Introduction
As the most common type of lung cancer, non-small
cell lung cancer (NSCLC) accounts for 80–85% of all lung cancer
types (1). Lung cancer was the
leading cause of cancer-related mortality in the United States in
2020 (2). The activation or
inactivation of signaling pathways and the physiological activities
activated by multiple gene alterations, including mutations,
amplifications, deletions and fusions, aggravate the disease
progression of NSCLC (3). Despite
significant advances in therapeutic strategies of NSCLC, early
diagnosis remains a challenge, and thus, most patients are
diagnosed with advanced lung cancer, and these patients exhibit
poor prognosis (4,5). The limited efficacy of conventional
chemotherapy and tumor metastasis are the predominant challenges in
the treatment of NSCLC (6), and so,
the identification of ideal diagnostic biomarkers for early
detection and elucidation of the mechanisms underlying NSCLC
metastasis are urgently required.
Circular RNA (circRNAs), a class of non-coding RNAs
(ncRNAs), can serve as competing endogenous RNAs (ceRNAs) or
microRNA (miRNA/miR) sponges, resulting in miRNA degradation or
translation inhibition (7).
Dysregulated circRNAs in cancer binds to the mRNA 3′untranslated
region (UTR) via miRNA response elements and affect the
characteristics of tumor cells (8,9).
hsa_circ_0003998 upregulation accelerates the
tumorigenesis of NSCLC by elevating the expression level of Notch
receptor 1 via sponging miR-326 (10). On the basis of the regulatory
mechanism of ceRNAs, hsa_circ_0023404 also induces tumor
progression of NSCLC by regulating the miR-217/zinc finger
E-box-binding homeobox 1 axis (11). Moreover, hsa_circ_0008305 inhibits
TGF-β-induced epithelial-mesenchymal transition (EMT) and
metastasis by regulating tripartite motif containing 24
(TRIM24) in NSCLC (12).
Therefore, the regulatory mechanism of ncRNAs/miRNAs serves an
important role during the progression of NSCLC. Although the role
of circRNAs on the development of NSCLC has been extensively
investigated, the underlying mechanisms via which circRNA mediate
oncogenic pathways that are involved in NSCLC progression remain
poorly understood.
AVL9 cell migration associated (AVL9) is a
crucial conserved protein in the late secretory pathway and is
required for the generation of secretory vesicles, as well as for
actin polarization and polarized growth (13). Depletion of AVL9 results in
secretory defects and the accumulation of Golgi-like membranes
(14). AVL9 is a key regulatory
factor in intracellular trafficking and cell cycle progression, and
AVL9 knockdown-induced abnormalities are derived from both aberrant
intracellular trafficking and defective mitosis, leading to an
elevated rate of cell apoptosis (15). In colorectal cancer, AVL9-mediated
epithelial cell polarity control may suppress colorectal tumor
progression (16). These data
suggest that AVL9 may be a cancer driver candidate gene. A
previous study reported that LINC00662, which belongs to the class
of ncRNAs with length of >200 nucleotides known as long ncRNAs
(lncRNAs), initiates colorectal cancer progression and metastasis
by competing with miR-497-5p, which subsequently represses the
expression of AVL9 (17). However,
whether the ncRNAs/miRNAs axis, especially circRNAs/miRNAs, affects
oncogenesis of NSCLC by targeting AVL9 remains poorly
understood.
Herein, the present study aimed to identify the
expression pattern of hsa_circ_0058357 [produced from the autophagy
related 9A (ATG9A) gene] in NSCLC. The current study further
investigated the underlying regulation mechanism of
hsa_circ_0058357 on the proliferation, migration, apoptosis and
tumor growth of NSCLC cells via sponging miR-24-3p and regulating
AVL9 expression. The present findings could highlight the role of
circ_0058357 in NSCLC and provide a novel drug target for treating
NSCLC.
Materials and methods
Subjects
A total of 20 fresh human NSCLC tissues and
paired-paracancerous tissues located 2 cm away from the lesion were
collected from patients with NSCLC (6 females and 14 males) at the
Affiliated Hangzhou First People's Hospital (Hangzhou, China)
between March 2015 and September 2018. The age of all patients
ranged from 45–58 years. All samples were analyzed retrospectively
in this study. All experiments were approved by the local Ethics
Committee of Affiliated Hangzhou First People's Hospital (ethical
approval no. 2016HZFPH-B072Z), and signed informed consent was
obtained from all the patients. All fresh samples were immediately
frozen in liquid nitrogen (−196°C) and maintained for subsequent
experimentations.
Cell culture and transfection
The human bronchial epithelial (HBE) cell line and
NSCLC cell lines, including H292, A549, H1299 and H460, were
obtained from the American Type Culture Collection. Cells were all
cultured in complete growth medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (cat. no. 10099-141; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
For cell transfection, A549 and H1299 cells were
seeded at a density of 5×105 in 6-well plates. After
incubation overnight at 37°C with 5% CO2, 2 µl
circ_0058357 small interfering (si)RNA (100 mM), circ_0058357
overexpression (OE) plasmid or 50 µM miR-24-3p mimic were
transfected into cells for 48 h using Lipofectamine®
2000 at room temperature (cat. no. 11668-019; Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacture's
introduction. The siRNA targeting circ_0058357 and the miR-24-3p
mimic were synthesized by Guangzhou RiboBio Co., Ltd. (Table I). The recombinant circ_0058357 OE
plasmid was constructed using PLCDH-ciR scrambled control vector
(Biovector Science Lab, Inc). Following 48 h of transfection, all
cells were used for subsequent experiments.
 | Table I.Sequences of siRNA circ_0058357. |
Table I.
Sequences of siRNA circ_0058357.
| Name | Sequence |
|---|
| siRNA1
circ_0058357 |
5′-GCTCATGCAGTTCCTCTTT-3′ |
| siRNA2
circ_0058357 |
5′-GGATCCACCGGCTTATCAA-3′ |
| siRNA3
circ_0058357 |
5′-CCAGATCTGCATCCACAAA-3′ |
| siRNA NC |
5′-TTCTCCGAACGTGTCACGT-3′ |
Cell Counting Kit (CCK)-8 assay
After transfection with circ_0058357 siRNA or
co-transfected with circ_0058357 OE plasmid and miR-24-3p mimic,
A549 and H1299 cells were plated at 96-well plate at a density of
3×103/well. After incubation for 24, 48, 72 and 96 h at
37°C with 5% CO2, 10 µl CCK-8 (cat. no. BS350B; Biosharp
Life Sciences) was added to each well for an additional 4 h
incubation at 37°C, according to the manufacturer's protocol. Then,
the absorption optical density value of cells was determined using
a microplate reader (MULTISKAN MK3; Thermo Fisher Scientific, Inc.)
at the wavelength of 450 nm.
Flow cytometry
A549 and H1299 cells transfected with circ_0058357
siRNA or the combination of circ_0058357 OE plasmid and miR-24-3p
mimic were digested using 0.25% trypsin (Gibco; Thermo Fisher
Scientific, Inc.) and resuspended in 500 µl Annexin V
binding-buffer. After adding 5 µl 7-AAD and 10 µl PI (Sigma
Aldrich; Merck KGaA), the mixture of the binding-buffer and cells
was incubated for 20 min in darkness at room temperature. Then, the
proportion of apoptotic cell was determined via flow cytometry
(CytoFLEX; Beckman Coulter, Inc.) and analyzed using ModFit LT
software (v3.3; Becton, Dickinson and Company). The apoptotic rate
was calculated based on the percentage of early and late apoptotic
cells. Each experiment was performed at least in triplicate.
Transwell and wound healing
assays
Migration assays of A549 and H1299 cells were
detected using Transwell chambers (cat. no. 353097; BD
Biosciences). Briefly, 800 µl complete medium containing 10% FBS
was added into the lower chambers. Then, 200 µl A549 and H1299
cells (3×105 cells/ml) resuspended in DMEM were
transferred to the upper chambers and allowed to migrate at 37°C in
5% CO2 for 28 h. Subsequently, cells in the upper
surface of top chambers were removed using cotton swab. Cells
migrating to the bottom surface were stained for 20 min at room
temperature with 0.5% crystal violet dye (Sigma-Aldrich; Merck
KGaA). The number of migrated cells was counted using a DMR
inverted microscope (magnification, ×40; OLYMP®S, IX51;
Olympus Corporation).
For the wound healing assay, cells were scratched to
create a linear wound with the pipette tip (200-µl) at the
confluence of 90–95%, which was followed by an incubation in
serum-free RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
for 24 h. Then, cells were imaged using a light microscope
(magnification, ×10) and the percent of wound closure was
calculated by analyzing the wound area using Image Pro Plus (v7.0;
Media Cybernetics, Inc.).
Western blot analysis
Total protein from cells and tumor tissues was
extracted using RIPA buffer (Beyotime Institute of Biotechnology).
Protein determination was performed using a BCA kit (Beyotime
Institute of Biotechnology). Equal amounts of protein (30 µg) were
subjected to 10% polyacrylamide gel electrophoresis for 2 h and
then all protein was transferred to a PVDF membrane. After blocking
with 5% skimmed milk for 1 h at room temperature, the activated
PVDF membrane was incubated with an antibody against AVL9 (cat. no.
GTX16209; 1:2,000; GeneTeX, Inc.) overnight at 4°C. GADPH (cat. no.
ab181602; 1:5,000; Abcam) served as the internal control. The
following day, the membrane was incubated with the HRP-conjugated
secondary antibody (1:5,000; cat. no. BA1054; Boster Biological
Technology) for 1 h at room temperature and the bands were
visualized using an ECL reagent (EMD Millipore). Densitometric
analysis was performed using Image Pro Plus software.
Luciferase assay
The target miRNAs of circ_0058357 were identified
using the online StarBase database (http://starbase.sysu.edu.cn/index.php). Subsequently,
the wild-type (WT) or mutant (MUT) 3′-UTR sequence of AVL9 and WT
or MUT circ_0058357 were amplified using genomic DNA and sub-cloned
into the region directly downstream of the stop codon in the
luciferase reporter vector (pGL4-Basic system; Addgene, Inc.). For
the analysis of the relationship between circ_0058357 and
miR-24-3p, A549 and H1299 cells were co-transfected with 50 µM
miR-24-3p mimic and 0.5 µg circ_0058357WT or
circ_0058357MUT plasmids using Lipofectamine 2000. For
the verification of the association between miR-24-3p and AVL9, the
two cell lines were co-transfected with miR-24-3p mimic and
AVL9WT or AVL9MUT plasmids. After
transfection for 48 h, firefly and Renilla luciferase
activities were measured using the dual-luciferase reporter assay
system (Promega Corporation) and the luciferase activities were
calculated based on the ration between firefly luciferase
activities and Renilla luciferase activities.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from tumor cells and tissues
and purified using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and an RNeasy Mini kit (Qiagen, Inc.)
according to the manufacturer's instructions. Then, 10 ng RNA was
reverse transcribed into cDNA using a TaqMan™ miRNA RT kit (cat.
no. 4366596; Thermo Fisher Scientific, Inc.) at 25°C for 5 min,
50°C for 15 min, 85°C for 5 min and 4°C for 10 min. qPCR was
subsequently performed using a SYBR Green Master mix (Vazyme
Biotech Co., Ltd.) on an ABI 7500 system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The following thermocycling
conditions were used for the qPCR: Initial denaturation at 50°C for
2 min and 95°C for 10 min; followed by 40 cycles of 95°C for 30 sec
and 60°C for 30 sec. U6 and GAPDH served as the internal control.
All primer information of miRNA and circRNA is shown in Table II. After the PCR reaction, the data
were normalized to the selected reference gene using U6 or GAPDH.
Relative gene expression levels were quantified using the
2−∆∆Cq method (18).
 | Table II.Primer sequences for reverse
transcription-quantitative PCR. |
Table II.
Primer sequences for reverse
transcription-quantitative PCR.
| Name | Primer | Sequence |
|---|
| GAPDH | Forward |
5′-TCAAGAAGGTGGTGAAGCAGG-3′ |
|
| Reverse |
5′-TCAAAGGTGGAGGAGTGGGT-3′ |
| circ_0058357 | Forward |
5′-GCACAGGGCTGAAGTCGC-3′ |
|
| Reverse |
5′-GACCTTGACGGGTTCAGTAGG-3′ |
| U6 | Forward |
5′-CGCTTCGGCAGCACATATAC-3′ |
|
| Reverse |
5′-AAATATGGAACGCTTCACGA-3′ |
| hsa-miR-24-3p | Forward |
5′-TGCGCGAGTTCAGCAGGAACAG-3′ |
|
| Loop |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGTTCCT-3′ |
Xenograft model
Male nude mice (BALB/c; age, 4 weeks; weight, 18–20
g) were purchased from Charles River Laboratories, Inc., and housed
in a specific-pathogen-free room under a controlled temperature
(20±2°C) and 45–55% humidity. All mice were exposed to a 12-h
light-dark cycle with free access to standard rodent chow and
water. All experiments were approved by the Animal Ethics Committee
of The Affiliated Hangzhou First People's Hospital (approval no.
2016HZFPH-AB19Z). After 1 week of adjustable feeding, ~0.2 ml
(1×107/ml) A549 cells transfected with circ_0058357
shRNA or negative control (NC) shRNA, or cells that were not
transfected were inoculated subcutaneously into the right flank of
mice (n=8/each group). After 1 week of injection, tumor size was
detected once a week for consecutive 6 weeks. Tumor growth rates
were determined by measuring the two orthogonal dimensional
diameters of each tumor. Tumor volumes were calculated according to
the formula V=½xa2xb (with a=short axis, and b=long
axis). At 7 weeks post-injection, mice were sacrificed and the
tumors were completely removed. After imaging and weighing, the
tumor tissues were stored at −80°C for the subsequent assays. Mice
were euthanasia via an overdose of CO2 (30% volume/min)
in the following situations: Weight loss of >15% of mouse body
weight, or mice continued to suffer pain from the tumor burden,
such as the tumor diameter reaching to >2.5 cm and ulcerated
tumors. In this research, the long and short diameters of the
largest tumor were as follows: Long diameter, 1.55 cm; and short
diameter, 1.12 cm.
Immunohistochemistry (IHC)
Tumor tissues were fixed with 4% paraformaldehyde
for >24 h at 4°C and embedded in paraffin. For IHC assay, after
heating in an oven for 30 min at 60°C, 5-µm thick slides of tumor
tissues were deparaffinized in xylene and rehydrated in gradient
ethanol, then retrieved by 0.01 M citrate salt solution (pH 6.0)
using a microwave method. Then, endogenous peroxidase was blocked
at room temperature using 3% H2O2 for 15 min.
After blocking with 5% BSA (Beijing Solarbio Science &
Technology Co., Ltd.) for 1 h at room temperature, a primary
antibody against Ki67 (1:200; cat. no. ab15580; Abcam) was
incubated with slides at 4°C overnight and the slides were then
washed thrice with PBS-0.05% Tween 20. Subsequently, a secondary
antibody (1:500; cat. no. PV-9001; OriGene Technologies, Inc.) was
incubated with slides for an additional 15 min at room temperature.
Antigen-antibody complexes were visualized using DAB staining
buffer (cat. no. ZLI-9017; OriGene Technologies, Inc.). The image
was observed under a biological inverted microscope (magnification,
×20; IX51; Olympus Corporation).
Statistical analysis
All independent experiments were performed at least
in triplicate. Data are presented as the mean ± SD. The data for
the luciferase assay results were examined using an unpaired
t-test. A paired Student's t-test was employed to compare the
difference between two groups. Moreover, a mixed two-way ANOVA and
a Bonferroni or Sidak post hoc test was used for analysis where
applicable. One-way ANOVA with Bonferroni test was conducted to
analyze the differences among multiple groups (>2 groups) using
GraphPad Prism 6.0 (GraphPad Software, Inc.) software. P<0.05
was considered to indicate a statistically significant
difference.
Results
Knockdown of circ_0058357 inhibits the
viability of NSCLC cells
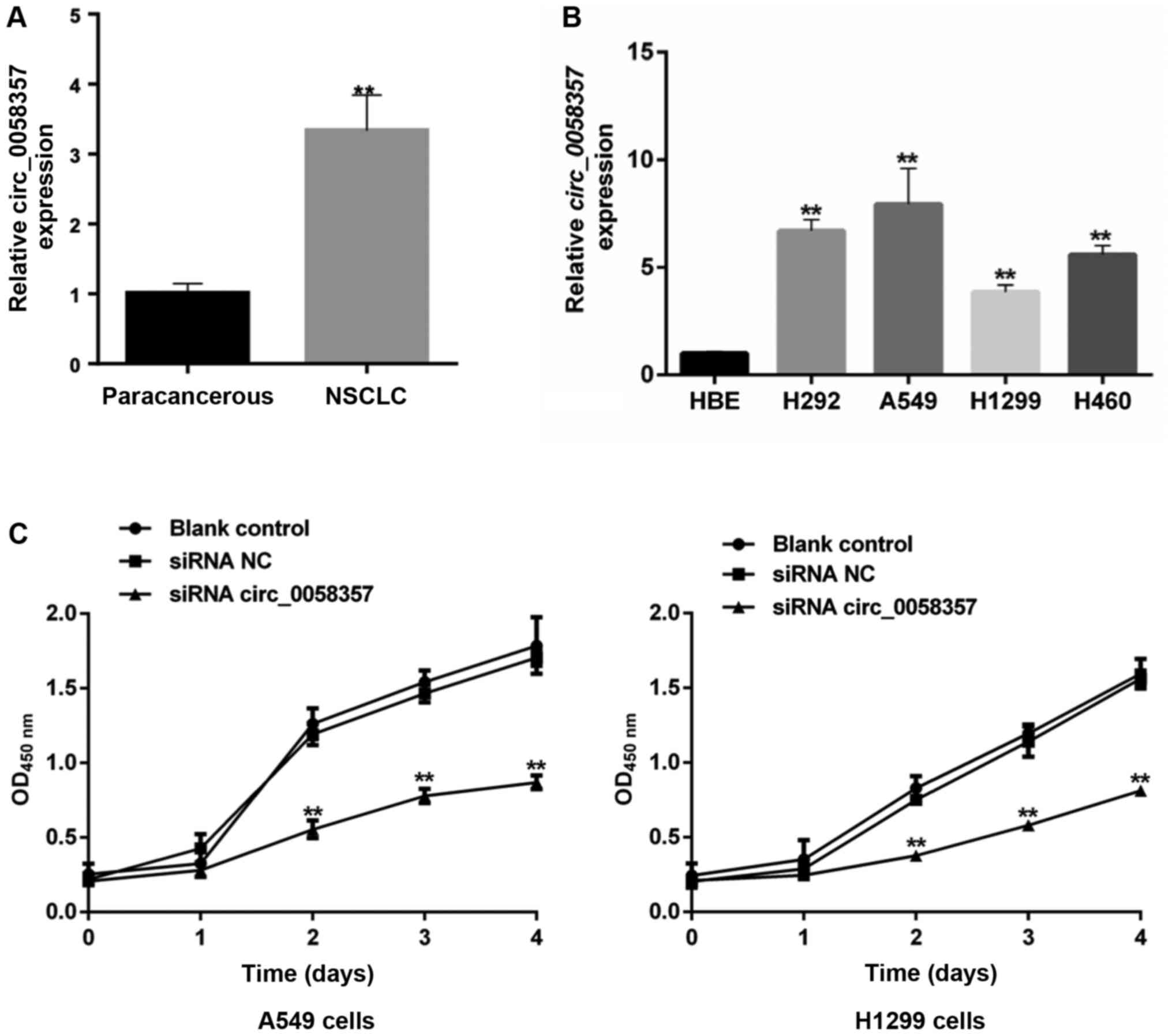
To investigate the role of circ_0058357 on NSCLC,
its expression pattern was firstly determined in human NSCLC
tissues and NSCLC cell lines. The relative expression level of hsa_
circ_0058357 was significantly elevated by ~3.5-fold in human NSCLC
tissues compared with that in the paired-paracancerous tissues
(Fig. 1A). Moreover, in comparison
with normal HBE cells, hsa_ circ_0058357 expression was upregulated
in the NSCLC cell lines, such as H292, A549, H1299 and H460, with
the highest level in A549 cells and a moderate increase in H1299
cells (Fig. 1B). Thus, A549 and
H1299 cell lines were selected to conduct the subsequent
experiments.
 | Figure 1.Effects of circ_0058357 on the
viability of NSCLC cells. (A) Relative expression level of
circ_0058357 in human NSCLC tissues and paired-paracancerous
tissues as determined by RT-qPCR, (n=20/group). **P<0.01 vs.
paracancerous. (B) Relative expression level of circ_0058357 in
NSCLC cells, including H292, A549, H1299, H460 and HBE cells as
detected by RT-qPCR. **P<0.01 vs. HBE cells. (C) A549 and H1299
cells were transfected with siRNA NC and circ_0058357 siRNA. Then,
cell viability was measured using a Cell Counting Kit-8 assay at
days 1, 2, 3 and 4 after incubation. **P<0.01 vs. A549
cells/H1299 cells transfected with siRNA NC. HBE, human bronchial
epithelial; RT-qPCR, reverse transcription-quantitative PCR; NC,
negative control; siRNA, small interfering RNA; OD, optical
density; circ, circular RNA; NSCLC, non-small cell lung cancer. |
Given the high expression level of circ_0058357, it
was suggested that it could be a tumor suppressor in NSCLC. First,
the specific siRNAs targeting circ_0058357 were screened. Among the
three candidate siRNAs, it was discovered that circ_0058357 siRNA-2
had the strongest inhibition efficiency on the expression of
circ_0058357. Thus, circ_0058357 siRNA-2 was selected to perform
the following experiments (Fig.
S1).
When knocking down circ_0058357 using specific
siRNA, the viability of both A549 and H1299 cells was significantly
repressed compared with that in cells transfected with NC siRNA
from days 2 to 4 (Fig. 1C).
However, there was no difference in cell viability between the
blank and NC siRNA groups. Therefore, the present data indicated
that circ_0058357 was highly expressed in NSCLC tissues and NSCLC
cells. Moreover, the knockdown of circ_0058357 contributed to the
proliferation inhibition of NSCLC cells.
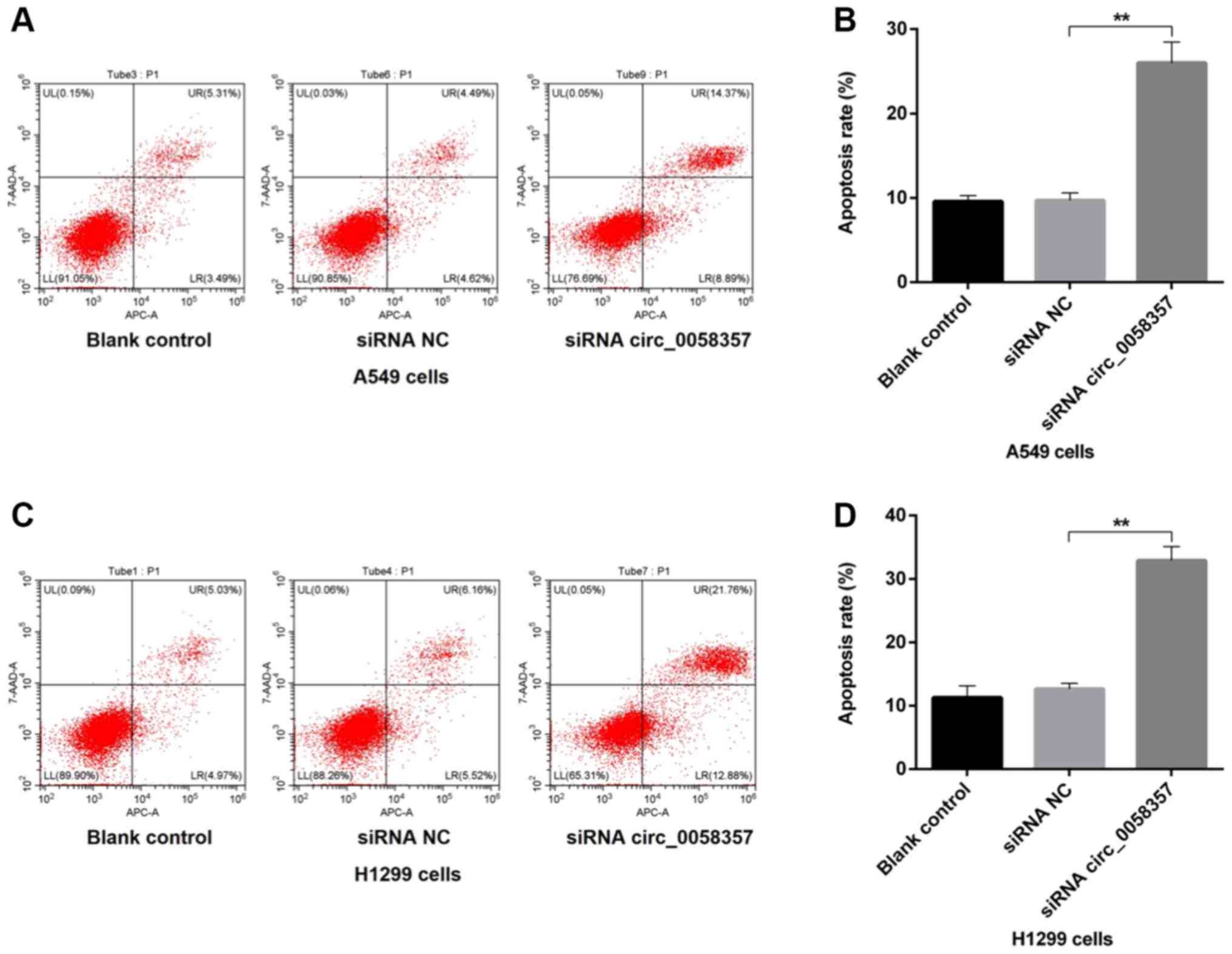
Knockdown of circ_0058357 accelerates
the apoptosis of NSCLC cells
Next, it was determined whether the circ_0058357
knockdown-mediated decrease of cell viability was associated with
programmed cell death in NSCLC cells. Compared with the blank
control A549 cells, the proportion of apoptotic cells, including
early and late apoptotic cells, was not affected by the
transfection of siRNA NC, while there was a 15% increase in the
number of apoptotic cells in circ_0058357-siRNA group compared with
the siRNA NC group (Fig. 2A and B).
Consistently, in H1299 cells, knockdown of circ_0058357 also
enhanced the population of apoptotic cells by ~20% when compared
with cells transfected with siRNA NC. Furthermore, exogenous siRNA
NC had no effect on apoptotic events in untransfected H1299 cells
(Fig. 2C and D). Thus, circ_0058357
knockdown-induced proliferation inhibition may be associated with
the elevation of programmed cell death.
circ_0058357 knockdown impedes the
migration of NSCLC cells
Then, the effect of circ_0058357 knockdown on the
metastasis of NSCLC cells was determined. Based on the Transwell
assay results, a 40% reduction in migrated cells was observed in
A549 cells transfected with circ_0058357-siRNA when compared with
the siRNA NC group (Fig. 3A and B).
In the wound healing assay, circ_0058357-siRNA transfection
significantly inhibited healing rate of A549 cells compared with
cells transfected with siRNA NC (Fig.
3C and D). circ_0058357 knockdown also significantly limited
the migratory ability of H1299 cells, showing a 50% decrease
compared with the siRNA NC group (Fig.
3E and F). Consistently, circ_0058357 knockdown decreased the
wound healing rate of H1299 cells after 24 h of transfection
compared with the siRNA NC group (Fig.
3G and H). In both cell lines, the addition of siRNA NC did not
affect the cellular migration (Fig.
3A-H). Collectively, circ_0058357 was suggested to be a key
regulator during the metastasis of NSCLC cells.
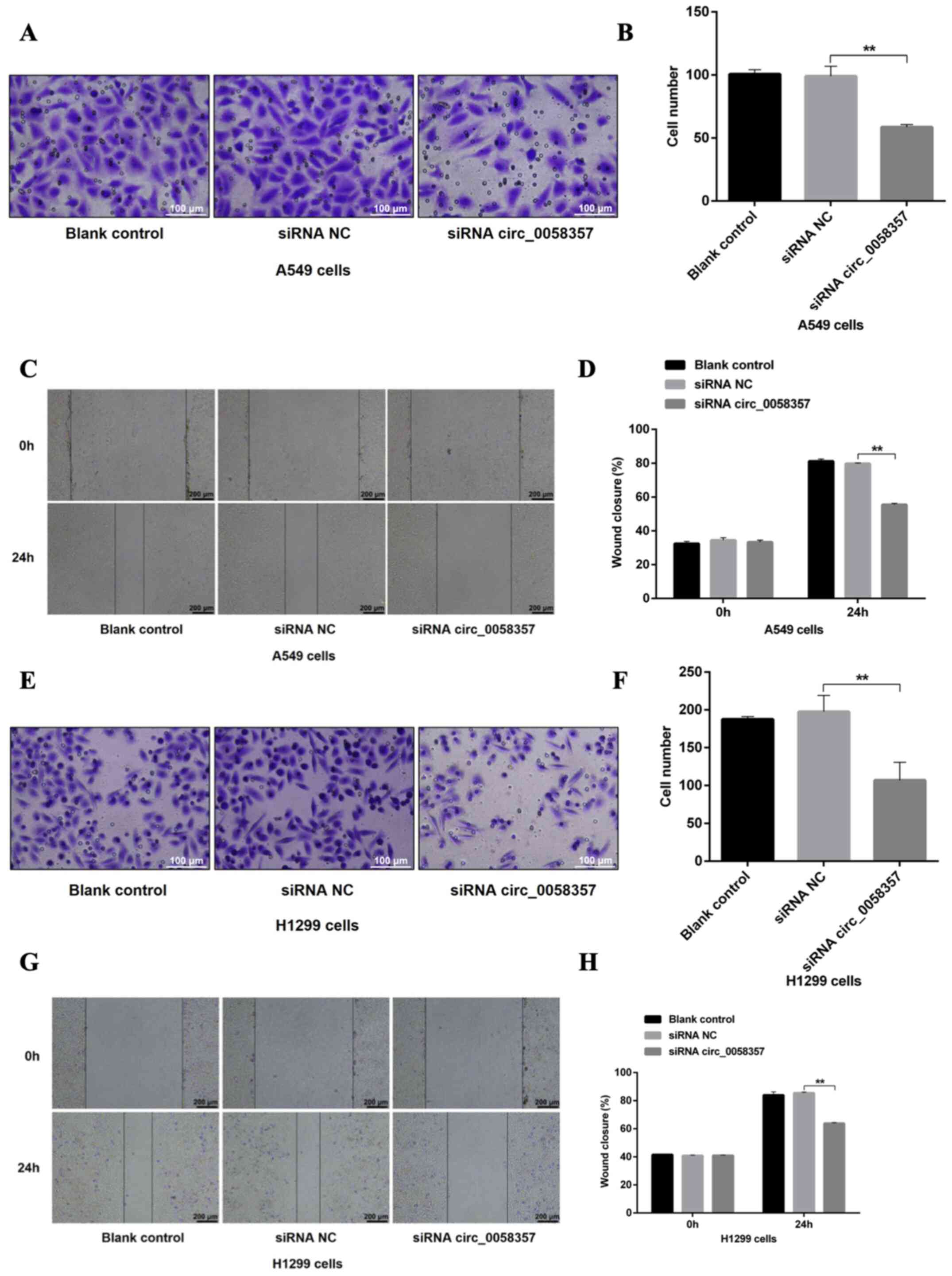 | Figure 3.Effects of circ_0058357 on the
migration of NSCLC cells. (A) A549 cells were transfected with
siRNA NC and circ_0058357 siRNA. The migrated A549 cells were
stained with crystal violet. Scale bar, 100 µm. (B) Quantitative
analysis of migration cells in panel A. (C) After transfection with
siRNA NC and circ_0058357 siRNA, A549 cells were wounded using a
pipette tip. Subsequently, the cells were imaged using a
microscope. Scale bar, 200 µm. (D) The healing rate was measured as
the relative percent of wound closure. (E) H1299 cells were
transfected with siRNA NC and circ_0058357 siRNA. The number of
migrated H1299 cells was determined via crystal violet staining.
Scale bar, 100 µm. (F) Quantitative analysis of migrated cells in
panel E. (G) After transfection with siRNA NC and circ_0058357
siRNA, H1299 cells were wounded using a pipette tip. Subsequently,
the cells were imaged using a microscope. Scale bar, 200 µm. (H)
The healing rate was measured as the relative percent of wound
closure. **P<0.01. NC, negative control; siRNA, small
interfering RNA; circ, circular RNA. |
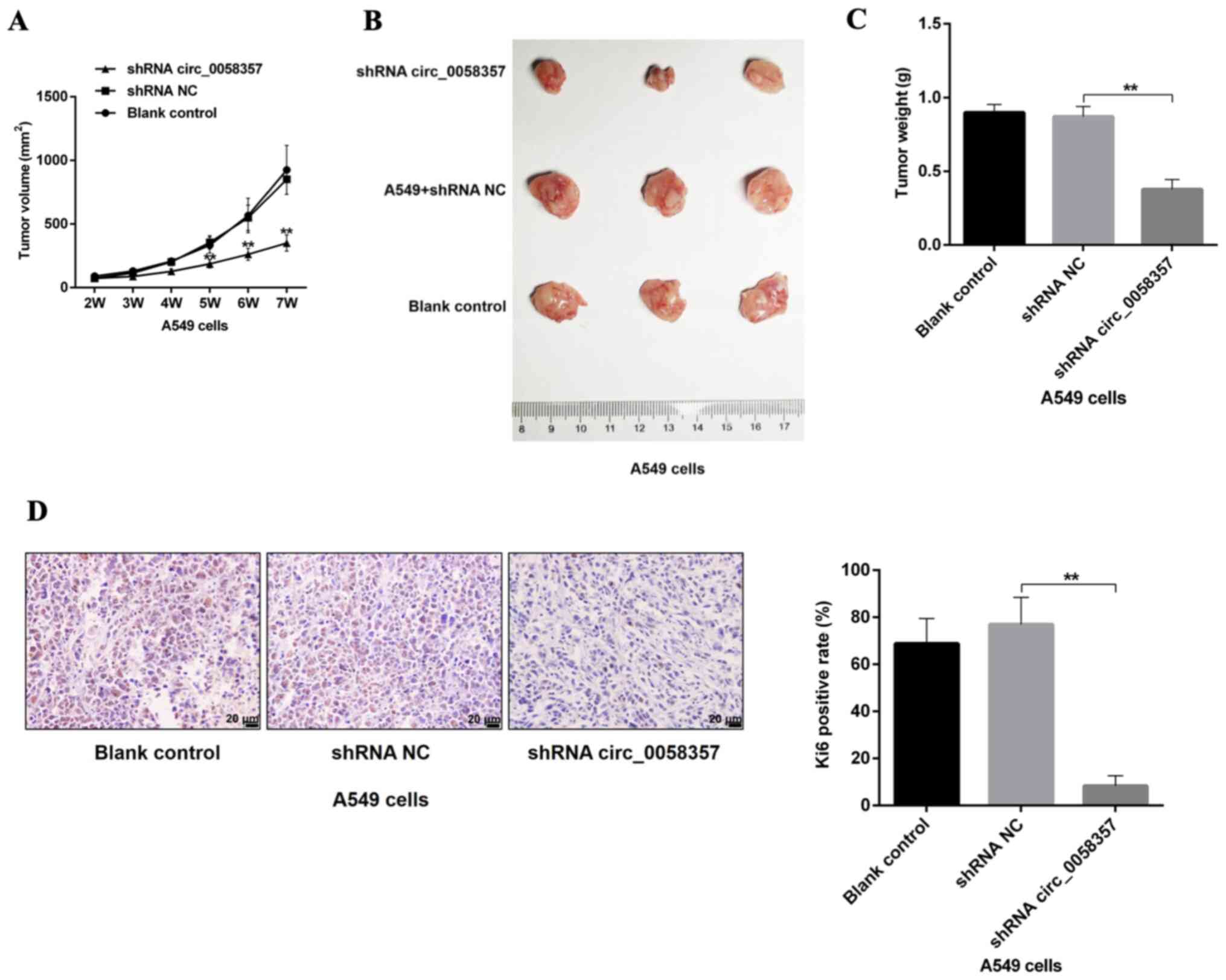
circ_0058357 knockdown restrains tumor
growth of NSCLC cells in vivo
Based on the tumor volumes at different time point,
it was identified that circ_0058357 shRNA transfection slowed down
the tumor growth rate, which presented a significant difference
compared with the shRNA NC group, starting 5 weeks after injection
(Fig. 4A). Additionally, knockdown
of circ_0058357 in A549 cells using shRNA significantly inhibited
tumor size and tumor weight by ~50% at the end of tumor xenograft
assay (Fig. 4B and C). Moreover,
the shRNA NC did not affect the tumor growth and tumor volume
compared with the blank control group. It was also found that both
knockdown of circ_0058357 and the shRNA NC in A549 cells did not
impact the body weight of the three groups (Fig. S2).
Next, the proliferative activity of tumor tissues in
different groups was assessed using an IHC staining assay with an
antibody against Ki67. A low expression level of Ki67 was observed
in tumor tissues from circ_0058357-shRNA group compared with that
in the shRNA NC group. Furthermore, Ki67-positive cells in
circ_0058357-shRNA group accounted for only 14% of the
Ki67-positive cells in tissues of shRNA NC group (Fig. 4D). Therefore, circ_0058357 knockdown
facilitated the tumor growth inhibition of NSCLC cell in
vivo.
hsa-miR-24-3p is sponged by
circ_0058357 in NSCLC cells
Using the online StarBase database, the potential
downstream sponges of circ_0058357 were identified. Among the
screened miRNAs, the alignScore 26 of miR-24-3p was ranked in the
top five, and autophagy events were associated with miR-24-3p and
circ_0058357 (Table SI).
Therefore, miR-24-3p was selected to predict the regulatory
mechanism of circ_0058357 on NSCLC.
A recent study revealed that hsa-miR-24-3p was lowly
expressed in lung adenocarcinomas (19). The present study also found that
miR-24-3p was lowly expressed in NSCLC tissues and NSCLC cells
compared with paracancerous tissues and HBE cells, respectively
(Fig. 5A and B). Interestingly, it
was predicted that circ_0058357 may be a ceRNA of miR-24-3p by
directly binding to the ‘GACTCGGT’ element of miR-24-3p (Fig. S3A). Next, A549 cells were
transfected with the miR-24-3p mimic, which significantly
upregulated the expression of miR-24-3p compared with mimic
NC-transfected cells (Fig. 5C). The
luciferase results indicated that miR-24-3p mimic transfection
significantly suppressed the luciferase activity of WT circ_0058357
both in A549 and H1299 cells, while the upregulation of miR-24-3p
did not affect the transcriptional activity of MUT circ_0058357
(Fig. 5D and E). It was also found
that knockdown of circ_0058357 promoted the expression level of
miR-24-3p, but miR-24-3p OE had no effect on the expression level
of circ_0058357 in both cells (Figs.
5F-H and S3B and C). These
data indicated that circ_0058357 can downregulate the expression
level of miR-24-3p by directly sponging it in NSCLC cells.
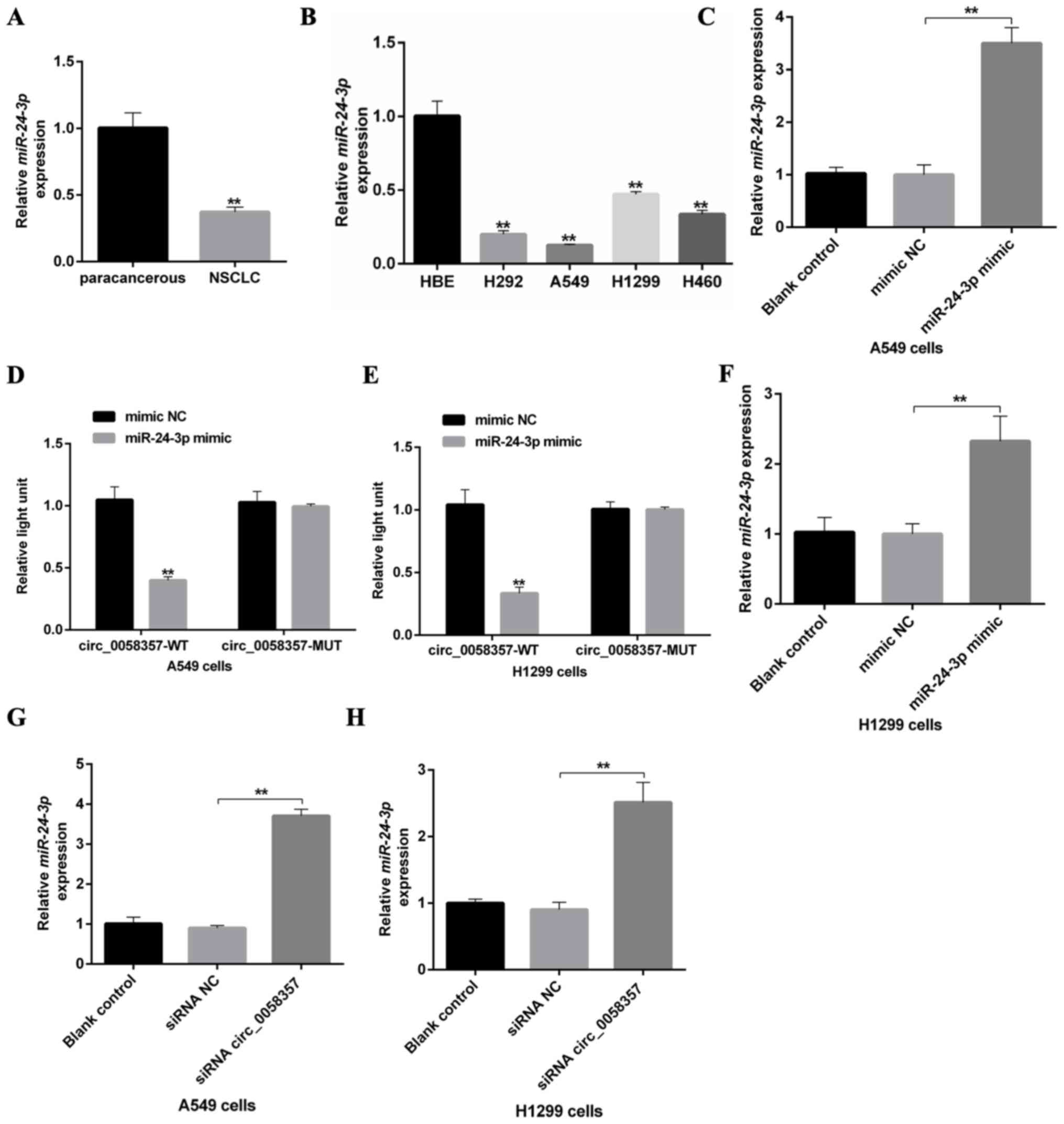 | Figure 5.An association between circ_0058357
and miR-24-3p in NSCLC cells. (A) Relative expression level of
miR-24-3p in human NSCLC tissues and paired-paracancerous tissues,
as determined via RT-qPCR, (n=20/group). (B) Relative expression
level of miR-24-3p in NSCLC cells, including H292, A549, H1299 and
H460, and HBE cells, as detected via RT-qPCR. (C) Transfection
efficiency of miR-24-3p mimics, as determined via RT-qPCR in A549
cells. (D) A549 and (E) H1299 cells were co-transfected with
miR-24-3p mimic and WT or MUT-circ_0058357, then luciferase
activity was detected and normalized with Renilla. (F)
Transfection efficiency of miR-24-3p mimics, as determined via
RT-qPCR in H1299 cells. (G) A549 and (H) H1299 cells were firstly
transfected with circ_0058357 siRNA, then the relative expression
of miR-24-3p was determined via RT-qPCR. **P<0.01. NC, negative
control; siRNA, small interfering RNA; circ, circular RNA; HBE,
human bronchial epithelial; RT-qPCR, reverse
transcription-quantitative PCR; WT, wild-type; MUT, mutant; miR,
microRNA. |
miR-24-3p mimic abolishes the
exogenous circ_0058357- mediated proliferation promotion of NSCLC
cells
Since knockdown of circ_0058357 inhibited
tumorigenesis of NSCLC cells, it was suggested that OE of
circ_0058357 could enhanced the oncogenesis of A549 and H1299
cells. The promotion efficiency of circ_0058357 OE plasmid was
confirmed (Fig. S4). It was
identified that the cell growth curve was accelerated and cell
proliferation was significantly enhanced from days 2 to 4 in A549
cells transfected with circ_0058357 OE plasmid compared with cells
transfected with vector (Fig. 6A and
B). Moreover, exogenous circ_0058357 induced proliferation in
H1299 cells after incubation for 2, 3 and 4 days (Fig. 6C and D). However, when
co-transfection with circ_0058357 OE and miR-24-3p mimic, the
exogenous circ_0058357-mediated promotion in cell proliferation was
notably rescued in both A549 and H1299 cells (Fig. 6A-D). Thus, miR-24-3p may act as a
negative regulator during circ_0058357-evoked tumorigenesis in
NSCLC.
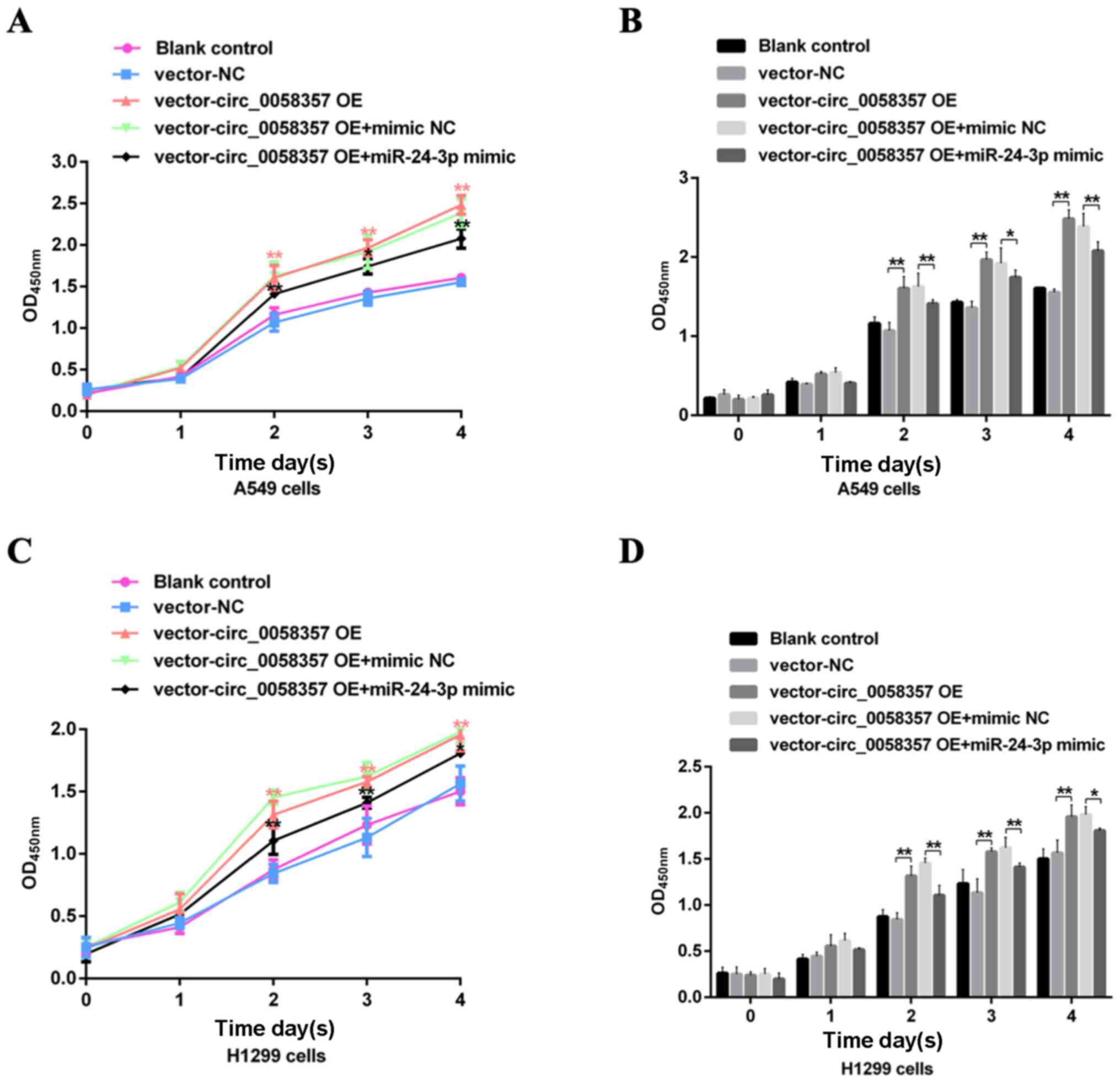 | Figure 6.Effects of miR-24-3p on the exogenous
circ_0058357-mediated viability of NSCLC cells. A549 and H1299
cells were transfected with vector and circ_0058357 OE plasmids
alone, or in combination with vector or circ_0058357 OE plasmids
and NC mimic or miR-24-3p mimic. Then, the (A) cell growth curve of
and (B) viability of A549 cells, and (C) growth curve and (D)
viability of H1299 cells were determined using Cell Counting Kit-8
assay at days 1, 2, 3 and 4 after incubation. *P<0.05,
**P<0.01. NC, negative control; siRNA, small interfering RNA;
circ, circular RNA; OE, overexpression; OD, optical density; miR,
microRNA. |
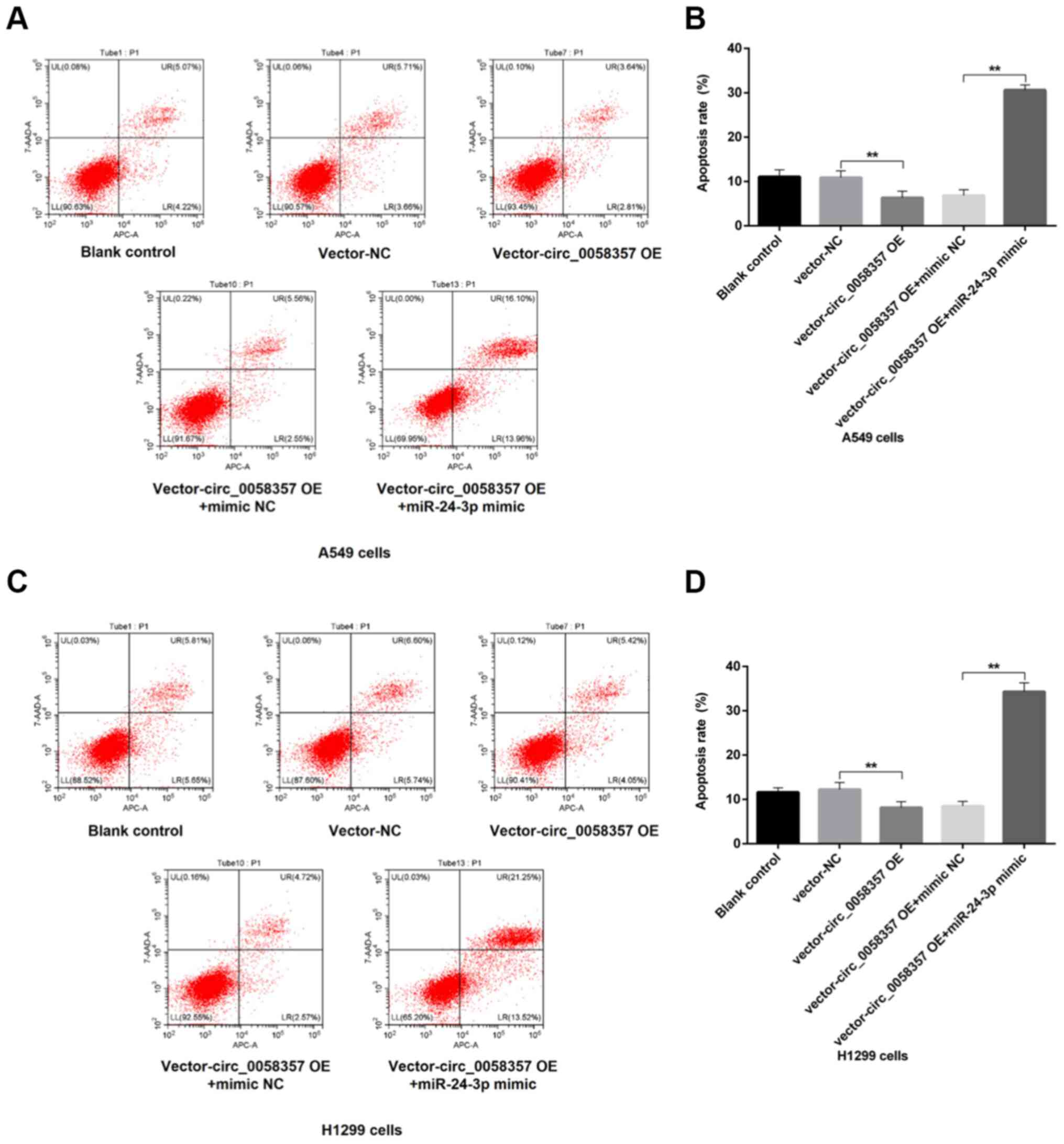
miR-24-3p mimic abrogates circ_0058357
OE-induced apoptosis inhibition and migration promotion
To further determine the regulatory role of
miR-24-3p on circ_0058357-initiated biological behaviors of NSCLC
cells, flow cytometry and Transwell assays were employed in cells
co-transfected with exogenous circ_0058357 and miR-24-3p. The
results demonstrated that OE of circ_0058357 repressed the number
of apoptotic cells by ~50% in A549 and H1299 cells (Fig. 7A-D). However, the co-administration
of miR-24-3p mimic not only abolished the inhibition role of
exogenous circ_0058357, but also significantly amplified the
proportion of apoptotic cells in both cell lines, showing a 5-fold
increase compared with cells co-transfected with circ_0058357 OE
and NC mimic, and a 3-fold increase compared with the blank cells
or cells transfected with vector-NC (Fig. 7A-D).
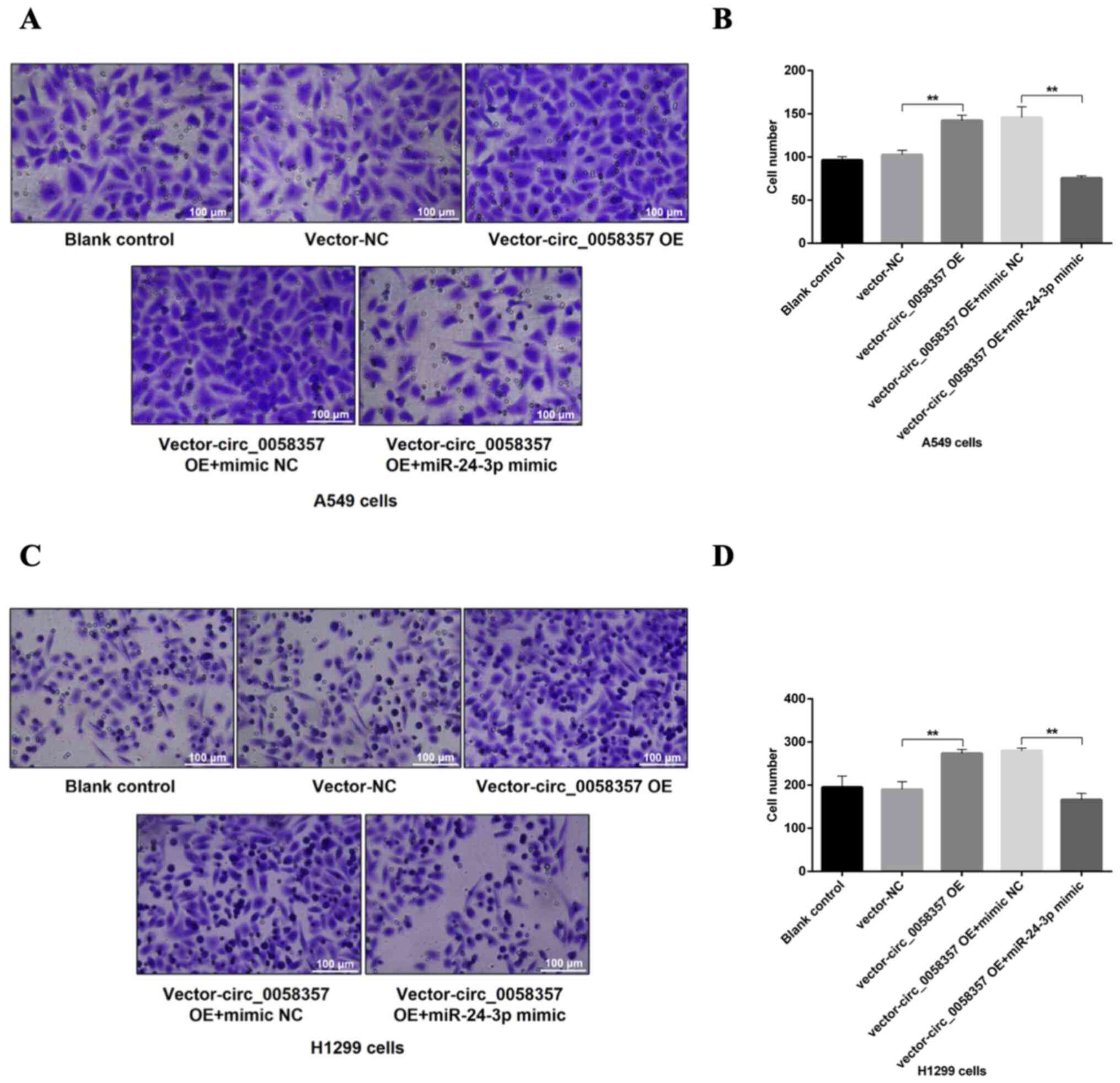
In the Transwell assay, after the OE of
circ_0058357, there was a 1.5-fold elevation in the number of
migrated A549 and H1299 cells compared with the vector-NC group
(Fig. 8A-D). In comparison with the
group of cells co-transfected with circ_0058357 OE and NC mimic,
the miR-24-3p mimic significantly reversed the increase of cell
metastasis induced by circ_0058357 OE in both cells, and returned
to a normal level (Fig. 8A-D).
Collectively, it was indicated that miR-24-3p negatively regulated
circ_0058357-induced cell migration or the inhibition in apoptotic
events.
miR-24-3p can inhibit AVL9 expression
by directly binding its' 3′-UTR domain
As well as being a crucial protein in the late
secretory pathway, AVL9 is also considered as a cancer
driver candidate gene (16).
Herein, it was found that the expression level of AVL9 protein was
higher in the tumor tissues from patients with NSCLC compared with
that in the paired-paracancerous tissues (Fig. 9A and B). In comparison with HBE
cells, there was a high expression level of AVL9 protein in NSCLC
cells (Fig. 9C).
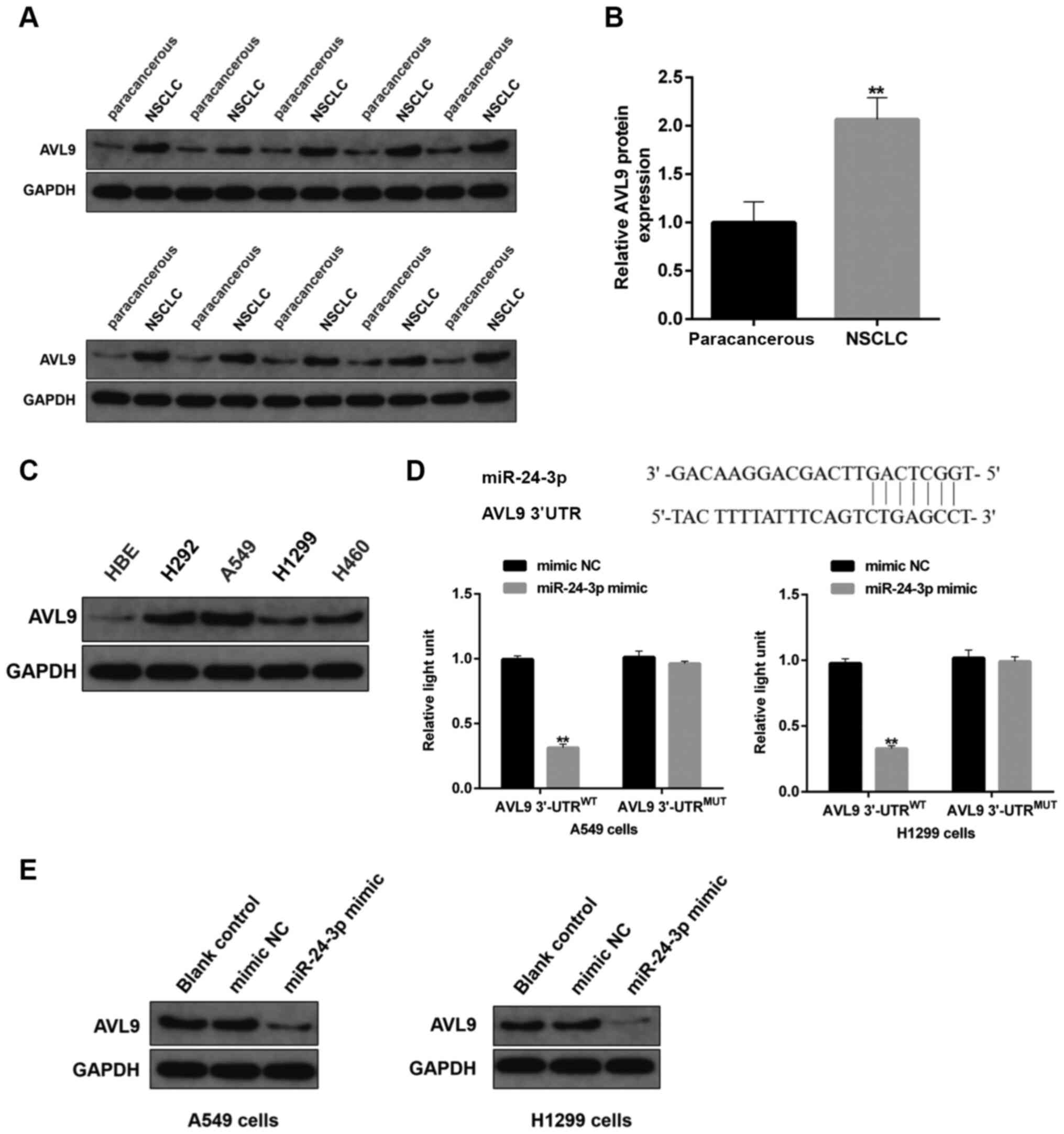 | Figure 9.Effects of miR-24-3p on protein
expression of AV9L. (A) Protein expression level of AVL9 in human
NSCLC tissues and paired-paracancerous tissues was determined via
immunoblotting, (n=20/group). (B) Semi-quantitative analysis of the
protein expression level of AVL9 in panel A. (C) Protein expression
level of AVL9 in NSCLC cells, including H292, A549, H1299 and H460,
and HBE cells, as detected by western blotting. (D) The binding
sites between miR-24-3p and circ_0058357 were identified and shown
in the top part of this panel. A549 and H1299 cells were
co-transfected with mimic NC/miR-24-3p mimic and WT/MUT-AVL9, then
the luciferase activity of AVL9 was detected and normalized with
Renilla. (E) A549 and H1299 cells were firstly transfected
with mimic NC or miR-24-3p mimic. Then, the protein expression
level of AVL9 was evaluated via western blotting in the two cell
lines. **P<0.01. NC, negative control; miR, microRNA; AVL9, AVL9
cell migration associated; NSCLC, non-small cell lung cancer; WT,
wild-type; MUT, mutant; UTR, untranslated region. |
Using the prediction analysis from the online
StarBase database, it was observed that there was direct binding
site of miR-24-3p in the AVL9 gene (Fig. 9D). Utilizing the luciferase assay,
the binding of miR-24-3p to AVL9 was also confirmed in both A549
and H1299 cells. The miR-24-3p mimic significantly reduced the
transcriptional activity of AVL9 in the WT 3′-UTR of AVL9 group,
while there was no effect on the luciferase activity in cells
administrated with MUT 3′-UTR of AVL9 (Fig. 9D). Of note, transfection with
exogenous miR-24-3p markedly decreased the protein expression level
of AVL9 in NSCLC cells (Fig. 9E).
These findings suggested that AVL9 was directly inhibited by
miR-24-3p in NSCLC cells, and that AVL9 may be the key switch in
the circ_0058357/miR-24-3p axis during the development of
NSCLC.
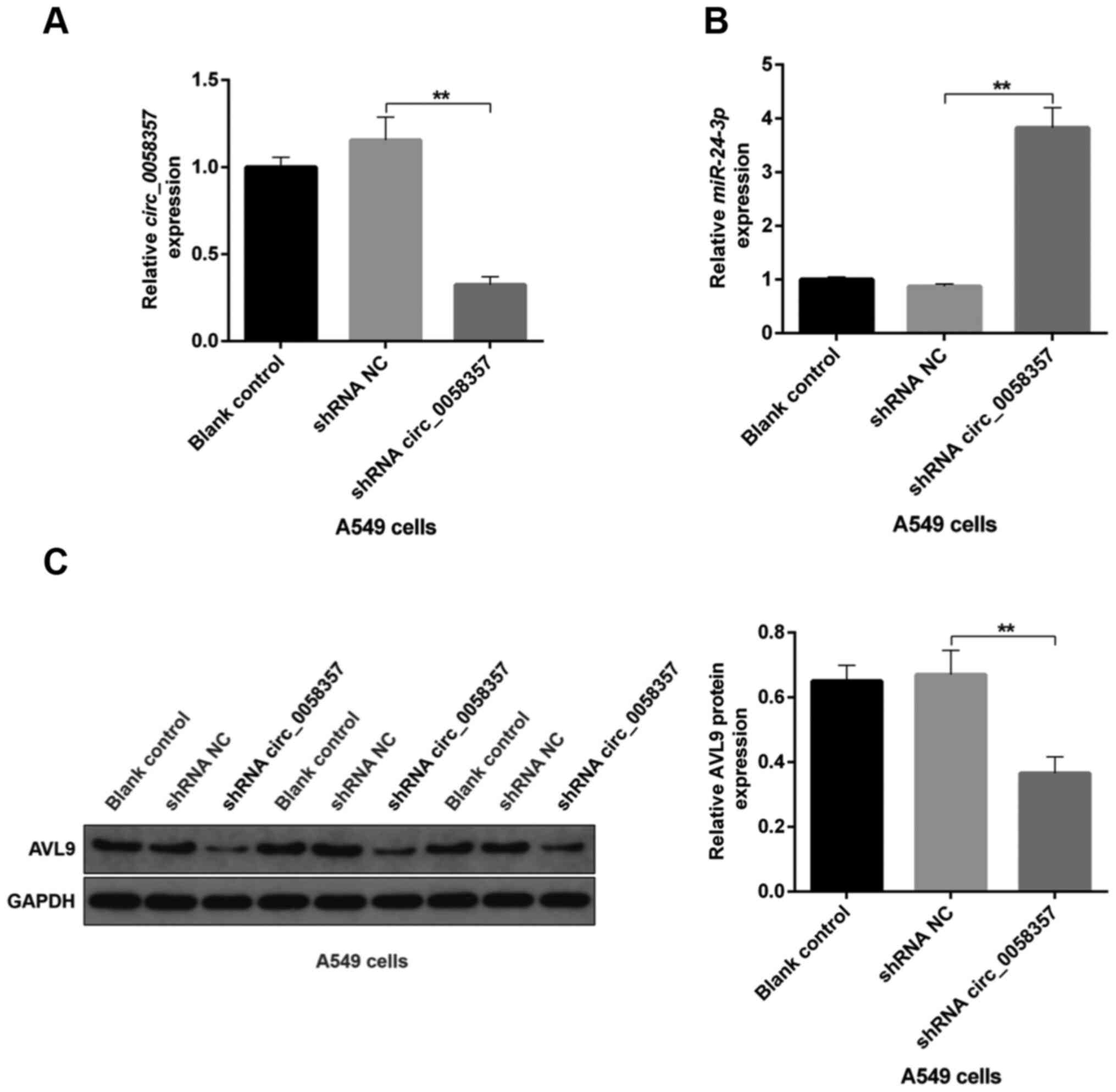
A circ_0058357/miR-24-3p/AVL9
signaling axis is involved in tumor development of NSCLC in
vivo
Next, the relationship between
circ_0058357/miR-24-3p axis and AVL9 was assessed in the tumor
tissues from xenograft model. Tumors from mice bearing A549 cells
transfected with circ_0058357 shRNA had a significant decline in
the expression level of circ_0058357 compared with the shRNA NC
group (Fig. 10A). On the contrary,
compared with the shRNA NC, miR-24-3p expression was significantly
elevated by ~4-fold in tumor tissues when circ_0058357 was knocked
down (Fig. 10B). Additionally, the
protein expression level of AVL9 was significantly decreased in
tumors of circ_0058357 shRNA group (Fig. 10C). It was also found that
transfection with shRNA NC of circ_0058357 in A549 cells did not
affect the circ_0058357/miR-24-3p/AVL9 signaling axis in tumor
tissues (Fig. 10A-C). In summary,
it was suggested that AVL9 may be an important downstream switch of
the circ_0058357/miR-24-3p axis in the process of tumorigenesis of
NSCLC.
Discussion
To date, as a tumor driver gene, whether and how
AVL9 contributes to the progression of NSCLC remains elusive. In
the present study, it was demonstrated that AVL9 was possibly
regulated by the circ_0058357/miR-24-3p signaling pathway, thereby
leading to the development of NSCLC. The present data firstly
indicated that the involvement of circ_0058357 could accelerate
tumor progress of NSCLC via a sponge-regulation mechanism, thereby
promoting the expression of AVL9.
Accumulating evidence has suggested that
dysregulated circRNAs contribute to carcinogenesis and tumor
progression. For instance, in comparison with healthy subjects,
both hsa_circ_0014130 and hsa_circ_0020123 are upregulated in NSCLC
tissues and have a significant positive correlation with TNM stage
and lymph node metastasis, which demonstrates good diagnostic
potential (20,21). hsa_circ_0013958 can also be
considered as a potential non-invasive biomarker for the early
diagnosis of patients with NSCLC (22). In the current study, circ_0058357
expression was notably enhanced in human NSCLC tissues, while it
was lowly expressed in paracancerous tissues. However, a larger
sample size is required to further investigate the application of
circ_0058357 in diagnosing NSCLC. It has been confirmed that
hsa_circ_0075930 is highly expressed in NSCLC, and its depletion
impairs cell proliferation, migration and invasion potential by
abolishing EMT progress (23).
Moreover, knockdown of hsa_circ_0007385 markedly reduces tumor
growth of NSCLC cells in vivo (24). These aberrant circRNAs may be
cancer-promoting genes and could be potential therapeutic targets
for NSCLC. In the present study, knockdown of circ_0058357
repressed cancer properties, including cell viability, cell
migration and tumor growth in vitro and in vivo.
These data suggested that circ_0058357 could be a novel oncogene of
NSCLC.
Previous research has shown that hsa_circ_0008305
[produced from protein tyrosine kinase 2 (PTK2) gene, termed
as circPTK2] inhibited TGF-β-induced EMT, and thus, blunts cancer
metastasis by controlling TRIM24 in NSCLC (12). However, circPTK2 did not influence
its parent gene PTK2 expression. Another previous study
reported that exon-intron or intron-derived circRNAs increased the
transcription of their parent gene, but exon-derived circRNAs do
not affect the expression of their parent genes (25). However, the present study did not
evaluate the regulatory role of circ_0058357 on its parent gene
ATG9A. ATG9A is an important target to develop new specific
cancer therapies, such as those for triple negative breast cancer
and colorectal cancer (26,27). Thus, if circ_0058357 can affect the
expression of its parent ATG9A gene, this finding would have
high significance in the clinic.
While circRNAs have a low abundance, they are
emerging as key oncogenic stimuli in cancer (28). circRNAs work as miRNA sponges
(29). Most recently, circ_0002483
has been confirmed to regulate the expression levels of
cancer-related genes by sponging miR-182-5p, thereby modulating
NSCLC progression (30).
Furthermore, hsa_circ_0004015 acts as a sponge for miR-1183 to
regulate its target gene 3-phosphoinositide dependent protein
kinase 1 and exerts oncogenic effects in NSCLC (31). It has demonstrated that miR-24-3p
may act as an oncogene by targeting chromodomain helicase
DNA-binding domain 5 in head and neck squamous cell carcinoma and
p27Kip1 in breast cancer, respectively (32,33).
Moreover, miR-24-3p can be regulated by several lncRNAs in numerous
types of cancer, including hepatocellular carcinoma (34), prostate cancer (35) and lung cancer (36). miR-24-3p is also sponged by
circ_0080425 in diabetic nephropathy, leading to cell proliferation
and fibrosis inhibition (37).
However, the sponge regulation of miR-24-3p by circRNAs in NSCLC
remains unknown. In the present study, it was observed that
miR-24-3p was directly sponged by circ_0058357 in NSCLC cells.
Moreover, exogenous miR-24-3p abolished the circ_0058357 OE-induced
tumorigenesis of NSCLC cells. These findings suggested that
circ_0058357-mediated carcinogenesis in NSCLC could be primarily
via sponging miR-24-3p.
As aforementioned, AVL9 may be considered as an
oncogene in colorectal cancer (16). Herein, it was discovered that AVL9
protein expression was higher in NSCLC tissues and cells compared
with in paracancerous tissues and HBE cells, suggesting that AVL9
could also be a cancer-driven gene in NSCLC. AVL9 can be directly
regulated by miRNA in cancer cells (17). The present study demonstrated that
AVL9 was the direct downstream target of miR-24-3p in NSCLC cells.
Additionally, in mouse tumor tissues, circ_0058357 knockdown
mediated the decline of miR-24-3p, and subsequently, the increase
of AVL9. It was indicated that circ_0058357 may positively regulate
the expression of AVL9 by sponging miR-24-3p in NSCLC cells.
Except for in colorectal cancer, the AVL9 protein is
also downregulated by lncRNA CRPAT4-siRNA, thereby inhibiting cell
migration and proliferation in the absence of hypoxia-inducible
factor 1α in clear cell renal cell carcinomas (38). Of note, AVL9 ablation notably
suppresses the migration of A549 cells (39). Therefore, AVL9 may be the key
downstream switch in the progress of the circ_0058357/miR-24-3p
signaling axis-induced excessive cell proliferation, metastasis and
tumor growth of NSCLC cells. Mechanistically, the generation of
secretory vesicles in late secretory pathway serves a crucial role
in the progression of several types of cancer, such as NSCLC,
breast cancer and colorectal cancer (40,41).
Rab GTPases define the vesicle trafficking pathways underpinning
cell polarization, proliferation and migration, which involves AVL9
(39,42). As a pivotal regulatory factor in the
late secretory pathway, it was suggested that the
circ_0058357/miR-24-3p pathway-induced carcinogenicity in NSCLC
cells by elevating AVL9 expression should be associated with
vesicle secretion events. However, the link between
hsa_circ_0058357 and hsa-miR-24-3p/AVL9 should be investigated
using the combination of AVL9 OE plasmids and circ_0058357 shRNA in
a xenograft model. Additionally, a larger sample size of specimens
should be included in a future study.
In conclusion, the present study identified a novel
circRNA, circ_0058357, as a potential therapeutic biomarker in
patients with NSCLC. The functional experiments suggested the
significant promotion role of circ_0058357 on cell migration,
migration and tumor growth by positively regulating the expression
level of AVL9 by sponging miR-24-3p. These findings encourage
further efforts to study the detailed regulatory mechanisms
underlying the circ_0058357/miR-24-3p/AVL9-mediated oncogenesis in
NSCLC progression.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Projects of Health
and Family Planning Commission of Zhejiang Province (grant no.
2018KY600), Zhejiang Provincial Natural Science Foundation of China
(grant no. LY21H010001), Zhejiang Provincial Science and Technology
of Traditional Chinese Medicine (grant no. 2021ZB219), Medical and
Health Planning Program of Zhejiang Province (grant no. 2020PY001),
Health Innovation Talent of Zhejiang Province (grant no. 2021PY038)
and Beijing Medical and Health Foundation (grant no.
YWJKJJHKYJJ-F2056G).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WF conceived the idea for the study. WF and LS
drafted the manuscript. DW and LS designed and performed the CCK-8,
Transwell, apoptosis and transfection assays. LS analyzed the data
and designed the figures. DW contributed to xenograft and western
blotting assays. WF provided the clinical breast cancer specimens.
WF and LS confirm the authenticity of all the raw data. All authors
discussed the results and edited this manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All patient experiments were approved by the local
Ethics Committee of Affiliated Hangzhou First People's Hospital
(ethical approval no. 2016HZFPH-B072Z). All animal experiments were
approved by the Animal Ethics Committee of The Affiliated Hangzhou
First People's Hospital (approval no. 2016HZFPH-AB19Z). Signed
informed consent was obtained from all the patients
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zheng HM, Zhan YT, Liu SL, Lu J, Luo J,
Feng J and Fan S: The roles of tumor-derived exosomes in non-small
cell lung cancer and their clinical implications. J Exp Clin Canc
Res. 37:2262018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu G, Pei F, Yang F, Li L, Amin AD, Liu
S, Buchan JR and Cho WC: Role of autophagy and apoptosis in
non-small-cell lung cancer. Int J Mol Sci. 18:3672017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rong B, Nan Y, Liu H and Gao W: Increased
stathmin correlates with advanced stage and poor survival of
non-small cell lung cancer. Cancer Biomark. 19:35–43. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu ZL, Zhu WR, Zhou WC, Ying HF, Zheng L,
Guo YB, Chen JX and Shen XH: Traditional Chinese medicinal herbs
combined with epidermal growth factor receptor tyrosine kinase
inhibitor for advanced non-small cell lung cancer: A systematic
review and meta-analysis. J Integr Med. 12:346–358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta GP and Massague J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cui J, Li W, Liu G, Chen X, Gao X, Lu H
and Lin D: A novel circular RNA, hsa_circ_0043278, acts as a
potential biomarker and promotes non-small cell lung cancer cell
proliferation and migration by regulating miR-520f. Artif Cells
Nanomed Biotechnol. 47:810–821. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie G: Circular RNA hsa-circ-0012129
promotes cell proliferation and invasion in 30 cases of human
glioma and human glioma cell lines U373, A172, and SHG44, by
targeting MicroRNA-661 (miR-661). Med Sci Monit. 24:2497–2507.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Z, Yu K, Liu O, Xiong Y, Yang X,
Wang S, Zhang S, Feng Y and Peng Y: Expression profile and
bioinformatics analyses of circular RNAs in keloid and normal
dermal fibroblasts. Exp Cell Res. 388:1117992020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu W, Jiang H, Zhang H and Li J:
Hsa_circ_0003998 promotes cell proliferation and invasion by
targeting miR-326 in non-small cell lung cancer. Onco Targets Ther.
11:5569–5577. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu C, Zhang Z and Qi D: Circular RNA
hsa_circ_0023404 promotes proliferation, migration and invasion in
non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco
Targets Ther. 12:6181–6189. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang L, Tong X, Zhou Z, Wang S, Lei Z,
Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al: Circular RNA
hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced
epithelial-mesenchymal transition and metastasis by controlling
TIF1γ in non-small cell lung cancer. Mol Cancer. 17:1402018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Nebane NM, Wennerberg K, Li Y,
Neubauer V, Hobrath JV, McKellip S, Rasmussen L, Shindo N, Sosa M,
et al: A high-throughput screen for chemical inhibitors of exocytic
transport in yeast. Chembiochem. 11:1291–1301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Harsay E and Schekman R: Avl9p, a member
of a novel protein superfamily, functions in the late secretory
pathway. Mol Biol Cell. 18:1203–1219. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Xu J, Xiong H, Ma Z, Wang Z, Kipreos
ET, Dalton S and Zhao S: Cancer driver candidate genes AVL9,
DENND5A and NUPL1 contribute to MDCK cystogenesis. Oncoscience.
1:854–865. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang J, Li Y, Lyon K, Camps J, Dalton S,
Ried T and Zhao S: Cancer driver-passenger distinction via sporadic
human and dog cancer comparison: A proof-of-principle study with
colorectal cancer. Oncogene. 33:814–822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Yu M, Hu W, Chen X, Luo Y, Lin X,
Zeng Y and Yao X: Linc00662 promotes tumorigenesis and progression
by regulating miR-497-5p/AVL9 axis in colorectal cancer. Front
Genet. 10:13852020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Olbromski M, Rzechonek A, Grzegrzolka J,
Glatzel-Plucinska N, Chachaj A, Werynska B, Podhorska-Okolow M and
Dziegiel P: Influence of miR-7a and miR-24-3p on the SOX18
transcript in lung adenocarcinoma. Oncol Rep. 39:201–208.
2018.PubMed/NCBI
|
|
20
|
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S
and Yuan H: Microarray profile of circular RNAs identifies
hsa_circ_0014130 as a new circular RNA biomarker in non-small cell
lung cancer. Sci Rep. 8:28782018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu D, Yan B, Xin R and Ma T: A novel
circular RNA hsa_circ_0020123 exerts oncogenic properties through
suppression of miR-144 in non-small cell lung cancer. Am J Cancer
Res. 8:1387–1402. 2018.PubMed/NCBI
|
|
22
|
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan
X, Han S and Wu G: hsa_circ_0013958: A circular RNA and potential
novel biomarker for lung adenocarcinoma. FEBS J. 284:2170–2182.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Wang J, Chen Z, Chen Y and Jin M:
Hsa_circ_0079530 promotes cell proliferation and invasion in
non-small cell lung cancer. Gene. 665:1–5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X,
Sun BH and Di QG: Microarray profiles reveal that circular RNA
hsa_circ_0007385 functions as an oncogene in non-small cell lung
cancer tumorigenesis. J Cancer Res Clin Oncol. 144:667–674. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cortés-López M and Miura P: Emerging
functions of circular RNAs. Yale J Biol Med. 89:527–537. 2016.
|
|
26
|
Gil J, Ramsey D, Pawlowski P, Szmida E,
Leszczynski P, Bebenek M and Sasiadek MM: Interdependence between
an expression of the ATG9A gene and the BAX gene in colorectal
cancer. J Biol Regul Homeost Agents. 33:183–185. 2019.PubMed/NCBI
|
|
27
|
Claude-Taupin A, Fonderflick L, Gauthier
T, Mansi L, Pallandre JR, Borg C, Perez V, Monnien F, Algros MP,
Vigneron M, et al: ATG9A is overexpressed in triple negative breast
cancer and its in vitro extinction leads to the inhibition of
pro-cancer phenotypes. Cells. 7:2482018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lyu DB and Huang SL: The emerging role and
clinical implication of human exonic circular RNA. Rna Biol.
14:1000–1006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, Yang B, Ren H, Xiao T, Zhang L, Li
L, Li M, Wang X, Zhou H and Zhang W: Hsa_circ_0002483 inhibited the
progression and enhanced the Taxol sensitivity of non-small cell
lung cancer by targeting miR-182-5p. Cell Death Dis. 10:9532019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou Y, Zheng X, Xu B, Chen L, Wang Q,
Deng H and Jiang J: Circular RNA hsa_circ_0004015 regulates the
proliferation, invasion, and TKI drug resistance of non-small cell
lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys
Res Commun. 508:527–535. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun X, Xiao D, Xu T and Yuan Y:
miRNA-24-3p promotes cell proliferation and regulates
chemosensitivity in head and neck squamous cell carcinoma by
targeting CHD5. Future Oncol. 12:2701–2712. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu K, Wang J, Song Y, Zhao S, Liu H, Tang
D, Pan B, Zhao H and Zhang Q: miRNA-24-3p promotes cell
proliferation and inhibits apoptosis in human breast cancer by
targeting p27Kip1. Oncol Rep. 34:995–1002. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fan JC, Zeng F, Le YG and Xin L: LncRNA
CASC2 inhibited the viability and induced the apoptosis of
hepatocellular carcinoma cells through regulating miR-24-3p. J Cell
Biochem. 119:6391–6397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Han X, Wei P, Yang J and Sun J:
Knockdown of lncRNA CCAT1 enhances sensitivity of paclitaxel in
prostate cancer via regulating miR-24-3p and FSCN1. Cancer Biol
Ther. 21:452–462. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Su C, Shi K, Cheng X, Han Y, Li Y, Yu D
and Liu Z: Long noncoding RNA LINC00472 inhibits proliferation and
promotes apoptosis of lung adenocarcinoma cells via regulating
miR-24-3p/DEDD. Technol Cancer Res Treat. 17:15330338187904902018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu H, Wang X, Wang ZY and Li L:
Circ_0080425 inhibits cell proliferation and fibrosis in diabetic
nephropathy via sponging miR-24-3p and targeting fibroblast growth
factor 11. J Cell Physiol. 235:4520–4529. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang W, Wang J, Chai R, Zhong G, Zhang C,
Cao W, Yan L, Zhang X and Xu Z: Hypoxia-regulated lncRNA CRPAT4
promotes cell migration via regulating AVL9 in clear cell renal
cell carcinomas. Onco Targets Ther. 11:4537–4545. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Linford A, Yoshimura S, Nunes Bastos R,
Langemeyer L, Gerondopoulos A, Rigden DJ and Barr FA: Rab14 and its
exchange factor FAM116 link endocytic recycling and adherens
junction stability in migrating cells. Dev Cell. 22:952–966. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang M and Petersen NO: Lipid-coated gold
nanoparticles promote lamellar body formation in A549 cells.
Biochim Biophys Acta. 1831:1089–1097. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schaaij-Visser TB, de Wit M, Lam SW and
Jiménez CR: The cancer secretome, current status and opportunities
in the lung, breast and colorectal cancer context. Biochim Biophys
Acta. 1834:2242–2258. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang L, Huang M and Harsay E: A chemical
genetic screen for modulators of exocytic transport identifies
inhibitors of a transport mechanism linked to GTR2 function.
Eukaryot Cell. 9:116–126. 2010. View Article : Google Scholar : PubMed/NCBI
|