Introduction
Glioma is the most prevalent histological subtype
among primary tumors of the central nervous system that originates
from normal glial cells (1). Glioma
has been estimated to account for ~75% of brain malignant tumors
worldwide (2). According to the
2016 World Health Organization classification, gliomas are
generally classified into astrocytoma, oligodendroglioma,
oligoastrocytoma, ependymoma and neuronal and mixed neuronal-glial
tumors (3). Currently, the most
definitive treatment modality for glioma is surgical resection of
the primary lesion coupled with postoperative radiotherapy and
chemotherapy (4). However, due to
the limitations of brain function and structure, and the formation
of chemical resistance of tumor cells, the recurrence rate after
surgery is extremely high, resulting in a 5-year survival rate that
is <5% (5).
Cancer stem cells (CSCs) have been confirmed to
serve an significant regulatory role in tumor metastasis,
recurrence and chemoresistance (6),
and thus, they may represent a highly valuable therapeutic target
in anti-cancer treatment. A study by Auffinger et al
(7) revealed that temozolomide
(TMZ)-treated glioma cells can interconvert from non-CSCs to CSCs,
thereby supplementing the original tumor population, and ultimately
enhancing its infiltration and TMZ resistance. More importantly, it
has been suggested that it is of profound significance to
investigate the molecular mechanism of alleviating the
chemoresistance caused by CSCs of gliomas.
Long non-coding (lnc)RNAs are a large and
functionally diverse class of non-RNA with a length of >200
nucleotides, which serve a crucial role in multiple diseases, such
as tumors (8), kidney diseases
(9) and cardiovascular diseases
(10). Moreover, several studies
have reported that the abnormal expression of lncRNA is closely
associated with the occurrence and development of malignant tumors,
including glioma (11–13). For instance, taurine upregulated
gene 1 (TUG1) acts as a tumor-suppress factor of human glioma by
promoting cell apoptosis (14).
Interestingly, Zhao et al (15) discovered that TUG1 acted as a
tumor-promoting factor for human glioma by promoting cell
proliferation and invasion, and inhibiting its apoptosis. This
indicates that TUG1 may be a tumor-promoting or a tumor suppressive
factor in glioma under different circumstances. However, there is a
lack of information regarding the mechanism of TUG1 regulating
TMZ-resistance in CSCs of gliomas.
The histone-lysine N-methyltransferase, enhancer of
Zeste homolog 2 polycomb repressive complex subunit 2 (EZH2), is an
indispensable catalytic enzyme for the methylation of histone H3
lysine 27 (H3K27me) and histone H3 lysine 9 in mammalian cells
(16). Furthermore, our previous
studies reported that EZH2 acted as a cancer-promoting factor in
glioma cells, and increased the proliferation, invasion and
migration of glioma cells (17).
However, the underlying mechanism of the interaction between TUG1
and EZH2 in the occurrence and development of glioma remains
elusive.
Therefore, the present study aimed to investigate
the underlying mechanism of TUG1 on the CSCs-like properties of
TMZ-resistant glioma cells via EZH2. The purpose of this study was
to provide basic experimental evidence and therapeutic targets for
the treatment of glioma resistance.
Materials and methods
Cell lines and cell culture
For this study, normal human astrocytes (NHAs; NHA2;
cat. no. CP-H122) were purchased from Procell Life Science &
Technology Co., Ltd. Human glioblastoma cells (U251, TJ861, U87MG,
T98G and A172; cat. nos. TCHu58, TCHu216, TCHu138, TCHu48 and
TCHu171, respectively) were acquired from The Cell Bank of the
Chinese Academy of Sciences. After short tandem repeat profiling,
it was noted that U87MG cells were glioblastoma cells of unknown
origin from the American Type Culture Collection. All cells were
cultured in DMEM (Hyclone; Cytiva), supplemented with 10% FBS
(Hyclone; Cytiva), 100 U/ml penicillin and 0.1 g/ml streptomycin,
and were cultured in a 5% CO2 incubator at 37°C for
routine use.
To establish TMZ-resistant glioma cells, A172 cells
were continuously exposed to 40–100 µM TMZ (Sigma-Aldrich; Merck
KGaA) for 6 months daily (18). The
expression level of drug resistance markers, glutathione
S-transferase (GST)-π and P-glycoprotein 1 (P-gp), in A172 cells
were assessed using western blot analysis. The obtained
TMZ-resistant cells were named A172/TMZ cells.
Cell Counting Kit (CCK)-8 assay (Dojindo Molecular
Technologies, Inc.) was used to evaluate the survival of A172/TMZ
cells and the parent A172 cell line following treatment with TMZ
(0, 5, 10, 20, 50, 100, 200, 500 and 1,000 µM). The IC50
calculator website (https://www.aatbio.com/tools/ic50-calculator)
calculated the IC50 (50% inhibiting concentration) of
TMZ in A172 cells and A172/TMZ cells. According to the
IC50 obtained, the resistance index (RI) was further
calculated using the following equation: IC50 of the
drug-resistant cell/IC50 of the parent cell.
Construction and cell
transfection
The cDNA strands of TUG1 and EZH2 were synthesized
and purified by Shanghai GeneChem Co., Ltd. and were cloned into
the expression vector pCDNA3.1 (Shanghai GeneChem Co., Ltd.).
Moreover, the short hairpin RNA (shRNA/sh) of TUG1 and EZH2 was
obtained from Shanghai GeneChem Co., Ltd. and was used to prepare
the plasmid vector for transfection using a DNA Midiprep kit
(Thermo Fisher Scientific, Inc.). Next, A172/TMZ cells treated with
IC50 TMZ (450 µM) were transfected with the plasmid
construct [40 nM sh-TUG1/EZH2 or sh-negative control (NC), 2 µg
pcDNA3.1-TUG1/EZH2 vector or pcDNA3.1-NC empty vector] using
Lipofectamine® 2000 reagent (Thermo Fisher Scientific,
Inc.), following the manufacturer's instructions at 37°C for 24 h.
Further experimentation was performed after cells were incubated at
37°C for 48 h. The shRNA sequences are as follows: sh-TUG1,
5′-CAGAAGAATGGTACAAATCCAAG-3′; sh-EZH2,
5′-CTGATGAAGTAAAGAGTATGTTT-3′; and sh-NC,
5′-TTCTCCGAACGTGTCACGTTT-3′.
Reverse transcription-quantitative
(RT-q)PCR assay
Total RNA from NHAs and human glioma cell lines was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Then, the concentration and purity of total RNA were determined
using the UV absorption method. According to the QuantiTect RT kit
(Qiagen, Inc.) instructions, 1 µg RNA was reverse transcribed into
cDNA. Subsequently, GAPDH was employed as an internal control to
detect the expression levels of TUG1 and EZH2 mRNA, as per the
instructions of Power SYBR Green (Takara Biotechnology Co., Ltd.).
The following thermocycling conditions were used for the qPCR:
Initial denaturation at 95°C for 30 sec; followed by 40 cycles of
95°C for 3 sec and 60°C for 30 sec. Relative expression levels of
each sample were calculated using the 2−ΔΔCq method
(19). The PCR primers for TUG1,
EZH2 and GAPDH were as follows: TUG1 forward,
5′-AGCGTGGGTGTACGTAAAGG-3′ and reverse, 5′-CCAAGGATTGGGGAACTGCT-3′;
EZH2 forward, 5′-GGACTCAGAAGGCAGTGGAG-3′ and reverse,
5′-CTTGAGCTGTCTCAGTCGCA-3′; and GAPDH forward,
5′-GCAACTAGGATGGTGTGGCT-3′ and reverse,
5′-TCCCATTCCCCAGCTCTCATA-3′.
Western blotting
After collecting the transfected cells, RIPA lysate
(Beyotime Institute of Biotechnology) was used to extract the total
protein in the cells, and a Bio-Rad protein (Bio-Rad Laboratories,
Inc.) assay kit was used to determine the concentration and purity
of the total protein. Then, 20 µg protein lysates were loaded onto
10% SDS-PAGE gels for electrophoresis, and separated proteins were
subsequently transferred onto PVDF membranes. After blocking with
5% BSA [Roche Diagnostics (Shanghai) Co., Ltd.] at room temperature
for 1 h, the membranes were incubated overnight at 4°C with primary
antibodies against: EZH2 (1:1,000; cat. no. ab228697), GST-π
(1:1,000; cat. no. ab138491), P-gp (1:1,000; cat. no. ab242104),
Oct4 (1:1,000; cat. no. ab109183), Nanog (1:500; cat. no. ab173368)
and SOX2 (1:1,000; cat. no. ab171380) (all from Abcam).
Subsequently, membranes were washed three time with 0.1%
TBS-Tween-20, 10 min each time, and further incubated with a
secondary antibody goat anti-mouse IgG H&L (1:5,000; cat. no.
ab97019; Abcam) or goat anti-rabbit IgG H&L (1:2,000; cat. no.
ab6721; Abcam) at room temperature for 2 h. After visualization
using an ECL kit (Thermo Fisher Scientific, Inc.), the images were
captured with an E-Gel Imager gel imager (Thermo Fisher Scientific,
Inc.), while the gray value of the protein band was semi-quantified
using ImageJ software (version 1.0; National Institutes of
Health).
CCK-8 assay
A172 and A172/TMZ cells were seeded in a 96-well
plate at a density of 5×103 cells/well and were
incubated at 37°C for 24 h. According to the manufacturer's
instructions, 10 µl CCK-8 solution was added to the medium at the
indicated time intervals (0, 24, 48, 72 or 96 h), followed by
incubation at 37°C for 2 h. Cells were subsequently detected using
a microplate reader (Beckman Coulter, Inc.) at a wavelength of 450
nm.
Annexin V-FITC/PI assay
Cell apoptosis was detected using flow cytometry
(FACSCalibur; BD Biosciences) using an Annexin V-FITC/PI kit (BD
Biosciences). The apoptotic rate was calculated using the
percentage of early + late apoptotic cells using CellQuest software
(version 5.0; BD Biosciences). Briefly, A172/TMZ cells grown to log
phase were collected, resuspended in pre-cooled PBS, centrifuged at
150 × g for 10 min at room temperature and washed. Following the
addition of 300 µl 1X binding buffer to suspend the cells, 500 µl
pre-cooling buffer and 5 µl Annexin V-FITC (BD Biosciences) were
added and then incubated at room temperature for 15 min in the
dark. Subsequently, 2.5 µl PI staining solution was added for 5 min
in the dark at room temperature. Finally, 200 µl 1X binding buffer
was added to each tube to detect cell apoptosis.
Colony formation assay
A172/TMZ cells were digested with 0.25% trypsin and
suspended in DMEM. Then, cells were plated at a density of 500
cells/well, containing 10 ml DMEM, and incubated at room
temperature for 2 weeks. When the clones were visible on the plate,
the culture was stopped. Subsequently, the cells were fixed in 5 ml
paraformaldehyde for 15 min and washed with PBS at room
temperature. Then, the cells were stained with GIMSA staining
solution for 20 min and then the staining solution was slowly
washed off with PBS at room temperature. The number of cells was
counted under a light microscope (Olympus Corporation;
magnification, ×100). The number of clones was counted using ImageJ
software (version 1.0; National Institutes of Health), and >50
cells was considered a colony.
Sphere formation assay
A172/TMZ cells (70–90% confluence) were digested and
centrifuged at 150 × g for 10 min at room temperature. Then, the
medium was removed. After washing twice with PBS, the cells were
resuspended in DMEM and counted. Cells were then seeded at a
concentration of 1×104 cells/well into a 6-well plate
for ultra-low adsorption cell culture, and were supplemented with 4
ml DMEM. After 10 days, the sphere state of cells was observed
under a light microscope (Olympus Corporation; magnification,
×100), and the number of sphere-forming cells was calculated by
dividing the total number of spheres by the number of cells
plated.
In vivo tumorigenicity
All experimental procedures involving animals were
performed following the guidelines of the Animal Experimental
Center of Kunming Medical University. Ethical approval was obtained
from The First Affiliated Hospital of Kunming Medical University
Ethics Committee (approval no. kmu-eac-2018056; date, 2018-05-01).
For this experiment, 12 male nude mice (age, 6 weeks; weight, 20–30
g) were purchased from Shanghai Experimental Animal Center of the
Chinese Academy of Sciences. All animal experiments in this study
were conducted strictly adhering to the Guidelines for the Care and
Use of Laboratory Animals proposed by the National Institutes of
Health (20). The mice were housed
in a specific pathogen-free environment at 22±2°C and 45–65%
humidity, with a 12-h light/dark cycle and access to food and water
ad libitum. Nude mice were subcutaneously injected with
2×105 A172/TMZ cells stably expressing TUG1
(pcDNA-lncRNA-TUG1) or the corresponding pcDNA3.1 NC vector. Then,
4 days after the injection, TMZ (10 mg/kg) was orally administered
to mice. A total of 36 days after injection, mice of each group
were euthanized by cervical dislocation. When tumors were palpable
and visible, the tumor volume was measured weekly in two dimensions
with a Vernier caliper and was calculated using the following
formula: Length × width 2 × 0.5. All mice were sacrificed at 5
weeks following injection. At the end of the study, the tumors were
isolated, weighed and imaged. Subsequently, the expression levels
of Ki67, Bcl2 and Bax were examined via immunohistochemistry.
Immunohistochemistry
Tumor tissues were fixed with 10% paraformaldehyde
at room temperature for 12 h, embedded in paraffin using an
automatic Biological Tissue Embedding machine (Leica Microsystems,
Inc.) and sliced into thin 4-µm thick sections. The sections were
incubated with 3% H2O2 for 30 min at 37°C to
block endogenous peroxidase activity and with 10% goat serum for 30
min at 37°C to block non-specific binding. The sections were then
incubated with Ki67 (1:200; cat. no. ab16667), Bcl-2 (1:250; cat.
no. ab32124) and Bax (1:50; cat. no. ab81083) primary antibodies
(all from Abcam) for 12 h at 4°C. Then, the sections were incubated
with goat anti-rabbit IgG H&L (1:1,000; cat. no. ab6721; Abcam)
for 30 min at 37°C. Sections were then stained with a DAB kit
(Thermo Fisher Scientific, Inc.) for 2 min at room temperature. The
results were observed under a light microscope (Olympus
Corporation; magnification, ×200). As previously described, the
expression levels of EZH2, GST-π, P-gp, Oct4, Nanog and SOX2 were
detected using western blotting.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 20.0; IBM Corp.), while GraphPad Prism 7
(GraphPad Software, Inc.) was used for drawing graphs. The
comparison between the two groups was performed using an unpaired
Student's t-test, whereas the comparison between multiple groups
was evaluated using a one-way ANOVA followed by Tukey's post hoc
test. Data are presented as the mean ± SD. P<0.05 was considered
to indicate a statistically significant difference. All experiments
were replicated three times.
Results
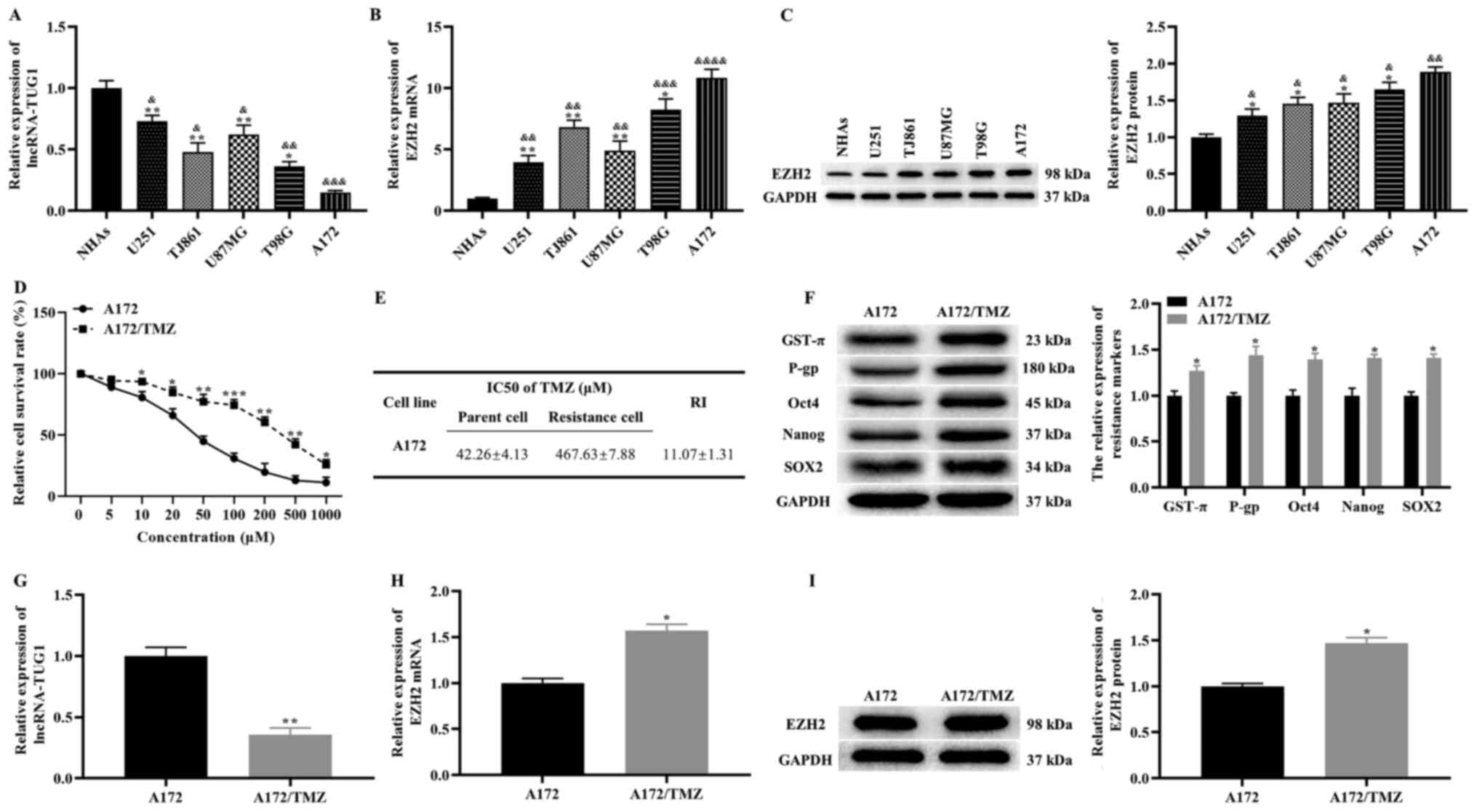
Expression levels of TUG1 and EZH2 in
glioma cell lines
The expression levels of TUG1 and EZH2 were detected
in NHA2 cells and glioma cell lines (U251, TJ861, U87MG, T98G and
A172). Compared with NHA2 cells, TUG1 was downregulated in glioma
cell lines (lowest in A172 cells), while the mRNA expression level
of EZH2 exhibited the opposite pattern, based on RT-qPCR results
(Fig. 1A and B). In addition, the
western blotting results demonstrated that the protein expression
level of EZH2 was significantly higher in glioma cell lines
(highest in A172 cells) compared with that in NHA2 cells (Fig. 1C). Taken together, these results
suggest that abnormal expression of TUG1 and EZH2 in glioma cell
lines may be closely associated with the occurrence and development
of glioma. Moreover, A172 cells were selected for further
experiments.
Expression levels of TUG1 and EZH2 in
A172/TMZ cells
Previous studies have reported that CSCs are closely
associated with the TMZ resistance of tumor cells (21,22).
Thus, A172/TMZ cells were established using increasing TMZ
concentrations. The CCK-8 results revealed that the inhibitory
effect of TMZ on the proliferation of A172/TMZ and A172 cells was
dose-dependent (Fig. 1D). The
IC50 and RI values of A172/TMZ cells were 467.63±7.88
and 11.07±1.31, respectively (Fig.
1E). Moreover, the expression levels of drug resistance
markers, GST-π and P-gp, were detected. The western blotting
results demonstrated that the expression levels of GST-π and P-gp
were significantly higher in A172/TMZ cells compared with those in
A172 cells (Fig. 1F). This finding
suggests that the drug resistance level of A172/TMZ cells met the
requirements of resistant strains.
Subsequently, the expression levels of CSCs markers,
including Oct4, Nanog and SOX2, were measured in A172/TMZ and A172
cells. As expected, the expression levels of Oct4, Nanog and SOX2
were markedly higher in A172/TMZ cells compared with those in A172
cells (Fig. 1F). Thus, it was noted
that A172/TMZ cells exhibited CSCs-like phenotypes.
The difference in TUG1 and EZH2 expression in the
parent A172 cells was subsequently measured. The findings of
RT-qPCR and western blotting demonstrated that the expression level
of TUG1 was significantly lower in A172/TMZ cells compared with
that in A172 cells, while opposite results were found for the mRNA
and protein expression levels of EZH2 (Fig. 1G-I). Collectively, these results
suggested that A172/TMZ cells displayed CSCs-like properties, and
that TUG1 and EZH2 contributed to these properties.
Effects of the knockdown of TUG1 on
the proliferation, apoptosis and CSCs-like properties of A172/TMZ
cells
To analyze the effect of the knockdown of TUG1 on
A172/TMZ cell survival and CSC-like properties, A172/TMZ cells were
treated with 450 µM TMZ. The RT-qPCR results indicated that after
knockdown of TUG1, the expression levels of TUG1 in A172/TMZ cells
were significantly downregulated (Fig.
2A). Additionally, the results of CCK-8 revealed that the
proliferative activity of A172/TMZ cells in the sh-lncRNA-TUG1
group was markedly higher compared with that in the sh-NC group
(Fig. 2B).
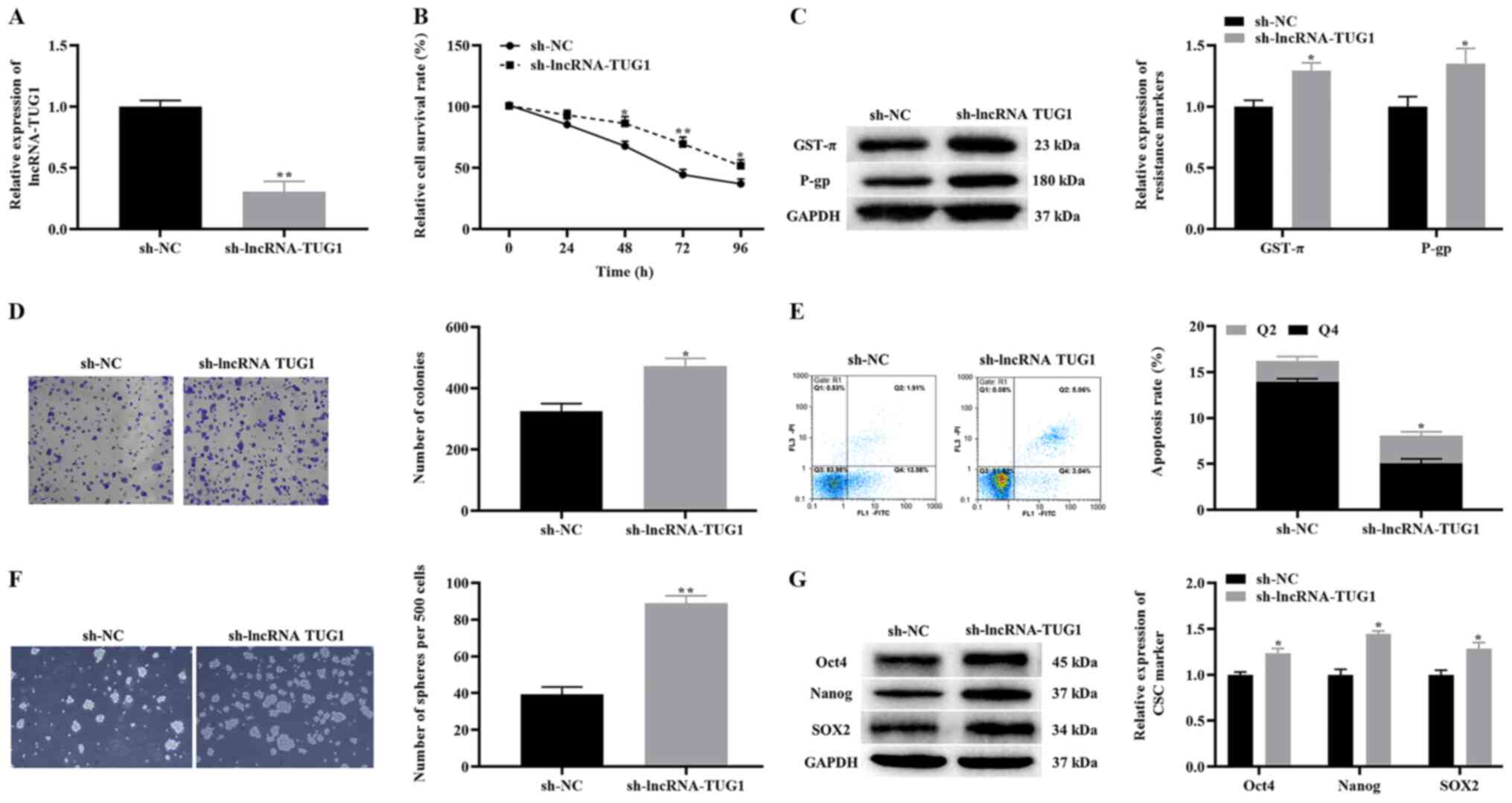 | Figure 2.Effect of TUG1 knockdown on the
proliferation, apoptosis and CSCs-like properties of A172/TMZ
cells. (A) Effect of knockdown of TUG1 on the expression levels of
TUG1 in A172/TMZ cells. (B) Cell Counting Kit-8 results of the
proliferation of A172/TMZ cells before and after knockdown of TUG1.
(C) Effects of TUG1 knockdown on the expression levels of drug
resistance markers. (D) Clone formation results showing the
proliferation of A172/TMZ cells in each group (magnification,
×100). (E) Annexin V FITC/PI apoptosis depicting the apoptotic
rates of A172/TMZ cells in each group. (F) Stem cell
spheroidization experiment showing the spheroidizing ability of
A172/TMZ cells (magnification, ×100). (G) Western blotting results
demonstrated the effect of TUG1 knockdown on the expression levels
of CSCs markers. The error bars represent the mean ± SD of at least
triplicate experiments. *P<0.05, **P<0.01 vs. sh-NC group.
sh, short hairpin RNA; NC, negative control; lncRNA, long
non-coding RNA; TMZ, temozolomide; TUG1, taurine upregulated gene
1; GST-π, glutathione S-transferase-π; P-gp, P-glycoprotein 1. |
The effect of the knockdown of TUG1 on drug
resistance markers was determined in A172/TMZ cells. The results
demonstrated that knockdown of TUG1 could significantly upregulate
the expression levels of GST-π and P-gp (Fig. 2C). Moreover, the proliferation of
A172/TMZ cells was examined using the clone formation experiment,
and the findings indicated that the proliferative activity of
A172/TMZ cells was significantly higher in the sh-lncRNA-TUG1 group
compared with that of the sh-NC group (Fig. 2D). It was also observed that the
apoptosis level of A172/TMZ cells was lower in the sh-lncRNA-TUG1
group compared with that of the sh-NC group, according to the
annexin V FITC/PI apoptosis experiment (Fig. 2E).
Next, the effect of the knockdown of TUG1 on the
CSC-like properties of A172/TMZ cells was investigated. The results
of the stem cell spheroidization experiment showed that, compared
with the sh-NC group, the number of stem cells formed was increased
in the sh-lncRNA-TUG1 group (Fig.
2F). The western blotting results revealed that, compared with
the sh-NC group, the expression levels of Oct4, Nanog and SOX2 were
significantly upregulated in the sh-lncRNA-TUG1 group (Fig. 2G). Based on these findings, it was
identified that knockdown of TUG1 promoted the proliferation and
CSC-like properties of A172/TMZ cells treated with TMZ, thereby
inhibiting their apoptosis. As a result, enhancing the TMZ
resistance of A172/TMZ cells.
Effects of the knockdown of EZH2 on
the proliferation, apoptosis and CSCs-like properties of A172/TMZ
cells
To test the effect of the knockdown of EZH2 on the
proliferation, apoptosis and CSC-like properties of A172/TMZ cells,
A172/TMZ cells were treated with 450 µM TMZ. The western blotting
results demonstrated that knockdown of EZH2 significantly
downregulated the expression level of EZH2 in A172/TMZ cells
(Fig. 3A). Then, CCK-8 and clone
formation experiments were used to examine the effects of EZH2
knockdown on the proliferation of A172/TMZ cells. The findings
indicated that compared with the sh-NC group, the proliferation
activity and the number of cell clones of A172/TMZ cells were
significantly lower in the sh-EZH2 group (Fig. 3B and D).
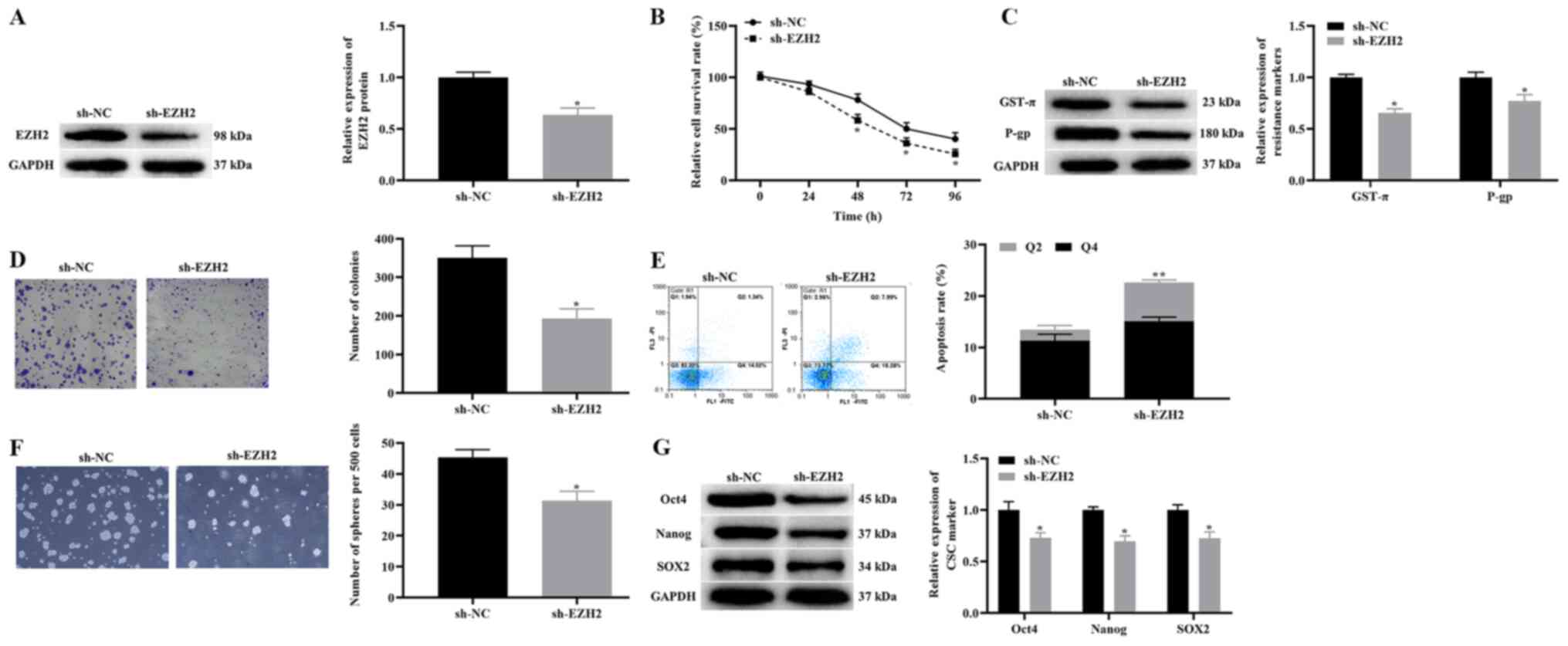 | Figure 3.Effect of TUG1 knockdown on the
proliferation, apoptosis and CSCs-like properties of A172/TMZ
cells. (A) Western blotting detected the change of EZH2 expression
in A172/TMZ cells after knockdown of EZH2. (B) Cell Counting Kit-8
results displaying the effect of the knockdown of EZH2 on the
proliferation of A172/TMZ cells. (C) Western blotting results
indicated the effect of EZH2 knockdown on the expression levels of
drug resistance markers. (D) Clone formation experiment detected
the effect of EZH2 knockdown on the clone formation of A172/TMZ
cells (magnification, ×100). (E) Annexin V FITC/PI apoptosis
experiment displaying the effect of EZH2 knockdown on the apoptosis
of A172/TMZ cells. (F) Stem cell spheroidization depicting the
effect of EZH2 knockdown on the spheroidization ability of A172/TMZ
cells (magnification, ×100). (G) Western blotting results
indicating the effect of EZH2 knockdown on the expression levels of
CSCs markers. The error bars represent the mean ± SD of at least
triplicate experiments. *P<0.05, **P<0.01 vs. sh-NC group.
sh, short hairpin RNA; NC, negative control; TMZ, temozolomide;
GST-π, glutathione S-transferase-π; P-gp, P-glycoprotein 1; EZH2,
enhancer of Zeste homolog 2 polycomb repressive complex subunit
2. |
Subsequently, the expression levels of drug
resistance markers were detected in A172/TMZ cells using western
blotting. The results suggested that knockdown of EZH2
significantly downregulated GST-π and P-gp expression in A172/TMZ
cells (Fig. 3C). It was
demonstrated that knockdown of EZH2 can enhance the sensitivity of
A172/TMZ cells to TMZ. Moreover, the Annexin V FITC/PI apoptosis
experiment revealed that knockdown of EZH2 significantly increased
the apoptotic rate of A172/TMZ cells (Fig. 3E). On the other hand, the findings
of stem cell spheroidization experiments and western blotting
demonstrated that knockdown of EZH2 significantly decreased the
number of sphere-forming A172/TMZ cells and downregulated the
expression levels of Oct4, Nanog, and SOX2 (Fig. 3F and G). In summary, these results
suggested that knockdown of EZH2 inhibited the proliferation and
CSC-like properties of A172/TMZ cells, as well as induced their
apoptosis, thereby alleviating the TMZ resistance of A172/TMZ
cells.
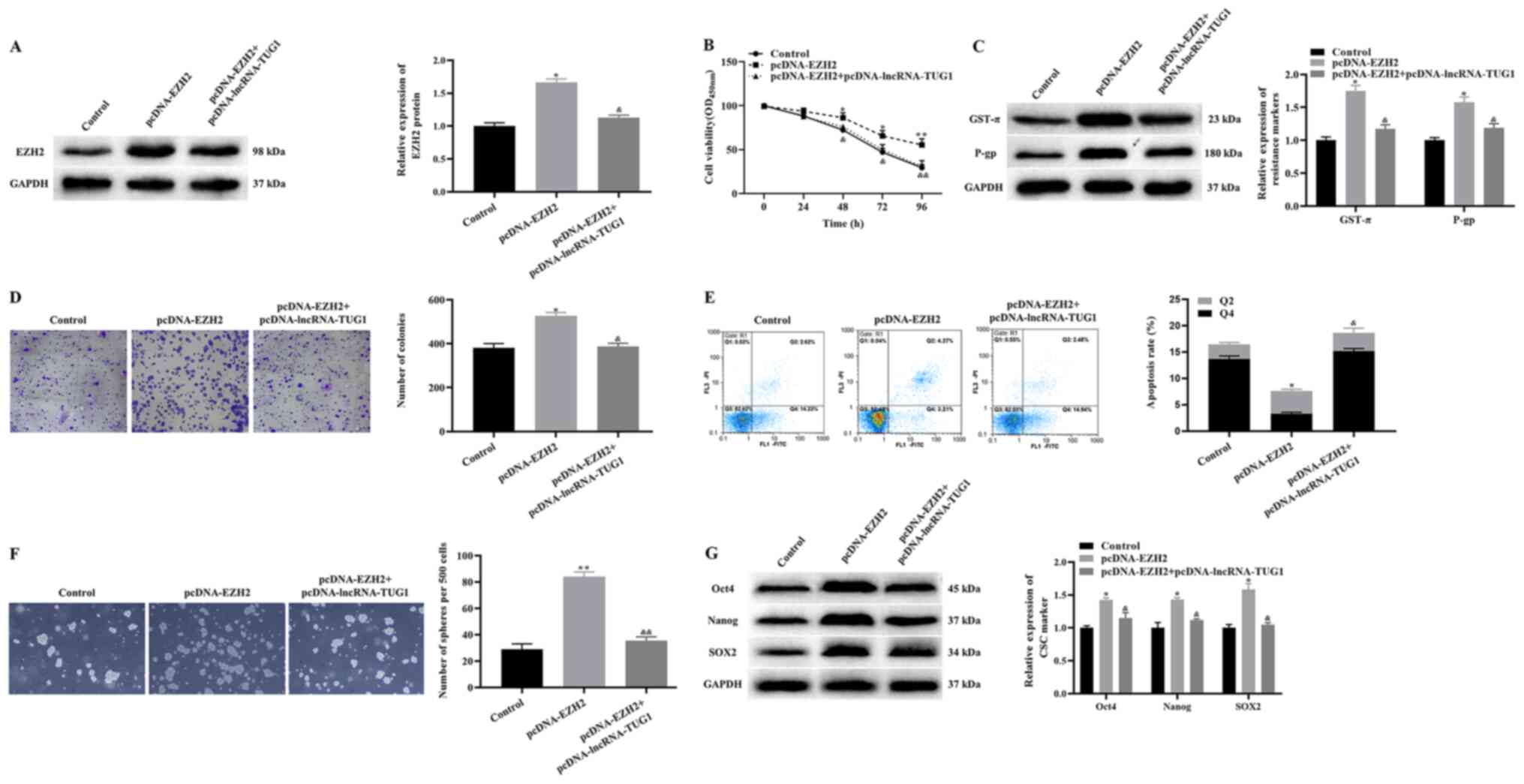
Effects of TUG1 on the proliferation,
apoptosis and CSCs-like properties of A172/TMZ cells via EZH2
Next, it was examined whether TUG1 may regulate the
proliferation, apoptosis and CSC-like properties of A172/TMZ cells
via EZH2. pcDNA-EZH2 and pcDNA-EZH2 + pcDNA-lncRNA-TUG1 were
transfected into A172/TMZ cells. Then, A172/TMZ cells were treated
with 450 µM TMZ. The western blotting results revealed that
overexpression of EZH2 significantly upregulated the expression
level of EZH2, while overexpression of EZH2 and TUG1 at the same
time had no significant effect on the expression level of EZH2
compared with the control group (Fig.
4A). The results of CCK-8 and clone formation experiments
indicated that, compared with the control group, the proliferation
and colony number of A172/TMZ cells was significantly increased in
the pcDNA-EZH2 group, while that in the pcDNA-EZH2 +
pcDNA-lncRNA-TUG1 group showed no significant change (Fig. 4B and D).
Subsequently, the current study determined the
expression levels of drug resistance markers. The results
demonstrated that overexpression of EZH2 significantly upregulated
the expression levels of GST-π and P-gp, while there was no
significant difference in these proteins between the control and
pcDNA-EZH2 + pcDNA-lncRNA-TUG1 groups (Fig. 4C). The annexin V FITC/PI apoptosis
experiment results demonstrated that, compared with the control
group, the apoptotic rate of A172/TMZ cells was decreased in the
pcDNA-EZH2 group, while that in the pcDNA-EZH2 + pcDNA-lncRNA-TUG1
group showed no significant change (Fig. 4E). In addition, the results of
western blotting and stem cell spheroidization experiments
identified that overexpression of EZH2 significantly increased the
expression levels of Oct4, Nanog and SOX2, as well as the
spheroidizing ability of A172/TMZ cells, while there was no
significant difference between the control and pcDNA-EZH2 +
pcDNA-lncRNA-TUG1 groups (Fig. 4F and
G). Notably, compared with the pcDNA-EZH2 group, the
expressions of EZH2, GST-π, P-gp, Oct4, Nanog and SOX2 were
significantly downregulated (Fig. 4A, C
and G), the proliferation, and number of colonies and spheres
were significantly reduced (Fig. 4B, D
and F) and the apoptosis rate was significantly increased
(Fig. 4E) in the pcDNA-EZH2 +
pcDNA-lncRNA-TUG1 group. Taken together, it was concluded that TUG1
inhibited the proliferation and CSC-like properties of A172/TMZ
cells and induced apoptosis by downregulating the expression level
of EZH2, thereby enhancing the TMZ sensitivity of A172/TMZ
cells.
Effect of the overexpression of TUG1
on the tumorigenicity of A172/TMZ cells
To evaluate the effect of the overexpression of TUG1
on the tumorigenicity of A172/TMZ cells in vivo, nude mouse
xenografts of A172/TMZ cells were constructed. The experimental
mice were divided into two groups, where one group was injected
with A172/TMZ cells transfected with pcDNA control, while the other
group was injected with A172/TMZ cells transfected with pcDNA-TUG1.
The RT-qPCR results demonstrated that the overexpression of TUG1
significantly increased the expression level of TUG1 of A172/TMZ
cells (Fig. 5A), and the same
results was observed in mice tumor tissue (Fig. 5B). The two nude mouse groups and
tumor bodies are presented in Fig.
5C. We also recorded the volume and weight of the tumor. The
data indicated that overexpression of TUG1 significantly reduced
the volume and weight of the tumor, while exhibiting no effect on
the body weight of the nude mice (Fig.
5D-F).
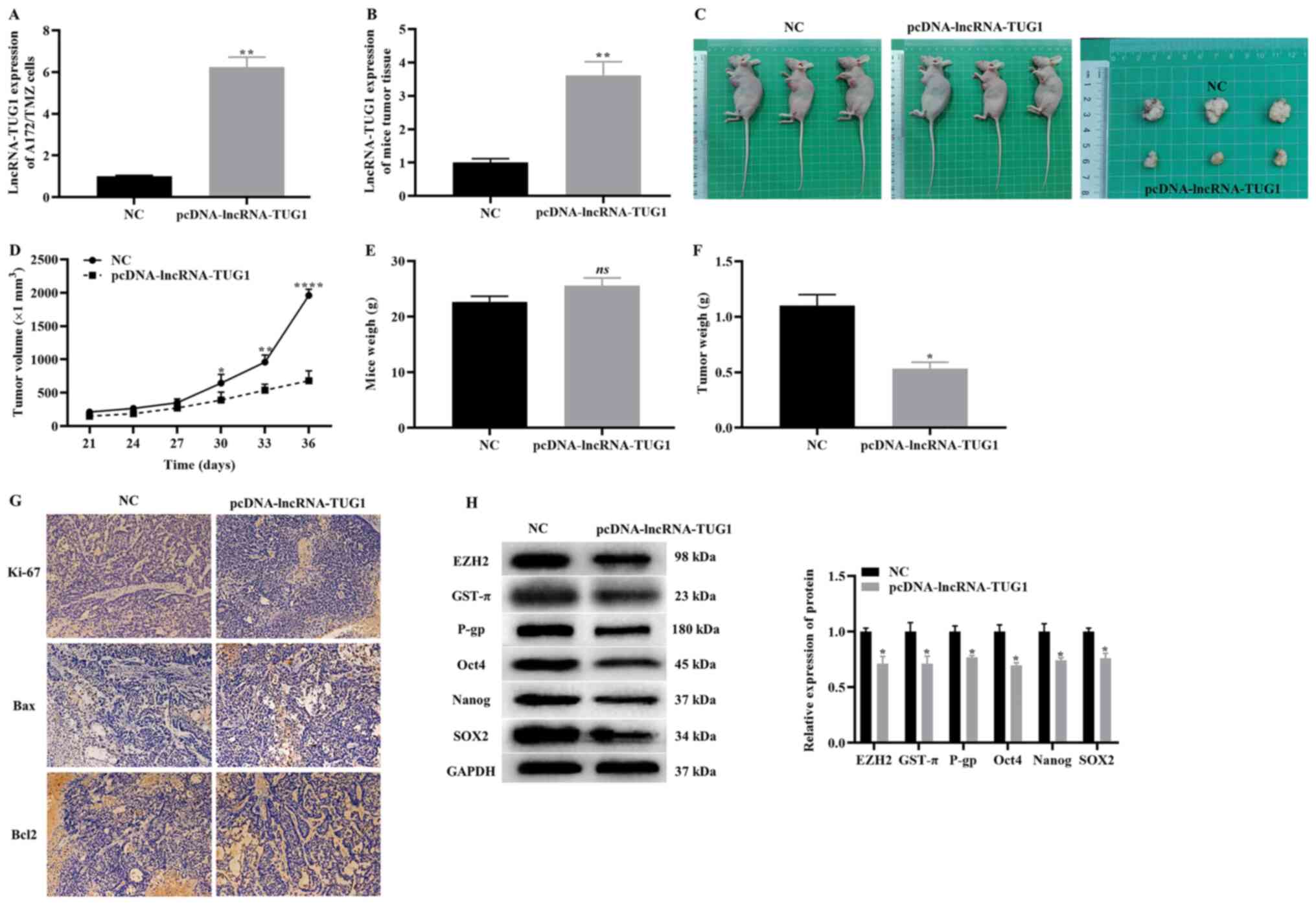 | Figure 5.Effect of overexpression TUG1 on the
tumorigenicity of A172/TMZ cells. Reverse
transcription-quantitative PCR indicating (A) the transfection
efficiency of pcDNA-TUG1 in A172/TMZ cells and (B) in mice tumor
tissue. (C) Images of two groups nude mice and tumors. Changes in
(D) tumor volume, (E) tumor mass and (F) body weight in the two
groups of nude mice. (G) Immunohistochemistry detected the
expression levels of cell proliferation and apoptosis markers.
Original magnification, ×200. (H) Western blotting results
displaying the expression levels of EZH2, markers of drug
resistance and CSCs. The error bars represent the mean ± SD of at
least triplicate experiments. *P<0.05, **P<0.01 and
****P<0.0001 vs. NC group. NC, negative control group; lncRNA,
long non-coding RNA; TMZ, temozolomide; TUG1, taurine upregulated
gene 1; GST-π, glutathione S-transferase-π; P-gp, P-glycoprotein 1;
EZH2, enhancer of Zeste homolog 2 polycomb repressive complex
subunit 2. |
Next, the markers of proliferation and apoptosis
were detected in tumors using immunohistochemistry. It was
identified that the overexpression of TUG1 notably downregulated
the expression levels of Ki67 and Bcl2, as well as upregulated the
expression levels of Bax (Fig. 5G).
Similarly, the western blotting results demonstrated that
overexpression of TUG1 significantly downregulated the expression
levels of EZH2, resistance markers (GST-π and P-gp) and CSCs
markers (Oct4, Nanog and SOX2) (Fig.
5H). Based on the aforementioned findings, it was suggested
that overexpression of TUG1 can inhibit the tumorigenicity of
A172/TMZ cells.
Discussion
At present, TMZ is the first-choice drug for the
treatment of glioma. However, patients with gliomas show poor
prognosis due to the TMZ resistance of gliomas, and thus require
aggressive treatment (23).
Previous studies have reported that only <10% of patients
survive for >5 years after treatment (24–26).
The mechanism of glioma resistance has been attributed to the fact
that chemical drugs targeting glioma cells exhibit a poor effect on
CSCs, which results in chemotherapy-resistant CSCs driving more
aggressive recurrent tumors (27).
Another plausible explanation is that glioma cells can transform
from non-CSCs to CSCs after chemotherapy, and subsequently, the
phenotypic plasticity of this tumor cell population can lead to
tumor regrowth (7,22). To the best of our knowledge, the
present study demonstrated for the first time that TUG1 inhibited
the CSCs-like properties of TMZ-resistant glioma cells via EZH2,
thereby alleviating TMZ resistance of gliomas.
TUG1 is a lncRNA located on chromosome 22q12, which
was initially found to be involved the formation of photoreceptors
by serving a key role in retinal development (28). The essential role of TUG1 in the
occurrence and development of various malignant tumors, including
glioma, has recently been confirmed. For example, Long et al
(29) discovered that TUG1 was
highly expressed in melanoma, where it upregulated the expression
level of astrocyte elevated gene-1 protein by competitively binding
miR-129-5p, which promoted the proliferation and invasion of
melanoma cells, as well as inhibiting their apoptosis. Moreover, a
study by Liao et al (30)
reported that TUG1 was significantly downregulated in gliomas, and
its overexpression could inhibit the proliferation of glioma cells,
along with their apoptosis. Interestingly, several studies have
highlighted that TUG1 can be utilized as a new therapeutic target
for glioma, whereas its downregulation may inhibit cell
proliferation and invasion, and promote the apoptosis of glioma
U251 cells (15,31,32).
However, the findings of Liao are inconsistent with the current
results.
The present study identified that TUG1 had a tumor
suppressor effect in gliomas, which was achieved by inhibiting
CSCs-like properties. Currently, the associated mechanism of TUG1
with CSCs-like properties has been rarely studied. Nevertheless,
there are recent multiple reports that have emphasized that TUG1
was involved in the regulation of drug resistance of a variety of
malignant tumors. For example, TUG1 has been noted to be markedly
downregulated in triple-negative breast cancer (TNBC), and at the
same time it inhibits Wnt signaling pathway activation by
regulating the miR-197/nemo like kinase molecular axis, thereby
increasing the chemosensitivity of TNBC cells (33). Moreover, Xie et al (34) have shown that TUG1 inhibited the
resistance of bladder urothelial carcinoma to adriamycin via the
Wnt/β-catenin pathway. Some studies have also concluded that the
Wnt signaling transcription factors, T-cell factor 1 and lymphoid
enhancer binding factor 1, are upregulated in malignant astrocytic
tumors (35). These reports suggest
that TUG1 may also regulate the drug resistance of gliomas via the
Wnt pathway. This may be the direction of future research.
Accumulating evidence has revealed that EZH2 is
involved in the abnormal transcriptome of cancer cells (36). In addition, it has been established
that EZH2, together with its enzymatic production of H3K27, are
closely associated with poor prognosis in a variety of malignant
tumors, such as prostate and breast cancer (37,38).
Likewise, our previous studies reported that EZH2 promoted the
malignant biological behavior of glioma cells. Moreover, EZH2 is
involved in regulating the chemical resistance of various malignant
tumors and the stemness of CSCs. It has been shown that EZH2 is
upregulated in glioma and glioma CSCs (39–41).
Furthermore, inhibiting both BMI1 and EZH2 enhances the
chemotherapy sensitivity of glioma stem cells (42). It has also been shown that
phosphorylation of EZH2 activates STAT3 signaling by methylating
STAT3, and promotes the tumorigenicity of glioblastoma CSC-like
cells (43). Moreover, the direct
transcriptional regulation of c-myc by EZH2 may constitute a novel
mechanism underlying glioma CSC maintenance (44), which suggests that EZH2 may be a
valuable new therapeutic target for glioma management. The present
study demonstrated that EZH2 could promote CSCs-like properties to
enhance the TMZ resistance of A172/TMZ cells.
Epigenetic disorders may induce tumorigenesis and
influence targeted therapy (45)
Carcinogenic pathways converge to the epigenome to maintain
tumorigenesis, and so, epigenetic regulators may be particularly
effective targets for cancer treatment (46). As a chromatin-modifying enzyme, EZH2
serves a vital role in the epigenetic silencing of genes via a
complex mechanism (47). Most
studies have demonstrated that 40% of gliomas harbor mutations in
epigenetic regulatory factors, such as EZH2 (48–50).
DNA methylation is a chemical modification of DNA, catalyzed by the
DNA methyltransferase (DNMT) enzyme, which serves a valuable role
in epigenetic modifications. In particular, the polycomb group
protein EZH2 directly controls DNA methylation (51), as EZH2 interacts with DNMTs to
methylate the EZH2-binding promoter (50,52,53),
which indicates that EZH2 is closely associated with the regulation
of gene expression via epigenetic transcription. In the current
study, EZH2 is likely to be involved in epigenetic regulation, but
the specific mechanism requires further investigation.
In conclusion, the present study demonstrated that
TUG1 inhibited CSCs-like properties and tumorigenicity, as well as
induced the apoptosis of A172/TMZ cells by downregulating the
expression level of EZH2, thereby alleviating the TMZ resistance of
A172/TMZ cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and YC were responsible for the conceptualization
and methodology; WC and YW performed the cell experiments, data
analysis and prepared the figure; HS, DT and DS were responsible
for animal experiments, data acquisition and data validation; JL
and YC prepared the original draft of the manuscript, and reviewed
and edited the manuscript. All authors confirm the authenticity of
all raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from The First
Affiliated Hospital of Kunming Medical University Ethics Committee
(approval no. kmu-eac-2018056; date, 2018-05-01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Modrek AS, Bayin NS and Placantonakis DG:
Brain stem cells as the cell of origin in glioma. World J Stem
Cells. 6:43–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lapointe S, Perry A and Butowski NA:
Primary brain tumours in adults. Lancet. 392:432–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wesseling P and Capper D: WHO 2016
Classification of gliomas. Neuropathol Appl Neurobiol. 44:139–150.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alifieris C and Trafalis DT: Glioblastoma
multiforme: Pathogenesis and treatment. Pharmacol Ther. 152:63–82.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun Q, Xu R, Xu H, Wang G, Shen X and
Jiang H: Extracranial metastases of high-grade glioma: The clinical
characteristics and mechanism. World J Surg Oncol. 15:1812017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chang JC: Cancer stem cells: Role in tumor
growth, recurrence, metastasis, and treatment resistance. Medicine
(Baltimore). 95 (Suppl 1):S20–S25. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Auffinger B, Tobias AL, Han Y, Lee G, Guo
D, Dey M, Lesniak MS and Ahmed AU: Conversion of differentiated
cancer cells into cancer stem-like cells in a glioblastoma model
after primary chemotherapy. Cell Death Differ. 21:1119–1131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ignarski M, Islam R and Müller RU: Long
non-coding RNAs in kidney disease. Int J Mol Sci. 20:32762019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang Y: The novel regulatory role of
lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med.
22:5768–5775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Buccarelli M, Lulli V, Giuliani A, Signore
M, Martini M, D'Alessandris QG, Giannetti S, Novelli A, Ilari R,
Giurato G, et al: Deregulated expression of the imprinted DLK1-DIO3
region in glioblastoma stemlike cells: Tumor suppressor role of
lncRNA MEG3. Neuro Oncol. 22:1771–1784. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu C, Wei Y, Wang X, Zhang Z, Yin J, Li W,
Chen L, Lyu X, Shi Z, Yan W and You Y: DNA-methylation-mediated
activating of lncRNA SNHG12 promotes temozolomide resistance in
glioblastoma. Mol Cancer. 19:282020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Voce DJ, Bernal GM, Wu L, Crawley CD,
Zhang W, Mansour NM, Cahill KE, Szymura SJ, Uppal A, Raleigh DR, et
al: Temozolomide treatment induces lncRNA MALAT1 in an NF-κB and
p53 codependent manner in glioblastoma. Cancer Res. 79:2536–2548.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Zhang M, An G and Ma Q: LncRNA TUG1
acts as a tumor suppressor in human glioma by promoting cell
apoptosis. Exp Biol Med (Maywood). 241:644–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao Z, Wang B, Hao J, Man W, Chang Y, Ma
S, Hu Y, Liu F and Yang J: Downregulation of the long non-coding
RNA taurine-upregulated gene 1 inhibits glioma cell proliferation
and invasion and promotes apoptosis. Oncol Lett. 15:4026–4032.
2018.PubMed/NCBI
|
|
16
|
Margueron R and Reinberg D: The Polycomb
complex PRC2 and its mark in life. Nature. 469:343–349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang XP, Shan C, Deng XL, Li LY and Ma W:
Long non-coding RNA PAR5 inhibits the proliferation and progression
of glioma through interaction with EZH2. Oncol Rep. 38:3177–3186.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krell A, Wolter M, Stojcheva N, Hertler C,
Liesenberg F, Zapatka M, Weller M, Malzkorn B and Reifenberger G:
MiR-16-5p is frequently down-regulated in astrocytic gliomas and
modulates glioma cell proliferation, apoptosis and response to
cytotoxic therapy. Neuropathol Appl Neurobiol. 45:441–458.
2019.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
National Research C: Guide for the Care
and Use of Laboratory Animals. 8th edition. The National Academies
Press; Washington, DC: 2011
|
|
21
|
Chumakova A and Lathia JD: Outlining
involvement of stem cell program in regulation of O6-methylguanine
DNA methyltransferase and development of temozolomide resistance in
glioblastoma: An Editorial Highlight for ‘Transcriptional control
of O6 -methylguanine DNA methyltransferase expression
and temozolomide resistance in glioblastoma’ on page 780. J
Neurochem. 144:688–690. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Safa AR, Saadatzadeh MR, Cohen-Gadol AA,
Pollok KE and Bijangi-Vishehsaraei K: Glioblastoma stem cells
(GSCs) epigenetic plasticity and interconversion between
differentiated non-GSCs and GSCs. Genes Dis. 2:152–163. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee SY: Temozolomide resistance in
glioblastoma multiforme. Genes Dis. 3:198–210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thakkar JP, Dolecek TA, Horbinski C,
Ostrom QT, Lightner DD, Barnholtz-Sloan JS and Villano JL:
Epidemiologic and molecular prognostic review of glioblastoma.
Cancer Epidemiol Biomarkers Prev. 23:1985–1996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ostrom QT, Gittleman H, Xu J, Kromer C,
Wolinsky Y, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical
report: Primary brain and other central nervous system tumors
diagnosed in the United States in 2009–2013. Neuro Oncol. 18 (Suppl
5):v1–v75. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee G, Auffinger B, Guo D, Hasan T,
Deheeger M, Tobias AL, Kim JY, Atashi F, Zhang L, Lesniak MS, et
al: Dedifferentiation of glioma cells to glioma stem-like cells by
therapeutic Stress-induced HIF signaling in the recurrent GBM
model. Mol Cancer Ther. 15:3064–3076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Long J, Menggen Q, Wuren Q, Shi Q and Pi
X: Long noncoding RNA taurine-upregulated gene1 (TUG1) promotes
tumor growth and metastasis through
TUG1/Mir-129-5p/astrocyte-elevated gene-1 (AEG-1) axis in malignant
melanoma. Med Sci Monit. 24:1547–1559. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao Y, Zhang B, Zhang T, Zhang Y and Wang
F: LncRNA GATA6-AS promotes cancer cell proliferation and inhibits
apoptosis in glioma by downregulating lncRNA TUG1. Cancer Biother
Radiopharm. 34:660–665. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Katsushima K, Natsume A, Ohka F, Shinjo K,
Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, et
al: Targeting the Notch-regulated non-coding RNA TUG1 for glioma
treatment. Nat Commun. 7:136162016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z,
Xi Z, Li Z, Bao M and Liu Y: Long non-coding RNA taurine
upregulated 1 enhances tumor-induced angiogenesis through
inhibiting microRNA-299 in human glioblastoma. Oncogene.
36:318–331. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang T, Cheng Y, She Q, Jiang Y, Chen Y,
Yang W and Li Y: Long non-coding RNA TUG1 sponges miR-197 to
enhance cisplatin sensitivity in triple negative breast cancer.
Biomed Pharmacother. 107:338–346. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xie D, Zhang H, Hu X and Shang C:
Knockdown of long non-coding RNA Taurine Up-Regulated 1 inhibited
doxorubicin resistance of bladder urothelial carcinoma via
Wnt/β-catenin pathway. Oncotarget. 8:88689–88696. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pećina-Šlaus N, Kafka A, Tomas D, Marković
L, Okštajner PK, Sukser V and Krušlin B: Wnt signaling
transcription factors TCF-1 and LEF-1 are upregulated in malignant
astrocytic brain tumors. Histol Histopathol. 29:1557–1564.
2014.
|
|
36
|
Kim KH and Roberts CW: Targeting EZH2 in
cancer. Nat Med. 22:128–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McCabe MT and Creasy CL: EZH2 as a
potential target in cancer therapy. Epigenomics. 6:341–351. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chase A and Cross NC: Aberrations of EZH2
in cancer. Clin Cancer Res. 17:2613–2618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Orzan F, Pellegatta S, Poliani PL, Pisati
F, Caldera V, Menghi F, Kapetis D, Marras C, Schiffer D and
Finocchiaro G: Enhancer of Zeste 2 (EZH2) is up-regulated in
malignant gliomas and in glioma stem-like cells. Neuropathol Appl
Neurobiol. 37:381–394. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu H, Zhao G, Zhang Y, Jiang H, Wang W,
Zhao D, Hong J, Yu H and Qi L: Mesenchymal stem cell-derived
exosomal microRNA-133b suppresses glioma progression via
Wnt/β-catenin signaling pathway by targeting EZH2. Stem Cell Res
Ther. 10:3812019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han B, Meng X, Wu P, Li Z, Li S, Zhang Y,
Zha C, Ye Q, Jiang C, Cai J and Jiang T: ATRX/EZH2 complex
epigenetically regulates FADD/PARP1 axis, contributing to TMZ
resistance in glioma. Theranostics. 10:3351–3365. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin X, Kim LJY, Wu Q, Wallace LC, Prager
BC, Sanvoranart T, Gimple RC, Wang X, Mack SC, Miller TE, et al:
Targeting glioma stem cells through combined BMI1 and EZH2
inhibition. Nat Med. 23:1352–1361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim E, Kim M, Woo DH, Shin Y, Shin J,
Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al: Phosphorylation of
EZH2 activates STAT3 signaling via STAT3 methylation and promotes
tumorigenicity of glioblastoma stem-like cells. Cancer Cell.
23:839–852. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Suvà ML, Riggi N, Janiszewska M,
Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino
D, Cironi L, et al: EZH2 is essential for glioblastoma cancer stem
cell maintenance. Cancer Res. 69:9211–9218. 2009. View Article : Google Scholar
|
|
45
|
Khani P, Nasri F, Khani Chamani F, Saeidi
F, Sadri Nahand J, Tabibkhooei A and Mirzaei H: Genetic and
epigenetic contribution to astrocytic gliomas pathogenesis. J
Neurochem. 148:188–203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mazor T, Pankov A, Johnson BE, Hong C,
Hamilton EG, Bell RJA, Smirnov IV, Reis GF, Phillips JJ, Barnes MJ,
et al: DNA methylation and somatic mutations converge on the cell
cycle and define similar evolutionary histories in brain tumors.
Cancer Cell. 28:307–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Eich ML, Athar M, Ferguson JE III and
Varambally S: EZH2-targeted therapies in cancer: Hype or a reality.
Cancer Res. 80:5449–5458. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Brennan CW, Verhaak RG, McKenna A, Campos
B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ,
Berman SH, et al: The somatic genomic landscape of glioblastoma.
Cell. 155:462–477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bian EB, Li J, Xie YS, Zong G, Li J and
Zhao B: LncRNAs: New players in gliomas, with special emphasis on
the interaction of lncRNAs With EZH2. J Cell Physiol. 230:496–503.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bian EB, Li J, He XJ, Zong G, Jiang T, Li
J and Zhao B: Epigenetic modification in gliomas: Role of the
histone methyltransferase EZH2. Expert Opin Ther Targets.
18:1197–1206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Viré E, Brenner C, Deplus R, Blanchon L,
Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden
JM, et al: The polycomb group protein EZH2 directly controls DNA
methylation. Nature. 439:871–874. 2006. View Article : Google Scholar
|
|
52
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang R and Liu X: Epigenetic regulation of
prostate cancer. Genes Dis. 7:606–613. 2019. View Article : Google Scholar : PubMed/NCBI
|