Introduction
Lung cancer is a major cause of mortality and has a
survival rate of only 15.6% in all affected patients (1), although this rate can reach up to
55–80% in early-stage cases (2). In
the United States, >225,000 new cases of lung cancer are
diagnosed each year and ~160,000 deaths occur annually (3). Most patients with lung cancer (~85%)
have non-small cell lung cancer (NSCLC) (4). Despite improvements in cancer
diagnosis and treatments, the 5-year survival rate has remained as
low as 15% since the 1970s (5).
Therefore, further investigations into the mechanisms of NSCLC
development and progression are of the utmost importance for the
diagnosis, prevention and treatment of this disease.
MicroRNAs (miRNAs/miRs), a class of endogenous
non-coding RNAs, can mediate various gene expression levels by
suppressing the translation or accelerating the decay of the target
mRNA (6,7). Previous data have revealed that miRNAs
can regulate 30% of mRNAs (8).
Furthermore, miRNAs have important effects on various pathological
processes, such as cancer cell proliferation, migration and
differentiation, as well as patient survival (9,10).
Numerous dysregulated miRNAs, including miR-21, miR-100, miR-138
and miR-847, have been verified to be involved in NSCLC cell
processes (11–14), which indicates that the aberrant
expression of mRNA is associated with NSCLC. Moreover, miR-205 is
significantly increased in NSCLC and has been demonstrated to serve
as a biological marker for NSCLC (15). Su et al (16) observed that abnormal expression of
miR-205 in endometrial cancer was closely associated with tumor
proliferation and invasion. Another study reported that upregulated
miR-205 expression could enhance the proliferation and the
migration of cervical cancer cells (17). Collectively, these previous findings
have suggested the potential promotive role of miR-205 on cancer
processes. Despite of these findings, the role and underlying
mechanisms of miR-205 in NSCLC remain largely unclarified.
Amyloid β precursor protein-binding family B member
2 (APBB2) is an adaptor protein that belongs to the β-amyloid
precursor protein-binding family B. APBB2 can bind to the
cytoplasmatic domain of the β-amyloid precursor protein (βAPP).
Thus, APBB2 may be involved in mediating APP processing (18). Lim et al (19) revealed that APBB2 overexpression
promoted the formation of APP and reduced the response to apoptotic
stimuli. Although the function of APBB2 is important, it remains
unclear whether APBB2 could be regulated at the transcriptional
level in NSCLC.
As stated in the aforementioned previous studies,
both miR-205-3p and APBB2 may play crucial roles in the development
of NSCLC, but the association between them and their regulatory
roles in NSCLC remain unclear. In the current study, the expression
of miR-205-3p in NSCLC was detected, followed by miR-205-3p
biofunction analyses with MTT, TUNEL and flow cytometry assays.
Following this, the potential downstream target of miR-205-3p,
APBB2, was predicted and further investigated using a
dual-luciferase reporter assay. In addition, the effect of the
miR-205-3p/APBB2 axis on the proliferation and apoptosis of NSCLC
was also explored. According to these investigation, the present
study aimed to clarify the underlying mechanism of the
miR-205-3p/APBB2 axis in NSCLC and provide some novel insights in
the treatment of NCSCLC.
Materials and methods
Cell culture and tissue samples
The NSCLC cell lines A549, H1299, 95-D and H460, and
the BEAS-2B normal human epithelial cell line were obtained from
The Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. The cells were cultured in Dulbecco's modified Eagle
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 100 IU/ml penicillin (Shanghai Sangong Pharmaceutical Co.,
Ltd.) and 100 mg/ml streptomycin (Shanghai Sangong Pharmaceutical
Co., Ltd.) at 37°C with 5% CO2.
NSCLC tissue samples and adjacent normal tissues (1
cm from tumor margin) were collected from 20 patients at the
Shaanxi Provincial People's Hospital (Xi'an, China) between January
and October 2019. All samples were snap-frozen in liquid nitrogen
and maintained at −80°C. None of the patients had undergone
chemotherapy or radiotherapy before tissue sampling. The NSCLC
samples were obtained from 8 patients with advanced-stage disease
(III and IV) and 12 patients with early-stage disease (I and II)
who had provided informed consent before collecting; all pertinent
diagnoses were conducted under the Revised International System for
Staging Lung Cancer (20). This
research was performed in accordance with the World Medical
Association Declaration of Helsinki and was approved by the
Institutional Review Board of the Shaanxi Provincial People's
Hospital (approval no. SXRMYY-2019-015). Written informed consent
was obtained from all patients.
Cell transfection
miR-205-3p inhibitor (50 nM;
5′-GAUUUCAGUGGAGUGAAGUUC-3′) and negative controls (miR-NC; 50 nM;
5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were obtained from Changzhou Ruibo
Bio-Technology Co., Ltd. Two specific small interfering (si)RNAs
against APBB2 (50 nM; APBB2-siRNA1, 5′-AUUUUCCAUCCAGCAAAUGUU-3′;
APBB2-siRNA2, 5′-UUGCUUAAAUUAUUGUCAGCC-3′) and a scrambled NC (50
nM; si-NC, 5′-CACUGAUUUCAAAUGGUGCUAUU-3′) were synthesized by
Shanghai GenePharma Co., Ltd. All transfections were performed
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol.
Briefly, 3×105 cells/well were seeded in a 6-well plate
and were incubated until they reached ≥70% confluence. Cells were
then transfected with the miR-205 inhibitor, APBB2 siRNA1, APBB2
siRNA2 and their respective NCs using Lipofectamine 2000 reagent.
After 72 h, the cells were harvested for further investigation. The
efficiency was validated by reverse transcription-quantitative PCR
(RT-qPCR) or western blotting.
RT-qPCR
Total RNA from tissues and cell lines was isolated
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was synthesized using the PrimeScript RT
reagent kit (Takara Bio, Inc.) according to the manufacturer's
protocol. qPCR was performed using the SYBR® Green
PrimeScript™ PLUS RT-PCR kit (Takara Bio, Inc.). The reaction
conditions were as follows: Initial denaturation at 95°C for 30
sec; followed by 40 cycles of denaturation at 95°C for 15 sec,
annealing at 60°C for 25 sec and extension at 72°C for 1 min; and a
final extension step at 72°C for 10 min. β-actin (mRNA) and U6
(miRNA) were used as the internal controls and the relative
expression of genes was calculated using 2−ΔΔCq method
(21). The following primers were
used: miR-205-3p forward, 5′-CTTGTCCTTCATTCCACCGGA-3′ and reverse,
5′-TGCCGCCTGAACTTCACTCC-3′; APBB2 forward,
5′-CACAGAGAAGAGTCTGGCCC-3′ and reverse, 5′-AGGTTGCTTGTGACAGGTCC-3′;
β-actin forward, 5′-GATGGACTCTGGTGATGGTGTGAC-3′ and reverse,
5′-TTTCTCTTTCGGCTGTGGTGGTG-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
MTT assay
An MTT assay was used to analyze cell viability.
Transfected cells were seeded into 96-well plates at a density of
1.0×104 per well and cultured at 37°C for 24, 48 and 72
h. After being mixed with 0.5 mg/ml MTT solution (Sigma; Merch
KGaA) for 4 h and co-culturing with 100 µl DMSO for 10 min at 37°C,
the optical density values were determined at 490 nm.
TUNEL staining
A DeadEnd Fluorometric TUNEL Detection system
(Promega Corporation) was used to evaluate apoptosis, according to
the manufacturer's protocol. Briefly, 3×105 cells were
seeded onto a cover slip in a six-well plate and attached
overnight. Then, cells were treated as indicated followed by 4%
formaldehyde-PBS fixation for 15 min at room temperature, after
which they were permeabilized with 0.2% Triton X-100 in PBS for
another 10 min under the same conditions. After being washed twice
with PBS, the cells were incubated with a fluorometric terminal
deoxytransferase mixture at 37°C for 1 h and then washed twice with
PBS buffer. Then, cells on slips were mounted using DAPI-containing
mounting medium (Vector Laboratories, Inc.) according to the
manufacturer's protocol. Fluorescence images were captured in at
least five views using a Nikon Eclipse Ti-E fluorescence microscope
(Nikon Corporation).
EdU assay
The effect of miR-205-5p on cell proliferation was
detected using EdU staining (Guangzhou RiboBio Co., Ltd.). Briefly,
3×105 cells were seeded onto a cover slip in a six-well
plate and maintained overnight followed by the indicated
treatments. Then, cells were incubated with 30 µM EdU contained in
serum-free DMEM. After washing in PBS three times, cells were fixed
with 4% polyformaldehyde in PBS at room temperature for 30 min in
the dark. Following this, cells were incubated with Apollo staining
solution and Hoechst 33342 at room temperature for 30 min. Finally,
staining images were obtained using a Nikon Eclipse Ti-E
fluorescence microscope (Nikon Corporation). Five random field of
views were chosen to count the number of positively stained cells,
and proliferation was assessed using the percentage of EdU-positive
cells relative to the total cell number.
Flow cytometric analysis of
apoptosis
Flow cytometry was performed to determine the
alteration of apoptosis. After harvesting, the cells were washed
with PBS solution twice, followed by staining with a Annexin
V-FITC/PI Kit in the dark (Sigma-Aldrich; Merck KGaA), according to
the manufacturer's instructions. Apoptosis was analyzed using a BD
FACSCalibur flow cytometer (BD Biosciences) with Cell Quest
software (version 5.0; BD Biosciences). The apoptotic rate was
counted as the percentage of early apoptosis plus the percentage of
late apoptosis.
Western blotting
After reaching 80–90% confluence, cells were lysed
in RIPA buffer (BioVision, Inc.) solution with protease inhibitor
and phosphatase inhibitor cocktail (Sigma-Aldrich; Merck KGaA),
followed by centrifugation at 10,000 × g at 4°C for 10 min. Then,
the supernatant of cell lysate was collected and quantified using
BCA method (Thermo Fisher Scientific, Inc.). Total protein (25 µg)
was separated via 10% SDS-PAGE, and subsequently transferred to a
nitrocellulose membrane (EMD Millipore). Then, the membrane was
blocked in 10% dried milk and 0.1% BSA (Fraction V; Sigma-Aldrich;
Merck KGaA) in PBS for 1 h at room temperature, followed by
incubation at 4°C overnight with primary antibodies against APBB2
(1:2,500; cat. no. PA5-54816; Thermo Fisher Scientific, Inc.), Bax
(1:2,500; cat. no. MA5-13994; Sigma-Aldrich; Merck KGaA), Bcl-2
(1:1,000; cat. no. 13–8800; Thermo Fisher Scientific, Inc.) and
β-actin (1:5,000; cat. no. MA1-744; Thermo Fisher Scientific,
Inc.). Then, membranes were incubated with HRP-conjugated secondary
goat anti-rabbit (1:20,000; cat. no. G-21234; Sigma-Aldrich; Merck
KGaA) or goat anti-mouse antibody (1:20,000; cat. no. G-21040;
Sigma-Aldrich; Merck KGaA) at room temperature for 1 h. Protein
bands on membranes were visualized using the enhanced
chemiluminescence (ECL) method (Pierce; Thermo Fisher Scientific,
Inc.). β-actin was used as the internal control and protein
expression levels were semi-quantified using Quantity One 4.6
software (Bio-Rad Laboratories, Inc.).
Prediction of miR-205-3p target
genes
The downstream target genes of miR-205-3p were
predicted using miRDB (http://mirdb.org/index.html) and TargetScan (version
7.0; http://www.targetscan.org/vert_72/). Cell cycle
arrest-associated genes were obtained from Gene Ontology (GO;
http://geneontology.org). Then, the overlapping
genes among miRDB target genes, TargetScan target genes and GO cell
cycle arrest-associated genes were isolated using Venn analysis
(http://bioinformatics.psb.ugent.be/webtools/Venn/).
Luciferase assay
The amplified wild-type (WT) APBB2-3′untranslated
region (UTR) and mutant (MUT) APBB2-3′UTR, which was produced using
a Q5 Site-Directed Mutagenesis kit (cat. no. E0554S; New England
BioLabs, Inc.), according to the manufacturer's protocol, were
cloned into pGL3 luciferase vectors (Promega Corporation). The
cells were co-transfected with 10 nM miR-205-3p inhibitor (or
miR-NC) and luciferase reporter vectors (200 ng/well) of the WT or
MUT type 3′UTR of the APBB2 gene using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Subsequently, the Dual-Luciferase
Reporter Assay kit (Promega Corporation) was used to detect the
relative luciferase activities 48 h after the transfections,
according to the manufacturer's protocols. Renilla
luciferase activity was used for normalization against firefly
luciferase activity.
In vivo tumor biology
The present study experiments were performed at the
Shaanxi Provincial People's Hospital; all experiments were
performed following the Guidelines of Animal Use and Care Committee
of the Shaanxi Provincial People's Hospital, and were approved by
the Institutional Animal Care and Use Committee of Shaanxi
Provincial People's Hospital. C57BL/6 mice (age, 6–8 weeks old;
weight, 20±2 g; n=10) were purchased from Shanghai SLAC Laboratory
Animal Co., Ltd., maintained in the animal facility of the Shaanxi
Provincial People's Hospital at room temperature (22±1°C) with
humidity of 55±2% and a 12/12-h light/dark cycle, with access to
food and water ad libitum. Briefly, 2×106 A549
cells in 0.2 ml PBS were inoculated subcutaneously in the right
flank of each mouse. After 7 days, the mice were intravenously
injected with 100 µl antagomir NC or miR-205-3p antagomir (5 nmol)
in the tail. Tumor volume was assessed using a slide caliper every
5 days and calculated using the following formula: Volume=length ×
width2/2. At 25 days after inoculation, all mice were
anaesthetized with an intramuscular injection of 50 mg/kg ketamine
mixed with 5 mg/kg xylazine (22).
Then, mice were euthanized by cervical dislocation, and the tumor
volume was measured. miR-205-3p expression levels were determined
by RT-qPCR and western blot analysis was used to detect the APBB2
protein expression levels in tumors. Ki67 was detected by
immunofluorescence.
Immunofluorescence
Fresh tumor samples were washed with PBS to remove
blood, and dehydrated with gradient sucrose solution. Then, samples
were embedded in opti-MEM cutting temperature compound and frozen
at −80°C to form hardened blocks. Following this, tumor samples
were cut into 6-µm thick slides, fixed in 100% acetone at 4°C for
5~10 min, permeabilized in 0.025% Triton X-100 with PBS, blocked by
PBS with 10% goat serum (cat. no. C0265; Beyotime Institute of
Biotechnology) and 1% BSA at room temperature for 1 h. Following
this, slides were incubated with rabbit anti-Ki67 antibody (1:500;
cat. no. ab92742; Abcam) at 4°C in a humid room overnight. After
rinsing with PBS three times, slides were incubated with goat
anti-rabbit secondary antibody (Alexa Fluor® 488;
1:1,000; cat. no. ab150077; Abcam) at room temperature for 1 h.
Then, slides were incubated with PI (100 µg/ml; Sigma-Aldrich;
Merck KGaA) at room temperature for 15 min. Subsequently, slides
were mounted with glycerin (Sangon Biotech Co., Ltd.) and stained
images were collected using a Nikon Eclipse Ti-E fluorescence
microscope (Nikon Corporation).
Statistical analysis
All data were collected in triplicate and presented
as the mean ± SD. A paired Student's t-test was used for
statistical analysis of RT-qPCR results of NSCLC tissues and their
adjacent tissues. An unpaired Student's t-test or one-way ANOVA
followed by Tukey's post hoc test was applied to the rest of
the data to evaluate statistical differences. Kaplan-Meier analysis
followed by a log-rank test was used to compare the overall
survival between the two patient groups. Statistical analysis was
performed using GraphPad Prism 6.0 (GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
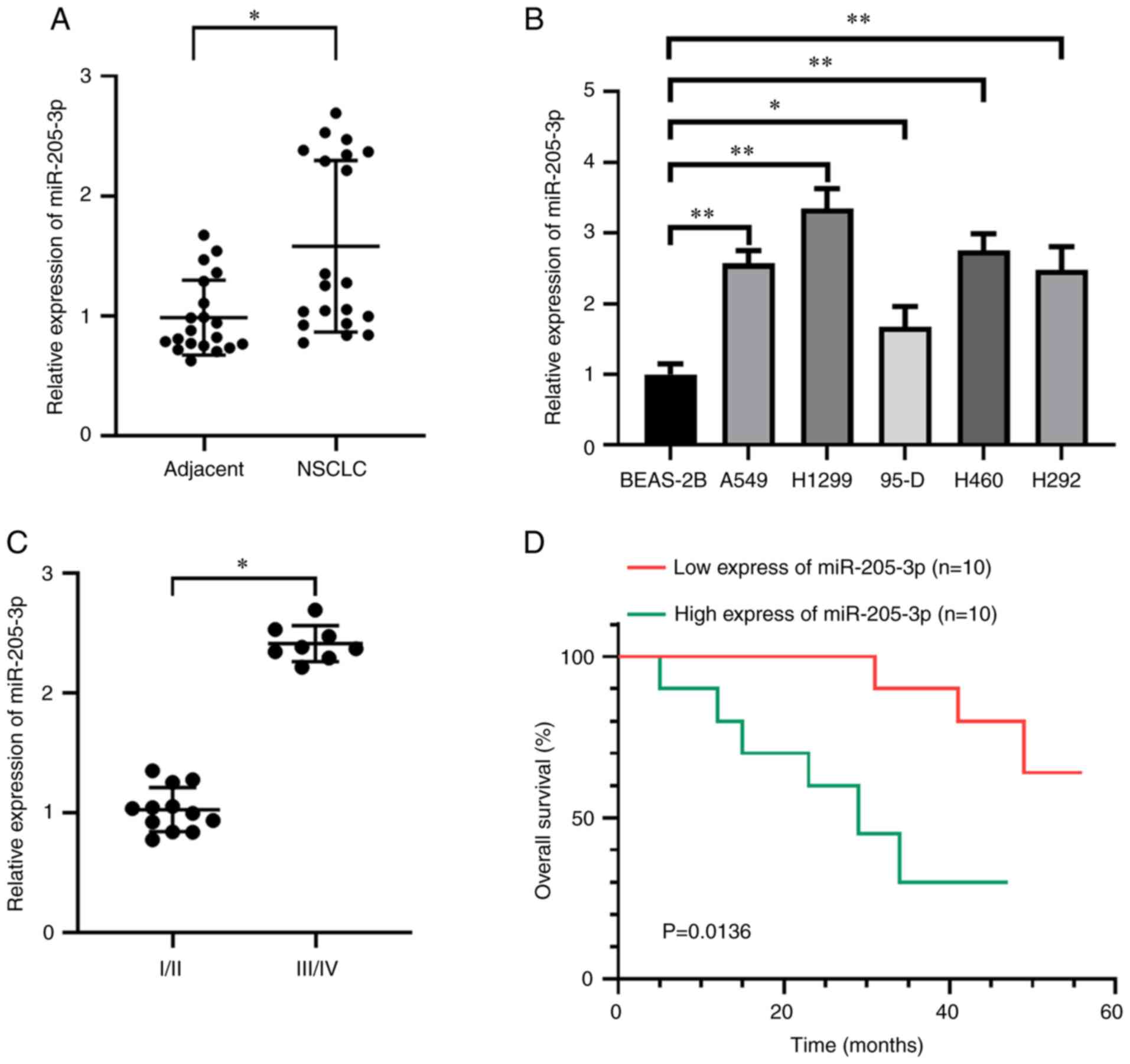
miR-205-3p is upregulated in NSCLC
tissues and cell lines
RT-qPCR was used to evaluate the expression levels
of miR-205-3p in normal adjacent lung tissues and NSCLC tissues
from patient samples. miR-205-3p expression was significantly
higher in the 20 NSCLC tissues compared with the normal adjacent
tissues (Fig. 1A). NSCLC cell
lines, including A549, H1299, 95-D, H460 and H292 cells, and the
BEAS-2B normal lung epithelial cells were used for further
investigation. As presented in Fig.
1B, miR-205-3p expression was significantly elevated in NSCLC
cell lines compared with that in the normal lung epithelial
cells.
To investigate the variations in miR-205-3p
expression, eight patients with advanced-stage disease (III and IV)
and 12 patients with early-stage disease (I and II) were examined.
It was revealed that miR-205-3p expression was significantly
elevated in advanced stages compared with early stages (Fig. 1C). The 20 patients with NSCLC were
divided into two groups based on their miR-205-3p expression levels
in relation to the median value (cut-off, 1.26) as follows: Low
expression (n=10) and high expression (n=10) miR-205-3p groups.
These groups were followed-up for 50 months to obtain survival
information. The results demonstrated that the patients in the low
miR-205-3p expression group displayed higher survival rates
compared with those in the high expression group (Fig. 1D). The characteristics of the
patients with NSCLC are listed in Table
I.
 | Table I.Clinicopathological characteristics
of patients with non-small cell lung cancer at the time of first
diagnosis. |
Table I.
Clinicopathological characteristics
of patients with non-small cell lung cancer at the time of first
diagnosis.
| Clinicopathological
characteristics | Patients
(n=20) |
|---|
| Age, years | 58.73±10.22 |
| BMI,
kg/m2 | 22.14±2.43 |
| Sex |
|
|
Male | 11 (55) |
|
Female | 9
(45) |
| TNM stage |
|
|
I–II | 12 (60) |
|
III–IV | 8
(40) |
| Tumor size, cm |
|
|
>5 | 13 (65) |
| ≤5 | 7
(35) |
| Smoking
history |
|
|
Yes | 14 (70) |
| No | 6
(30) |
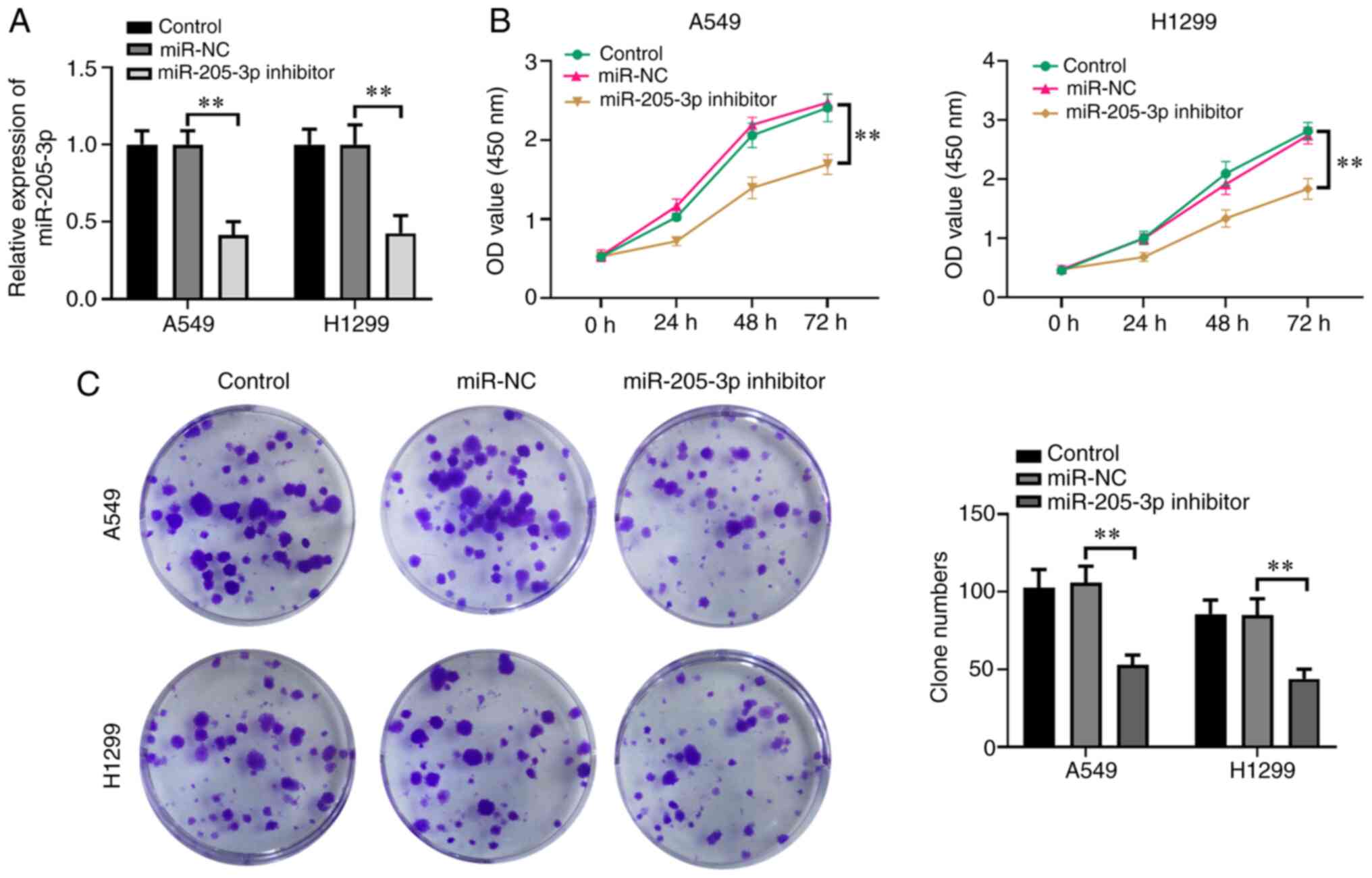
miR-205-3p knockdown reduces the
proliferation of NSCLC cells
A miR-205-3p inhibitor was used to investigate the
effects of miR-205-3p on H1299 and A549 NSCLC cell proliferation.
A549 and H1299 cells were chosen for this assay as they showed the
highest expression of miR-205-3p among the NSCLC cell lines.
Compared with the control and miR-NC treatment groups, miR-205-3p
expression was significantly lower in the miR-205-3p
inhibitor-transfected group (Fig.
2A). The MTT assay was performed to examine the effects of
miR-205 on H1299 and A549 cell proliferation. As presented in
Fig. 2B, the miR-205-3p
inhibitor-mediated knockdown of miR-205-3p significantly decreased
cell viability at 72 h post-transfection. A colony formation assay
was also conducted to analyze the possible effects of miR-205-3p
knockdown on cell proliferation. The results indicated that the
miR-205-3p inhibitor led to fewer clones compared with the control
groups (Fig. 2C).
To further validate the function of miR-205-3p,
BEAS-2B normal human lung epithelial cells were transfected with
miR-205-3p mimics or the inhibitor (Fig. S1A and B). It was found that neither
the miR-205-3p mimics or inhibitors altered cell proliferation in
BEAS-2B cells (Fig. S1C).
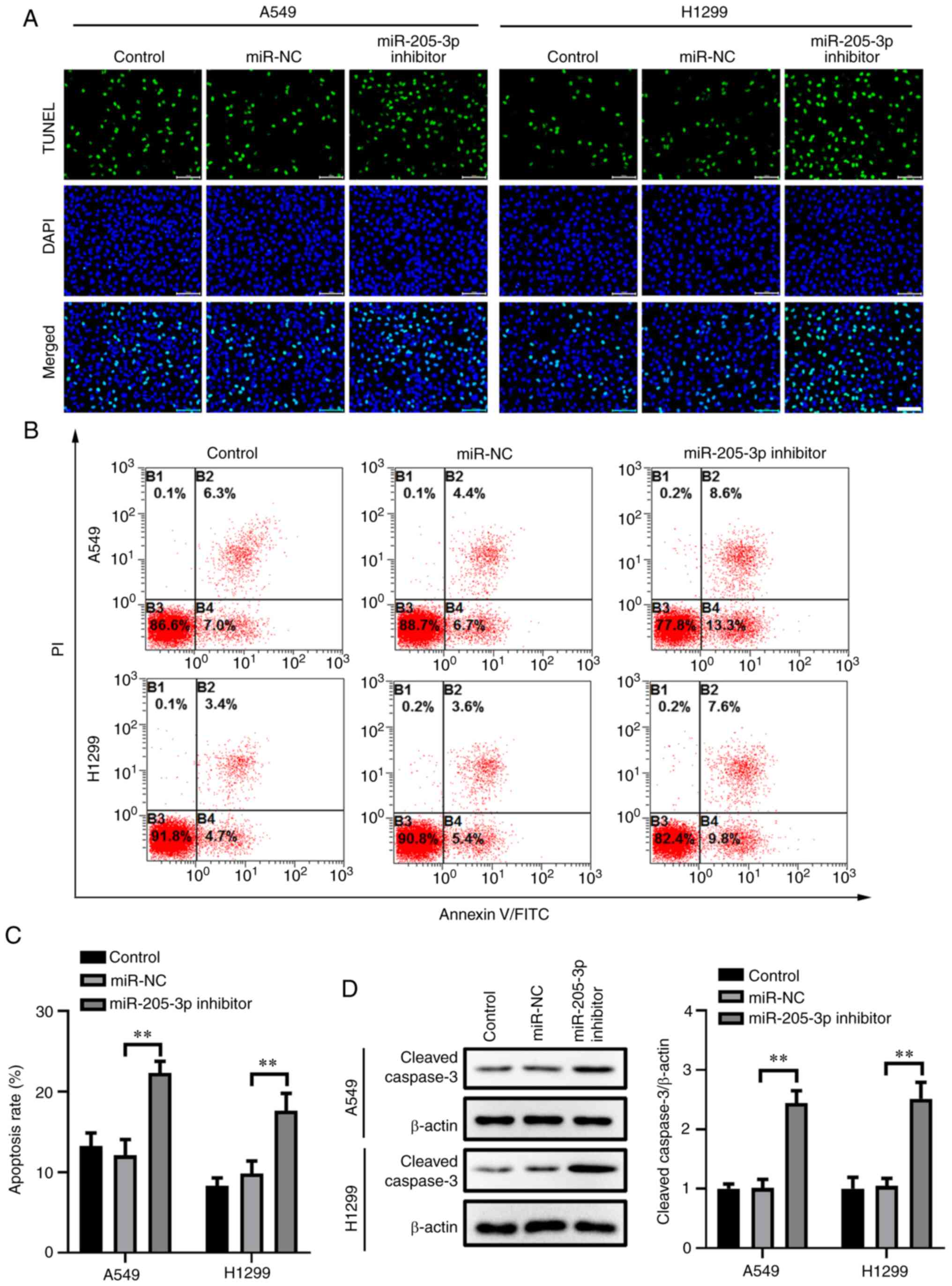
miR-205-3p knockdown accelerates the
apoptosis of NSCLC cells
TUNEL staining was used to determine the potential
impact of miR-205-3p knockdown on apoptosis in A549 and H1299
cells. Compared with the control and miR-NC groups, A540 and H1299
cells transfected with the miR-205-3p inhibitor had a high number
of cells undergoing apoptosis, as identified by TUNEL staining
(Fig. 3A). After being transfected
for 48 h, the apoptotic rates in the A549 and H1299 cells were also
analyzed with PI and Annexin V/FITC staining (Fig. 3B and C). This assessment revealed
that miR-205-3p knockdown significantly increased the apoptotic
rates in the miR-205-3p inhibitor-transfected group compared with
the controls.
Subsequently, the protein expression levels of
cleaved caspase-3, which is closely associated with apoptosis, were
investigated by western blotting. As shown in Fig. 3D, the results suggested that
miR-305-3p knockdown could significantly increase cleaved caspase-3
expression in both NSCLC cell lines, which confirmed the outcomes
of the TUNEL staining and Annexin V/PI staining.
APBB2 is a target gene of
miR-205-3p
miRNAs can inhibit the biological functions of
target genes by regulating gene expression (23). Moreover, each miRNA can target
thousands of genes. The potential targets of miR-205-3p were
predicted using miRDB and TargetScan. Then, cell cycle-associated
genes targeted by miR-205-3p were isolated using Venn analysis
based on miRDB target genes, target genes predicted using
TargetScan, and cell cycle-associated genes found using GO
(GO:0007050, ‘cell cycle arrest’). As shown in Fig. S2, a single target gene in the
center was identified among the three programs, APBB2. Thus, it was
hypothesized that miR-205-3p may inhibit APBB2 expression by
binding to the 3′UTR. Moreover, APBB2 was found to have potential
binding ability with miR-205-3p at the 3′UTR site (Fig. 4A). Therefore, the WT or MUT 3′UTR of
APBB2 was inserted downstream of the firefly luciferase gene to
confirm this hypothesis using a dual-luciferase assay (Fig. 4B). miR-205-3p-transfected A549 and
H1299 cells were co-transfected with WT 3′UTR or MUT 3′UTR of APBB2
plasmid, and relative luciferase expression data were obtained
after 48 h. It was found that the miR-205-3p inhibitor could
significantly increase WT 3′UTR luciferase activity compared with
the miR-NC group, and no significant differences were identified
between miR-NC and MUT 3′UTR (Fig.
4B).
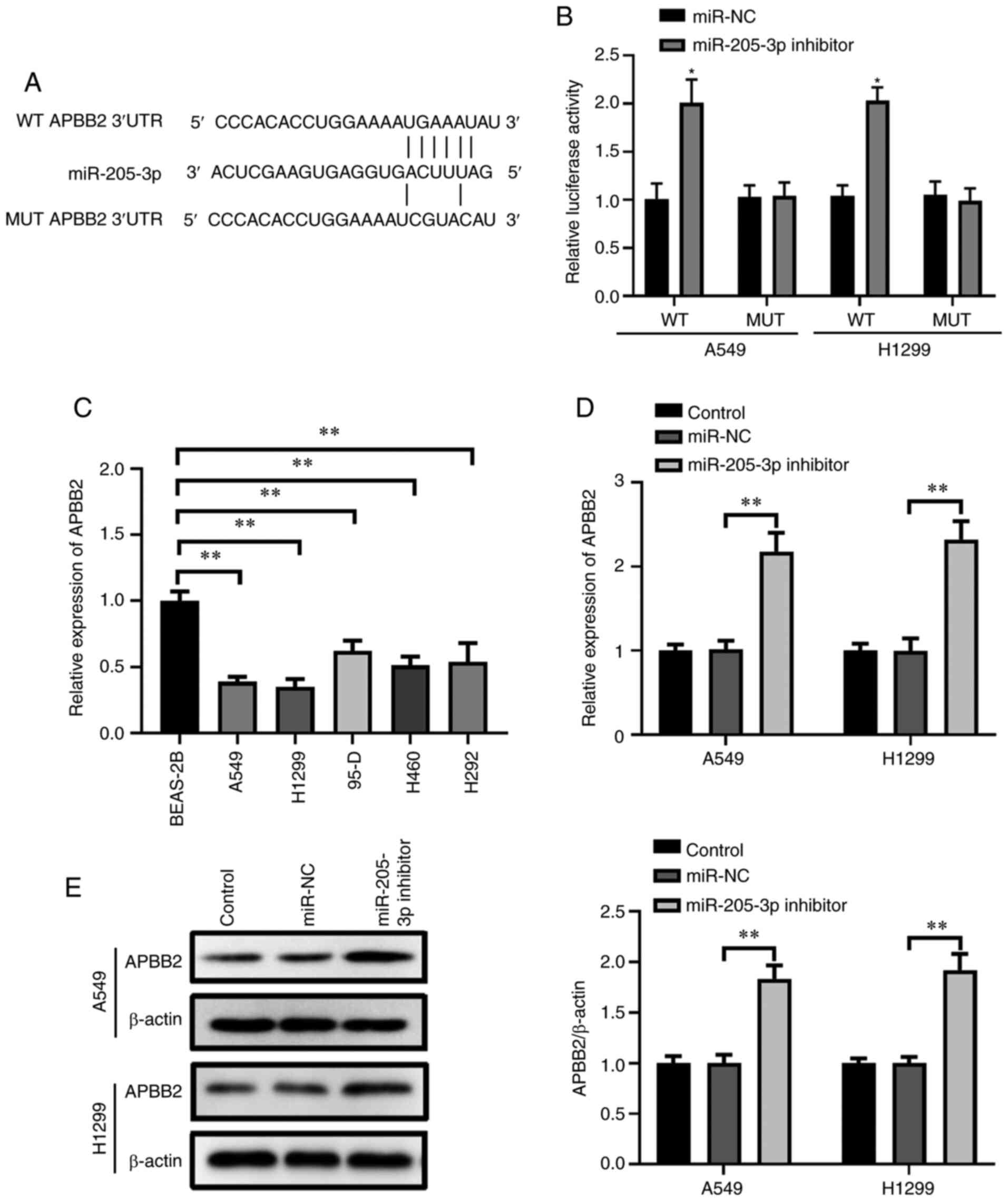 | Figure 4.APBB2 is a target gene of miR-205-3p.
(A) Predicted miR-205-3p target site in the 3′UTR of APBB2. (B)
A549 and H1299 cells were co-transfected with miR-NC or miR-205-3p
inhibitor and luciferase reporter vectors containing the WT or the
MUT 3′UTR of APBB2. Relative luciferase activity was analyzed 48 h
later. *P<0.05 vs. miR-NC. (C) Reverse
transcription-quantitative PCR results of APBB2 mRNA expression in
NSCLC cell lines (95-D, A549, H1299, H292 and H460) and normal
human lung epithelial cells (BEAS-2B). A549 and H1299 cells were
transfected with the miR-205-3p inhibitor, miR-NC or left
untreated, and APBB2 (D) mRNA and (E) protein expression levels
were analyzed. Data are presented as the mean ± SD of three
independent experiments. **P<0.01. miR, microRNA; NC, negative
control; WT, wild-type; MUT, mutant; APBB2, amyloid β precursor
protein-binding family B member 2; UTR, untranslated region. |
Based on the aforementioned findings, RT-qPCR was
conducted to further examine the potential alteration of APBB2
expression between normal human lung epithelial cells (BEAS-2B) and
NSCLC cell lines (95-D, A549, H1299, H292 and H460). The data
revealed that APBB2 mRNA expression levels in the NSCLC cell lines
were significantly lower compared with that in BEAS-2B cells
(Fig. 4C). As shown in Fig. 4D and E, after being transfected with
the miR-205-3p inhibitor, the mRNA and protein expression levels of
APBB2 were significantly increased in the A549 and H1299 cells.
These results indicated that APBB2 may be a direct target of
miR-205-3p in NSCLC.
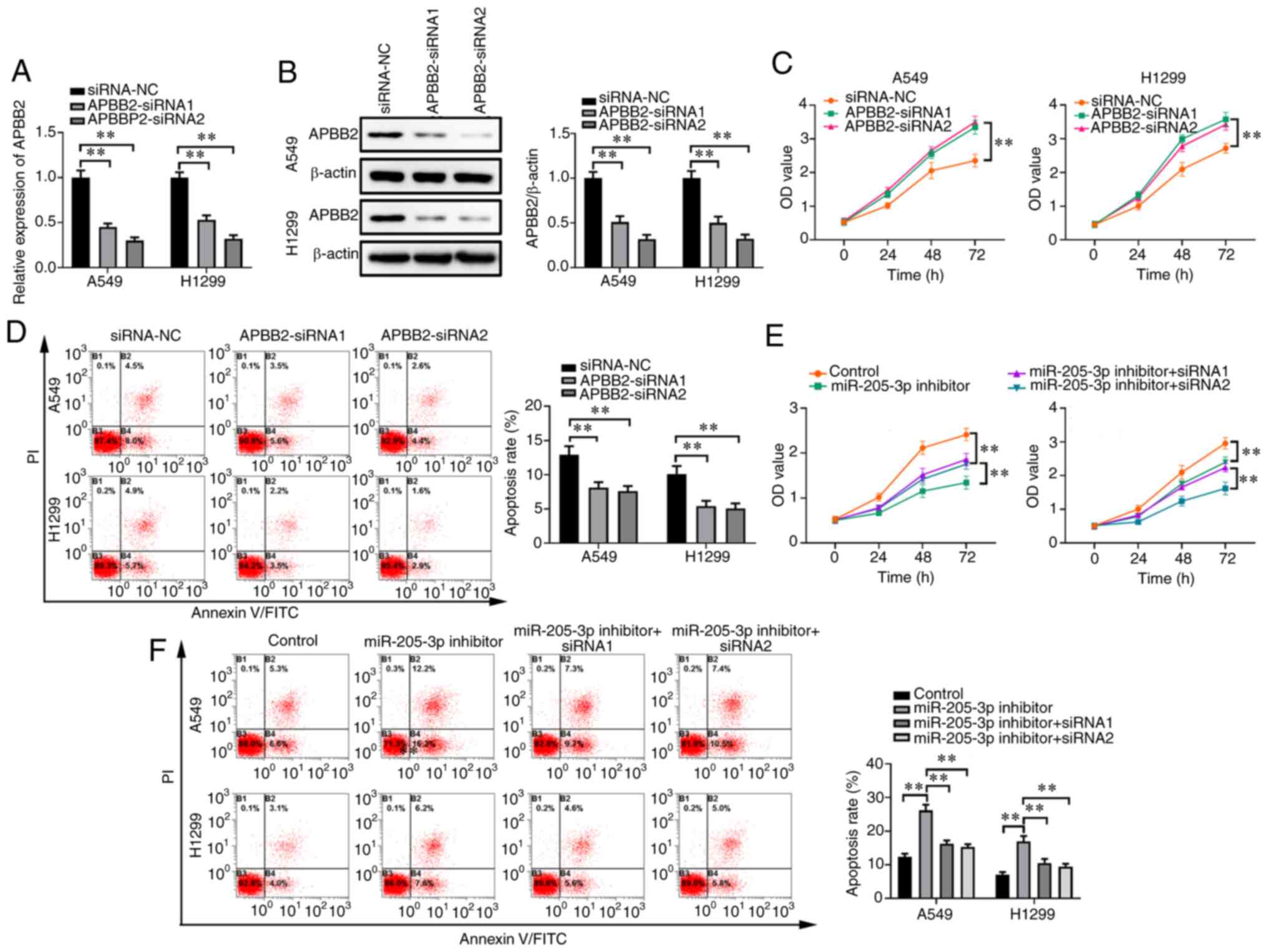
Knockdown of APBB2 promotes cell
proliferation
APBB2 genes were knocked down through transfection
with APBB2-siRNA1 or APBB2-siRNA2, which were found to
significantly decrease APBB2 mRNA and protein expression levels
(Fig. 5A and B, respectively).
After being transfected with either APBB2-siRNA, proliferation of
the A549 and H1299 cells was significantly enhanced, whereas the
apoptotic rates were decreased, compared with that of the siRNA-NC
group (Fig. 5C and D). Furthermore,
A549 and H1299 cells were co-transfected with either APBB2-siRNA
and the miR-205-3P inhibitor. It was found that APBB2-siRNA
partially alleviated miR-205-3p inhibitor-induced suppression of
cell proliferation and, to a certain degree, reversed the promotion
of apoptosis in both cell lines (Fig.
5E and F).
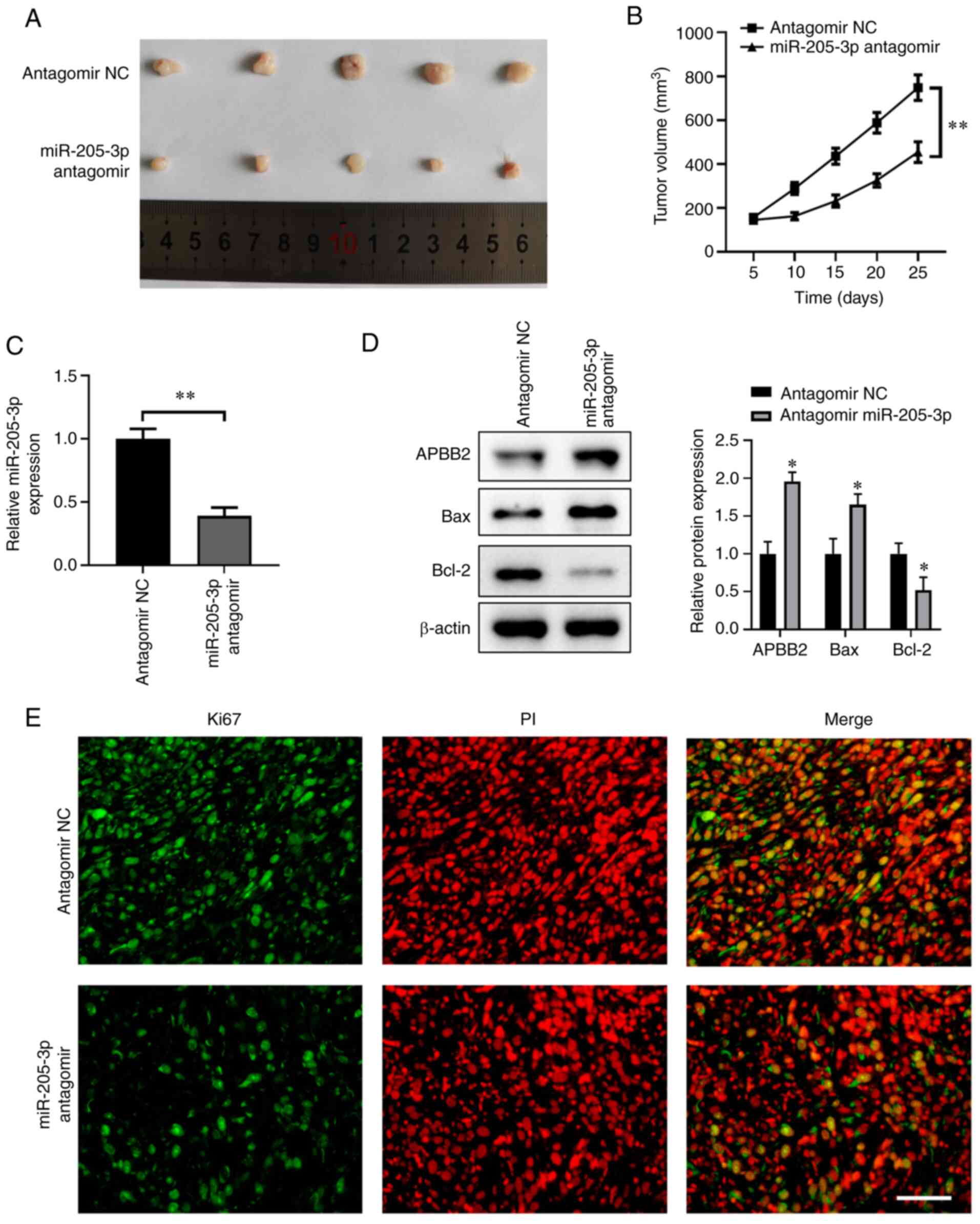
miR-205-3p antagomir inhibits lung
cancer growth and promotes apoptosis in mouse xenografts
miR-205-3p-medidated tumor suppression at the tissue
level was investigated using a miR-205-3p antagomir administered to
mouse xenograft models subcutaneously injected with A549 cells. As
shown in Fig. 6A and B, tumors
harvested from mice treated with miR-205-3p antagomir were smaller
in size and weight compared with those of the antagomir NC group.
RT-qPCR results demonstrated that the miR-205-3p antagomir could
significantly decrease miR-205-3p expression (Fig. 6C), as well as promote APBB2
expression in tumors. Moreover, the miR-205-3p antagomir induced a
significant decrease in Bcl-2 and a significant increase in Bax
protein expression levels (Fig.
6D). It was also identified that APBB2 protein expression was
upregulated after injection with miR-205-3p antagomir in the same
tumor issues (Fig. 6D).
Furthermore, the miR-205-3p antagomir could markedly inhibit cell
proliferation in the tumor, with Ki67 as the index (Fig. 6E).
Discussion
Previous studies have investigated the potential
effects of the aberrant expression levels of miRNAs on tumor
development (24–26). Given the close association between
miRNAs and tumors, accumulating evidence has revealed that
modulating miRNA expression could be an effective novel therapeutic
management of tumors (27). The
present results indicated that miR-205-3p knockdown could
significantly inhibit tumor growth and the viability of NSCLC
cells, as well as promote the apoptotic rate, at least partially,
by targeting APBB2, a tumor suppressor.
miR-205, which is located on the second intron of
the LOC642587 locus in chromosome 1, serves an important role in
various types of cancer (28–30).
In brief, as a tumor suppressor, miR-205 overexpression can notably
inhibit tumor development (31).
Moreover, the expression level of miR-205 is closely associated
with the presence of fewer melanoma tissues, and a lower expression
results in worse clinical outcomes (32,33).
Conversely, miR-205 can promote cancer formation in some cancer
types. For example, Niu et al (34) reported that miR-205 was upregulated
in ovarian cancer, and its expression level was positively
correlated with advanced clinical stages. The different roles of
miR-205 may be associated with the targets in different cancer
microenvironments. Thus, the aim of the present research was to
determine the potential relative expression levels of miR-205-3p in
cancer and normal adjacent tissues from patients with NSCLC. The
present study initially revealed that miR-205-3p expression in lung
tumor tissues was significantly higher compared with that in
adjacent normal tissues. Moreover, miR-205-3p overexpression was
associated with advanced stages (III and V) and a shorter overall
survival for patients in the clinic. Conversely, the knockdown of
miR-205-3p using a miR-205-3p inhibitor lead to increased apoptosis
in H1299 and A549 cells and a smaller tumor volume in mice.
Collectively, the current data suggested that miR-205-3p served an
essential role in NSCLC tumor development and could accelerate lung
cancer progression.
The present study further investigated the
underlying mechanism of the promotion function of miR-205-3p on
tumor development. APBB2 is an adaptor protein that binds to the
cytoplasmatic domain of βAPP (35),
and it was predicted to be the target of miR-205-3p through binding
at the 3′UTR. To verify this prediction, the present study detected
the altered expression level of APBB2 by transfecting the
miR-205-3p inhibitor into NSCLC cells and normal human lung
epithelial cells. The current data revealed that there were
significant differences in APBB2 expression between the NSCLC cells
and normal cells. Moreover, APBB2 mRNA and protein expression was
significantly elevated by the miR-205-3p inhibitor in both cellular
and tissue models compared with the control-treated group.
Collectively, these data suggested that APBB2 was a target gene of
miR-205-3p at the 3′UTR sites, and this finding was supported by
the luciferase reporter assay results.
However, there are some limitations of the present
study. First, although it was documented that miR-205-3p could
regulate the process of lung cancer via targeting APBB2, the
detailed mechanisms of how APBB2 modulates cancer cell behaviors
need further exploration. Furthermore, due to the low number of
clinical samples, the expression of APBB2 in NSCLC, and the
association between APBB2 expression and clinical features and
prognosis of patients with NSCLC remains unknown. The underlying
mechanisms of APBB2 in NSCLC will be further explored in future
studies. Despite these limitations, the findings of the current
study also provided some novel insights into our knowledge of
NSCLC.
In conclusion, the present study demonstrated that
miR-205-3p was significantly increased in NSCLC tissue and
associated with the poor prognosis of patients with NSCLC.
Mechanically, miR-205-3p promoted lung cancer progression by
targeting APBB2. These results suggested that miR-205-3p may play a
critical role in the development of NSCLC and it is of importance
to further explore the biofunction of the miR-205-3p/APBB2 axis in
NSCLC in future studies.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by The Shaanxi Provincial
Key Research and Development Project (grant no. 2017SF-023).
Availability of data and materials
The datasets used and/or analyzed during the current
study available from the corresponding author on reasonable
request.
Authors' contributions
LBX and MQ conceived and designed the experiments.
JX, YHZ, YD and XPR performed the experiments. JX, YJR, DH and SHW
analyzed and interpreted the data. JX wrote the manuscript. LBX and
MQ revised the manuscript. LBX, JX and MQ confirm the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and the study protocol was approved by the Ethics
Committee of Shaanxi Provincial People's Hospital (Shaanxi, China).
Animal care and study were approved by the Institutional Animal
Care and Use Committee of Shaanxi Provincial People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bade BC and Dela Cruz CS: Lung cancer
2020: Epidemiology, etiology, and prevention. Clin Chest Med.
41:1–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grimminger J, Richter M, Tello K, Sommer
N, Gall H and Ghofrani HA: Thin Air resulting in high pressure:
Mountain sickness and hypoxia-induced pulmonary hypertension. Can
Respir J. 2017:83816532017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ardila D, Kiraly AP, Bharadwaj S, Choi B,
Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, et al:
End-to-end lung cancer screening with three-dimensional deep
learning on low-dose chest computed tomography. Nat Med.
25:954–961. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sutendra G and Michelakis ED: The
metabolic basis of pulmonary arterial hypertension. Cell
Metabolism. 19:558–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parra V, Bravo-Sagua R, Norambuena-Soto I,
Hernández-Fuentes CP, Gómez-Contreras AG, Verdejo HE, Mellado R,
Chiong M, Lavandero S and Castro PF: Inhibition of mitochondrial
fission prevents hypoxia-induced metabolic shift and cellular
proliferation of pulmonary arterial smooth muscle cells. Biochim
Biophys Acta Mol Basis Dis. 1863:2891–2903. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farber HW and Loscalzo J: Pulmonary
arterial hypertension. N Engl J Med. 351:1655–1665. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aydin S, Kuloglu T, Aydin S, Kalayci M,
Yilmaz M, Cakmak T, Albayrak S, Gungor S, Colakoglu N and Ozercan
IH: A comprehensive immunohistochemical examination of the
distribution of the fat-burning protein irisin in biological
tissues. Peptides. 61:130–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bueno MJ, Pérez de Castro I and Malumbres
M: Control of cell proliferation pathways by microRNAs. Cell Cycle.
7:3143–3148. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu F, Song H, Zhang Y, Zhang Y, Mu Q,
Jiang M, Wang F, Zhang W, Li L, Li H, et al: Irisin induces
angiogenesis in human umbilical vein endothelial cells in vitro and
in zebrafish embryos in vivo via activation of the ERK signaling
pathway. PLoS One. 10:e01346622015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chalmers S, Saunter C, Wilson C, Coats P,
Girkin JM and McCarron JG: Mitochondrial motility and vascular
smooth muscle proliferation. Arterioscler Thromb Vasc Biol.
32:3000–3011. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li DJ, Li YH, Yuan HB, Qu LF and Wang P:
The novel exercise-induced hormone irisin protects against neuronal
injury via activation of the Akt and ERK1/2 signaling pathways and
contributes to the neuroprotection of physical exercise in cerebral
ischemia. Metabolism. 68:31–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szarka RJ, Wang N, Gordon L, Nation PN and
Smith RH: A murine model of pulmonary damage induced by
lipopolysaccharide via intranasal instillation. J Immunol Methods.
202:49–57. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duan B, Guo T, Sun H, Cai R, Rui Q and Xi
Z: miR-205 as a biological marker in non-small cell lung cancer.
Biomed Pharmacother. 91:823–930. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Su N, Qiu H, Chen Y, Yang T, Yan Q and Wan
X: miR-205 promotes tumor proliferation and invasion through
targeting ESRRG in endometrial carcinoma. Oncol Rep. 29:2297–2302.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao H, Pratt N, Mattison J, Zhao Y and
Chang NS: Characterization of an apoptosis inhibitory domain at the
C-temini of FE65-like protein. Biochem Biophys Res Commun.
276:843–850. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goodman RB, Pugin J, Lee JS and Matthay
MA: Cytokine-mediated inflammation in acute lung injury. Cytokine
Growth Factor Rev. 14:523–535. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lim JH, Lee HJ, Ho Jung M and Song J:
Coupling mitochondrial dysfunction to endoplasmic reticulum stress
response: A molecular mechanism leading to hepatic insulin
resistance. Cell Signal. 21:169–177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lim W, Ridge CA, Nicholson AG and
Mirsadraee S: The 8th lung cancer TNM classification and clinical
staging system: Review of the changes and clinical implications.
Quant Imaging Med Surg. 8:709–718. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kumar G, Goldberg SN, Wang Y, Velez E,
Gourevitch S, Galun E and Ahmed M: Hepatic radiofrequency ablation:
Markedly reduced systemic effects by modulating periablational
inflammation via cyclooxygenase-2 inhibition. Eur Radiol.
27:1238–1247. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer-a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
EI-Awady RA, Hersi F, AI-Tunaiji H, Saleh
EM, Abdel-Wahab AH, Al Homssi A, Suhail M, El-Serafi A and Al-Tel
T: Epigenetics and miRNAs as predictive markers and targets for
lung cancer chemotherapy. Cancer Biol Ther. 16:1056–1070. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Markou A, Zacridou M and Lianidou ES:
miR-21 as a novel therapeutic target in lung cnacer. Lung Cancer
(Auckl). 7:19–27. 2016.PubMed/NCBI
|
|
27
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang M, Wang J, Liu Y, Wang J, Nie Y, Si
B, Liu Y, Wang X, Chen S, Hei TK, et al: Subcellular targets of
zinc oxide nanoparticles during the aging process: Role of
cross-talk between mitochondrial dysfunction and endoplasmic
reticulum stress in the genotoxic response. Toxicol Sci. Jun
7–2019.doi: 10.1093/toxsci/kfz132 (Epub ahead of print). View Article : Google Scholar
|
|
29
|
Mazur-Bialy AI: Irisin acts as a regulator
of macrophages host defense. Life Sci. 176:21–25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park DW, Jiang S, Liu Y, Siegal GP, Inoki
K, Abraham E and Zmijewski JW: GSK3β-dependent inhibition of AMPK
potentiates activation of neutrophils and macrophages and enhances
severity of acute lung injury. Am J Physiol Lung Cell Mol Physiol.
307:L735–L745. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Batty D, Rapic'-Otrin V, Levine AS and
Wood RD: Stable binding of human XPC complex to irradiated DNA
confers strong discrimination for damaged sites. J Mol Biol.
300:275–290. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hanna JA, Hahn L, Agarwal S and Rimm DL:
In situ measurement of miR-205 in malignant melanoma tissue
supports its role as a tumor suppressor microRNA. Lab Invest.
92:1390–1397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rubenfeld GD, Caldwell E, Peabody E,
Weaver J, Martin DP, Neff M, Stern EJ and Hudson LD: Incidence and
outcomes of acute lung injury. N Engl J Med. 353:1685–1693. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Niu K, Shen W, Zhang Y, Zhao Y and Lu Y:
MiR-205 promotes motility of ovarian cancer cells via targeting
ZEB1. Gene. 574:330–336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Golanska E, Sieruta M, Gresner SM, Pfeffer
A, Chodakowska-Zebrowska M, Sobow TM, Klich I, Mossakowska M,
Szybinska A, Barcikowska M and Liberski PP: APBB2 genetic
polymorphisms are associated with severe cognitive impairment in
centenarians. Exp Gerontol. 48:391–394. 2013. View Article : Google Scholar : PubMed/NCBI
|