Introduction
Colorectal cancer (CRC) is one of the commonest
types of cancer and a leading cause of cancer mortality worldwide
(1). It is well established that
the pathogenesis of CRC involves genetic and epigenetic changes and
an imbalance in oncogenes and tumor suppressor genes (2). With the improvement of early screening
strategies and CRC treatment methods, such as surgery,
chemotherapy, or pre-operative neoadjuvant chemoradiotherapy plus
surgery, the survival rate of patients with CRC has improved.
However, the prognosis for advanced CRC remains poor.
Chemoresistance is the main reason for treatment failure and
disease progression (3). The
survival of patients is significantly reduced because of consequent
tumor recurrence and distant metastasis (4). Therefore, further identification of
key molecules, which not only regulate the development and
progression of CRC but may also be used as early detection
biomarkers or novel therapeutic targets, would be of great benefit
to patients with CRC.
MicroRNAs (miRNAs/miRs) belong to a group of
non-coding single-stranded small RNA molecules that contain ~22
nucleotides and are highly conserved (5). miRNAs perform modulatory functions in
a series of biological processes (6,7).
Increasing evidence indicates that miRNA may be promising
biomarkers for disease diagnosis and prognosis (8,9) and
dysregulation of miRNA may contribute to the development and
progression of cancer (10–12). As miRNAs exist stably in the blood
and blood samples are easy to obtain, the detection of circulating
miRNAs in human bodily fluids such as serum has become more broadly
studied in research (13). As serum
miRNAs expression profiles in tumor patients differ significantly
from that of healthy individuals (14,15),
serum miRNAs may be considered promising biomarkers for tumor
detection (16,17).
As a tumor suppressor gene, miR-451 inhibits the
survival, proliferation and invasion of cancer cells and serves a
significant role in tumor development (18–20).
Furthermore, studies focused on multiple solid tumor types have
shown that miR-451 may be a meaningful biomarker for cancer
diagnosis, treatment and drug resistance (21–23).
Several studies have focused on the decreased expression of
miR-451, which is closely associated with CRC progression (24,25).
However, the underlying molecular mechanisms remain
understudied.
The present study was designed to detect the
expression of miR-451 in patients with CRC and CRC cells and to
determine the role of miR-451 in CRC cells. The targets of miR-451
that induce the related signaling pathway changes were explored.
The findings demonstrated that miR-451 is essential to blocking
tumor growth via targeting SAMD4B.
Materials and methods
Cell culture and human blood serum
specimens
CRC cells (HT29, SW480 and HCT116) and normal
colorectal mucosal cell lines (FHC and HCoEpiC) were purchased from
the American Type Culture Collection in March 2018 and grown in
DMEM media (Sigma-Aldrich; Merck KGaA) with 10% fetal bovine serum
(HyClone; Cytiva). The HT-29 cell line was authenticated by STR
profiling. Cells were cultured in a humidified incubator at 37°C
with 5% CO2. Oxaliplatin was purchased from Jiangsu
Hengrui Pharmaceutical Co., Ltd.; 5-Fluorouracil (5-FU) was
purchased from Shanxi Pude Pharmaceutical Co., Ltd.
Serum was collected and pre-processed in the
Departments of Gastrointestinal Surgery and Oncology of Jiangjin
Central Hospital of Chongqing prior to surgical treatment. The
process of extracting serum was performed as described by Allen
et al (14). A total of 50
diagnostic patient specimens were used in this study (Table I). Serum from 50 healthy subjects
served as the control group (Table
II). The Ethics Committee of the Jiangjin Central Hospital of
Chongqing approved the research protocol (approval no. 20190611-28)
and informed written consent was received from each patient.
 | Table I.Clinical features of patients
enrolled in the present study. |
Table I.
Clinical features of patients
enrolled in the present study.
| Characteristic | n= |
|---|
| Age |
|
|
≥60 | 33 |
|
<60 | 17 |
| Sex |
|
|
Male | 31 |
|
Female | 19 |
| Pathology |
|
|
High-moderately differentiated
adenocarcinoma | 28 |
| Poorly
differentiated adenocarcinoma | 22 |
| Stage |
|
|
I–II | 15 |
|
III–IV | 35 |
| Type of
therapy |
|
|
Adjuvant chemotherapy | 39 |
|
Neoadjuvant chemotherapy | 11 |
 | Table II.Clinical features of patients and
controls enrolled in the present study. |
Table II.
Clinical features of patients and
controls enrolled in the present study.
| Characteristic | CRC patients
(n=50) | Control (n=50) |
χ2/t | P-value |
|---|
| Age (mean ±
standard deviation) | 62.0±10.4 | 60.8±14.2 | 0.457 | 0.648 |
| Sex |
|
| 0.167 | 0.683 |
|
Male | 31 | 29 |
|
|
|
Female | 19 | 21 |
|
|
Transfection experiments
For cell function research, negative control (NC),
synthetic miR-451 mimics and an miR-451 inhibitor were purchased
from Guangzhou RiboBio Co., Ltd. The SAMD4B overexpression plasmid
and the scramble plasmid were purchased from Shanghai GeneChem Co.,
Ltd. The process of cell transfection was performed as described by
Fan and Zhao (26). Transfection
was performed using Lipofectamine® 3000 Reagent (Thermo
Fisher Scientific, Inc.) in accordance with the manufacturer's
protocols. In brief, HCT 116 cells were cultured in 6-well plates
at 37°C until they reached 60–70% confluence. Then ~4 µg
transfectant [negative control (NC), synthetic miR-451 mimics,
miR-451 inhibitor, SAMD4B overexpression plasmid, or scramble
plasmid] were added. Cells were collected for subsequent
experiments after 24 h co-culture at 37°C. The oligonucleotide
sequences were as follows: hsa-miR-451 mimics,
5′-AAACCGUUACCAUUACUGAGUU-3′; mimics NC, 5′-TTCTCCGAACGTGTCACGT-3′;
miR-451 inhibitor, 5′-AACUCAGUAAUGGUAACGGUUU-3′; and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′ (all Guangzhou RiboBio Co., Ltd.). The
plasmid vector was pcDNA 3.1, and the over expression sequences of
SAMD 4B was 5′-ACTGGAGGACCGCAACGCAC-3′ (Shanghai GeneChem Co.,
Ltd.).
In vivo experiments
A total of six female athymic (nu/nu) mice per group
were purchased from the Laboratory Animal Center of Chongqing
Medical University [certificate. SCXK(YU)2018-0003]. A total of 24
mice (age, 6 weeks) were maintained in polycarbonate cages
(temperature, 21–23°C; humidity, 40–60%; 5 animals per cage) on a
12-h light/dark cycle. All of the animal procedures were approved
by the Ethics Committee of the Chongqing Medical University
(approval no. 2019–256). Mice were utilized to generate a xenograft
tumor model and each group included six mice. They were injected
subcutaneously with 1×106 HCT116 cells that were stably
transfected with i) negative control (NC), ii) lentiviral vectors
with miR-451, iii) lentiviral vectors with SAMD4B, or iv)
lentiviral vectors with miR-451 plus SAMD4B. The lentiviral vector
system included the pCDH-CMV-EF1-copGFP-T2A-SAMD4B-puro and the
pcDNA3.1 plasmids, which were purchased from Shanghai GeneChem Co.,
Ltd. The process of transfection was performed as described by Zhu
et al (27). In brief, HCT
116 cells were cultured in 6-well plates to 60–70% confluence. A
total of 100 µl opti-MEM (Gibco; Thermo Fisher Scientific, Inc.)
was used to dissolve 2.5 µg plasmid; the same volume was also added
to 5 µl Lipofectamine 3000 (Thermo Fisher Scientific, Inc.), then
incubated at room temperature for 5 min. Lastly, the two solutions
were mixed and incubated at room temperature for 15 min, then added
to the cells. After 72 h transfection at 37°C, neomycin (450 µg/ml)
was used for selection. Following transfection, 1×106
HCT116 cells in 100 µl serum-free medium were injected
subcutaneously into each mouse (right back). The subcutaneous tumor
size of each mouse was measured weekly with an electronic caliper.
Mice were sacrificed when the tumor size reached 1,500
mm3, the largest diameter of a tumor was 19 mm and the
mice were sacrificed using cervical dislocation and the xenograft
tumor tissues were excised and weighed to determine the tumor
weights. Tumor volume was measured as following: 1/2× a (the
largest diameter) × b2 (the perpendicular diameter).
Cell viability analysis
Cells were transfected with the indicated plasmid or
drugs (Oxaliplatin was purchased from Jiangsu Hengrui
Pharmaceutical Co., Ltd., and 5-FU). Oxaliplatin was used at a
concentration gradient of 2, 4, 8, 12, 24 and 48 µM. 5-FU was used
at a concentration gradient of 1, 2, 4, 8, 16 and 32 µg/ml at 8,
24, 48 and 72 h after miR-451mimics and inhibitors transfection and
evaluation of cell proliferation via cell viability was determined
by a Cell Counting Kit-8 (CCK-8; MedChemExpress). The cell
viability was measured using optical density (OD) at 450 nm.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Blood samples were collected in BD Vacutainer Serum
Separation Tubes, incubated for 1 h at room temperature and
centrifuged at 1,300 × g for 10 min at 4°C. The serum supernatant
was transferred to new tubes, centrifuged at 12,000 × g for 10 min
at 4°C to remove any residual cells and debris and stored at −80°C.
The serum was collected from fasting patients and controls and, for
patients, the serum was collected before the first cycle of
chemotherapy. RNA was extracted from cells or human blood serum
specimens using TRIzol® reagent (Thermo Fisher
Scientific, Inc.) and RNA concentration was measured on a NanoDrop
spectrophotometer (Thermo Fisher Scientific, Inc.). RT-qPCR was
conducted according to the manufacturer's protocol. miR-451, U6,
SAMD4B and GAPDH expression were measured by RT-qPCR using the
primer set from Guangzhou RiboBio Co., Ltd. The primer sequences
for these genes are given in Table
III. miRNA was reverse transcribed using the polyA tailing
method (All-in-One™ miRNA RT-qPCR detection kit), while mRNA was
reverse transcribed using the All-in-One™ First-Strand cDNA
Synthesis kit (both from GeneCopoeia, Inc.) and then the DNAse
digestion on RNA extracts. The following temperature protocol was
used: Incubation at 37°C for 60 min and 72°C for 5 min; the samples
were stored at 4°C. The following thermocycling conditions were
used: Initial denaturation at 95°C for 10 min, followed by 40
cycles at 95°C for 10 sec, 58°C for 20 sec and 72°C for 15 sec. The
relative expression levels of miR-451 and SAMD4B were normalized to
RNU6 and GAPDH, respectively. The 2−ΔΔCq method
(28) was used to analyze the
relative fold changes.
 | Table III.The primer and sequence information
used for reverse transcription-quantitative PCR. |
Table III.
The primer and sequence information
used for reverse transcription-quantitative PCR.
| Gene | Sequence |
|---|
| SAMD4B | F:
5′-CTCACTGGCGGACTGCAAT-3′ |
|
| R:
5′-GTAGCCTCATGTACTCCGACTT-3′ |
| GAPDH | F:
5′-GTCGATGGCTAGTCGTAGCATCGAT-3′ |
|
| R:
5′-TGCTAGCTGGCATGCCCGATCGATC-3′ |
| miR-451 | F:
5′-TCCGATTGAGTCATTACCAT-3′ |
|
| R:
5′-GTGCAGGGTCCGAGGT-3′ |
| U6 | F:
5′-TAAGATCGTGAAGCGTTC-3′ |
|
| R:
5′-GTGCAGGGTCCGAGGT-3′ |
| miR-451 mimics | F:
5′-AAACCGUUACCAUUACUGAGUU-3′ |
|
| R:
3′-CUCAGUAAUGGUAACGGUUUUU-5′ |
The Cancer Genome Atlas (TCGA)
database
Using data from TCGA the top 100 patients with high
expression and the bottom 100 patients with low expression were
selected for analysis.
Apoptosis analysis
HCT116 cells were seeded in 6-well cell culture
plates after 6 h of transfection. After 24 h, the cells were
harvested and then stained with Annexin V and PE according to the
manufacturer's protocol (Biotium, Inc.). The cells were then
analyzed using flow cytometry. Briefly, HCT-116 cells were seeded
in 6-well plates at a density of 5×105 cells/ml. After
12 h incubation at 37°C, the HCT-116 cells in the experimental
group were treated with miR-451 mimics, miR-451 inhibitor or
miR-451 mimics + SAMD4B, and the cells were collected following 24
h incubation at 37°C. A total of 5 µl Annexin V-FITC and PE were
added. Samples were left to stand in the dark at room temperature
for 15 min and then analyzed for apoptosis by flow cytometry. The
experiments were repeated three times. The total percentage of
apoptotic cells is presented as the sum of the apoptotic cell
populations in the early and late stages. CytoFLEX flow cytometer
was used for detection and CytExpert 2.4 software (both Beckman
Coulter, Inc.) was used for data analysis.
Transwell assays
Cells were stained with a 0.1% crystal violet
solution. The assay uses 8.0 µm Transwell inserts (Corning, Inc.).
The Transwell assay protocol was performed as described by Zhu
et al (29). In brief, cells
were digested and collected after 24 h transfection, then
resuspended in DMEM (Sigma-Aldrich; Merck KGaA) containing 1% FBS.
The cell density was adjusted density to 1×105/ml; then
200 µl cell suspension were added to the upper chamber, and 600 µl
10% FBS was added to the lower chamber before incubation at 37°C.
After 24 h, cells on the upper layer of the Transwell cell filter
membrane were removed using absorbent cotton; the filter membrane
was fixed in 4% paraformaldehyde for 30 min at room temperature and
stained with 1% crystal violet for 15 min at room temperature. The
slides were viewed and images captured under a light microscope
(Olympus Corporation) at ×100 magnification.
Luciferase reporter assay
A dual-luciferase assay system (Promega Corporation)
was applied to measure luciferase activity. The primer sequences
for amplifying the 3′-UTR of SAMD4B were: SAMD4B-wild-type (WT)
forward (F): 5′-CTCACTGGCGGACTGCAAT-3; reverse(R):
5′-GTAGCCTCATGTACTCCGACTT-3′. Mutation primers were designed based
on miR-451 and the SAMD4B gene 3 ′UTR sequence binding mutation
site. SAMD4B mutant (Mut) F: 5′-CTCTCCCTGCCACTGTCTTG-3′; R:
5′-ATTCTGCAAGGACAGGAGCC-3′. The 3′-UTR segments of SAMD4B that were
predicted to interact with miR-451 were amplified from human
genomic DNA via PCR and inserted into the HindIII and
SacI sites of the miR-451 expression reporter vector. The
dual luciferase assay (Promega Corporation) was performed according
to the manufacturer's protocol. The Luciferase reporter vector was
pGL3-Basic (Youbio) and Renilla luciferase was used for
normalization. Lipofectamine 3000 (Thermo Fisher Scientific, Inc.)
was used to transfect the plasmid and mimics. After 72 h
transfection, dual luciferase assay (Promega Corporation) was
performed according to the manufacturer's protocol.
Western blot analysis
Cells were lysed in ice-cold RIPA buffer (cat. no.
P0013B; Beyotime Institute of Biotechnology). The cells lysates
were centrifuged at 12,000 × g for 15 min at 4°C and the
supernatants were stored at −80°C. The protein concentration was
determined via BCA method. The protein extracts were fractionated
by electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide
gel, transferred onto PVDF membranes and blocked with 5% non-fat
milk powder for 2 h at room temperature. Then the membranes were
incubated overnight at 4°C with primary antibodies as follows:
Anti-SAMD4B (ProteinTech Group, Inc.; 1:800; cat. no. 17723-1-AP),
anti-Bax (ProteinTech Group, Inc.; 1:2,000; cat. no. 50599-2-Ig),
anti-Bcl2 (Abcam; 1:2,000; cat. no. ab182858) and anti-GAPDH
(ProteinTech Group, Inc.; 1:10,000; cat. no. 10494-1-AP). After
washing with 1X TBST (0.1% Tween) buffer four times, the blots were
incubated with horseradish peroxidase-conjugated Goat Anti-Rabbit
IgG(H+L; ProteinTech Group, Inc.; 1:2,000; cat. no. SA00001-2) at
4°C for 1 h. Exposure imaging was performed using the Bio-Rad
chemiluminescence imaging system (Bio-Rad Laboratories, Inc.). The
blot optical density was determined by Image Lab software 5.2.1
(Bio-Rad Laboratories, Inc.).
Immunohistochemistry and
Immunofluorescence Staining
The IHC assays were performed as described by
Yamadera et al (30). In
brief, the implanted xenograft tumor tissues were excised and fixed
with 4% paraformaldehyde for 24 h at room temperature, dehydrated
using a graded alcohol series of 60, 70, 80, 90, 95 and 100%
ethanol and finally embedded in paraffin. The 5-µm-thick paraffin
sections were dewaxed and dehydrated, incubated with sealed serum
and human rabbit ki-67 primary antibodies (1:400; cat. no. 9027;
CST), were added in a wet box at 4°C overnight. After washing with
PBS, sections were incubated with secondary antibodies (ProteinTech
Group, Inc.; 1:1,000; cat. no. SA00004-2) at room temperature for 1
h and positive expression was detected with a DAB kit (Beyotime
Institute of Biotechnology) at room temperature for 30 min. The
nuclei were stained with hematoxylin at room temperature for 8 min.
The slides were viewed and images captured under a light microscope
(Olympus Corporation) at 200× magnification.
When conducting immunofluorescence experiments,
HCT116 cells were seeded in 24-well cell culture plates after 6 h
of transfection. The culture medium was removed after 24 h, cells
were fixed with 4% paraformaldehyde at room temperature for 20 min
and permeabilized with 0.5% Triton X-100 at room temperature for 20
min and then blocked with 5% blocking solution at room temperature
for 2 h. Following a phosphate-buffered saline (PBS) wash, the
cells were then incubated with an anti-SAMD4B (ProteinTech Group,
Inc.; 1:50; cat. no. 17723-1-AP) antibody overnight at 4°C. The
cells were then incubated in the dark with a goat anti-rabbit
IgG-FITC antibody (ProteinTech Group, Inc.; 1:50; cat. no.
SA00003-2) at room temperature for 1 h and cell nuclei were
counterstained with DAPI at room temperature for 5 min. Cells were
reviewed and images captured with a fluorescence microscope
(Olympus Corporation) at ×400 magnification
Statistical analysis
The data were expressed as the means ± SD and
analyzed using SPSS 23 statistical software (IBM Corp.). Unpaired
Student's t-tests or one-way ANOVA with Dunnett's or Bonferroni
post hoc tests were performed to determine statistical
significance. Categorical variables were analyzed using
χ2 test or Fisher's exact test. The Kaplan-Meier method
was applied to assess the survival rate of patients with CRC.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Differential expression of miR-451 in
patients with CRC and cell lines
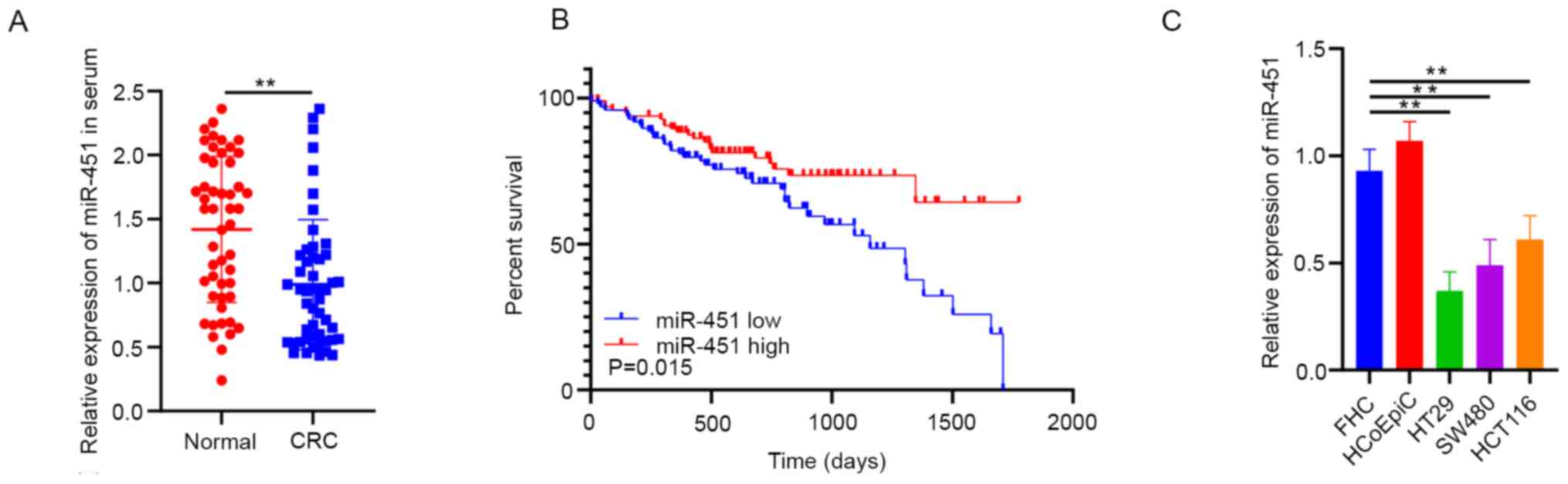
miR-451 expression was significantly decreased in
patients with CRC when compared with serum from normal individuals
(Fig. 1A). In addition, data from
TCGA demonstrated that decreased miR-451 expression was notably
associated with shorter overall survival rate of patients with CRC
(Fig. 1B). In addition, miR-451
expression was lower in CRC cell lines (HT29, SW480 and HCT116)
compared with the normal colorectal mucosal cell lines (FHC and
HCoEpiC) (Fig. 1C).
miR-451 suppresses CRC cell
proliferation and migration, increases sensitivity to chemotherapy
and promotes apoptosis
To explore the role of miR-451 in the tumorigenesis
of CRC, the HCT116 cell line, which exhibits intermediate
expression of miR-451, was selected for further study. Mimics and
inhibitors specific to miR-451 were transfected into HCT116 cells
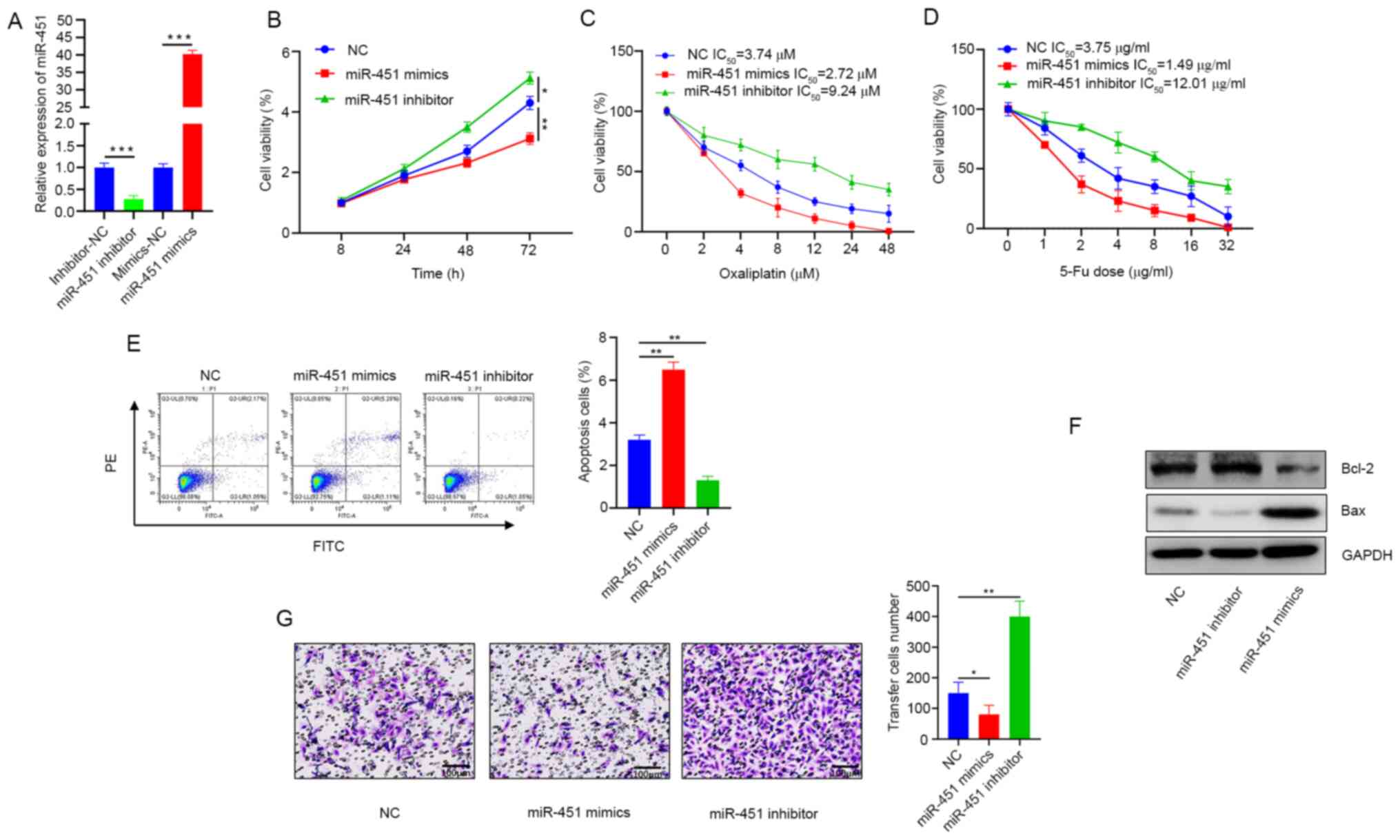
and miR-451 expression detected 72 h post transfection (Fig. 2A). Cell viability was notably
decreased in the miR-451mimic groups but increased in the
miR-451-inhibited cells (Fig. 2B).
Overexpression of miR-451 increased oxaliplatin and 5-FU-induced
inhibition of CRC cell growth and increased sensitivity to
chemotherapy (Fig. 2C and D). When
miR-451 was overexpressed, the number of apoptotic cells increased
significantly, whereas less apoptosis was noted in the inhibitor
group (Fig. 2E). Western blotting
results demonstrated that expression of the apoptosis-inhibitor
protein Bcl-2 was downregulated but the apoptosis-promoting protein
Bax was upregulated, supporting the conclusion that miR-451
promoted apoptosis (Fig. 2F).
Additionally, results from the Transwell assay also demonstrated a
lower migration rate in cells where miR-451 was overexpressed, but
higher migration rates were observed when miR-451 was knocked down
(Fig. 2G).
SAMD4B is a target of miR-451
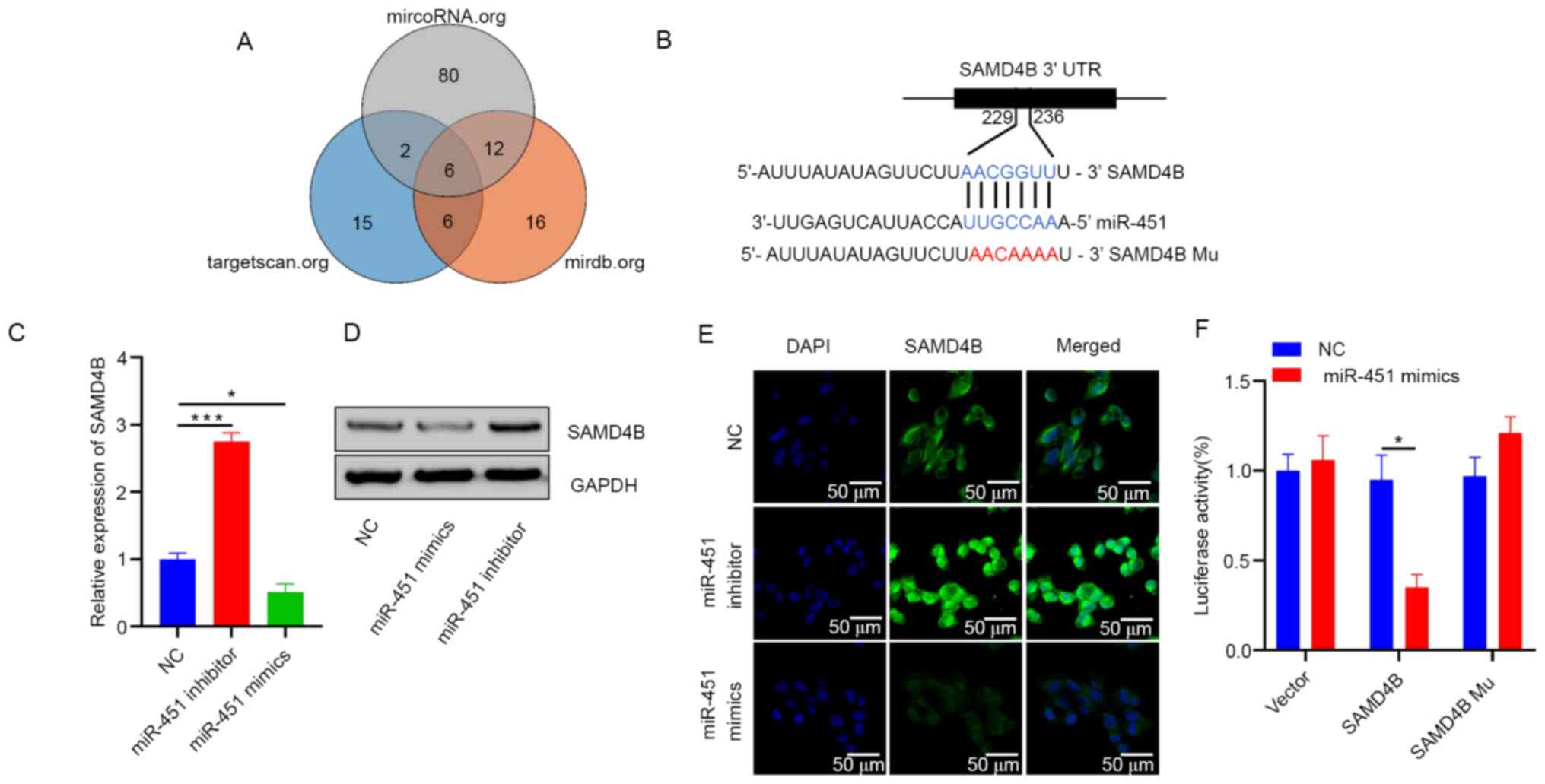
To investigate how miR-451 regulates the malignant
biological behavior of CRC cells, target gene candidates of miR-451
were searched for (TargetScan, targetscan.org/; microRNA, microRNA.org/ and mirdb, mirdb.org/) and
SAMD4B was identified as a potential candidate (Fig. 3A and B). To explore the regulatory
effect of miR-451 on SAMD4B, HCT116 cells were transfected with the
miR-451 mimics or inhibitors. After 72 h transfection, SAMD4B
expression was measured by RT-qPCR and western blotting. Results
demonstrated that overexpression of miR-451 significantly
downregulated the expression of SAMD4B at the mRNA and protein
levels and inhibition of miR-451 expression had the opposite effect
(Fig. 3C and D). These results were
confirmed using immunofluorescence (Fig. 3E). Furthermore, miR-451 directly
targeted the 3′-UTR of SAMD4B as determined by luciferase reporter
assay (Fig. 3F). These findings
indicated that miR-451 inhibited SAMD4B mRNA and protein expression
by directly targeting its 3′-UTR.
miR-451 regulates growth and migration
via SAMD4B in CRC
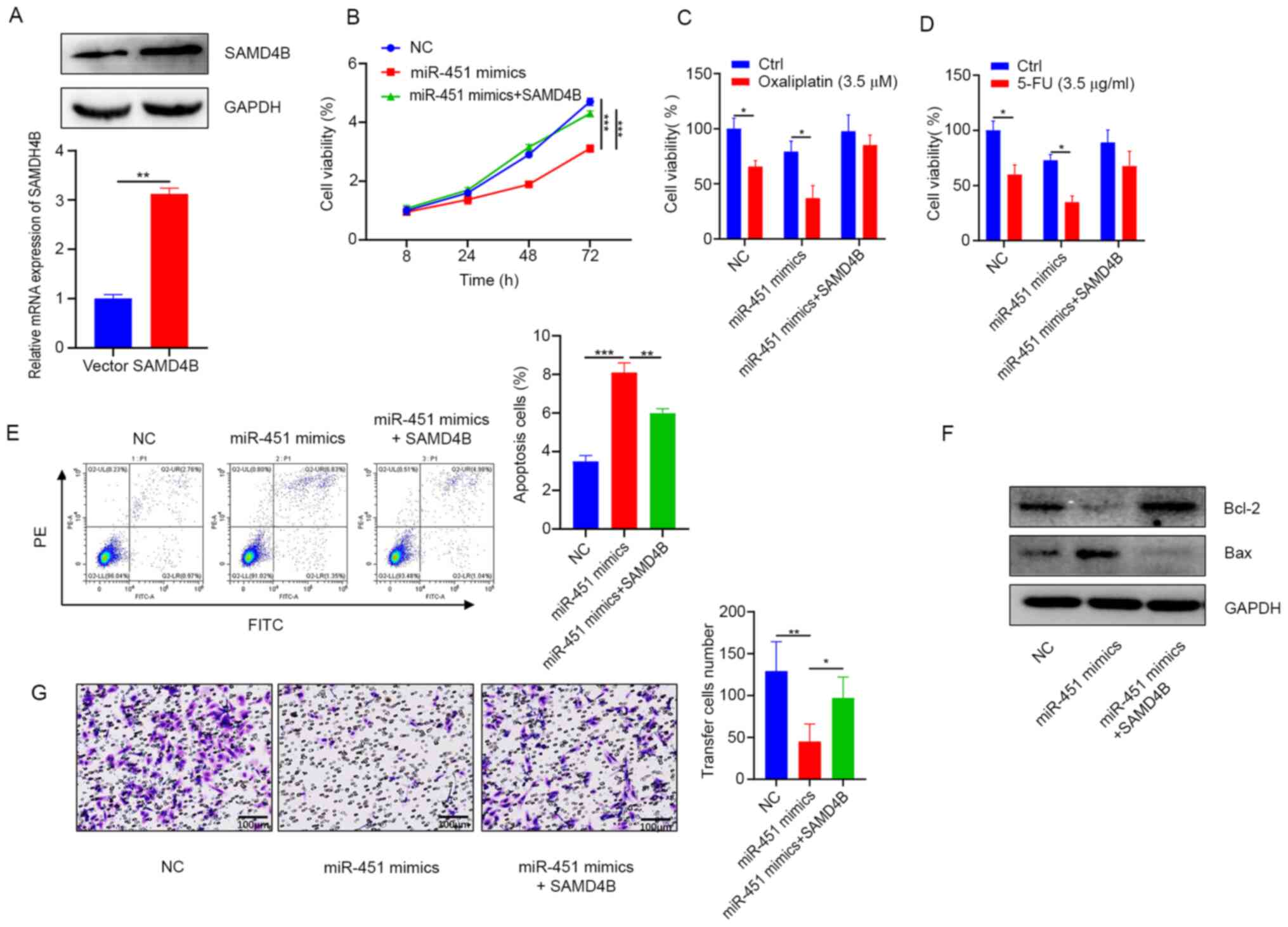
Migration and malignant proliferation are important
factors in the progression and recurrence of CRC. Plasmids specific
to SAMD4B were transfected into HCT116 cells and SAMD4B mRNA and
protein levels analyzed 72 h post transfection (Fig. 4A). The present study then explored
whether miR-451 was closely linked to the regulation of CRC
proliferation and migration via SAMD4B. The cell viability analysis
results demonstrated that overexpression of SAMD4B restored the
cell proliferation inhibited by miR-451 (Fig. 4B) and overexpression of SAMD4B
decreased the oxaliplatin and 5-FU chemosensitivity induced by
miR-451 (Fig. 4C and D). Flow
cytometric analysis revealed that the overexpression of SAMD4B
attenuated miR-451-induced apoptosis (Fig. 4E). Consistent with these results,
analysis of apoptotic protein expression via western blotting
further confirmed the above results (Fig. 4F). In addition, Transwell
experiments suggested that the overexpression of SAMD4B inhibited
miR-451-induced effects on cell migration (Fig. 4G). Taken together, these results
illustrate that miR-451 regulated proliferation and migration
through SAMD4B in CRC cells.
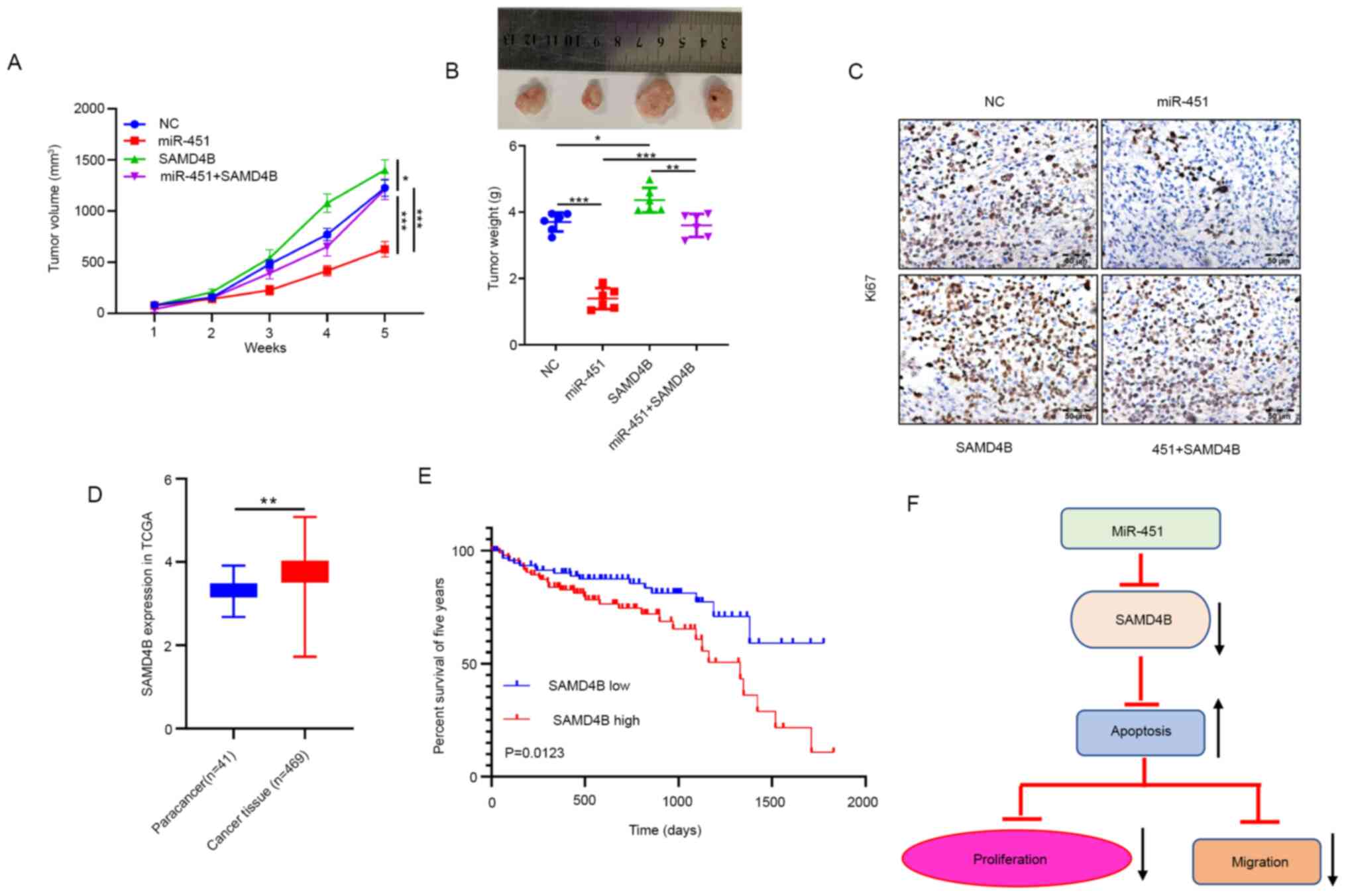
miR-451 inhibits CRC growth through
SAMD4B in vivo
In the animal experiment, it was observed that
miR-451 overexpression resulted in a significant decrease in tumor
volume (Fig. 5A) and tumor weight
(Fig. 5B). When both miR-451 and
SAMD4B were overexpressed, SAMD4B overexpression reversed
inhibition of tumor growth induced by miR-451. The IHC experiment
demonstrated that the Ki-67 protein was dramatically decreased in
the miR-451 group but overexpressed in the SAMD4B group and
co-transfection of miR-451 and SAMD4B reversed inhibition of Ki-67
expression compared with miR-451 expression alone (Fig. 5C). The TCGA database was used to
identify the expression of SAMD4B in CRC and paracancer tissues and
found that SAMD4B is highly expressed in both types of tumor
tissues (Fig. 5D). SAMD4B is
significantly related to poor prognosis in patients with CRC
(Fig. 5E). In summary, these
findings supported the hypothesis that miR-451 is essential to
inhibition of malignant behavior via targeting SAMD4B (Fig. 5F).
Discussion
CRC is a common malignant tumor of the digestive
system. The pathological development of CRC is a long, multi-step
process progressing from benign precancerous lesions (adenoma) to
malignant tumors (adenocarcinoma) (31). The treatment of CRC involves
surgical resection of the primary lesion combined with
chemotherapy, radiotherapy and/or molecular targeted therapy
(32). Chemotherapeutic drugs based
on 5-FU and oxaliplatin do not sufficiently control the progression
of advanced CRC (33). Molecular
targeted drugs such as EGFR/VEGFR inhibitors offer hope for
patients with advanced CRC (34),
but the prognosis of advanced patients with CRC remains poor.
Conventional tumor markers such as CEA and CA19-9 cannot suitably
predict the occurrence and development of tumors due to poor
sensitivity (1). In recent years,
researchers have been searching for valuable markers for the
diagnosis, treatment and prediction of CRC (35).
In 2005, Altuvia et al (36) searched miRNA precursor sequences
near known miRNA genes. He used bioinformatics to first identify an
miRNA adjacent to miR-144 and named it miR-451 (36). Subsequently, a growing number of
studies focused on miR-451 in living organisms have been published.
Regarding physiological processes, miR-451 is involved in
hematopoietic differentiation (37–39),
embryo maturation (40) and human
nervous system development (41).
In pathological processes, miR-451 has been shown to improve
cardiac hypertrophy (42) and
attenuate ischemic brain infarction (43).
In oncology, miR-451 has significance in different
types of tumors such as lung cancer, pancreatic cancer and glioma
(19,20,44).
In CRC, miR-451 serves an important role. Mamoori et al
(24) found that elevated
expression of miR-451 contributed to increased apoptosis, reduced
proliferation and changed cell cycle distribution. Vidal et
al (45) evaluated miRNA
profiles in embryonic samples and cancer cell lines. They concluded
that miR-451 is highly expressed in placental samples but is
significantly lower in cancer cell lines. Transfection of miR-451
significantly improves the malignant biological behavior of the
HT-29 CRC cell line by decreasing proliferation, migration,
invasion and colony formation (45). Consistent with these studies, the
present study also demonstrated overexpression of miR-451
sensitized CRC cells to oxaliplatin and 5-FU. Expression of the
apoptosis-inhibitory protein Bcl-2 was downregulated whereas the
apoptosis-promoting protein Bax was upregulated, supporting the
hypothesis that miR-451 promoted apoptosis. miR-451 also reduced
cell migration. In animal experiments, miR-451 reduced the growth
of xenograft tumors, decreased tumor volume and tumor weight and
IHC staining indicated a decrease in Ki-67 expression. These
results suggest that the proliferative capacity of the tumors was
suppressed.
The present study further explored the internal
tumor suppressor mechanisms of miR-451 in CRC. In vitro and
in vivo functional experiments confirmed that miR-451
suppressed the malignant biological behavior of CRC cells by
targeting SAMD4B.
In conclusion, the present study identified miR-451
as a predictive miRNA that is closely associated with longer
survival rates in patients with CRC. Through gain and
loss-of-function studies, the present study concluded that miR-451
inhibited proliferation and migration, enhanced the sensitivity of
CRC cells to chemotherapy and probably mediated its effects by
targeting SAMD4B. The present study illustrated the potential value
of miR-451 and its target gene SAMD4B in predicting the progression
and prognosis of patients with CRC. The miR-451/SAMD4B axis may
serve as a novel therapeutic target in patients with CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science
Foundation of Chongqing, China (grant no. cstc2020jcyj-msxmX0183),
District Science and Technology Commission Project of Chongqing
Jiangjin (grant nos. Y2019067 and Y2020050) and Health and Family
Planning Commission Medical Research Project of Chongqing (grant
no. 2017MSXM171).
Availability of data and materials
The data of this study are available from the
corresponding author on reasonable request.
Authors' contributions
CW and XL contributed equally to the present study.
CW and XL designed the study and drafted the manuscript; BL
interpreted the data and performed statistical analysis; GS and CP
participated in the coordination of the study; and DX supervised
the study and helped to draft the manuscript. All authors
contributed toward data analysis and critically revising the paper
and agree to be accountable for all aspects of the work. CW and XL
confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the Jiangjin Central
Hospital of Chongqing approved the research protocol (approval no.
20190611-28) and informed and written consent was received from
each patient. Animal procedures were approved by the Ethics
Committee of Chongqing Medical University (approval no.
2019-256).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Das V, Kalita J and Pal M: Predictive and
prognostic biomarkers in colorectal cancer: A systematic review of
recent advances and challenges. Biomed Pharmacother. 87:8–19. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang FW, Cao CH, Han K, Zhao YX, Cai MY,
Xiang ZC, Zhang JX, Chen JW, Zhong LP, Huang Y, et al:
APC-activated long noncoding RNA inhibits colorectal carcinoma
pathogenesis through reduction of exosome production. J Clin
Invest. 129:727–743. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dy GK, Hobday TJ, Nelson G, Windschitl HE,
O'Connell MJ, Alberts SR, Goldberg RM, Nikcevich DA and Sargent DJ:
Long-term survivors of metastatic colorectal cancer treated with
systemic chemotherapy alone: A North Central Cancer Treatment Group
review of 3811 patients, N0144. Clin Colorectal Cancer. 8:88–93.
2009. View Article : Google Scholar
|
|
4
|
Skarkova V, Kralova V, Vitovcova B and
Rudolf E: Selected aspects of chemoresistance mechanisms in
colorectal carcinoma-a focus on epithelial-to-mesenchymal
transition, autophagy, and apoptosis. Cells. 8:2342019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–96. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee SD, Yu D, Lee DY, Shin HS, Jo JH and
Lee YC: Upregulated microRNA-193a-3p is responsible for cisplatin
resistance in CD44(+) gastric cancer cells. Cancer Sci.
110:662–673. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ding HX, Lv Z, Yuan Y and Xu Q: miRNA
polymorphisms and cancer prognosis: A systematic review and
meta-analysis. Front Oncol. 8:5962018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rodríguez-Martínez A, de Miguel-Pérez D,
Ortega FG, García-Puche JL, Robles-Fernández I, Exposito J,
Martorell-Marugan J, Carmona-Sáez P, Garrido-Navas MDC, Rolfo C, et
al: Exosomal miRNA profile as complementary tool in the diagnostic
and prediction of treatment response in localized breast cancer
under neoadjuvant chemotherapy. Breast Cancer Res. 21:212019.
View Article : Google Scholar
|
|
9
|
Vychytilova-Faltejskova P and Slaby O:
MicroRNA-215: From biology to theranostic applications. Mol Aspects
Med. 70:72–89. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang R, Sun Y, Yu W, Yan Y, Qiao M, Jiang
R, Guan W and Wang L: Downregulation of miRNA-214 in
cancer-associated fibroblasts contributes to migration and invasion
of gastric cancer cells through targeting FGF9 and inducing EMT. J
Exp Clin Cancer Res. 38:202019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou GL, Xu WF, Jin YB, Wu J, Pan Y and
Zhou F: miRNA-217 accelerates the proliferation and migration of
bladder cancer via inhibiting KMT2D. Biochem Biophys Res Commun.
519:747–753. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu AN, Qu HJ, Gong WJ, Xiang JY, Yang MM
and Zhang W: LncRNA AWPPH and miRNA-21 regulates cancer cell
proliferation and chemosensitivity in triple-negative breast cancer
by interacting with each other. J Cell Biochem. 120:14860–14866.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shivapurkar N, Weiner LM, Marshall JL,
Madhavan S, Deslattes Mays A, Juhl H and Wellstein A: Recurrence of
early stage colon cancer predicted by expression pattern of
circulating microRNAs. PLoS One. 9:e846862014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Allen B, Schneider A, Victoria B, Nunez
Lopez YO, Muller M, Szewczyk M, Pazdrowski J, Majchrzak E, Barczak
W, Golusinski W, et al: Blood serum from head and neck squamous
cell carcinoma patients induces altered microRNA and target gene
expression profile in treated cells. Front Oncol. 8:2172018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao L, Yang S, You X, He W and Xue J:
Novel miRNA-based biomarker panel for detection β2-agonists in
goats. Food Chem. 288:15–21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Usuba W, Urabe F, Yamamoto Y, Matsuzaki J,
Sasaki H, Ichikawa M, Takizawa S, Aoki Y, Niida S, Kato K, et al:
Circulating miRNA panels for specific and early detection in
bladder cancer. Cancer Sci. 110:408–419. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng D, Ding Y, Ma Q, Zhao L, Guo X, Shen
Y, He Y, Wei W and Liu F: Identification of serum MicroRNAs as
novel biomarkers in esophageal squamous cell carcinoma using
feature selection algorithms. Front Oncol. 8:6742018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen Y, Gong JM, Zhou LL and Sheng JH:
Correction: miR-451 as a new tumor marker for gastric cancer.
Oncotarget. 10:63962019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Li H, Li LH, Tang JB and Sheng YL:
Mir-451 inhibits proliferation and migration of non-small cell lung
cancer cells via targeting LKB1/AMPK. Eur Rev Med Pharmacol Sci. 23
(Suppl 3):S274–S280. 2019.
|
|
20
|
Guo R, Gu J, Zhang Z, Wang Y and Gu C:
miR-451 promotes cell proliferation and metastasis in pancreatic
cancer through targeting CAB39. Biomed Res Int. 2017:23814822017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kong W, Feng L, Yang M, Chen Q, Wang H,
Wang X and Hou J: Prognostic value of microRNA-451 in various
cancers: A meta-analysis. Pathol Res Pract. 215:1527262019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khordadmehr M, Jigari-Asl F, Ezzati H,
Shahbazi R, Sadreddini S, Safaei S and Baradaran B: A comprehensive
review on miR-451: A promising cancer biomarker with therapeutic
potential. J Cell Physiol. 234:21716–21731. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bai H and Wu S: miR-451: A novel biomarker
and potential therapeutic target for cancer. Onco Targets Ther.
12:11069–11082. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mamoori A, Wahab R, Vider J, Gopalan V and
Lam AK: The tumour suppressor effects and regulation of cancer stem
cells by macrophage migration inhibitory factor targeted miR-451 in
colon cancer. Gene. 697:165–174. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mamoori A, Gopalan V, Lu CT, Chua TC,
Morris DL, Smith RA and Lam AK: Expression pattern of miR-451 and
its target MIF (macrophage migration inhibitory factor) in
colorectal cancer. J Clin Pathol. 70:308–312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fan X and Zhao Y: miR-451a inhibits cancer
growth, epithelial-mesenchymal transition and induces apoptosis in
papillary thyroid cancer by targeting PSMB8. J Cell Mol Med.
23:8067–8075. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu D, Fang C, He W, Wu C, Li X and Wu J:
MicroRNA-181a inhibits activated B-cell-like diffuse large B-cell
lymphoma progression by repressing CARD11. J Oncol.
2019:98329562019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu G, Cheng Z, Huang Y, Zheng W, Yang S,
Lin C and Ye J: MyD88 mediates colorectal cancer cell
proliferation, migration and invasion via NF-κB/AP-1 signaling
pathway. Int J Mol Med. 45:131–140. 2020.PubMed/NCBI
|
|
30
|
Yamadera M, Shinto E, Kajiwara Y,
Mochizuki S, Okamoto K, Shimazaki H, Hase K and Ueno H:
Differential clinical impacts of tumour budding evaluated by the
use of immunohistochemical and haematoxylin and eosin staining in
stage II colorectal cancer. Histopathology. 74:1005–1013. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mármol I, Sánchez-de-Diego C, Pradilla
Dieste A, Cerrada E and Rodriguez Yoldi MJ: Colorectal carcinoma: A
general overview and future perspectives in colorectal cancer. Int
J Mol Sci. 18:1972017. View Article : Google Scholar
|
|
33
|
To KK, Tong CW, Wu M and Cho WC: MicroRNAs
in the prognosis and therapy of colorectal cancer: From bench to
bedside. World J Gastroenterol. 24:2949–2973. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yalcin S, Trad D, Kader YA, Halawani H,
Demir OG, Mall R, Meshcheryakov A, Nasr F, Nosworthy A, Osinsky D,
et al: Personalized treatment is better than one treatment fits all
in the management of patients with mCRC: A consensus statement.
Future Oncol. 10:2643–2657. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Sun XF, Cao Y, Ye B, Peng Q, Liu
X, Shen B and Zhang H: CBD: A biomarker database for colorectal
cancer. Database (Oxford). 2018:bay0462018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Altuvia Y, Landgraf P, Lithwick G, Elefant
N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T and Margalit H:
Clustering and conservation patterns of human microRNAs. Nucleic
Acids Res. 33:2697–2706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu P, Palmer LE, Lechauve C, Zhao G, Yao
Y, Luan J, Vourekas A, Tan H, Peng J, Schuetz JD, et al: Regulation
of gene expression by miR-144/451 during mouse erythropoiesis.
Blood. 133:2518–2528. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shokri G, Kouhkan F, Nojehdehi S,
Soleimani M, Pourfathollah AA, Nikougoftar Zarif M, Tamaddon M and
Obeidi N: Simultaneous regulation of miR-451 and miR-191 led to
erythroid fate decision of mouse embryonic stem cell. Iran J Basic
Med Sci. 224:432–438. 2019.PubMed/NCBI
|
|
39
|
Yao H, Ma Y and Huang LJ: Deletion of
miR-451 curbs JAK2(V617F)-induced erythrocytosis in polycythemia
vera by oxidative stress-mediated erythroblast apoptosis and
hemolysis. Haematologica. 105:e153–e156. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Zhang W, Fu J, Xu Y, Gu R, Qu R, Li
L, Sun Y and Sun X: MicroRNA-451 is downregulated in the follicular
fluid of women with endometriosis and influences mouse and human
embryonic potential. Reprod Biol Endocrinol. 17:962019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Z, Chang H, Li Y, Zhang T, Zou J,
Zheng X and Wu J: MicroRNAs: Potential regulators involved in human
anencephaly. Int J Biochem Cell Biol. 42:367–374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gan M, Zheng T, Shen L, Tan Y, Fan Y,
Shuai S, Bai L, Li X, Wang J, Zhang S and Zhu L: Genistein reverses
isoproterenol-induced cardiac hypertrophy by regulating
miR-451/TIMP2. Biomed Pharmacother. 112:1086182019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fu C, Chen S, Cai N, Liu Z, Wang P and
Zhao J: Potential neuroprotective effect of miR-451 against
cerebral ischemia/reperfusion injury in stroke patients and a mouse
model. World Neurosurg. 130:e54–e61. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ogawa D, Ansari K, Nowicki MO, Salińska E,
Bronisz A and Godlewski J: MicroRNA-451 inhibits migration of
glioblastoma while making it more susceptible to conventional
therapy. Noncoding RNA. 5:252019.PubMed/NCBI
|
|
45
|
Vidal DO, Ramão A, Pinheiro DG, Muys BR,
Lorenzi JCC, de Pádua Alves C, Zanette DL, de Molfetta GA, Duarte G
and Silva WA Jr: Highly expressed placental miRNAs control key
biological processes in human cancer cell lines. Oncotarget.
9:23554–23563. 2018. View Article : Google Scholar : PubMed/NCBI
|