherapy and chemotherapy, have been used to
treat OC; however, these treatments do not achieve the expected
results (4–6). To enhance the prognosis of patients
with OC, more treatment approaches should be explored.
Laminin subunit γ2 (LAMC2) serves an important role
in OC, and the human LAMC2 gene is located on chromosome 1q25-q31
(7). More specifically, this gene
has been reported to be involved in head and neck cancer (8). Laminin γ2, which belongs to the
laminin protein family, is a basal lamina glycoprotein encoded by
the LAMC2 gene (9). As a specific
biomarker, LAMC2 is expressed in several types of malignant cancer,
including gastric cancer (10),
esophageal squamous cell carcinoma (11) and pancreatic ductal adenocarcinoma
(12). Furthermore, cell-surface
receptors combine with LAMC2 to guide tumor migration and invasion,
thus making it a possible effective cancer target (7,11,13,14).
To the best of our knowledge, the present study was the first to
investigate the expression levels and effects of LAMC2 in OC.
With a length of 18–25 nucleotides, microRNAs
(miRNAs/miRs) were previously regarded as junk RNAs that have no
effects (15–17). However, previous studies revealed
that when miRNAs are involved in the development of tumors, they
can inhibit the translation of mRNAs and shorten the half-life of
mRNAs (15–17). In tumors, most downregulated miRNAs
function as tumor suppressors, whereas upregulated miRNAs function
as cancer-promoting factors (18–21).
Efficient diagnosis and prognosis of OC may become possible, with
various tumors exhibiting a signature miRNA profile associated with
tumor progression. Small non-coding RNA molecule miR-5580 was once
identified along with another 84 miRNAs using the miRDeep 2
algorithm (22). The gene that
encodes miR-5580 is located on chromosome14q22.2, according to the
National Center for Biotechnology Information genome database
(https://www.ncbi.nlm.nih.gov/gene/100847076). However,
to the best of our knowledge, no previous study has explored the
effects of miR-5580-3p in human cancer types.
Therefore, the roles of miR-5580-3p and LAMC2 in OC
deserve further investigation. The purpose of the present study was
to investigate whether and how miR-5580-3p affected OC cell
phenotypes by regulating LAMC2.
Materials and methods
Bioinformatics analysis
Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo), a public functional
database, stores mRNA and non-coding RNA expression profiles. Three
mRNA expression profiles, GSE19089 (23), GSE23558 (24) and GSE138206 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138206),
which included the mRNA expression profiling in OC were analyzed
using the GEO2R built-in algorithm by R software 4.0.4 (https://www.r-project.org/) on the GEO database with
the criteria of logFC≥1.5 and adjusted P<0.05. The intersected
genes from the three mRNA expression profiles were uploaded to the
Search Tool for the Retrieval of Interacting Genes/Proteins
database (https://string-db.org/) for enrichment
analysis. The Gene Expression Profiling Interactive Analysis
(GEPIA) survival analysis tool (http://gepia2.cancer-pku.cn/#survival) was used to
study the prognostic effects of the genes of interest in human head
and neck squamous cell carcinoma (HNSCC), and to obtain the
expression of LAMC2 in HNSCC. The target miRNAs of LAMC2 predicted
by TargetScan Human 7.2 (25) were
eventually overlapped with the downregulated miRNAs in the GSE98463
dataset (a miRNA microarray dataset) (26) with logFC<-1.5 and P<0.05.
Clinical samples collection from
patients with OC
A total of 40 patients diagnosed with OC at Puai
Hospital, Tongji Medical College of Huazhong University of Science
and Technology (Wuhan, China) participated in the present study.
The inclusion criteria were patients with OC without radiotherapy,
chemotherapy or other treatments, and the exclusion criteria were
the patients with OC and with other diseases. The participants were
divided into a training set (n=20) and a validation set (n=20; the
requirement of Mann-Whitney test). The cancer tissues and
corresponding adjacent healthy tissues (<3 cm from tumor
tissues) were collected between January 2020 and March 2020, frozen
and embedded in paraffin until they were used in the present study.
The clinical characteristics of the 40 patients are listed in
Table I. The present study was
approved by the Ethics Committee of Puai Hospital, Tongji Medical
College, Huazhong University of Science and Technology (approval
no. KY2020-501-01). All patients who participated in the present
study provided written informed consent. It was ensured that all
experimental procedures, including the use and collection of
tissues, followed the ethical standards set out in the Declaration
of Helsinki.
 | Table I.Clinical characteristics of patients
with oral cancer in the training (n=20) and validation (n=20)
sets. |
Table I.
Clinical characteristics of patients
with oral cancer in the training (n=20) and validation (n=20)
sets.
| Pathological
characteristics | Training set, n
(%) | Validation set, n
(%) |
|---|
| Age, years |
|
|
|
>50 | 12 (60) | 10 (50) |
|
≤50 | 8
(40) | 10 (50) |
| Sex |
|
|
|
Male | 11 (55) | 12 (60) |
|
Female | 9
(45) | 8
(40) |
| Site |
|
|
| Buccal
mucosa | 6
(30) | 5
(25) |
|
Tongue | 6
(30) | 6
(30) |
|
Alveolus | 8
(40) | 9
(45) |
| T
classification |
|
|
|
T1-T2 | 11 (55) | 10 (50) |
| T3 | 6
(30) | 6
(30) |
| T4 | 3
(15) | 4
(20) |
| N
classification |
|
|
| N0 | 9
(45) | 8
(40) |
| N1 | 7
(35) | 8
(40) |
| N2 | 4
(20) | 4
(20) |
| Metastasis |
|
|
| No | 20
(100) | 20
(100) |
|
Yes | 0 | 0 |
| Histological
differentiation |
|
|
|
Well | 6
(30) | 8
(40) |
|
Moderate | 9
(45) | 7
(35) |
|
Poor | 5
(25) | 5
(25) |
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from tissues and
cells, according to the manufacturer's protocols. miRNAs were
purified using the miRcute miRNA isolation kit (cat. no. DP501;
Tiangen Biotech Co., Ltd.). Prior to the RNA reverse transcription
process, RNA was quantified using gel electrophoresis. miR-5580-3p
reverse transcription (cat. no. KR211; Tiangen Biotech Co., Ltd.)
and LAMC2 mRNA reverse transcription (cat. no. RR037A; Takara Bio,
Inc.) were then performed, according to the manufacturer's
protocols. Subsequently, the 7500 Fast Dx Real-Time PCR Instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to
analyze the expression levels of miR-5580-3p using a miRcute Plus
miRNA qPCR kit (SYBR Green) (cat. no. FP411; Tiangen Biotech Co.,
Ltd.) with the thermocycling conditions: 95°C 15 min, 40 cycles of
94°C 20 sec and 60°C 34 sec, and the mRNA expression levels of
LAMC2 were analyzed using a TB Green® Premix Ex Taq™ kit
(cat. no. RR420A; Takara Bio, Inc.) with the thermocycling
conditions: 95°C 30 sec, 40 cycles of 95°C 5 sec and 60°C 30 sec.
U6 and GAPDH were used as reference genes for miR-5580-3p and LAMC2
mRNA quantification using 2−ΔΔCq method (27), respectively. All primers are listed
in Table II.
 | Table II.Primer sequences included in the
present study. |
Table II.
Primer sequences included in the
present study.
| Genes | Primer sequences
(5′→3′) |
|---|
| miR-5580-3p | F:
CACAGTTGAAGAGAGCCAGCAC |
|
| R:
CAGTGCGTGTCGTGGAGT |
| LAMC2 | F:
GGAGCTGGAGTTTGACACGA |
|
| R:
GAGCATGGAGCTGGAAGGTT |
| GAPDH | F:
ATGGAGAAGGCTGGGGCTC |
|
| R:
AAGTTGTCATGGATGACCTTG |
| U6 | F:
TGCGGGTGCTCGCTTCGGCAGC |
|
| R:
CCAGTGCAGGGTCCGAGGT |
Cell lines and transfection
The human OC cell lines (SCC-4, SCC-9 and Cal-27),
as well as the human oral epithelial cell line (HOEC; cat. no.
BNCC340217), were purchased from BeNa Culture Collection; Beijing
Beina Chuanglian Biotechnology Research Institute. SCC-4, SCC-9,
Cal-27 and HOEC cells were cultured in DMEM (cat. no. C11965500BT;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(cat. no. 10439024; Gibco; Thermo Fisher Scientific, Inc.). These
cells were subsequently placed in a humidified incubator at 37°C
with 5% CO2. All plasmids, including small interfering
(si)RNA-LAMC2 (cat. no. siG000003918A-1-5), overexpression
(OE)-LAMC2, miR-5580-3p mimic (cat. no. miR10022274-1-5),
miR-5580-3p inhibitor and negative controls (NC), such as si-NC and
OE-NC, were obtained from Guangzhou RiboBio Co., Ltd. SCC-4 and
Cal-27 cells were seeded (1×106 cells/well) into plates
for 24 h. Subsequently, the samples were transfected with 50 nM
plasmids using Lipofectamine® 2000 reagent (cat. no.
11668027; Thermo Fisher Scientific, Inc.) for 48 h at 37°C. The
following sequences were used in the present study: miR-5580-3p
mimic,
5′-UGCUGGCUCAUUUCAUAUGUGUGCUGAGAAAAUUCACACAUAUGAAGUGAGCCAGCAC-3′;
miR-5580-3p inhibitor,
5′-ACGACCGAGUAAAGUAUACACACGACUCUUUUAAGUGUGUAUACUUCACUCGGUCG-3′;
miR-5580-3p mimic NC, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′; and inhibitor
NC, 5′-CAGTACTTTTGTGTAGTACAA-3′. The NC for LAMC2 overexpression
was an empty pEXP-RB-Mam vector (Guangzhou RiboBio Co., Ltd.), and
the NC for LAMC2 knockdown was an empty pRNAT-U6.1 vector
(Guangzhou RiboBio Co., Ltd.). After 48 h transfection, the
transfected cells were used for the subsequent experiments.
Luciferase reporter assay
SCC-4 and Cal-27 cells were seeded into 96-well
plates at a density of 3×105 cells/well. The wild-type
(Wt) pEZX-MT05-LAMC2-3′ untranslated region (3′UTR) constructs and
the mutant-type (Mut) pEZX-MT05-LAMC-3′UTR constructs were
purchased from GeneCopoeia, Inc. The 3′UTR constructs were
co-transfected with miR-5580-3p mimic, miR-5580-3p inhibitor, mimic
control or inhibitor control into SCC-4 and Cal-27 cells using
Lipofectamine 2000 reagent. Subsequently, the Secrete-Pair Dual
Luminescence assay kit (cat. no. LF031; GeneCopoeia, Inc.) was used
to detect the firefly and secreted alkaline phosphatase (SEAP)
activities. Cells were collected after 72 h of transfection and the
fluorescence intensity was measured. Relative luciferase activity
was normalized to SEAP.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was determined using a CCK-8 assay
(cat. no. HY-K0301; MedChemExpress). Briefly, 1,500 transfected
SCC-4 and Cal-27 cells were seeded into the wells of 96-well plates
and cultured at 37°C with 5% CO2 for 24 h. From the next
day, at 0, 24, 48 and 72 h, 10 µl CCK-8 solution was added to the
corresponding wells of each group. Next, the plate was incubated
for 4 h. Finally, the optical density (OD) at 450 nm of each well
was determined using a microplate reader.
5-Bromo-2′-deoxyuridine (BrdU) cell
proliferation assay
A BrdU incorporation assay was used to evaluate cell
proliferation. After the transfected SCC-4 and Cal-27 cells
(3×105 cells/well) were seeded into 96-well plates for 1
day, the old medium was replaced with DMEM without FBS.
Subsequently, 10 µl BrdU (cat. no. ab126556; Abcam) was added to
each well for an incubation period of 4 h to allow proliferating
cells to incorporate BrdU into their DNA. Then, the cells were
fixed using the fixing solution included in the kit. Primary
antibody against BrdU (prediluted, ready-to-use, provided in the
kit) and secondary HRP-conjugated antibody (1:2,000, provided in
the kit) were sequentially added to the wells and incubated at room
temperature. 3,3′,5,5′-tetramethylbenzidine solution was added to
develop the color. Finally, the OD at 450 nm was measured using a
scanning multi-well spectrophotometer immediately after the stop
solution was added.
Wound healing migration assay
A wound healing assay was performed to examine cell
migration. Briefly, SCC-4 and Cal-27 cells (3×106
cells/well) were seeded into 12-well plates for 24 h. Once the cell
density reached 80%, the fused cell monolayer was scratched in the
center using a 20-µl micropipette tip. Subsequently, the
non-adherent cells were washed off with PBS. The DMEM medium was
then replaced with serum-free DMEM medium, and the cells were
cultured for another 24 h. At 0 and 24 h, images were captured
under a light microscope at 100× magnification to observe the wound
width. The migration rate was calculated as: (wound width at 0
h-wound width at 24 h)/wound width at 0 h.
Western blotting
RIPA buffer (cat. no. 20-188; Sigma-Aldrich; Merck
KGaA) with 5 mM EDTA (cat. no. V900106; Sigma-Aldrich; Merck KGaA)
and PMSF (cat. no. 78830; Sigma-Aldrich; Merck KGaA) was used to
extract and lyse protein from SCC-4 and Cal-27 cells. After
determining protein concentration by BCA Protein Assay Kit (Thermo
Fisher Scientific, Inc.), 30 µg protein was separated via SDS-PAGE
on 10% gel. The separated proteins were then transferred to PVDF
membranes (cat. no. ISEQ00010; EMD Millipore). Next, the membranes
were blocked with 5% milk in TBS with 0.1% Tween-20 at room
temperature for 2 h. Subsequently, the membranes were incubated
with primary antibodies against LAMC2 (1:1,000; cat. no. ab210959;
Abcam) and GAPDH (1:5,000; cat. no. ab181602; Abcam) at 4°C
overnight. The following day, the membranes were incubated with the
Goat Anti-Rabbit IgG H&L (HRP) antibody (1:10,000; cat. no.
ab97051; Abcam) for 2 h at room temperature. Finally, ECL reagent
(cat. no. 1705062; Bio-Rad Laboratories, Inc.) was used to
visualize the protein signals. Image Lab v3.0 software (Bio-Rad
Laboratories, Inc.) was used for densitometry.
Statistical analysis
The experiments were repeated three times, and the
data were shown as mean ± standard deviation. SPSS v23.0 (IBM
Corp.) was used to analyze all data collected in the present study.
The Wilcoxon test was used to analyze the differences between tumor
and healthy groups for tissue data. Unpaired Student's t-test was
used to analyze the differences between two groups for cell
experiments. One-way ANOVA with Dunnett's post hoc test was used to
evaluate the differences among multiple groups in cell experiments.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-5580-3p/LAMC2 as a potential
biomarker and therapeutic target in OC
By analyzing three mRNA expression profiles
(GSE19089, GSE23558 and GSE138206), 35 common differentially
expressed genes were obtained using the criteria of logFC≥1.5 and
adjusted P<0.05 (Fig. 1A and B).
The mRNA expression analysis results for the GSE19089, GSE23558 and
GSE138206 datasets are shown in Tables
SI–III. The 35 genes were
uploaded to the Search Tool for the Retrieval of Interacting
Genes/Proteins database for enrichment analysis. The results
revealed that the ‘extracellular matrix organization’ biological
process and the ‘extracellular matrix component’ were significantly
enriched terms, in which LAMC2, collagen type IV α6 chain and
laminin subunit α3 were involved (Fig.
1C). Subsequently, the GEPIA survival analysis tool (http://gepia.cancer-pku.cn/detail.php)
was used to study the prognostic effects of the three genes in
human HNSCC. The results revealed that LAMC2 was a prognostic
marker (Fig. 1D). By analyzing
GEPIA expression data, it was also identified that LAMC2 expression
was markedly upregulated in HNSCC (Fig.
1E). Therefore, LAMC2 was considered to be the gene of
interest. The target miRNAs of LAMC2 predicted by TargetScan Human
7.2 (25) were overlapped with the
downregulated miRNAs in the GSE98463 dataset (a miRNA microarray
dataset) (26) with logFC<-1.5
and P<0.05. The results indicated that miR-5580-3p was a
potential cancer suppressor in OC (Fig.
1F). The targets of LAMC2 predicted by TargetScan Human 7.2 are
listed in Table SIV. The analysis
results for the GSE98463 dataset are shown in Table SV.
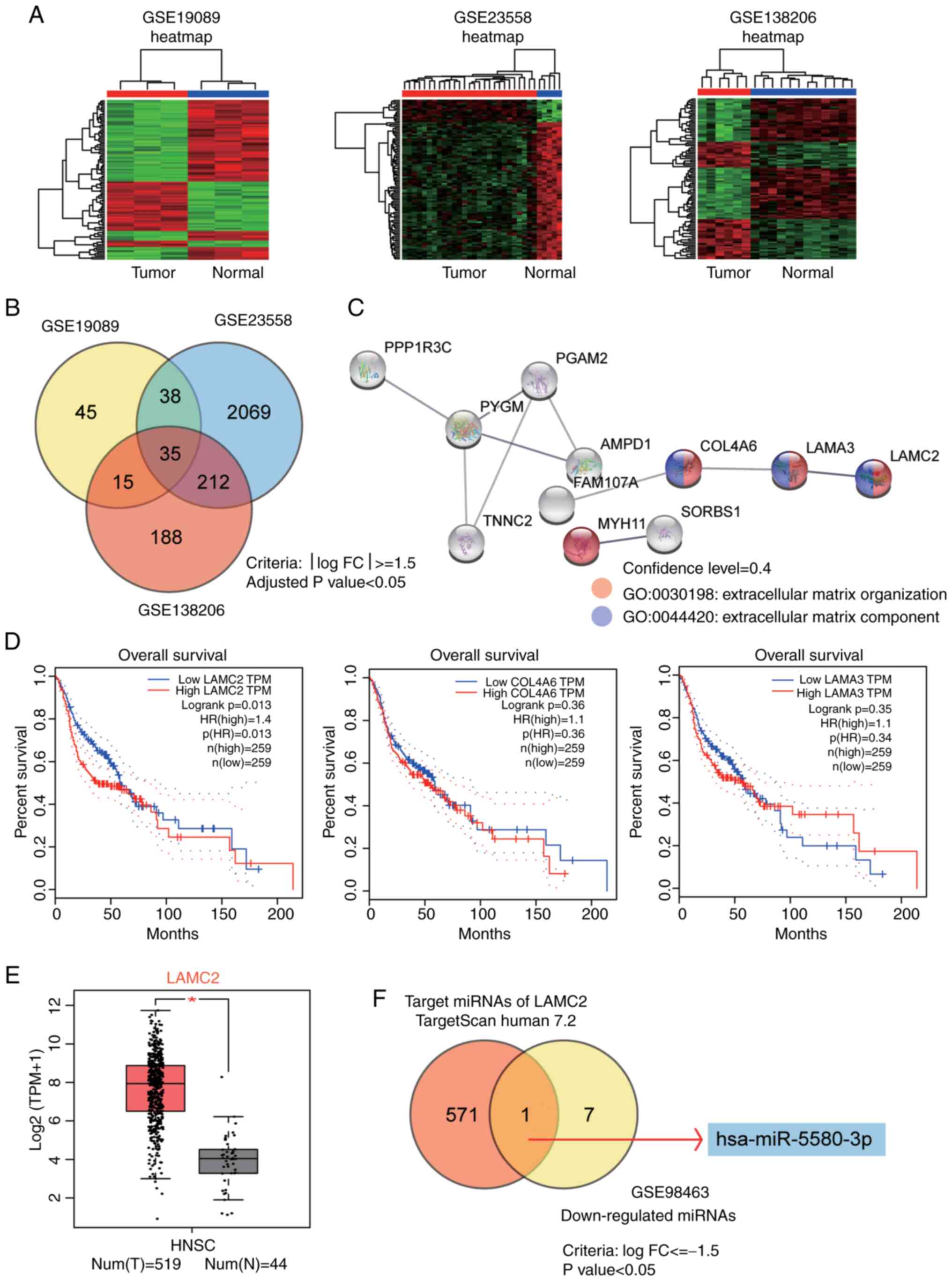 | Figure 1.Identification of miR-5580-3p and
LAMC2 as potential biomarkers and therapeutic targets in OC. (A)
Heatmaps of DEGs in three OC Gene Expression Omnibus datasets
(GSE19089, GSE23558 and GSE138206) using R software 4.0.4. (B) Venn
diagram revealing that 35 common DEGs were identified using the
criteria of logFC≥1.5 and adjusted P<0.05. (C) The 35 DEGs were
analyzed using the Search Tool for the Retrieval of Interacting
Genes/Proteins database, and the extracellular matrix organization
biological process and extracellular matrix component were
identified as enriched terms. Genes that were involved in the two
included LAMC2, COL4A6 and LAMA3. (D) GEPIA survival analysis was
employed to study the prognostic effects of the three genes in
HNSCC. (E) By interrogating GEPIA expression data, it was also
revealed that LAMC2 expression was upregulated in HNSCC.
*P<0.01. (F) Intersection of the target miRNAs of LAMC2
identified using TargetScan Human 7.2 and the downregulated miRNAs
in the GSE98463 dataset (criteria, logFC<-1.5 and adjusted
P<0.05). COL4A6, collagen type IV α6 chain; DEGs, differentially
expressed genes; FC, fold-change; GEPIA, Gene Expression Profiling
Interactive Analysis; HNSCC, head and neck squamous cell carcinoma;
LAMA3, laminin subunit α3; LAMC2, laminin subunit γ2; miR-5580-3p,
microRNA-5580-3p; miRNAs, microRNAs; num (T), number of the tumor
group; n (N), number of the normal group; OC, oral cancer. |
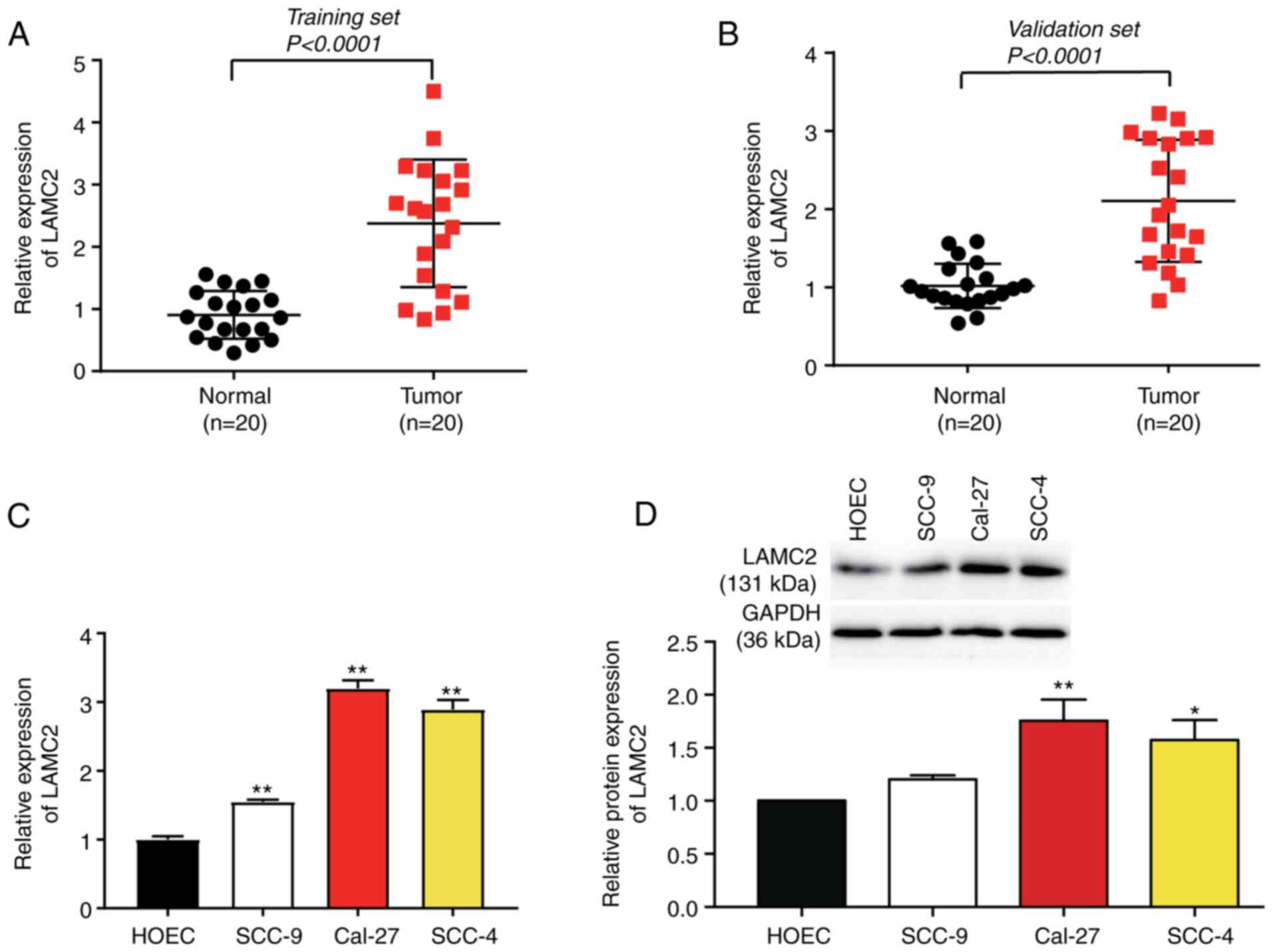
Upregulation of LAMC2 in OC
Compared with that in adjacent healthy tissues,
LAMC2 mRNA expression was upregulated ~2-fold in cancerous tissues
in the training and validation sets (Fig. 2A and B). LAMC2 mRNA and protein
expression levels were significantly upregulated in cancer cell
lines, particularly in Cal-27 and SCC-4 cells, compared with in the
HOEC cell line (Fig. 2C and D).
Therefore, SCC-4 and Cal-27 cells were selected for the follow-up
experiments.
LAMC2 enhances the viability,
proliferation and migration of OC cells
The successful transfection of si-LAMC2, si-NC,
OE-LAMC2 and OE-NC into SCC-4 and Cal-27 cells was demonstrated.
LAMC2 mRNA expression in the OE-LAMC2 group was increased 3-fold
compared with that in the OE-NC group. Furthermore, LAMC2 mRNA
expression was decreased by 0.7-fold in the si-LAMC2 group compared
with that in the si-NC group (Fig.
3A). Additionally, it was observed that the cell viability in
the OE-LAMC2 group was 0.5-fold higher compared with that in the
OE-NC group; however, that in the si-LAMC2 group was ~0.3-fold
lower compared with that in the si-NC group in SCC-4 and Cal-27
cells at 72 h (Fig. 3B). Cells in
the OE-LAMC2 group exhibited a 0.3-fold increase in cell
proliferation compared with cells in the OE-NC group. Nevertheless,
cells in the si-LAMC2 group had a 0.3-fold decline in the cell
proliferation level compared with cells in the si-NC group
(Fig. 3C). Furthermore, it was
observed that LAMC2 overexpression accelerated the wound healing
process by 0.3-fold, whereas LAMC2 knockdown decreased it by
~0.3-fold. This observation suggested that LAMC2 overexpression
enhanced OC cell migration (Fig.
3D). Overall, LAMC2 improved the viability, proliferation and
migration of OC cells.
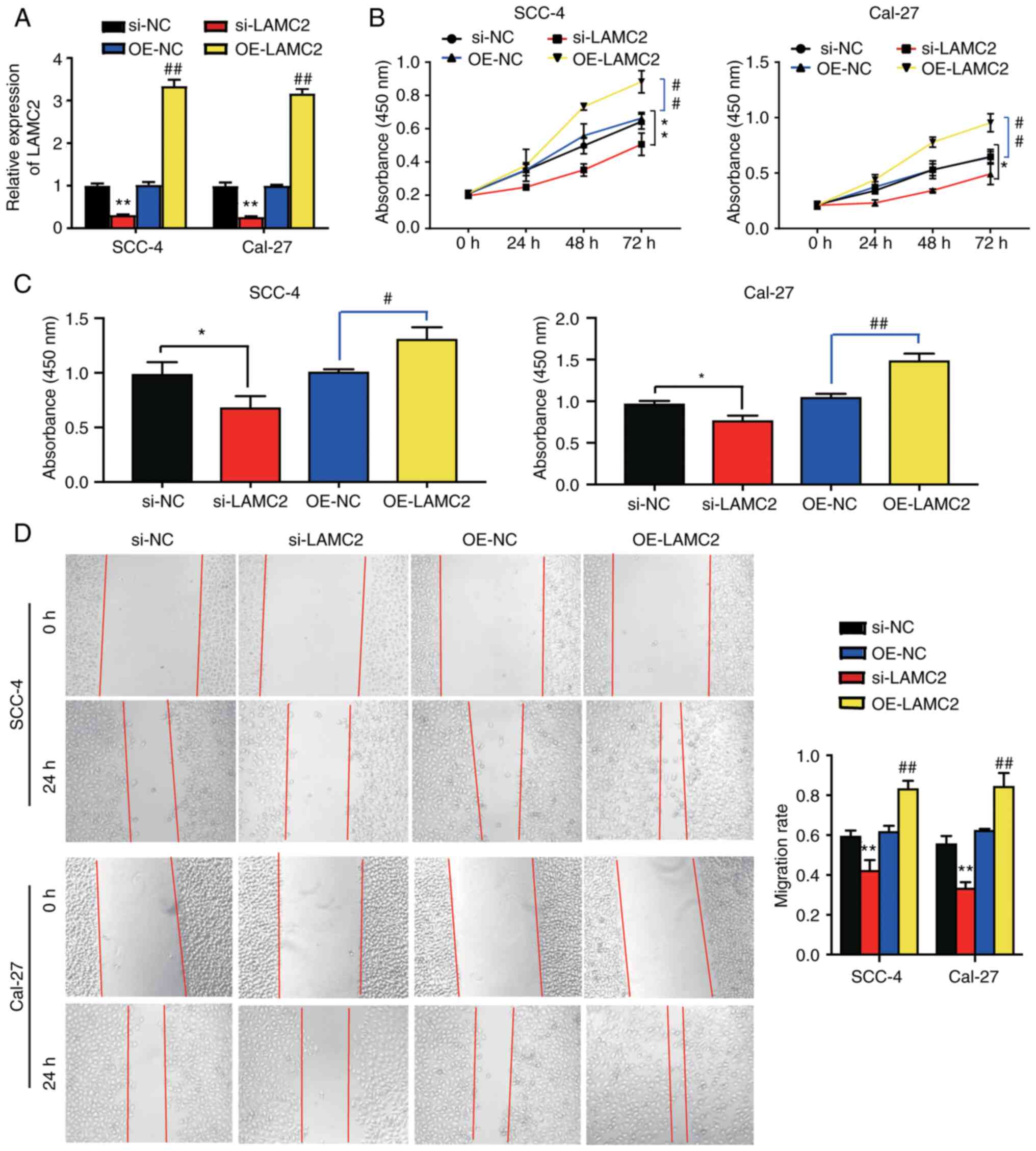 | Figure 3.LAMC2 affects the viability,
proliferation and migration of oral cancer cells. (A) Transfection
efficiency in oral tumor cell lines following transfection with
si-LAMC2, si-NC, OE-LAMC2 and OE-NC was detected by reverse
transcription-quantitative PCR. **P<0.01 vs. si-NC group;
##P<0.01 vs. OE-NC group. (B) Results of the Cell
Counting Kit-8 assay in SCC-4 and Cal-27 cells transfected with
si-LAMC2, si-NC, OE-LAMC2 and OE-NC. **P<0.01 vs. si-NC group;
##P<0.01 vs. OE-NC group at 72 h. (C) Results of the
5-bromo-2′-deoxyuridine incorporation assay in SCC-4 and Cal-27
cells transfected with si-LAMC2, si-NC, OE-LAMC2 and OE-NC.
*P<0.05 vs. si-NC group; #P<0.05 and
##P<0.01 vs. OE-NC group. (D) Wound healing assay
results in SCC-4 and Cal-27 cells. **P<0.01 vs. si-NC group;
##P<0.01 vs. OE-NC group. si-LAMC2, LAMC2 siRNA;
si-NC, siRNA-negative control; OE-LAMC2, overexpression of LAMC2;
OE-NC, negative control of overexpression; LAMC2, laminin subunit
γ2; si/siRNA, small interfering RNA. |
LAMC2 is a downstream target gene of
miR-5580-3p
The complementary relationship between miR-5580-3p
and LAMC2 mRNA is shown in Fig. 4A.
Luciferase activity in the cells co-transfected with Wt LAMC2 mRNA
3′UTR and miR-5580-3p mimic was decreased by 0.5-fold compared with
that in the cells co-transfected with Wt LAMC2 mRNA 3′UTR and
miR-5580-3p mimic-NC plasmids in both cell lines. On the other
hand, the luciferase activity in cells co-transfected with Wt LAMC2
mRNA 3′UTR and miR-5580-3p inhibitor was markedly increased by
~2-fold compared with that in the cells co-transfected with Wt
LAMC2 mRNA 3′UTR and miR-5580-3p inhibitor-NC (Fig. 4B). The expression levels of
miR-5580-3p were 0.5-fold lower in OC tissues compared with in the
healthy tissues in both the training and validation sets (Fig. 4C and D). The results of RT-qPCR
revealed that miR-5580-3p expression was significantly
downregulated in OC cell lines, particularly in Cal-27 and SCC-4
cells, compared with in the HOEC cell line (Fig. 4E). miR-5580-3p was significantly
upregulated in the mimic group, whereas LAMC2 mRNA was
significantly downregulated in the mimic group (Fig. S1). In addition, miR-5580-3p
expression was markedly increased (~2-fold) in the mimic + OE-NC
and mimic + OE-LAMC2 groups, compared with in the mimic-NC + OE-NC
group in Cal-27 and SCC-4 cells. LAMC2 expression was decreased by
~0.8-fold in the mimic + OE-NC group, but increased ~4-fold in the
mimic-NC + OE group compared with in the mimic-NC + OE-NC group in
Cal-27 and SCC-4 cells (Fig. 4F).
These results demonstrated that miR-5580-3p directly targeted
LAMC2.
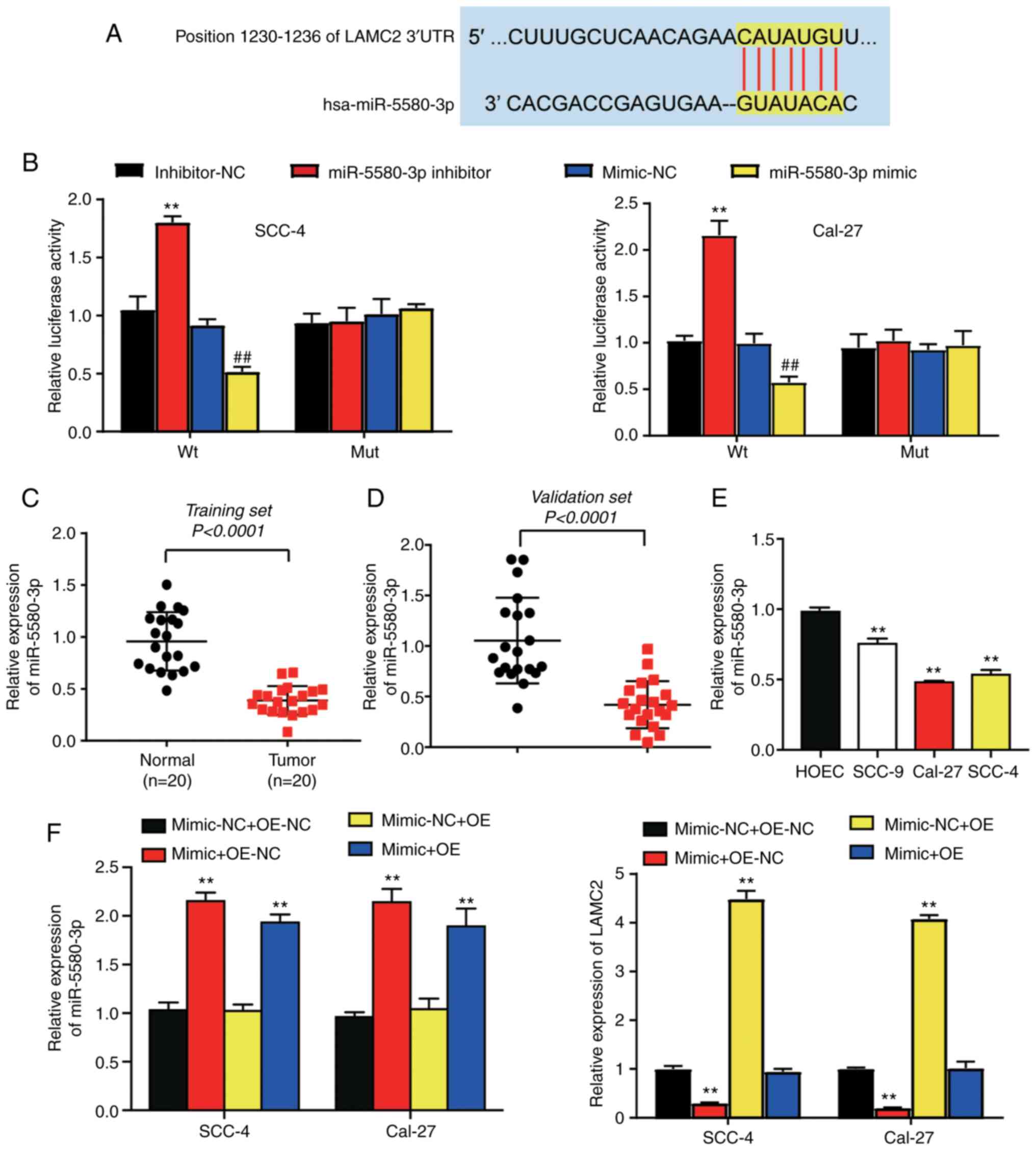 | Figure 4.LAMC2 is a downstream target gene of
miR-5580-3p. (A) Predicted binding sites of miR-5580-3p in the
LAMC2 mRNA 3′-UTR according to TargetScan Human 7.2. (B) A
luciferase reporter assay in SCC-4 and Cal-27 cells suggested that
LAMC2 is a downstream target of miR-5580-3p. **P<0.01 vs.
inhibitor-NC group; ##P<0.01 vs. mimic-NC group. (C)
miR-5580-3p expression was lower in oral tumor tissues than in
healthy tissues in the training set. P<0.001 (Wilcoxon test).
(D) miR-5580-3p expression was lower in oral tumor tissues compared
with that in healthy tissues in the validation set. P<0.001
(Wilcoxon test). (E) miR-5580-3p expression was significantly lower
in SCC-4 and Cal-27 cells than in HOEC cells. **P<0.01 vs. HOEC
cell line. (F) Expression levels of miR-5580-3p and LAMC2 in
co-transfection groups (miR-5580-3p mimic + OE-NC; mimic-NC +
OE-NC; miR-5580-3p mimic + OE-LAMC2; mimic-NC + OE-LAMC2) was
detected by reverse transcription-quantitative PCR. **P<0.01 vs.
mimic-NC + OE-NC group. Mimic-NC + OE-NC, miR-5580-3p mimic
negative control + LAMC2 overexpression negative control; mimic +
OE-NC, miR-5580-3p mimic + LAMC2 overexpression negative control;
mimic-NC + OE, miR-5580-3p mimic negative control + LAMC2
overexpression; mimic + OE, miR-5580-3p mimic + LAMC2
overexpression; LAMC2, laminin subunit γ2; miR-5580-3p,
microRNA-5580-3p; Wt, wild-type; Mut, mutant; UTR, untranslated
region; HOEC, human oral epithelial cell line. |
miR-5580-3p inhibits OC cell
viability, proliferation and migration by suppressing LAMC2
To further investigate whether miR-5580-3p inhibited
the viability, proliferation and migration of OC cells by
regulating LAMC2 mRNA, rescue experiments were designed. The
results of the CCK-8 assay revealed that the cell viability at 72 h
was decreased in the miR-5580-3p mimic + OE-NC group, whereas the
cell viability in the mimic-NC + OE-LAMC2 group was increased,
compared with that in the mimic-NC + OE-NC group. The viability of
cells in the miR-5580-3p mimic + OE-LAMC2 group was similar to that
of cells in the mimic-NC + OE-NC group, suggesting that LAMC2 could
reverse the effect of miR-5580-3p on OC cell viability (Fig. 5A). Furthermore, cell proliferation
was decreased by ~0.2-fold in the miR-5580-3p mimic + OE-NC group,
while that in the mimic-NC + OE-LAMC2 group was increased by nearly
0.5-fold compared with that in the mimic-NC + OE-NC group. In
addition, in the miR-5580-3p mimic + OE-LAMC2 group, the effect of
miR-5580-3p on OC cell proliferation was reversed (Fig. 5B). Furthermore, it was observed that
the migration rate was decreased by 0.2-fold in the miR-5580-3p
mimic + OE-NC group, while that in the mimic-NC + OE-LAMC2 group
was increased by 0.3-fold compared with that in the mimic-NC +
OE-NC group. Additionally, in the miR-5580-3p mimic + OE-LAMC2
group, the effect of miR-5580-3p on OC cell migration was reversed
(Fig. 5C). Overall, the results
demonstrated that miR-5580-3p inhibited OC cell viability,
proliferation and migration by suppressing LAMC2.
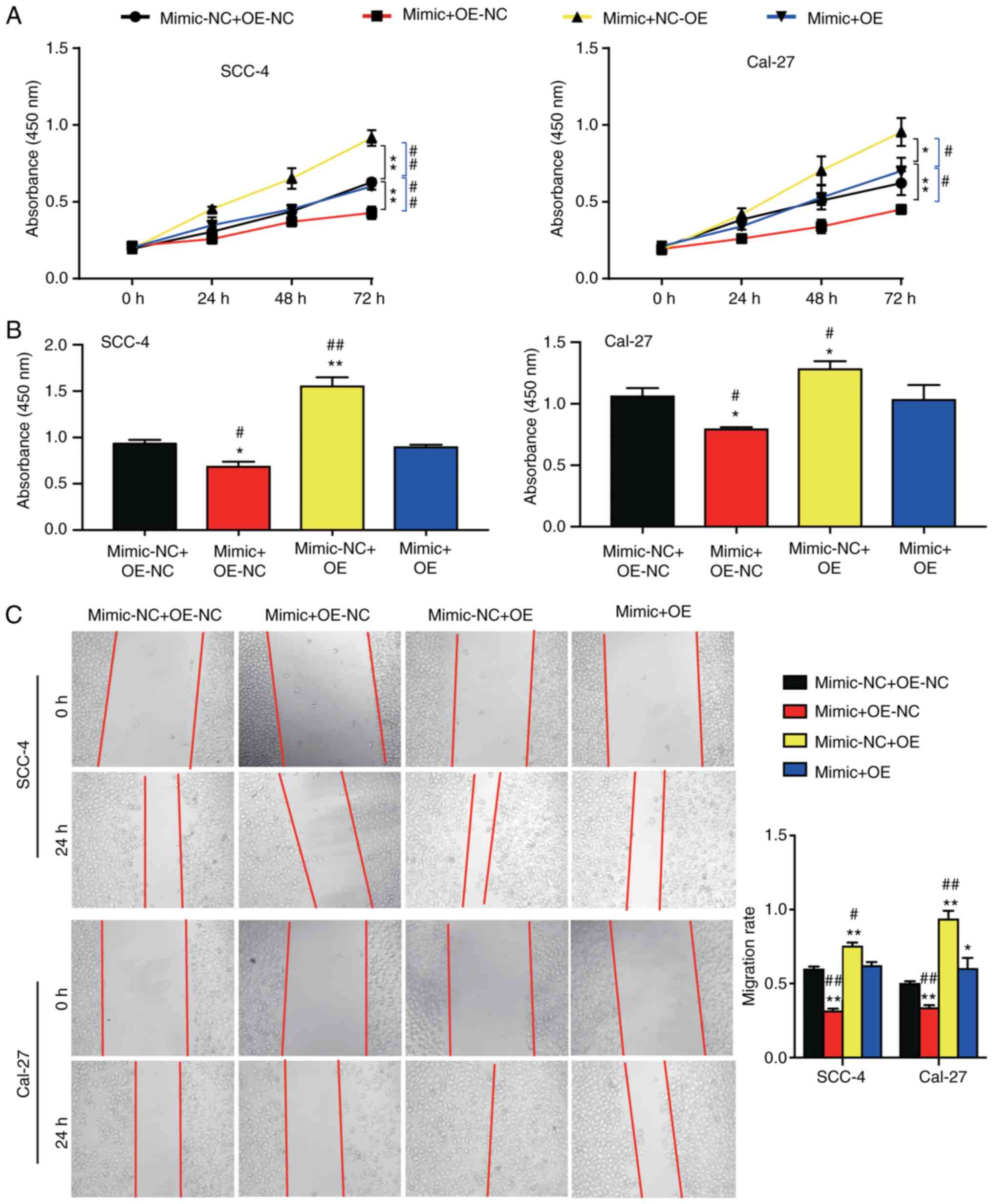 | Figure 5.Effects of miR-5580-3p on cell
viability, proliferation and wound healing via targeting of LAMC2.
(A) Cell Counting Kit-8 assay results revealed that miR-5580-3p
inhibited cell viability, while LAMC2 reversed its effect. (B)
5-bromo-2′-deoxyuridine incorporation assay results demonstrated
that miR-5580-3p inhibited cell proliferation, while LAMC2 reversed
its effect. (C) miR-5580-3p increased the wound healing rate, while
LAMC2 reversed its effect. *P<0.05 and **P<0.01 vs. NC +
OE-NC group; #P<0.05 and ##P<0.01 vs.
mimic + OE group. Mimic-NC + OE-NC, miR-5580-3p mimic negative
control + LAMC2 overexpression negative control; mimic + OE-NC,
miR-5580-3p mimic + LAMC2 overexpression negative control; mimic-NC
+ OE, miR-5580-3p mimic negative control + LAMC2 overexpression;
mimic + OE, miR-5580-3p mimic + LAMC2 overexpression; LAMC2,
laminin subunit γ2; miR-5580-3p, microRNA-5580-3p. |
Discussion
The present experiments demonstrated that LAMC2
expression was upregulated in OC tissues and cell lines, whereas
miR-5580-3p expression was downregulated in OC tissues and cells.
Following overexpression of LAMC2, cell viability, proliferation
and migration were increased notably in SCC-4 and Cal-27 cells.
Whereas upregulation of miR-5580-3p led to the opposite results.
Following co-transfection of miR-5580-3p mimic in
LAMC2-overexpressing cell lines, the effects on cell viability,
proliferation and migration abilities were reversed.
Increasing reports have revealed that LAMC2
expression is upregulated in various types of cancer, including
gastric carcinoma, lung carcinoma, colorectal carcinoma, pancreas
carcinoma, cervix carcinoma, oral carcinoma and melanoma (7,28–34).
These previous studies demonstrated that LAMC2 enhanced tumor
aggressiveness and was associated with shorter survival time, and
high recurrence or metastasis rate in patients (7,28–34).
Additionally, in a study by Lindberg et al (35), which included 20 LAMC2-positive
tumors, in 11 grade one cases of incipient carcinoma, LAMC2 had a
similar expression pattern to plasminogen activator inhibitor-1
(PAI-1). These results suggested that PAI-1 and LAMC2 with
coordinated expression sustained the early-phase features of
invading cancer cells in OSCC (35). Another study reported that LAMC2
expression was upregulated in OSCC tissues with α-smooth muscle
actin positivity, increasing to participate in the vascular
basement membrane reorganization in tumor angioneogenesis (36). The results of the present study
revealed that LAMC2 expression was upregulated in OC tissues and
cell lines to promote cell viability, proliferation and
migration.
Several miRNAs have been determined to function as
tumor suppressors or promoters during OC genesis (37,38).
In 2016, the results of a microarray study demonstrated that
let-7a, let-7d, let-7f and miR-16 were expressed at low levels in
OSCC tissues, whereas the expression levels of miR-29b, miR-142-3p,
miR-144, miR-203 and miR-223 were increased in OSCC tissues
(37). However, one study
demonstrated that miR-196 was associated with lymph node metastasis
when highly overexpressed in OC tissues, thus resulting in enhanced
cell migration and invasion (39).
A study published in 2014 revealed that miR-99a, one of the most
significantly downregulated miRNAs in OSCC, decreased the migration
and invasion of OSCC cells (40).
In the present study, it was observed that miR-5580-3p expression
was downregulated in both OC tissues and cell lines, and that it
inhibited cell viability, proliferation and migration.
The present study provided additional insights into
how miRNAs regulate OC. It has been widely hypothesized that a
miRNA can bind to the 3′UTR of its target mRNA (41–43).
For instance, miR-31-5p regulates the expression of extracellular
PEG2 antagonistically, thereby enhancing prostaglandin E receptor
1-ERK-MMP9 (44). Furthermore,
miR-125b, which is expressed at low levels in OSCC tissues, is a
direct upstream gene of peroxiredoxin like 2A, and its
overexpression in OSCC cells increases oxidative stress and
inhibits the activity of OSCC cells (45). Additionally, LAMC2 has been reported
to be targeted by other miRNAs and to affect OC cell phenotypes.
For instance, LINC00511 is highly expressed in tongue squamous cell
carcinoma, and it acts as a competing endogenous RNA sponging
miR-765, finally upregulating LAMC2 expression to enhance cell
proliferation and invasion (46).
In HNSCC, LAMC2 is regulated by miR-29s to promote cell migration
and invasion (47). In the present
study, miR-5580-3p was demonstrated to be an upstream miRNA of
LAMC2. By binding to the 3′UTR of LAMC2, miR-5580-3p decreased cell
viability, proliferation and migration in OSCC.
Another study revealed that laminin-5 protein could
co-deposit with large un-spliced tenascin-C in the extracellular
matrix to promote invasion and metastasis in OSCC (48). Therefore, further experiments should
be performed to study how LAMC2 enters the cell nucleus to remodel
the extracellular matrix and thus influence tumor invasion. To
further validate the current results, animal experiments could also
be conducted in the future. Clinically, the log-rank test may not
be applicable for survival plots where late stage crossover is
present in LAMC2 prognostic analysis, so further verification
should be performed using a weighted test, such as Renyi or
Cramer-von Mises.
In summary, the present study demonstrated that
miR-5580-3p suppressed cell viability, proliferation and migration
by decreasing the expression levels of LAMC2. This means that, in
the future, miR-5580-3p and LAMC2 may be used as potential
biomarkers or even therapeutic targets for OC diagnosis and
therapies.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and or/analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BX and QL designed the study. RF performed most of
the experiments. QL performed the data analysis and wrote the
paper. BX and QL confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Puai Hospital, Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China; approval no.
KY2020-501-01). All patients who participated in the present study
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
OC
|
oral cancer
|
|
LAMC2
|
laminin subunit γ2
|
|
OSCC
|
oral squamous cell carcinoma
|
|
miRNA
|
microRNA
|
|
CCK-8
|
Cell Counting Kit-8
|
|
BrdU
|
5-bromo-2′-deoxyuridine
|
|
NC
|
negative control
|
References
|
1
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: circRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carnielli CM, Macedo CCS, De Rossi T,
Granato DC, Rivera C, Domingues RR, Pauletti BA, Yokoo S, Heberle
H, Busso-Lopes AF, et al: Combining discovery and targeted
proteomics reveals a prognostic signature in oral cancer. Nat
Commun. 9:35982018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sophia J, Kowshik J, Dwivedi A, Bhutia SK,
Manavathi B, Mishra R and Nagini S: Nimbolide, a neem limonoid
inhibits cytoprotective autophagy to activate apoptosis via
modulation of the PI3K/Akt/GSK-3β signalling pathway in oral
cancer. Cell Death Dis. 9:10872018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kang CJ, Lin CY, Yang LY, Ho TY, Lee LY,
Fan KH, Wang HM, Huang SF, Chang KP, Fang KH, et al: Positive
clinical impact of an additional PET/CT scan before adjuvant
radiotherapy or concurrent chemoradiotherapy in patients with
advanced oral cavity squamous cell carcinoma. J Nucl Med. 56:22–30.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jardim JF, Francisco AL, Gondak R,
Damascena A and Kowalski LP: Prognostic impact of perineural
invasion and lymphovascular invasion in advanced stage oral
squamous cell carcinoma. Int J Oral Maxillofac Surg. 44:23–28.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hafiji J, Hussain W and Salmon P:
Reconstruction of perioral defects post-Mohs micrographic surgery:
A dermatological surgeon's approach. Br J Dermatol. 172:145–150.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garg M, Kanojia D, Okamoto R, Jain S,
Madan V, Chien W, Sampath A, Ding LW, Xuan M, Said JW, et al:
Laminin-5gamma-2 (LAMC2) is highly expressed in anaplastic thyroid
carcinoma and is associated with tumor progression, migration, and
invasion by modulating signaling of EGFR. J Clin Endocrinol Metab.
99:E62–E72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cotterill SJ: Chromosome 1. Cancer
Genetics. http://www.cancer-genetics.org/clinkc01.htmApril. 12,
2021
|
|
9
|
Koshikawa N, Minegishi T, Nabeshima K and
Seiki M: Development of a new tracking tool for the human monomeric
laminin-gamma 2 chain in vitro and in vivo. Cancer Res. 68:530–536.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu L, Hou Y, Tu G, Chen Y, Du YE, Zhang H,
Wen S, Tang X, Yin J, Lang L, et al: Nuclear Drosha enhances cell
invasion via an EGFR-ERK1/2-MMP7 signaling pathway induced by
dysregulated miRNA-622/197 and their targets LAMC2 and CD82 in
gastric cancer. Cell Death Dis. 8:e26422017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shou JZ, Hu N, Takikita M, Roth MJ,
Johnson LL, Giffen C, Wang QH, Wang C, Wang Y, Su H, et al:
Overexpression of CDC25B and LAMC2 mRNA and protein in esophageal
squamous cell carcinomas and premalignant lesions in subjects from
a high-risk population in China. Cancer Epidemiol Biomarkers Prev.
17:1424–1435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kosanam H, Prassas I, Chrystoja CC, Soleas
I, Chan A, Dimitromanolakis A, Blasutig IM, Rückert F, Gruetzmann
R, Pilarsky C, et al: Laminin, gamma 2 (LAMC2): A promising new
putative pancreatic cancer biomarker identified by proteomic
analysis of pancreatic adenocarcinoma tissues. Mol Cell Proteomics.
12:2820–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Zoltan M, Riquelme E, Xu H, Sahin
I, Castro-Pando S, Montiel MF, Chang K, Jiang Z, Ling J, et al:
Immune cell production of interleukin 17 induces stem cell features
of pancreatic intraepithelial neoplasia cells. Gastroenterology.
155:210–223.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin Y, Ge X, Zhang X, Wu Z, Liu K, Lin F,
Dai C, Guo W and Li J: Protocadherin-8 promotes invasion and
metastasis via laminin subunit γ2 in gastric cancer. Cancer Sci.
109:732–740. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Xu R and Li N: MicroRNAs from plants
to animals, do they define a new messenger for communication? Nutr
Metab (Lond). 15:682018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reinhart BJ, Weinstein EG, Rhoades MW,
Bartel B and Bartel DP: MicroRNAs in plants. Genes Dev.
16:1616–1626. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu X, Wang Y, Mojumdar K, Zhou Z, Jeong
KJ, Mangala LS, Yu S, Tsang YH, Rodriguez-Aguayo C, Lu Y, et al:
A-to-I-edited miRNA-379-5p inhibits cancer cell proliferation
through CD97-induced apoptosis. J Clin Invest. 129:5343–5356. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhatia V, Yadav A, Tiwari R, Nigam S, Goel
S, Carskadon S, Gupta N, Goel A, Palanisamy N and Ateeq B:
Epigenetic silencing of miRNA-338-5p and miRNA-421 drives
SPINK1-positive prostate cancer. Clin Cancer Res. 25:2755–2768.
2019.PubMed/NCBI
|
|
20
|
Croset M, Pantano F, Kan CWS, Bonnelye E,
Descotes F, Alix-Panabieres C, Lecellier CH, Bachelier R, Allioli
N, Hong SS, et al: miRNA-30 family members inhibit breast cancer
invasion, osteomimicry, and bone destruction by directly targeting
multiple bone metastasis-associated genes. Cancer Res.
78:5259–2573. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marcuello M, Duran-Sanchon S, Moreno L,
Lozano JJ, Bujanda L, Castells A and Gironella M: Analysis of A
6-mirna signature in serum from colorectal cancer screening
participants as non-invasive biomarkers for advanced adenoma and
colorectal cancer detection. Cancers (Basel). 11:15422019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Friedlander MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tuch BB, Laborde RR, Xu X, Gu J, Chung CB,
Monighetti CK, Stanley SJ, Olsen KD, Kasperbauer JL, Moore EJ, et
al: Tumor transcriptome sequencing reveals allelic expression
imbalances associated with copy number alterations. PLoS One.
5:e93172010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bhosale PG, Cristea S, Ambatipudi S, Desai
RS, Kumar R, Patil A, Kane S, Borges AM, Schäffer AA, Beerenwinkel
N and Mahimkar MB: Chromosomal alterations and gene expression
changes associated with the progression of leukoplakia to advanced
gingivobuccal cancer. Transl Oncol. 10:396–409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chamorro-Petronacci C, Perez-Sayáns M,
Padín-Iruegas ME, Marichalar-Mendia X, Gallas-Torreira M and García
García A: Differential expression of snoRNAs in oral squamous cell
carcinomas: New potential diagnostic markers. J Enzyme Inhib Med
Chem. 33:424–427. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Soini Y, Maatta M, Salo S, Tryggvason K
and Autio-Harmainen H: Expression of the laminin gamma 2 chain in
pancreatic adenocarcinoma. J Pathol. 180:290–294. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takahashi S, Hasebe T, Oda T, Sasaki S,
Kinoshita T, Konishi M, Ochiai T and Ochiai A: Cytoplasmic
expression of laminin gamma2 chain correlates with postoperative
hepatic metastasis and poor prognosis in patients with pancreatic
ductal adenocarcinoma. Cancer. 94:1894–901. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koshikawa N, Moriyama K, Takamura H,
Mizushima H, Nagashima Y, Yanoma S and Miyazaki K: Overexpression
of laminin gamma2 chain monomer in invading gastric carcinoma
cells. Cancer Res. 59:5596–5601. 1999.PubMed/NCBI
|
|
31
|
Hlubek F, Jung A, Kotzor N, Kirchner T and
Brabletz T: Expression of the invasion factor laminin gamma2 in
colorectal carcinomas is regulated by beta-catenin. Cancer Res.
61:8089–8093. 2001.PubMed/NCBI
|
|
32
|
Kagesato Y, Mizushima H, Koshikawa N,
Kitamura H, Hayashi H, Ogawa N, Tsukuda M and Miyazaki K: Sole
expression of laminin gamma 2 chain in invading tumor cells and its
association with stromal fibrosis in lung adenocarcinomas. Jpn J
Cancer Res. 92:184–192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Skyldberg B, Salo S, Eriksson E, Aspenblad
U, Moberger B, Tryggvason K and Auer G: Laminin-5 as a marker of
invasiveness in cervical lesions. J Natl Cancer Inst. 91:1882–1887.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oku N, Sasabe E, Ueta E, Yamamoto T and
Osaki T: Tight junction protein claudin-1 enhances the invasive
activity of oral squamous cell carcinoma cells by promoting
cleavage of laminin-5 gamma2 chain via matrix metalloproteinase
(MMP)-2 and membrane-type MMP-1. Cancer Res. 66:5251–5257. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lindberg P, Larsson A and Nielsen BS:
Expression of plasminogen activator inhibitor-1, urokinase receptor
and laminin gamma-2 chain is an early coordinated event in
incipient oral squamous cell carcinoma. Int J Cancer.
118:2948–2956. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Franz M, Wolheim A, Richter P, Umbreit C,
Dahse R, Driemel O, Hyckel P, Virtanen I, Kosmehl H and Berndt A:
Stromal laminin chain distribution in normal, hyperplastic and
malignant oral mucosa: Relation to myofibroblast occurrence and
vessel formation. J Oral Pathol Med. 39:290–298. 2010.PubMed/NCBI
|
|
37
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Park NJ, Zhou H, Elashoff D, Henson BS,
Kastratovic DA, Abemayor E, Abemayor E and Wong DT: Salivary
microRNA: Discovery, characterization, and clinical utility for
oral cancer detection. Clin Cancer Res. 15:5473–5477. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu YC, Chang JT, Liao CT, Kang CJ, Huang
SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al:
OncomiR-196 promotes an invasive phenotype in oral cancer through
the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 13:2182014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yen YC, Shiah SG, Chu HC, Hsu YM, Hsiao
JR, Chang JY, Hung WC, Liao CT, Cheng AJ, Lu YC and Chen YW:
Reciprocal regulation of microRNA-99a and insulin-like growth
factor I receptor signaling in oral squamous cell carcinoma cells.
Mol Cancer. 13:62014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen
Z, Dinglin X, Ma S, Li D, Wu Y, et al: N6-methyladenosine induced
miR-143-3p promotes the brain metastasis of lung cancer via
regulation of VASH1. Mol Cancer. 18:1812019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Buonfiglioli A, Efe IE, Guneykaya D,
Ivanov A, Huang Y, Orlowski E, Krüger C, Deisz RA, Markovic D, Flüh
C, et al: let-7 MicroRNAs regulate microglial function and suppress
glioma growth through toll-like receptor 7. Cell Rep.
29:3460–3471,e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu L, Wu D, Gao H, Balic JJ, Tsykin A, Han
TS, Liu YD, Kennedy CL, Li JK, Mao JQ, et al: Clinical utility of a
STAT3-regulated miRNA-200 family signature with prognostic
potential in early gastric cancer. Clin Cancer Res. 24:1459–1472.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lai YH, Liu H, Chiang WF, Chen TW, Chu LJ,
Yu JS, Chen SJ, Chen HC and Tan BC: MiR-31-5p-ACOX1 axis enhances
tumorigenic fitness in oral squamous cell carcinoma via the
promigratory prostaglandin E2. Theranostics. 8:486–504. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen YF, Wei YY, Yang CC, Liu CJ, Yeh LY,
Chou CH, Chang KW and Lin SC: miR-125b suppresses oral oncogenicity
by targeting the anti-oxidative gene PRXL2A. Redox Biol.
22:1011402019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ding J, Yang C and Yang S: LINC00511
interacts with miR-765 and modulates tongue squamous cell carcinoma
progression by targeting LAMC2. J Oral Pathol Med. 47:468–476.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kinoshita T, Nohata N, Hanazawa T, Kikkawa
N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto
Y and Seki N: Tumour-suppressive microRNA-29s inhibit cancer cell
migration and invasion by targeting laminin-integrin signalling in
head and neck squamous cell carcinoma. Br J Cancer. 109:2636–2645.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Franz M, Hansen T, Richter P, Borsi L,
Bohmer FD, Hyckel P, Schleier P, Katenkamp D, Zardi L, Kosmehl H
and Berndt A: Complex formation of the laminin-5 gamma2 chain and
large unspliced tenascin-C in oral squamous cell carcinoma in vitro
and in situ: Implications for sequential modulation of
extracellular matrix in the invasive tumor front. Histochem Cell
Biol. 126:125–131. 2006. View Article : Google Scholar : PubMed/NCBI
|