Introduction
Primary liver cancer refers to the primary malignant
cancer located in the hepatocytes or bile duct cells in the liver
(1). In China, liver cancer has the
fifth highest incidence rate (accounting for 9% of all
cancer-related incidences) and is the second leading cause of death
(accounting for 13% of all cancer-related mortalities); therefore,
liver cancer has a significant impact on the life and health of
individuals (2). Amongst the
subtypes of liver cancer, hepatocellular carcinoma (HCC) has the
highest incidence and accounts for >80% of primary liver cancer
types (3). Currently, there are
various therapeutic approaches available for HCC, including
surgical resection, cryotherapy, laser therapy and interventional
surgery; however, due to the highly invasive and metastatic nature
of HCC, the median survival time of patients with advanced HCC is
only 3–6 months, and the 5-year survival rate remains at 12.5%
(4,5). Therefore, the pathological mechanisms
of HCC at the molecular level should be further investigated to
determine effective treatment targets for HCC.
Circular RNAs (circRNAs/circs) are a type of
non-coding RNA formed by the covalent bonding between the 5′ and 3′
ends of linear precursor RNA through a reverse splicing mechanism,
which is associated with gene transcription and
post-transcriptional gene expression regulation (6). The expression levels of circ-protein
kinase C iota (PRKCI) have been discovered to be upregulated in
gastric cancer and promoted the proliferation, invasion and
migration of gastric cancer cells (7). circ-PRKCI was also found to bind with
microRNA (miRNA/miR) as a competitive RNA to promote the
progression of esophageal squamous cell carcinoma (8). Furthermore, the upregulation of
circ-PRKCI expression levels was discovered to enhance the
proliferation and migration of HCC cells. The upregulated
expression levels were also positively associated with an increase
in infiltration depth and tumor node metastasis classification
(9). In the present study, StarBase
was used to predict the target genes of circ-PRKCI.
Notably, the upregulation of miR-1294 expression
levels was demonstrated to inhibit the proliferation and promote
the apoptosis of HCC cells (10).
miR-186-5p expression levels were reported to be downregulated in
HCC tissues and cells, which promoted the tumorigenesis and
metastasis of HCC, while the overexpression of miR-186-5p
suppressed the viability and increased the apoptosis and autophagy
of the cancer cells (11,12). In addition, multiple previous
studies have suggested that both miR-1294 and miR-186-5p could
regulate the expression levels of forkhead box K1 (FOXK1) (13,14).
FOXK1 expression levels were discovered to be upregulated in liver
cancer cells, which regulated glycolysis and subsequently affected
the viability of liver cancer cells (15).
However, whether circ-PRKCI can regulate FOXK1
through miR-1294 and miR-186-5p to affect glycolysis in HCC cells
remains to be determined.
Materials and methods
Plasmids
For the stable overexpression (Ov) of circ-PRKCI and
FOXK1, sequences were cloned into pcDNA3.1 vectors (Invitrogen;
Thermo Fisher Scientific, Inc.); empty plasmids were used as the
negative control (NC) for the overexpression plasmids. The
following small interfering RNAs (siRNA/si) and miRNA
oligonucleotides were obtained from Guangzhou RiboBio Co., Ltd.:
Si-NC (cat. no. siB06525141922-1-5), si-circ-PRKCI-1 (cat. no.
siB0804170942011-1-5), si-circ-PRKCI-2 (cat. no.
siB0804170942012-1-5), si-FOXK1-1 (cat. no. siG151012011528-1-5),
si-FOXK1-2 (cat. no. siG151012011536-1-5), mimic-NC (cat. no.
miR1N0000001-1-5), miR-1294 mimic (cat. no. miR10005884-1-5),
miR-186-5p mimic (cat. no. miR10000456-1-5), miR-1294 inhibitor
(cat. no. miR20005884-1-5) and miR-186-5p inhibitor (cat. no.
miR20000456-1-5).
Cell culture
The human hepatic epithelial cell line, THLE-3, was
obtained from the American Type Culture Collection and the HCC cell
line, HCCLM3, was purchased from Procell Life Science &
Technology Co., Ltd. THLE-3 and HCCLM3 cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and
100 µg/ml streptomycin, and maintained in a humidified atmosphere
at 37°C with 5% CO2.
Cell transfection
HCCLM3 cells were cultured until reaching the
logarithmic stage and then seeded at a cell density of 1×105
cells/ml into a 6-well plate. The cells were subsequently incubated
in an incubator at 37°C for 24 h until the cells adhered to the
wall and the confluence reached 70%. The HCCLM3 cells were
subsequently transfected with plasmids, siRNAs or oligonucleotides
using Lipofectamine® 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h, and
cells then were used for subsequent experimentation.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) A total of 2 µg RNA was reverse transcribed into cDNA using a
Transcriptor First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. qPCR
was subsequently performed using a SYBR-Green Real-time PCR kit
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The following thermocycling conditions
were used for the qPCR: Initial denaturation at 95°C for 5 min;
followed by 40 cycles of denaturation at 95°C for 15 sec,
annellation at 60°C for 15 sec and extension at 72°C for 45 sec.
The following primer sequences were used for the qPCR: circ-PRKCI
forward, 5′-CGGAGGTTCCAGCTCGTTAGTC-3′ and reverse,
5′-GCAACCGGAATGTGGAATTGA-3′; FOXK1 forward,
5′-GCCACAAAGGCTGGCAGAATT-3′ and reverse,
5′-TGGCTTCAGAGGCAGGGTCTAT-3′; GAPDH forward,
5′-TCGACAGTCAGCCGCATCTT-3′ and reverse,
5′-GAGTTAAAAGCAGCCCTGGTG-3′; miR-1294 forward,
5′-TATGATCTCACCGAGTCCT-3′ and reverse, 5′-CACCTTCCTAATCCTCAGTT-3′;
miR-186-5p forward, 5′-AAGAATTCTCCTTTTGGGCT-3′ and reverse,
5′-GTGCGTGTCGTGGAGTCG-3′; and U6 forward,
5′-AGGTGTGTGCTGCTATGAAC-3′ and reverse, 5′-CAGGGCATTTGTCATTATTG-3′.
GAPDH or U6 were used as the internal reference genes for mRNA and
miRNA, respectively, and the 2-∆∆Cq method (16) was used to quantify the relative
expression levels of circ-PRKCI, miR-1294, miR-186-5p and
FOXK1.
Dual luciferase reporter assay
circ-PRKCI-wild-type (WT), circ-PRKCI-mutant (MUT),
FOXK1-WT and FOXK1-MUT 3′-untranslated region (UTR) sequences were
cloned into pmirGLO plasmids (Promega Corporation). The pmirGLO
plasmids were subsequently co-transfected into HCCLM3 cells with
the miR-1294, miR-186-5p mimic or miR-NC using Lipofectamine 2000
reagent. After transfection for 48 h, the relative luciferase (Luc)
activity was measured using a Dual Luciferase Reporter assay system
(Promega Corporation) and the Luc value was normalized to the
Renilla (R)-Luc value.
Cell Counting Kit-8 (CCK-8) assay
HCCLM3 cells from each group were plated into a
96-well plate (5×103 cells/well) and cultured overnight at 37°C.
Following incubation for 24 h, the medium was removed and HCCLM3
cells were rinsed with PBS. Subsequently, 10 µl CCK-8 solution
(Beyotime Institute of Biotechnology) was added/well and incubated
at 37°C with 5% CO2 for 2 h. The absorbance of each
group at 24 h was measured at a wavelength of 450 nm with a
microplate reader to reflect the change in the cell viability.
Wound healing assay
HCCLM3 cells from each group were seeded into
12-well plates (5×104 cells/well) and cultured in DMEM with 5% FBS
at 37°C. After reaching 100% confluence, a 200-µl sterile pipette
tip was used to scratch the confluent monolayer; the cells were
then washed with PBS to remove the non-adherent cells. The cells
were subsequently cultured with serum-free DMEM at 37°C for 24 h.
The wound area at 0 and 24 h was imaged using a light microscope
(magnification, ×100).
Transwell assay
A total of 3×104 cells from each group were seeded
in serum-free DMEM into the upper chamber of Transwell plates
precoated with Matrigel at 37°C for 30 min and then cultured at
37°C for 24 h. DMEM supplemented with 10% FBS was plated into the
lower chamber. Following incubation for 24 h at 37°C, the cells
remaining on the upper surface of the membrane were removed. The
invasive cells in the lower chamber were fixed with 4%
paraformaldehyde at room temperature for 30 min and stained with
0.2% crystal violet solution for 10 min at room temperature.
Finally, the membrane was washed with PBS once and visualized under
a light microscope (magnification, ×100).
Western blotting
HCCLM3 cells from each group were collected and
total protein was extracted using RIPA lysis buffer (Beyotime
Institute of Biotechnology) at 3,000 × g at 4°C for 15 min. Total
protein was quantified using a BCA assay kit (Pierce; Thermo Fisher
Scientific, Inc.) and 20 µg protein/lane was separated via 12%
SDS-PAGE. The separated proteins were transferred onto PVDF
membranes and blocked at room temperature with 5% skimmed milk for
30 min. The membranes were then incubated overnight at 4°C with the
following primary antibodies: Anti-FOXK1 (1:1,000; cat. no.
ab18196; Abcam), anti-hexokinase-2 (HK2; 1:1,000; cat. no.
ab209847; Abcam), anti-glucose transporter 1 (GLUT1; 1:200; cat.
no. ab150299; Abcam), anti-lactate dehydrogenase A (LDHA; 1:1,000;
cat. no. ab101562; Abcam) and anti-GAPDH (1:1,000; cat. no. ab8245;
Abcam). Following the primary antibody incubation, the membranes
were incubated with an anti-rabbit IgG HRP-conjugated antibody
(1:2,000; cat. no. ab6721; Abcam) or anti-mouse IgG HRP-conjugated
antibody (1:1,000; cat. no. 7076; Cell Signaling Technology, Inc.)
for 1 h at room temperature. Protein bands were visualized in a
darkroom using ECL reagent (Bio-Rad Laboratories, Inc.).
Densitometric analysis was performed using Image-Pro Plus software
(version 6.0; Media Cybernetics, Inc.)
Detection of glucose and lactic acid
levels
Following transfection for 24 h, HCCLM3 cells were
centrifuged at 3,000 × g at 4°C for 15 min to obtain the
supernatant. The levels of glucose and lactic acid in the
supernatant of each group were analyzed with a Glucose Assay kit
(cat. no. E-BC-K234-S; Elabscience) and a L-Lactic Acid
Colorimetric Assay kit (cat. no. E-BC-K044-S; Elabscience),
according to the manufacturers' protocols, respectively, as
previously described (17,18).
Statistical analysis
All experiments were repeated three times.
Statistical analysis was performed using SPSS 18.0 software (SPSS,
Inc.) and data are presented as the mean ± SD. Statistical
differences between two groups were determined using an unpaired
Student's t-test, while a one-way ANOVA with a Tukey's post hoc
test was used to determine the statistical differences between
multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
circ-PRKCI regulates the expression
levels of miR-1294 and miR-186-5p in HCC cells
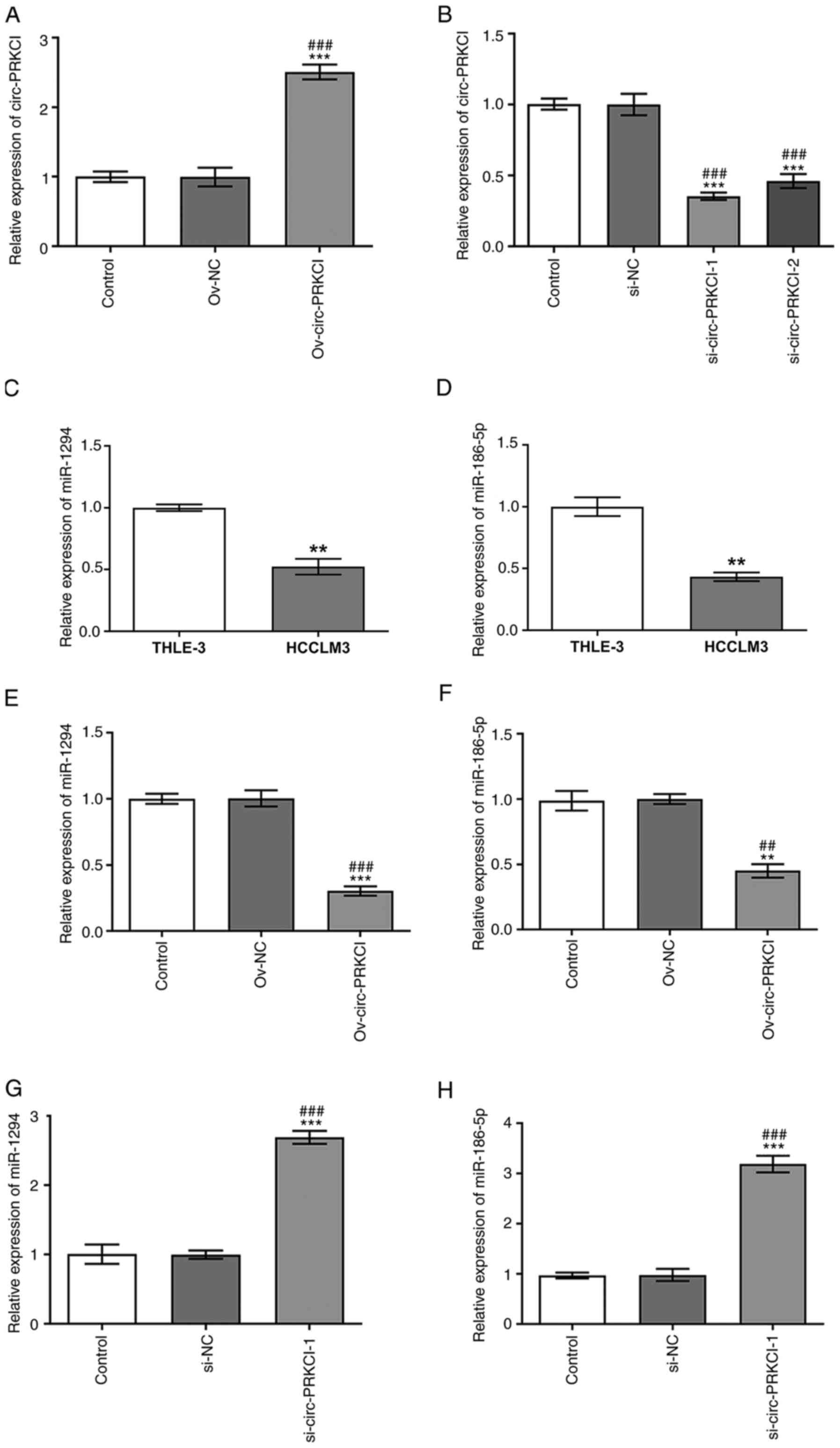
circ-PRKCI expression levels were significantly
upregulated in HCCLM3 cells transfected with Ov-circ-PRKCI vector
compared with the control and Ov-NC groups (Fig. 1A), while circ-PRKCI expression
levels were downregulated in HCCLM3 cells transfected with
si-circ-PRKCI-1 and si-circ-PRKCI-2 compared with the control and
si-NC groups (Fig. 1B). circ-PRKCI
expression levels were lower in HCCLM3 cells transfected with
si-circ-PRKCI-1, thus si-circ-PRKC1-1 was selected for use in
subsequent experiments. The expression levels of miR-1294 (Fig. 1C) and miR-186-5p (Fig. 1D) in HCCLM3 cells were downregulated
compared with THLE-3 cells. circ-PRKCI overexpression significantly
downregulated the expression levels of miR-1294 compared with the
Ov-NC and control groups (Fig. 1E)
and miR-186-5p (Fig. 1F), while the
knockdown of circ-PRKCI upregulated the expression levels of
miR-1294 (Fig. 1G) and miR-186-5p
(Fig. 1H) in HCCLM3 cells compared
with the control and si-NC groups.
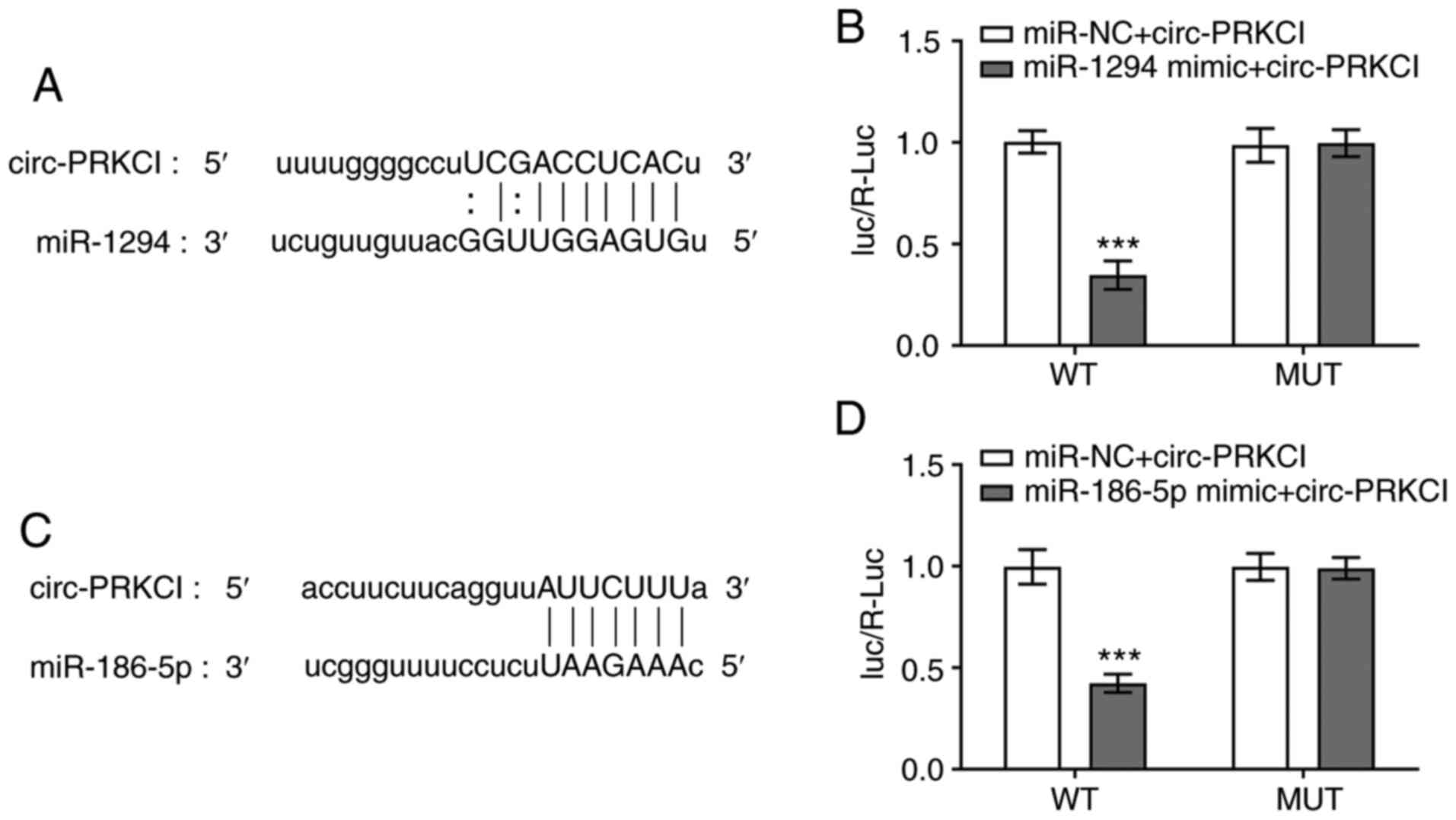
circ-PRKCI targets miR-1294 and
miR-186-5p
The predicted binding sites between circ-PRKCI and
miR-1294 are shown in Fig. 2A. The
relative Luc activity of cells was significantly decreased
following the co-transfection of cells with miR-1294 mimic and
circ-PRKCI-WT compared with cells co-transfected with miR-NC and
circ-PRKCI-WT. There were no significant differences observed in
the relative luciferase activity of MUT reporter plasmids between
cells transfected with the miRNA mimics and miR-NCs (Fig. 2B). The predicted binding sites
between circ-PRKCI and miR-186-5p are presented in Fig. 2C. The relative Luc activity was
significantly decreased following the co-transfection of cells with
miR-186-5p mimic and circ-PRKCI-WT compared with cells
co-transfected with miR-NC and circ-PRKCI-WT (Fig. 2D).
circ-PRKCI promotes the viability,
invasion and migration of HCC cells by targeting miR-1294 and
miR-186-5p
Following the transfection of HCCLM3 cells with the
miR-1294 mimic or miR-186-5p mimic, the expression levels of
miR-1294 (Fig. 3A) and miR-186-5p
(Fig. 3B), respectively, were
upregulated in HCCLM3 cells compared with cells transfected with
the mimic-NC or the control group. Following the transfection of
HCCLM3 cells with the miR-1294 inhibitor or miR-186-5p inhibitor,
the expression levels of miR-1294 (Fig.
3C) and miR-186-5p (Fig. 3D),
respectively, were downregulated in HCCLM3 cells compared with
cells transfected with inhibitor-NC or the control group.
circ-PRKCI overexpression significantly promoted the viability of
HCCLM3 cells compared with the Ov-NC and control groups, which was
subsequently reversed following the overexpression of miR-1294 or
miR-186-5p (Fig. 3E). The
inhibition of circ-PRKCI also significantly suppressed the
viability of HCCLM3 cells compared with the si-NC and control
groups, which was partially reversed by miR-1294 inhibition or
miR-186-5p inhibition (Fig. 3F). As
shown in Fig. 3G-J, circ-PRKCI
overexpression significantly promoted the migration and invasion of
HCCLM3 cells compared with the Ov-NC and control groups, which was
subsequently reversed following the overexpression of miR-1294 or
miR-186-5p. Conversely, the inhibition of circ-PRKCI significantly
suppressed the migration and invasion of HCCLM3 cells compared with
the Ov-NC and control groups, which was subsequently reversed
following the knockdown of miR-1294 or miR-186-5p (Fig. 3K-N)
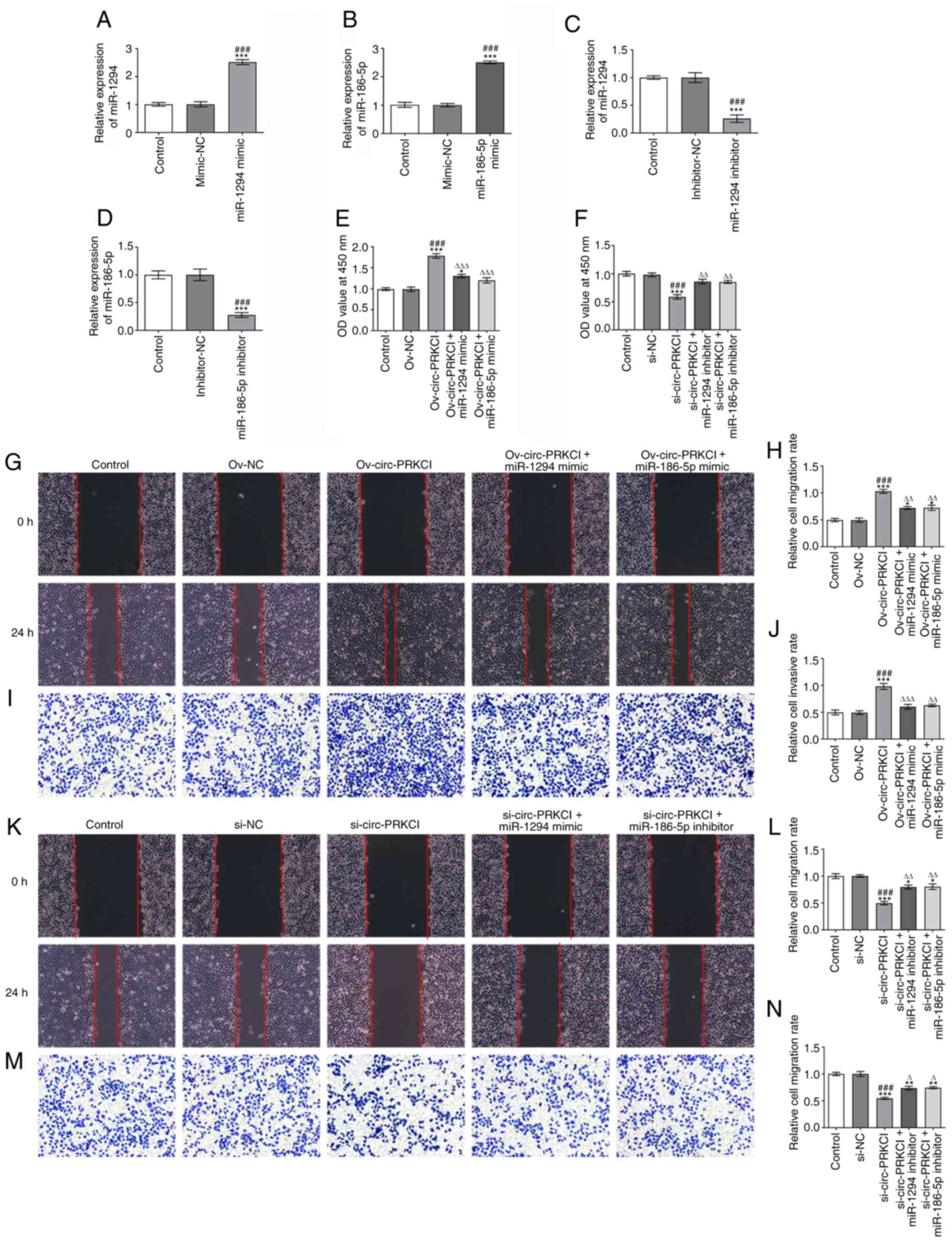 | Figure 3.circ-PRKCI promotes the viability,
invasion and migration of hepatocellular carcinoma cells by
targeting miR-1294 and miR-186-5p. Expression levels of (A)
miR-1294 and (B) miR-186-5p in HCCLM3 cells following the
transfection with miR-1294 mimic or miR-186-5p mimic, respectively,
were analyzed using RT-qPCR. ***P<0.001 vs. Control;
###P<0.001 vs. mimic-NC. Expression levels of (C) miR-1294 and
(D) miR-186-5p in HCCLM3 cells following the transfection with
miR-1294 inhibitor or miR-186-5p inhibitor were analyzed using
RT-qPCR. ***P<0.001 vs. Control; ###P<0.001 vs. inhibitor-NC.
(E) Viability of HCCLM3 cells following the transfection with
Ov-circ-PRKCI and/or miR-1294 mimic or miR-186-5p mimic was
analyzed using a CCK-8 assay. *P<0.05, ***P<0.001 vs.
Control; ###P<0.001 vs. Ov-NC; ∆∆∆P<0.001 vs.
Ov-circ-PRKCI. (F) Viability of HCCLM3 cells following the
transfection with si-circ-PRKCI and/or miR-186-5p inhibitor or
miR-186-5p inhibitor was analyzed using a CCK-8 assay.
***P<0.001 vs. Control; ###P<0.001 vs. si-NC;
∆∆P<0.01 vs. si-circ-PRKCI. (G and H) Migration of
HCCLM3 cells following the transfection with Ov-circ-PRKCI and/or
miR-1294 mimic or miR-186-5p mimic was determined using a wound
healing assay (magnification, ×100). *P<0.05, ***P<0.001 vs.
Control; ###P<0.001 vs. Ov-NC; ∆∆P<0.01 vs.
Ov-circ-PRKCI. (I and J) Invasion of HCCLM3 cells following the
transfection with Ov-circ-PRKCI and miR-1294 mimic or miR-186-5p
mimic was determined using a Transwell assay (magnification, ×100).
***P<0.001 vs. Control; ###P<0.001 vs. Ov-NC;
∆∆P<0.01, ∆∆∆P<0.001 vs. Ov-circ-PRKCI.
(K and L) Migration of HCCLM3 cells following the transfection with
si-circ-PRKCI and miR-1294 inhibitor or miR-186-5p inhibitor was
determined using a wound healing assay (magnification, ×100).
*P<0.05, ***P<0.001 vs. Control; ###P<0.001 vs. si-NC;
∆∆P<0.01 vs. si-circ-PRKCI. (M and N) Invasion of
HCCLM3 cells following the transfection with si-circ-PRKCI and
miR-1294 inhibitor or miR-186-5p inhibitor was determined using a
Transwell assay (magnification, ×100). **P<0.01, ***P<0.001
vs. Control; ###P<0.001 vs. si-NC; ∆P<0.05 vs.
si-circ-PRKCI. circ, circular RNA; PRKC1, protein kinase C iota;
miR, microRNA; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; CCK-8, Cell Counting Kit-8; si,
small interfering RNA; Ov, overexpression; OD, optical density. |
circ-PRKCI regulates FOXK1 expression
levels by targeting miR-1294 and miR-186-5p
The overexpression of circ-PRKCI upregulated the
expression levels of FOXK1 compared with the control and Ov-NC
groups, while the promoting effect of circ-PRKCI overexpression on
FOXK1 expression levels was significantly suppressed by the
overexpression of miR-1294 or miR-186-5p (Fig. 4A). The knockdown of circ-PRKCI
expression significantly downregulated FOXK1 expression levels
compared with the si-NC and control groups, which was partially
reversed following the knockdown by miR-1294 or miR-186-5p
(Fig. 4B). The predicted binding
sites between FOXK1 and miR-1294 are shown in Fig. 4C. The Luc/R-Luc value was
significantly decreased in HCCLM3 cells co-transfected with the
miR-1294 mimic and FOXK1-WT vector compared with the cells
co-transfected with miR-NC and FOXK1-WT. There were no significant
differences observed in the relative luciferase activity of MUT
reporter plasmids between cells transfected with the miRNA mimics
and miR-NCs (Fig. 4D). The binding
sites between FOXK1 and miR-186-5p are presented in Fig. 4E. The Luc/R-Luc value was also
significantly decreased in HCCLM3 cells co-transfected with the
miR-186-5p mimic and FOXK1-WT compared with the cells
co-transfected with miR-NC and FOXK1-WT (Fig. 4F).
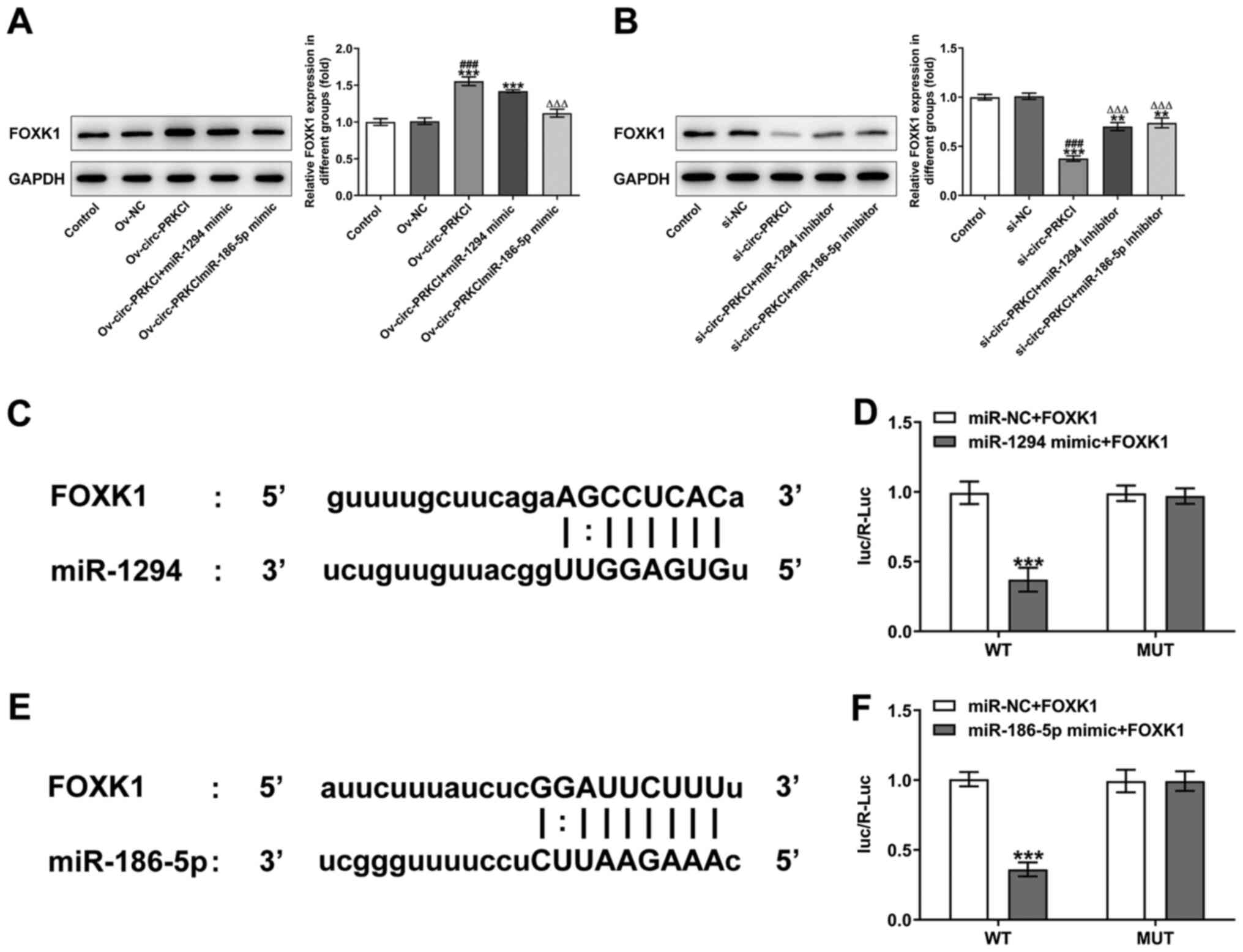 | Figure 4.circ-PRKCI regulates FOXK1 expression
levels by targeting miR-1294 and miR-186-5p. (A) FOXK1 expression
levels in HCCLM3 cells following the transfection with
Ov-circ-PRKCI and miR-1294 mimic or miR-186-5p mimic were analyzed
using western blotting. ***P<0.001 vs. Control; ###P<0.001
vs. Ov-NC; ∆∆∆P<0.001 vs. Ov-circ-PRKCI. (B) FOXK1
expression levels in HCCLM3 cells following the transfection with
si-circ-PRKCI and miR-1294 inhibitor or miR-186-5p inhibitor were
analyzed using western blotting. **P<0.01, ***P<0.001 vs.
Control; ###P<0.001 vs. si-NC; ∆∆∆P<0.001 vs.
si-circ-PRKCI. (C) Binding sites between FOXK1 and miR-1294 are
shown. (D) Dual luciferase reporter assay was used to validate the
direct binding relationship between FOXK1 and miR-1294.
***P<0.001 vs. miR-NC + FOXK1 WT. (E) Binding sites between
FOXK1 and miR-186-5p are shown. (F) Dual luciferase reporter assay
was used to validate the direct binding relationship between FOXK1
and miR-186-5p. ***P<0.001 vs. miR-NC + FOXK1 WT. circ, circular
RNA; PRKCI, protein kinase C iota; FOXK1, forkhead box K1; miR,
microRNA; Ov, overexpression; NC, negative control; WT, wild-type;
MUT, mutant; R, Renilla; Luc, luciferase. |
circ-PRKCI suppresses glycolysis in
HCC cells by regulating FOXK1 expression
FOXK1 expression levels were significantly
upregulated in HCCLM3 cells transfected with Ov-FOXK1 compared with
the Ov-NC and control groups (Fig.
5A). The mRNA expression levels of FOXK1 were also
significantly downregulated in HCCLM3 cells transfected with
si-FOXK1-1 and si-FOXK1-2 compared with the si-NC and control
groups (Fig. 5B). The
overexpression of circ-PRKCI significantly increased the levels of
glucose (Fig. 5C) and lactic acid
(Fig. 5E) compared with Ov-NC and
control groups, which were subsequently reversed following the
knockdown of FOXK1. Conversely, the knockdown of circ-PRKCI
expression levels significantly decreased the levels of glucose
(Fig. 5D) and lactic acid (Fig. 5F) compared with the Ov-NC and
control groups, which was subsequently reversed following the
overexpression of FOXK1. As shown in Fig. 5G, circ-PRKCI overexpression
upregulated the expression levels of HK2, GLUT1 and LDHA compared
with the Ov-NC and control groups, which were subsequently reversed
by FOXK1 inhibition. Conversely, the knockdown of circ-PRKCI
expression significantly downregulated the expression levels of
HK2, GLUT1 and LDHA compared with the si-NC and control groups,
which were subsequently reversed by the overexpression of FOXK1
(Fig. 5H).
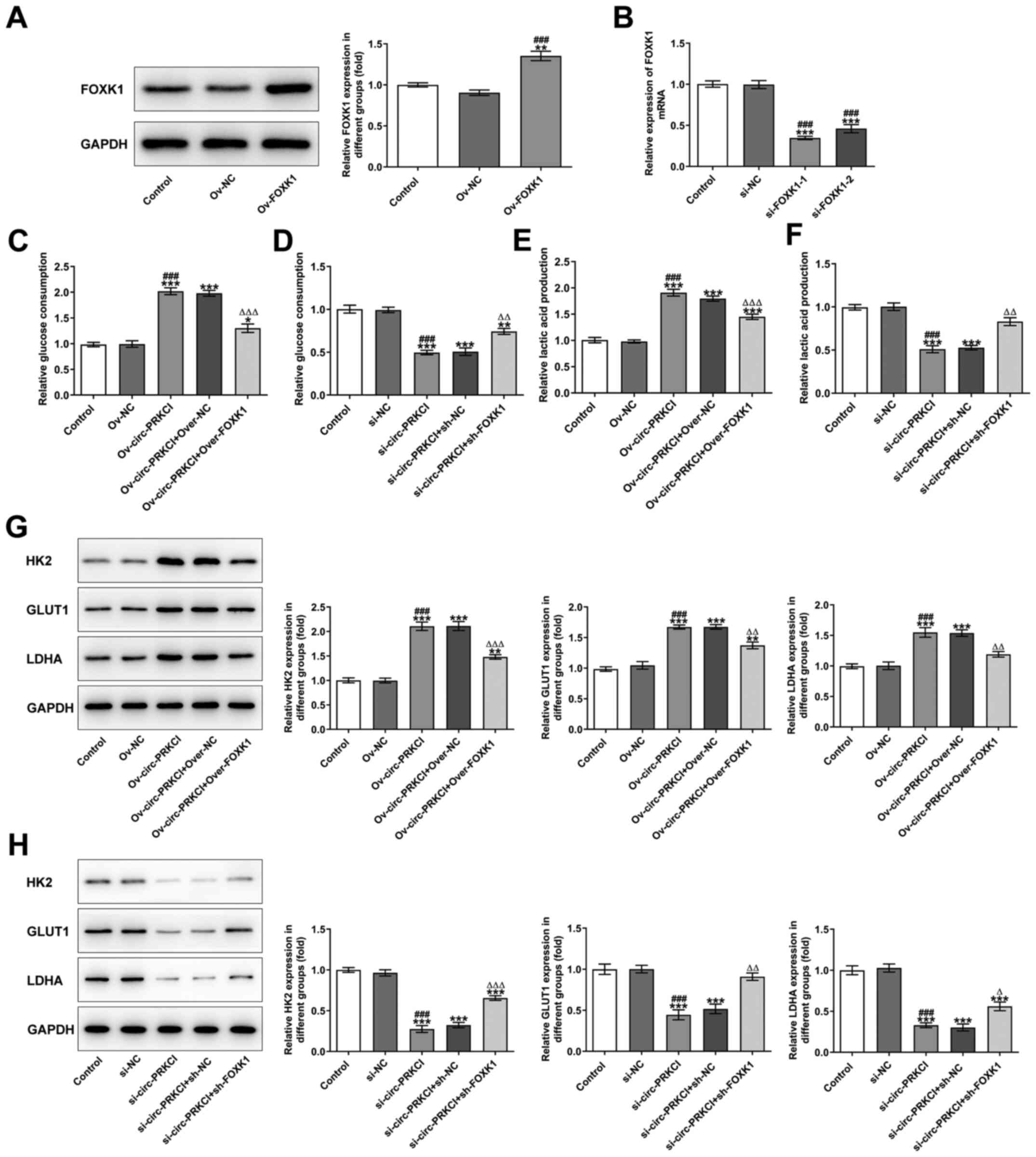 | Figure 5.circ-PRKCI suppresses glycolysis in
hepatocellular carcinoma cells by targeting miR-1294 and
miR-186-5p. (A) FOXK1 expression levels in HCCLM3 cells following
the transfection with Ov-FOXK1 were analyzed using western
blotting. **P<0.01 vs. Control; ###P<0.001 vs. Ov-NC. (B)
FOXK1 mRNA expression levels in HCCLM3 cells following the
transfection with si-FOXK1-1 and si-FOXK1-2 were analyzed using
reverse transcription-quantitative PCR. ***P<0.001 vs. Control;
###P<0.001 vs. si-NC. (C) Glucose levels in the supernatant of
HCCLM3 cells following the transfection with (C) Ov-circ-PRKCI and
si-FOXK1 and (D) si-circ-PRKCI and Ov-FOXK1 were determined using a
commercially available kit. *P<0.05, **P<0.01, ***P<0.001
vs. Control; ###P<0.001 vs. Ov-NC/si-NC; ∆∆P<0.01,
∆∆∆P<0.001 vs. Ov-circ-PRKCI/si-circ-PRKCI. Lactic
acid levels in the supernatant of HCCLM3 cells following the
transfection with (E) Ov-circ-PRKCI and si-FOXK1 and (F)
si-circ-PRKCI and ov-FOXK1 were determined using a commercially
available kit. ***P<0.001 vs. Control; ###P<0.001 vs.
Ov-NC/si-NC; ∆∆P<0.01, ∆∆∆P<0.001 vs.
Ov-circ-PRKCI/si-circ-PRKCI. Expression levels of HK2, GLUT1 and
LDHA in HCCLM3 cells following the transfection with (G)
Ov-circ-PRKCI and si-FOXK1 and (H) si-circ-PRKCI and Ov-FOXK1 were
analyzed using western blotting. **P<0.01, ***P<0.001 vs.
Control; ###P<0.001 vs. Ov-NC/si-NC; ∆P<0.05,
∆∆P<0.01, ∆∆∆P<0.001 vs.
Ov-circ-PRKCI/si-circ-PRKCI. Circ, circular RNA; PRKCI, protein
kinase C iota; miR, microRNA; FOXK1, forkhead box K1; Ov,
overexpression; si, small interfering RNA; NC, negative control;
HK2, hexokinase-2; GLUT1, glucose transporter 1; LDHA, lactate
dehydrogenase A. |
Discussion
It is estimated that ~50% of ATP in tumor cells is
synthesized through glycolysis (19). Although aerobic glycolysis occurs at
low levels, it is a rapid process. The intermediate products can
provide nutrients for cell proliferation and protect tumor cells,
thereby promoting the excessive proliferation and metastasis of
malignant tumor cells (20,21). Therefore, the effective inhibition
of glycolysis may suppress the progression of liver cancer.
Cancer is increasingly being regarded as a
metabolism-related disease, and tumor-related energy regulation has
been investigated as a biochemical pathway and drug target for
tumor therapy (22). In the past
decade, molecular and cellular studies have highlighted the
association between oncogenes, tumor suppressors and cancer
glycolysis (23). For example, Guo
et al (24) reported that
miR-199a-5p directly targeted HK2 to suppress glycolysis and the
tumorigenesis of liver cancer cells. The expression levels of
pyruvate kinase M1/2 (PKM2) were also found to be highly expressed
in HCC, which was essential for the aerobic glycolysis of HCC cells
(25). Liang et al (26) demonstrated that the expression
levels of miR-122 were closely associated with numerous glycolytic
genes, such as PKM2, hexokinase, LDHA and GLUT1, through the
comprehensive gene expression analysis of 94 liver cancer tissues.
The same study also revealed that miR-122 targeted PKM2, which
affected the metabolism of HCC. Numerous previous studies have
reported the role of miR-186 in mediating glycolysis in several
types of cancer; for example, miR-186 inhibited aerobic glycolysis
in osteosarcoma cells by decreasing hypoxia-inducible factor 1
expression levels (27);
circ-nuclear receptor interacting protein 1 promoted glycolysis and
gastric cancer progression by downregulating miR-186-5p expression
levels (28); and FOXK1 and/or
FOXK2 were discovered to regulate aerobic glycolysis (29). In the present study, bioinformatics
analysis predicted that circ-PRKCI targeted miR-186-5p and in turn,
miR-186-5p targeted FOXK1. circ-PRKCI was revealed to suppress
glycolysis in HCC cells by upregulating FOXK1 expression levels via
miR-186-5p.
Previous studies reported that miR-1294 expression
levels were downregulated in gastric, esophageal, epithelial
ovarian and cervical cancers, while miR-1294 overexpression
inhibited the cell proliferation and cell cycle progression of
cancer cells (13,30–32).
The results of the current study revealed that the expression
levels of miR-1294 and miR-186-5p were downregulated in HCC cells.
circ-PRKCI promoted the viability, migration and invasion of HCC
cells by downregulating the expression levels of miR-1294 and
miR-186-5p. In addition, circ-PRKCI was discovered to target
miR-1294 and miR-1294 was identified to target FOXK1. Inhibition of
circ-PRKCI also suppressed glycolysis in the HCC cells by
downregulating FOXK1 expression levels via miR-1294. A previous
study also showed that the knockdown of FOXK1 suppressed glycolysis
in liver cancer cells (15).
In conclusion, the findings of the present study
suggested that circ-PRKCI may promote the viability, migration and
invasion and glycolysis of HCC cells by upregulating FOXK1
expression levels by targeting miR-1294 and miR-186-5p. These
results supported that circ-PRKCI may represent a novel target for
the treatment of HCC. Due to the present study being limited by
only performing in vitro cell experiments, which do not
mimic the in vivo environment, the expression levels of
circ-PRKCI, miR-1294 and miR-186-5p will be further investigated in
tumor tissues and the present results will be further verified in
in vivo animal experiments.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GW conceived and designed the study. WC performed
the experiments. YL, JZ and WC analyzed and interpreted the data.
WC, YL and JZ confirmed the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for participation
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gingold JA, Zhu D, Lee DF, Kaseb A and
Chen J: Genomic profiling and metabolic homeostasis in primary
liver cancers. Trends Mol Med. 24:395–411. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Invenizzi F, Iavarone M, Donato MF,
Mazzucco A, Torre M, Conforti S, Rimessi A, Zavaglia C, Schiavon M,
Comacchio G, et al: Pulmonary resection for metastasis of
hepatocellular carcinoma recurring after liver transplant: An
Italian multicenter experience. Front Oncol. 10:3812020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wei CY, Chen PC, Chau GY, Lee RC, Chen PH,
Huo TI, Huang YH, Su YH, Hou MC, Wu JC, et al: Comparison of
prognosis between surgical resection and transarterial
chemoembolization for patients with solitary huge hepatocellular
carcinoma. Ann Transl Med. 8:2382020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu L, Li Y, Xu XM and Zhu X: Circular RNA
circ-PRKCI promotes cell proliferation and invasion by binding to
microRNA-545 in gastric cancer. Eur Rev Med Pharmacol Sci.
23:9418–9426. 2019.PubMed/NCBI
|
|
8
|
Shi N, Shan B, Gu B, Song Y, Chu H and
Qian L: Circular RNA circ-PRKCI functions as a competitive
endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p
in esophageal squamous cell carcinoma. J Cell Biochem.
120:10021–10030. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qi SX, Sun H, Liu H, Yu J, Jiang ZY and
Yan P: Role and mechanism of circ-PRKCI in hepatocellular
carcinoma. World J Gastroenterol. 25:1964–1974. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai X, Yu L, Chen Z, Ye F, Ren Z and Jin
P: Arsenic trioxide-induced upregulation of miR-1294 suppresses
tumor growth in hepatocellular carcinoma by targeting TEAD1 and
PIM1. Cancer Biomark. 28:221–230. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shan Y and Li P: Long intergenic
non-protein coding RNA 665 regulates viability, apoptosis, and
autophagy via the MiR-186-5p/MAP4K3 axis in hepatocellular
carcinoma. Yonsei Med J. 60:842–853. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lan T, Yan X, Li Z, Xu X, Mao Q, Ma W,
Hong Z, Chen X and Yuan Y: Long non-coding RNA PVT1 serves as a
competing endogenous RNA for miR-186-5p to promote the
tumorigenesis and metastasis of hepatocellular carcinoma. Tumour
Biol. 39:10104283177053382017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Liu G, Sun S and Qin J: miR-1294
alleviates epithelial-mesenchymal transition by repressing FOXK1 in
gastric cancer. Genes Genomics. 42:217–224. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Z, Zhang W, Mao J, Xu Z and Fan M:
miR-186-5p functions as a tumor suppressor in human osteosarcoma by
targeting FOXK1. Cell Physiol Biochem. 52:553–564. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cui H, Gao Q, Zhang L, Han F and Wang L:
Knockdown of FOXK1 suppresses liver cancer cell viability by
inhibiting glycolysis. Life Sci. 213:66–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kawauchi K, Araki K, Tobiume K and Tanaka
N: p53 regulates glucose metabolism through an IKK-NF-kappaB
pathway and inhibits cell transformation. Nat Cell Biol.
10:611–618. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi D, Zhao D, Niu P, Zhu Y, Zhou J and
Chen H: Glycolysis inhibition via mTOR suppression is a key step in
cardamonin-induced autophagy in SKOV3 cells. BMC Complement Altern
Med. 18:3172018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gillies RJ and Gatenby RA: Hypoxia and
adaptive landscapes in the evolution of carcinogenesis. Cancer
Metastasis Rev. 26:311–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding Z, Yang L, Xie X, Xie F, Pan F, Li J,
He J and Liang H: Expression and significance of hypoxia-inducible
factor-1 alpha and MDR1/P-glycoprotein in human colon carcinoma
tissue and cells. J Cancer Res Clin Oncol. 136:1697–1707. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liberti MV and Locasale JW: The Warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benjamin DI, Cravatt BF and Nomura DK:
Global profiling strategies for mapping dysregulated metabolic
pathways in cancer. Cell Metab. 16:565–577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Orang AV, Petersen J, McKinnon RA and
Michael MZ: Micromanaging aerobic respiration and glycolysis in
cancer cells. Mol Metab. 23:98–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen
T, Chen Z, Huang S, Gu J, Li J, et al: MiR-199a-5p is negatively
associated with malignancies and regulates glycolysis and lactate
production by targeting hexokinase 2 in liver cancer. Hepatology.
62:1132–1144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iansante V, Choy PM, Fung SW, Liu Y, Chai
JG, Dyson J, Del Rio A, D'Santos C, Williams R, Chokshi S, et al:
PARP14 promotes the Warburg effect in hepatocellular carcinoma by
inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat
Commun. 6:78822015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang Y, Zhang D, Zheng T, Yang G, Wang J,
Meng F, Liu Y, Zhang G, Zhang L, Han J, et al: lncRNA-SOX2OT
promotes hepatocellular carcinoma invasion and metastasis through
miR-122-5p-mediated activation of PKM2. Oncogenesis. 9:542020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao Q, Wei Z, Li Y, Zhou X, Chen J, Wang
T, Shao G, Zhang M and Zhang Z: miR-186 functions as a tumor
suppressor in osteosarcoma cells by suppressing the malignant
phenotype and aerobic glycolysis. Oncol Rep. 39:2703–2710.
2018.PubMed/NCBI
|
|
28
|
Liu Y, Jiang Y, Xu L, Qu C, Zhang L, Xiao
X, Chen W, Li K, Liang Q and Wu H: circ-NRIP1 promotes glycolysis
and tumor progression by regulating miR-186-5p/MYH9 axis in gastric
cancer. Cancer Manag Res. 12:5945–5956. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sukonina V, Ma H, Zhang W, Bartesaghi S,
Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, et al:
FOXK1 and FOXK2 regulate aerobic glycolysis. Nature. 566:279–283.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Z, Lin W, Gao L, Chen K, Yang C,
Zhuang L, Peng S, Kang M and Lin J: Hsa_circ_0004370 promotes
esophageal cancer progression through miR-1294/LASP1 pathway.
Biosci Rep. 39:BSR201823772019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo TY, Xu HY, Chen WJ, Wu MX and Dai X:
Downregulation of miR-1294 associates with prognosis and tumor
progression in epithelial ovarian cancer. Eur Rev Med Pharmacol
Sci. 22:7646–7652. 2018.PubMed/NCBI
|
|
32
|
Kan XQ, Li YB, He B, Cheng S, Wei Y and
Sun J: MiR-1294 acts as a tumor inhibitor in cervical cancer by
regulating FLOT1 expression. J Biol Regul Homeost Agents. Apr
24–2020.(Epub ahead of print).
|