Introduction
Uterine leiomyoma, also known as uterine fibroid, is
the most common benign tumor, affecting ~70% of women by the age of
50 (1). These lesions disrupt the
function of the uterus and cause several health complications, such
as irregular bleeding, recurrent pregnancy loss, and pelvic
discomfort (2). Although
pathogenetic factors such as genetics, epigenetics, microRNA,
ovarian steroids, and growth factors have been implicated in the
development of leiomyoma (1), the
underlying pathogenesis remains poorly understood.
Telomeres protect chromosome ends from unnecessary
DNA repair and nucleolytic degradation (3). Human somatic cells lose telomeres
progressively over the course of cell divisions, and cells with
critically short telomeres stop dividing and enter senescence
(4). Telomere shortening is thought
to be a tumor suppressor mechanism (5), and immortal cells, such as stem cells
and transformed cells, express telomerase, thereby maintaining
their telomere length (6).
Telomerase is a ribonucleoprotein consisting of two
essential components, telomerase reverse transcriptase (TERT) and
telomerase RNA component (TERC), and several accessary proteins.
TERT, the catalytic protein component, is expressed in stem cells
and most cancer cells (7), whereas
TERC functions as a template for the synthesis of telomeric
repeats, and is ubiquitously expressed in both primary and immortal
cells (8). The telomere contains
tandem TTAGGG repeats that are associated with a multiprotein
complex known as shelterin (3).
Telomeric repeat-binding factor (TRF)1 and TRF2 are components of
shelterin, which directly bind TTAGGG repeats, protect telomeres,
and contribute to telomere maintenance by acting as negative
regulators of telomere length (9,10).
Altered expression of TRF1 and TRF2 is
frequently observed in cancer. In humans, TRF1 and
TRF2 are induced in hepatitis-induced carcinoma (11), renal cell carcinoma (12), and lung cancer (13). Recently, upregulation of TRF1
and TRF2 was also detected in patients with obesity
(14). By contrast, these genes are
downregulated in breast cancer (15). In addition, TRF1 is highly
expressed in adult stem cells and pluripotent stem cells (16), where it plays a crucial role in
maintenance of tissue homeostasis and pluripotency by protecting
telomeres regardless of telomere length (17). PIN2-interacting telomerase inhibitor
1 (PINX1), a TRF1-interacting protein, is a potent telomerase
inhibitor and tumor suppressor that is essential for maintaining
telomerase activity and chromosome stability (18). Expression of PINX1 is reduced
in several types of cancers, including breast (18) and colorectal tumors (19), and correlates with poor prognosis in
patients with ovarian cancer (20).
Meanwhile, PINX1 is upregulated in esophageal squamous cell
carcinoma, cervical squamous cell carcinoma (21), and glioma (22). Abnormal regulation of PINX1
is complex, and the function of this protein in tumorigenesis
remains unresolved.
Although uterine leiomyoma are benign tumors, some
exhibit histopathological traits that mimic malignancy, including
hypercellularity, active mitotic activity, and abnormal nuclei
(23). Telomere shortening has been
reported in uterine leiomyoma (24,25).
However, the expression levels of telomeric repeat-binding proteins
and telomerase, as well as their relationship with telomere length,
have not been evaluated in uterine leiomyoma. To address this,
telomere length and mRNA levels of genes involved in telomere
function were analyzed in normal myometrium and uterine leiomyoma,
and their relationships were examined.
Materials and methods
Tissue samples
Uterine leiomyoma tissue samples were obtained from
18 female patients (mean age, 45.7±3.03 years; age range, 37–50
years) who underwent myomectomy or hysterectomy for uterine
leiomyoma at Hanyang Hospital in Seoul (South Korea) between
October 2017 and June 2019; all patients had multiple leiomyoma.
Normal myometrium tissues were obtained from 13 of 18 patients.
Inclusion criteria included the presence of a symptomatic myoma.
Patients with organ dysfunction, for example, in the liver, kidney
and lungs, were excluded. Samples taken from the centers of
leiomyoma and from adjacent normal myometrium were immediately
snap-frozen and stored in liquid nitrogen. This study was approved
by the Institutional Review Board of Hanyang Hospital, Hanyang
University College of Medicine (approval no. 201707012), and the
requirement for informed consent was waived.
Isolation of genomic DNA
Fresh tissue homogenized using a disposable
homogenizer (BioMasher; Nippi, Inc.) was incubated with 20 µg
DNase- and protease-free RNase A (Thermo Fisher Scientific, Inc.)
at 37°C for 1 h in 500 µl lysis buffer (10 mM Tris-HCl; pH 8.0; 100
mM EDTA; 0.5% (w/v) SDS), then further digested at 50°C overnight
with 50 µg proteinase K (Invitrogen; Thermo Fisher Scientific,
Inc.). Genomic DNA was extracted using phenol/chloroform/isoamyl
alcohol (PCI; Bioneer Corporation) a total of three times. MaXtract
High-Density tubes (Qiagen GmbH) were used for PCI extractions to
prevent carryover of organic solvent, proteins, and other
contaminants. The supernatant was supplemented with ammonium
acetate to a final concentration of 2.5 M and one volume of
isopropanol. Genomic DNA spooled on a pipette tip was washed with
70% ethanol three times, air-dried and resuspended in water.
Quantification of DNA was performed using a NanoDrop™ 2000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.).
Preparation of digoxigenin
(DIG)-labeled probes
Oligonucleotides were labeled at the 3′-end by
incorporation of a single DIG-labeled ddUTP (Roche Diagnostics).
Briefly, 100–200 pmol oligonucleotides were incubated at 37°C for
60 min with 1 µl of 1 mM DIG-ddUTP and 20 units terminal
deoxynucleotidyl transferase (Roche Diagnostics) in 1X reaction
buffer and 5 mM CoCl2 in a total volume of 20 µl, and
the reaction was stopped by addition of 2 µl of 0.2 M EDTA (pH
8.0).
Southern blot analysis of terminal
restriction fragment lengths
Southern hybridization was performed to measure
telomere length. Briefly, 5 µg HinFI-cut DNA was
fractionated on an 0.8% (w/v) agarose gel and blotted by capillary
transfer onto a nylon membrane (Hybond™ N+; GE Healthcare
Biosciences) in 10X saline-sodium citrate (SSC) buffer. Blots were
UV-crosslinked (Spectronics), pre-hybridized in DIG Easy Hyb (Roche
Diagnostics) at 45°C for 1 h, and hybridized with DIG-labeled
d(TTAGGG)4 at the same temperature overnight. The
membrane washing and antibody reaction were carried out at room
temperature as follows. The membranes were washed in 2X SSC with
0.1% SDS for 15 min twice, in 0.5X SSC with 0.1% SDS for 15 min
twice, and then in 1X maleic acid buffer (MAB; 0.1 M maleic acid;
0.15 M NaCl; pH 7.5) for 5 min. The blots were immersed in 1X
Blocking Solution (Roche Diagnostics; diluted in 1X MAB) for 30
min, then incubated with an anti-DIG antibody conjugated to
alkaline phosphatase (cat. no. 11093274910; 1:10,000; Roche;
MilliporeSigma; Merck KGaA) in 1X Blocking Solution for 60 min. The
membranes were washed in 1X washing buffer (0.3% Tween-20 in 1X
MAB) for 15 min twice, then in detection buffer (0.1 M NaCl; 0.1 M
Tris-HCl, pH 9.5) at room temperature for 5 min. CDP-Star (Roche
Diagnostics) was used as a chemiluminescence substrate, and the
hybridization signal was detected by scanning using a ChemiDoc XRS+
image analysis system (Bio-Rad Laboratories, Inc.). For the next
round of hybridization, the membrane was rinsed thoroughly with
sterile water, washed at 37°C in stripping buffer (0.2 N NaOH; 0.1%
SDS) for 15 min twice, then rinsed with 2X SSC buffer for 5 min.
The de-probed blot was rehybridized with the DIG-labeled
d(CAC)8 probe. Telomere length (TL) was calculated as
previously described (26).
Briefly, the telomere signal in each lane was quantified in a grid
object defined as a single column with 20 rows (1×20 grid over the
lane ranging from 3.5 to 21 kb), using the Image Lab software
(version 6; Bio-Rad Laboratories, Inc.), and the TL was defined as
∑(MWi × ODi)/∑(ODi), where
ODi is the optical density and MWi is the
molecular weight of the DNA at the ith position.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues using Tri-RNA
(Favorgen) or TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Then, 1 µg RNA was incubated with 2 units
RNase-free DNase I (New England Biolabs, Inc.) and 40 units RNase
inhibitor (Takara Bio, Inc.) with 2 mM DTT in a total volume of 1
µl at 37°C for 30 min, heat-inactivated at 75°C for 10 min, then
subjected to cDNA synthesis. First-strand cDNA synthesis was
carried out in 20-µl volumes containing 6 µM random hexamers
(Takara Bio, Inc.) and 200 units Moloney murine leukemia virus
(M-MLV) reverse transcriptase (Thermo Fisher Scientific, Inc.) at
37°C for 50 min. To ensure that genomic DNA was completely removed,
the RT reaction mixture without M-MLV reverse transcriptase was
subjected to PCR with TERC-specific primers. PCR was
performed in duplicate with 1 µl cDNA template, 0.2 µM primers
(Macrogen), and 1X LightCycler® 480 SYBR-Green I Master
mix (Roche Diagnostics) using the LightCycler® 480
system (Roche Diagnostics). Thermocycling conditions were as
follows: i) 95°C for 5 min; ii) 40 cycles of 95°C for 15 sec and
60°C for 1 min; iii) a dissociation stage of 95°C for 5 sec; 65°C
for 1 min; and iv) 40°C for 30 sec. GAPDH was used as the
reference gene, and sequences of the primers used in the present
study are listed in Table I
(27–30). Expression levels were quantified
using the 2−∆∆Cq method (31).
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| First author,
year | Primer | Sequence (5–3) | (Refs.) |
|---|
| Scheibe et
al, 2013 |
TRF1-forward |
TGCTTTCAGTGGCTCTTCTG | (27) |
| Scheibe et
al, 2013 |
TRF1-reverse |
ATGGAACCCAGCAACAAGAC | (27) |
| Scheibe et
al, 2013 |
TRF2-forward |
TTGTGGGGTCCTTGGACATA | (27) |
| Scheibe et
al, 2013 |
TRF2-reverse |
CCAGTAGAAAACTGGTCAAGGAA | (27) |
| Present study |
PINX1-forward |
CACTCCAGAGGAGAACGAAACC | – |
| Present study |
PINX1-reverse |
CACCGGCTTGGCAAAGTACT | – |
| Park et al,
2008 |
TERT-forward |
TGTGCACCAACATCTACAAG | (28) |
| Park et al,
2008 |
TERT-reverse |
GCGTTCTTGGCTTTCAGGAT | (28) |
| Lundberg et
al, 2002 |
TERC-forward |
TCTAACCCTAACTGAGAAGGGCGTAG | (29) |
| Lundberg et
al, 2002 |
TERC-reverse |
GTTTGCTCTAGAATGAACGGTGGAAG | (29) |
| Deng et al,
2012 |
GAPDH-forward |
AGCCACATCGCTCAGACAC | (30) |
| Deng et al,
2012 |
GAPDH-reverse |
GCCCAATACGACCAAATCC | (30) |
Statistical analysis
The data were obtained from a single experiment and
data are presented as the mean ± standard deviation. Statistical
analysis was performed using SPSS software (v. 24, IBM, Corp.) and
assessed using an unpaired Student's t-test and the Pearson
correlation. P<0.05 was considered to indicate a statistically
significant difference.
Results
Telomere shortening in uterine
leiomyoma
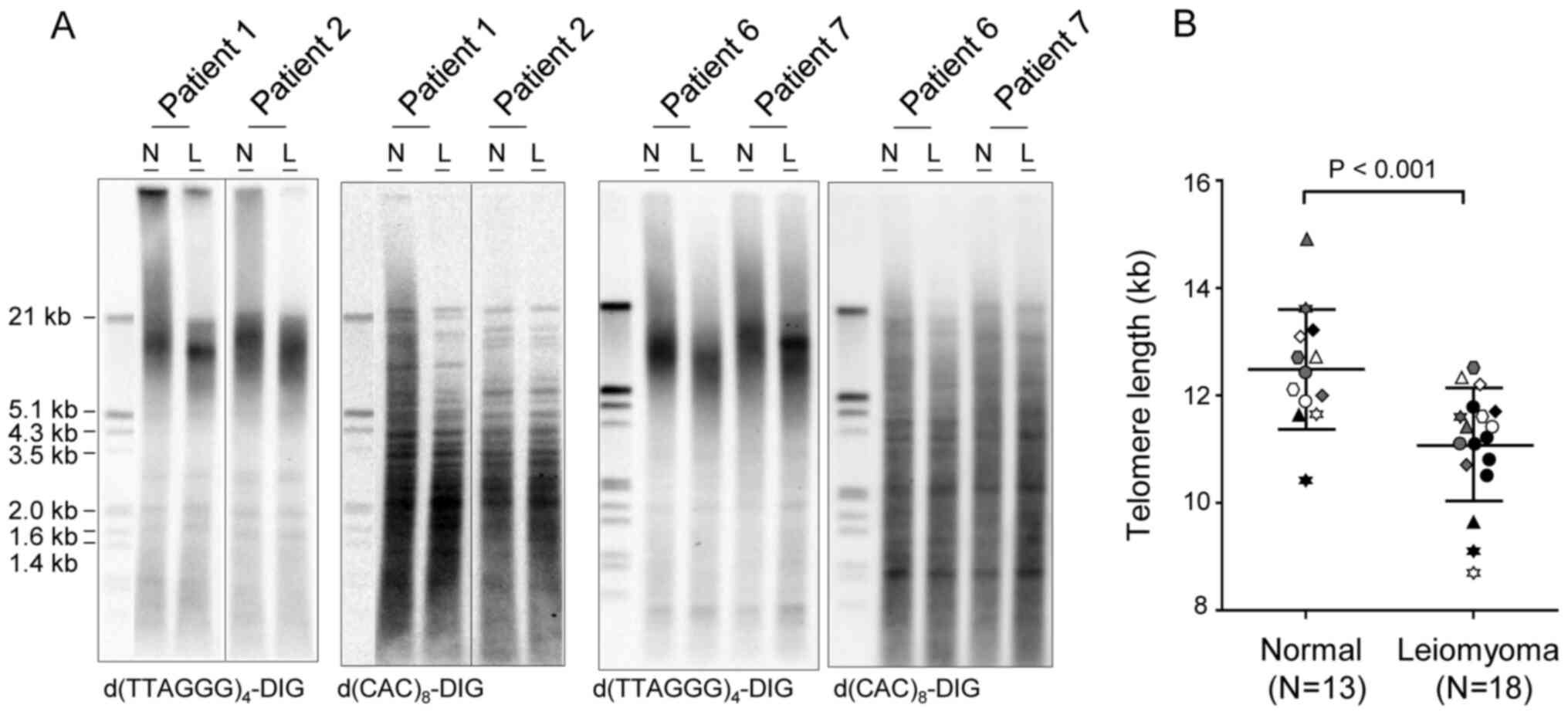
In this study, terminal restriction fragment lengths
were measured using Southern blotting in leiomyoma (n=18) and
adjacent normal myometrium (n=13) tissue. Of note, five of the
leiomyoma did not have matched normal tissues (Table II). Hybridization with the
telomere-specific 3′-DIG-labeled d(TTAGGG)4 probe
indicated that telomere length ranged from 10.4 to 14.9 kb in
normal myometrium and from 8.7 to 12.5 kb in leiomyoma (Fig. 1 and Table II). Leiomyoma samples presented
shorter telomeres than the normal myometrium (Fig. 1B). Mean telomere lengths in normal
and leiomyoma tissue samples were 12.5±1.11 (n=13) and 11.1±1.04 kb
(n=18), respectively (P<0.001; Fig.
1B and Table II). There were
no significant differences in telomere length related to patient
age, possibly due to the narrow range of patient ages (37–50 years)
(data not shown). Telomere lengths in multiple leiomyoma samples
from a single patient (Table II;
patients no. 7, 8, 9) were also measured, but no obvious variations
in lengths was observed among measurements. Membranes re-probed
with a 3′-DIG-labeled d(CAC)8 probe for minisatellite
DNA confirmed the integrity of genomic DNA. A minisatellite is a
tract of repetitive DNA sequences that are present throughout the
entire genome and often used for the fingerprinting of DNA
(32). Indeed, hybridization with
d(CAC)8 probe showed different hybridization patterns by
patients (Fig. 1A).
 | Table II.Telomere length in normal myometrium
and leiomyoma tissue. |
Table II.
Telomere length in normal myometrium
and leiomyoma tissue.
|
| Telomere length,
kb |
|
|
|---|
|
|
|
|
|
|---|
| Patient no. | Normal | Leiomyoma | Telomere
shorteninga, kb | Age, years |
|---|
| 1 | 13.6 | 11.6 | −2.0 | 44 |
| 2 | 13.1 | 12.2 | −0.9 | 45 |
| 3 | 11.6 | 8.7 | −2.9 | 47 |
| 4 | 12.4 | 11.1 | −1.3 | 37 |
| 5 | 12.7 | 12.5 | −0.2 | 48 |
| 6 | 11.6 | 9.6 | −2.0 | 47 |
| 7 | 12.0 | 10.7 | −1.3 | 48 |
|
| 12.3* | 10.1* |
| 8 | 12.7 | 12.2 | −0.5 | 50 |
|
| 12.1* | 11.7* |
|
|
|
|
| 11.4* |
|
|
| 9 | 11.9 | 11.4 | −0.5 | 49 |
|
| 12.5* | 11.8* |
|
|
|
|
| 12.2* |
|
|
| 10 | 10.4 | 9.1 | −1.3 | 46 |
| 11 | 12.1 | 11.6 | −0.5 | 48 |
| 12 | 14.9 | 11.4 | −3.5 | 45 |
| 13 | 13.2 | 11.7 | −1.5 | 44 |
| 14 |
| 11.1 |
| 43 |
| 15 |
| 10.5 |
| 43 |
| 16 |
| 11.2 |
| 44 |
| 17 |
| 10.8 |
| 48 |
| 18 |
| 11.8 |
| 47 |
| Total |
12.5±1.11b |
11.1±1.04b |
−1.4±0.98b |
45.7±3.03b |
|
| (n=13) | (n=18) | (n=13) | (n=18) |
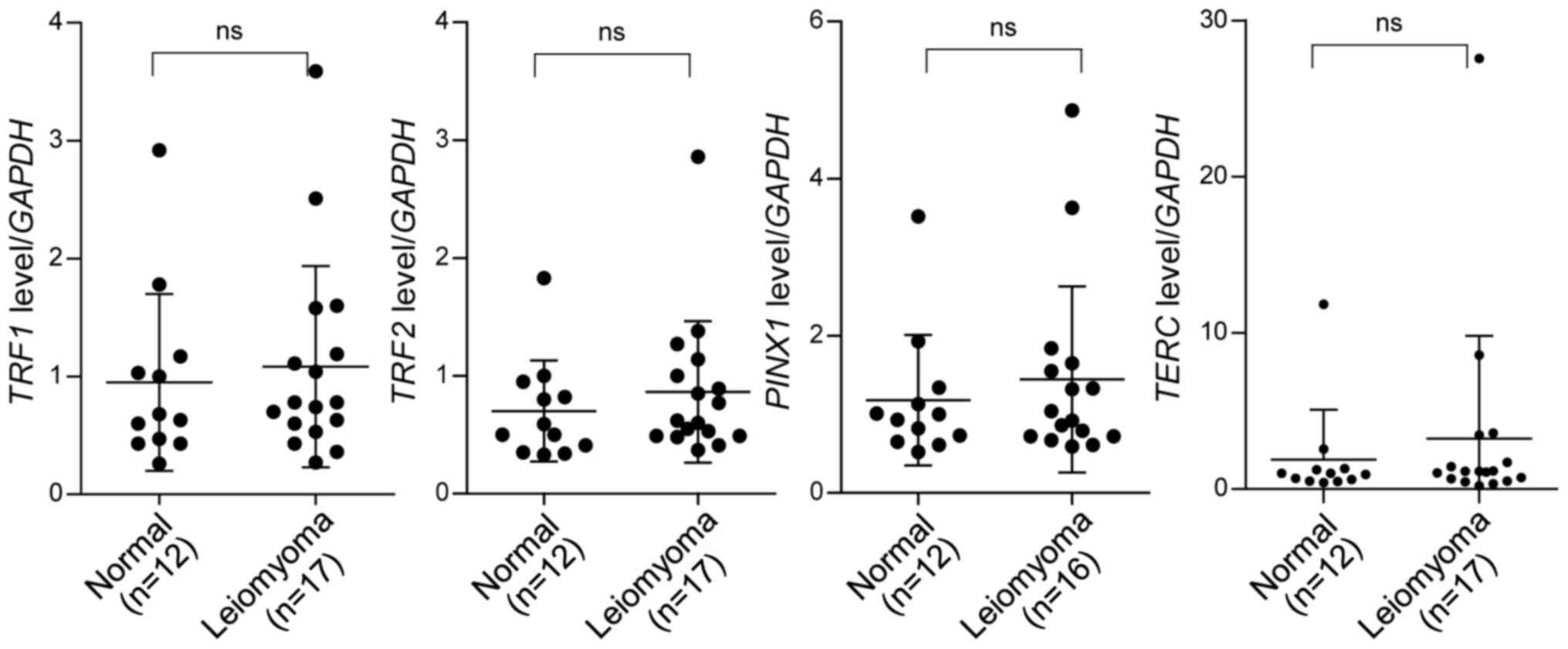
Expression of TRF1, TRF2 and PINX1 in
leiomyoma
TRF1 and TRF2, components of shelterin, protect
telomeres and regulate telomere length. Depletion or deletion of
these genes induces a persistent DNA damage response at telomeres,
resulting in cessation of cell division and induction of apoptosis
or senescence (16,33–35).
Meanwhile, overexpression of TRF1, TRF2 and PINX1
results in telomere shortening in telomerase-positive cells
(9,10,36),
ultimately leading to induction of cell crisis which is
characterized by the reduction in growth rate. To determine whether
the expression of TRF1, TRF2 and PINX1 varied during
the development of leiomyoma, the mRNA levels of these genes in
leiomyoma (n=17) and normal tissues (n=12) were determined. RT-qPCR
results suggested that these genes were expressed at similar levels
in normal and leiomyoma nodules (Fig.
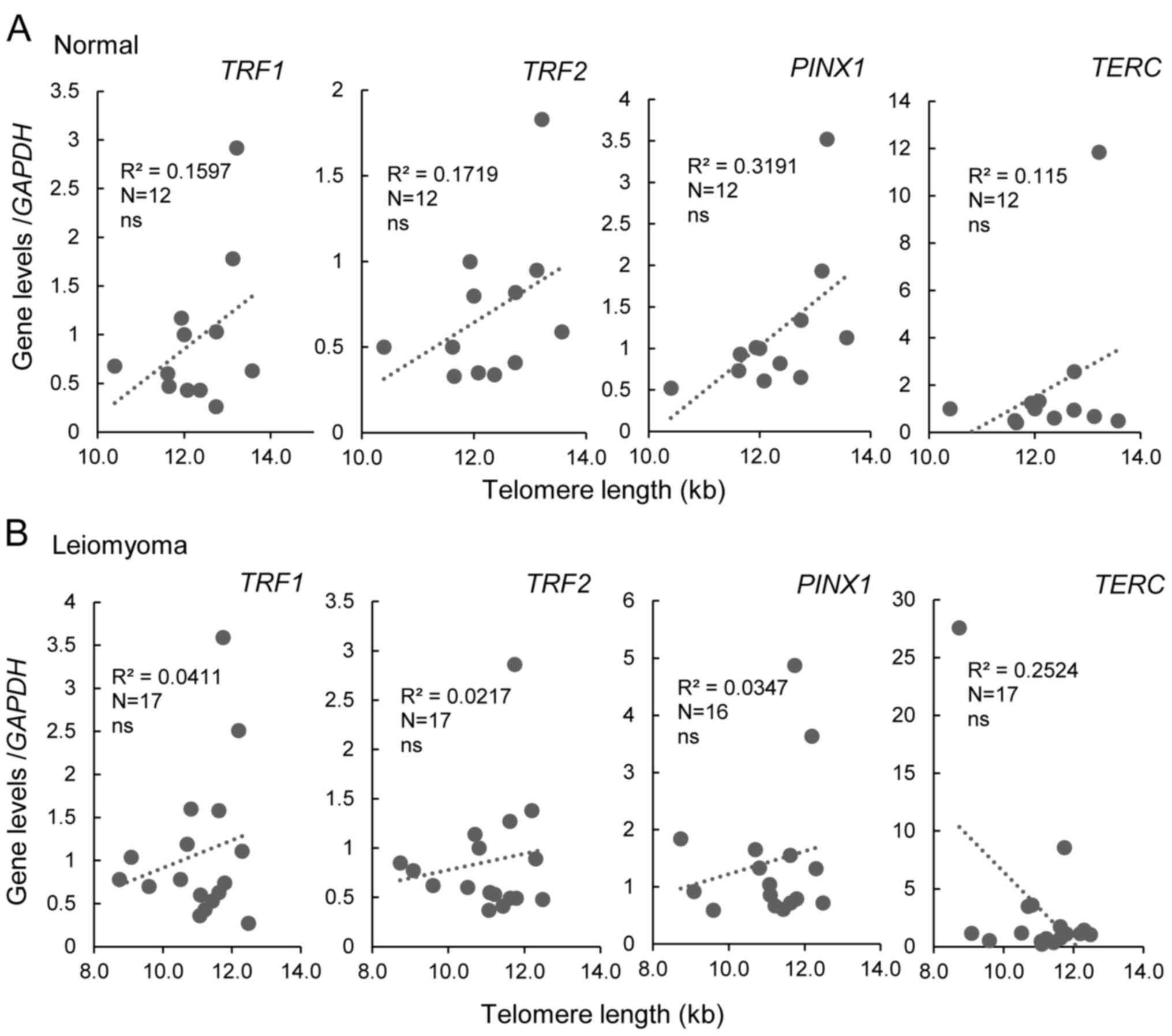
2). To determine whether there was a possible association
between gene expression and telomere length, Pearson's correlation
was used (Fig. 3). The expression
of TRF1, TRF2 and PINX1 did not correlate with
telomere length in either normal myometrium or leiomyoma (Fig. 3). Moreover, the mRNA levels of
TERT and TERC, the essential components of
telomerase, were also measured. TERC was expressed at
comparable levels in leiomyoma and normal tissue samples,
independently of telomere length (Figs.
2 and 3), whereas TERT
expression was negative in all samples tested (data not shown).
Discussion
Uterine leiomyoma accounts for the majority of
hysterectomies and is associated with substantial morbidity, such
as excessive uterine bleeding, anemia, pelvic discomfort and
recurrent pregnancy loss, in women of reproductive age (1). Development of leiomyoma proceeds
through a multistep process involving the transformation of normal
myocytes into abnormal ones and their growth into tumors (1). Accumulating evidence suggests that
some intrinsic abnormalities of the myometrium, abnormal myometrial
receptors for estrogen, and hormonal changes or altered responses
to ischemic damage during the menstrual period may be responsible
for the initiation of (epi)genetic changes found in uterine myoma
(37,38). However, the pathogenesis of
leiomyoma remains largely unclear. In this study, telomere
shortening was clearly identified as a key event associated with
leiomyoma. Leiomyoma samples displayed shorter telomeres, ranging
from a few hundred bases to several kilobases, than adjacent normal
tissues. However, most leiomyoma had telomeres of ≥10 kb. In fact,
critically short telomeres <5 kb, which are frequently found in
malignant tumors (11), were not
detected in leiomyoma. These observations suggested that
significant levels of cell division occur during progression to
leiomyoma, but telomeres are long enough to maintain their
integrity within the tumors.
Southern blot analysis was employed in this study to
measure telomere length, and the use of this technique is limited
in studies involving large numbers of samples. The quantitative PCR
technique which provides relative telomere length (RTL) and
requires a small number of cells is widely used in epidemiological
studies (39,40). In fact, the RTLs have been
successfully measured in circulating serum DNA from patients with
endometrial cancer (41,42). Further study is needed to assess the
RTL in the serum of a large number of uterine leiomyoma patients,
which will provide a better understanding of whether telomere
length is a diagnostic marker for early detection of uterine
leiomyoma.
There were no significant alterations in the levels
of TRF1, TRF2 or PINX1 in uterine leiomyoma, and the
expression of these genes did not correlate with telomere length.
Our findings are in accordance with a previous study that reported
low but constant levels of TRF1 and TRF2 expression
at early stages in human hepatocarcinogenesis (normal, chronic
hepatitis, liver cirrhosis, and large regenerative nodule) and
shortening of telomeres with disease progression (11). It may be hypothesized that the
steady-state expression of telomere protection genes such as
TRF1, TRF2 and PINX1 is important for the maintenance
of telomere integrity in benign tumors, which may impede tumor
progression to malignancy. In addition, unsurprisingly, leiomyoma
remained telomerase-negative as revealed by RT-qPCR for
TERT, confirming that cells in this tumor did not acquire
immortality. Several studies have demonstrated that TERC
levels are upregulated in early preneoplastic stages (43–45).
This phenomenon, however, was not detected in leiomyoma
progression. TERC is essential for telomere maintenance in
telomerase-positive cells, but the telomerase- and
telomere-independent functions of TERC remain elusive.
TERC may function as a noncoding RNA that prevents apoptosis
in normal telomerase-negative cells (46).
In conclusion, expression levels of genes essential
for telomere protection were maintained during neoplastic
transformation of myometrium to leiomyoma, and telomere shortening
was evident during this process. Persistent expression of telomere
protection genes may lead to maintenance of telomere integrity in
leiomyoma. These results provide insight into the progression of
normal tissue to benign tumors.
Acknowledgements
We thank Dr Baikseol Cho (Department of Obstetrics
and Gynecology, Hanyang University College of Medicine, Seoul,
South Korea) for her valuable input on Institutional Review Board
approval.
Funding
This work was supported by the Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education (grant no. 2019R1I1A1A01041367)
and the Ministry of Science and Information and Communication
Technology (grant no. 2017R1A2B4004721).
Availability of data and materials
The data are available from the corresponding author
on reasonable request.
Authors' contributions
BKO and JSC conceived and designed the study and BKO
drafted the manuscript. BKO and YC performed the experiments and
analyzed the data. All authors confirm the authenticity of all the
raw data, and read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional Review
Board of Hanyang Hospital, Hanyang University College of Medicine
(approval no. 201707012). The requirement for informed consent was
waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bulun SE: Uterine fibroids. N Engl J Med.
369:1344–1355. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Al-Hendy A, Myers ER and Stewart E:
Uterine Fibroids: Burden and Unmet Medical Need. Semin Reprod Med.
35:473–480. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Lange T: Shelterin-Mediated Telomere
Protection. Annu Rev Genet. 52:223–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim NW, Piatyszek MA, Prowse KR, Harley
CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL and Shay
JW: Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shay JW and Wright WE: Role of telomeres
and telomerase in cancer. Semin Cancer Biol. 21:349–353. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Counter CM, Hirte HW, Bacchetti S and
Harley CB: Telomerase activity in human ovarian carcinoma. Proc
Natl Acad Sci USA. 91:2900–2904. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakayama J, Tahara H, Tahara E, Saito M,
Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T and Ishikawa F:
Telomerase activation by hTRT in human normal fibroblasts and
hepatocellular carcinomas. Nat Genet. 18:65–68. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Avilion AA, Piatyszek MA, Gupta J, Shay
JW, Bacchetti S and Greider CW: Human telomerase RNA and telomerase
activity in immortal cell lines and tumor tissues. Cancer Res.
56:645–650. 1996.PubMed/NCBI
|
|
9
|
van Steensel B and de Lange T: Control of
telomere length by the human telomeric protein TRF1. Nature.
385:740–743. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Steensel B, Smogorzewska A and de
Lange T: TRF2 protects human telomeres from end-to-end fusions.
Cell. 92:401–413. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oh BK, Kim YJ, Park C and Park YN:
Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is
related to telomere shortening during human multistep
hepatocarcinogenesis. Am J Pathol. 166:73–80. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pal D, Sharma U, Singh SK, Kakkar N and
Prasad R: Over-expression of telomere binding factors (TRF1 &
TRF2) in renal cell carcinoma and their inhibition by using SiRNA
induce apoptosis, reduce cell proliferation and migration invitro.
PLoS One. 10:e01156512015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nakanishi K, Kawai T, Kumaki F, Hiroi S,
Mukai M, Ikeda E, Koering CE and Gilson E: Expression of mRNAs for
telomeric repeat binding factor (TRF)-1 and TRF2 in atypical
adenomatous hyperplasia and adenocarcinoma of the lung. Clin Cancer
Res. 9:1105–1111. 2003.PubMed/NCBI
|
|
14
|
Grun LK, Teixeira ND Jr, Mengden LV, de
Bastiani MA, Parisi MM, Bortolin R, Lavandoski P, Pierdoná V, Alves
LB, Moreira JC, et al: TRF1 as a major contributor for telomeres'
shortening in the context of obesity. Free Radic Biol Med.
129:286–295. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saito K, Yagihashi A, Nasu S, Izawa Y,
Nakamura M, Kobayashi D, Tsuji N and Watanabe N: Gene expression
for suppressors of telomerase activity (telomeric-repeat binding
factors) in breast cancer. Jpn J Cancer Res. 93:253–258. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schneider RP, Garrobo I, Foronda M,
Palacios JA, Marión RM, Flores I, Ortega S and Blasco MA: TRF1 is a
stem cell marker and is essential for the generation of induced
pluripotent stem cells. Nat Commun. 4:19462013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bejarano L, Schuhmacher AJ, Méndez M,
Megías D, Blanco-Aparicio C, Martínez S, Pastor J, Squatrito M and
Blasco MA: Inhibition of TRF1 Telomere Protein Impairs Tumor
Initiation and Progression in Glioblastoma Mouse Models and
Patient-Derived Xenografts. Cancer Cell. 32:590–607.e4. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou XZ, Huang P, Shi R, Lee TH, Lu G,
Zhang Z, Bronson R and Lu KP: The telomerase inhibitor PinX1 is a
major haploinsufficient tumor suppressor essential for chromosome
stability in mice. J Clin Invest. 121:1266–1282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deng W, Jiao N, Li N, Wan X, Luo S and
Zhang Y: Decreased expression of PinX1 protein predicts poor
prognosis of colorectal cancer patients receiving 5-FU adjuvant
chemotherapy. Biomed Pharmacother. 73:1–5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai MY, Zhang B, He WP, Yang GF, Rao HL,
Rao ZY, Wu QL, Guan XY, Kung HF, Zeng YX, et al: Decreased
expression of PinX1 protein is correlated with tumor development
and is a new independent poor prognostic factor in ovarian
carcinoma. Cancer Sci. 101:1543–1549. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qian D, Zhang B, He LR, Cai MY, Mai SJ,
Liao YJ, Liu YH, Lin MC, Bian XW, Zeng YX, et al: The
telomere/telomerase binding factor PinX1 is a new target to improve
the radiotherapy effect of oesophageal squamous cell carcinomas. J
Pathol. 229:765–774. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bai J, Chen YS, Mei PJ, Liu QH, Du Y and
Zheng JN: PinX1 is up-regulated and associated with poor patients'
survival in gliomas. Int J Clin Exp Pathol. 8:6952–6959.
2015.PubMed/NCBI
|
|
23
|
Oliva E, Carcangiu M, Carinelli SG, Ip P,
Loening T and Longacre TA: Mesenchymal tumours. WHO Classification
of Tumours of Female Reproductive Organs. 4th edition. Kurman RJ,
Carcangiu ML, Herrington CS and Young RH: IARC Press; Lyon: pp.
135–138. 2014
|
|
24
|
Bonatz G, Frahm SO, Andreas S, Heidorn K,
Jonat W and Parwaresch R: Telomere shortening in uterine
leiomyomas. Am J Obstet Gynecol. 179:591–596. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rogalla P, Rohen C, Hennig Y, Deichert U,
Bonk U and Bullerdiek J: Telomere repeat fragment sizes do not
limit the growth potential of uterine leiomyomas. Biochem Biophys
Res Commun. 211:175–182. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kruk PA, Rampino NJ and Bohr VA: DNA
damage and repair in telomeres: Relation to aging. Proc Natl Acad
Sci USA. 92:258–262. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scheibe M, Arnoult N, Kappei D, Buchholz
F, Decottignies A, Butter F and Mann M: Quantitative interaction
screen of telomeric repeat-containing RNA reveals novel TERRA
regulators. Genome Res. 23:2149–2157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park IH, Zhao R, West JA, Yabuuchi A, Huo
H, Ince TA, Lerou PH, Lensch MW and Daley GQ: Reprogramming of
human somatic cells to pluripotency with defined factors. Nature.
451:141–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lundberg AS, Randell SH, Stewart SA,
Elenbaas B, Hartwell KA, Brooks MW, Fleming MD, Olsen JC, Miller
SW, Weinberg RA, et al: Immortalization and transformation of
primary human airway epithelial cells by gene transfer. Oncogene.
21:4577–4586. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deng Z, Wang Z, Stong N, Plasschaert R,
Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei
MS, et al: A role for CTCF and cohesin in subtelomere chromatin
organization, TERRA transcription, and telomere end protection.
EMBO J. 31:4165–4178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeffreys AJ, Wilson V and Thein SL:
Hypervariable ‘minisatellite’ regions in human DNA. Nature.
314:67–73. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karlseder J, Broccoli D, Dai Y, Hardy S
and de Lange T: p53- and ATM-dependent apoptosis induced by
telomeres lacking TRF2. Science. 283:1321–1325. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Celli GB and de Lange T: DNA processing is
not required for ATM-mediated telomere damage response after TRF2
deletion. Nat Cell Biol. 7:712–718. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martínez P, Thanasoula M, Muñoz P, Liao C,
Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M
and Blasco MA: Increased telomere fragility and fusions resulting
from TRF1 deficiency lead to degenerative pathologies and increased
cancer in mice. Genes Dev. 23:2060–2075. 2009. View Article : Google Scholar
|
|
36
|
Zhou XZ and Lu KP: The
Pin2/TRF1-interacting protein PinX1 is a potent telomerase
inhibitor. Cell. 107:347–359. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Laganà AS, Vergara D, Favilli A, La Rosa
VL, Tinelli A, Gerli S, Noventa M, Vitagliano A, Triolo O,
Rapisarda AM, et al: Epigenetic and genetic landscape of uterine
leiomyomas: A current view over a common gynecological disease.
Arch Gynecol Obstet. 296:855–867. 2017. View Article : Google Scholar
|
|
38
|
Yang Q, Mas A, Diamond MP and Al-Hendy A:
The mechanism and function of epigenetics in uterine leiomyoma
development. Reprod Sci. 23:163–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cawthon RM: Telomere measurement by
quantitative PCR. Nucleic Acids Res. 30:e472002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tarik M, Ramakrishnan L, Sachdev HS,
Tandon N, Roy A, Bhargava SK and Pandey RM: Validation of
quantitative polymerase chain reaction with Southern blot method
for telomere length analysis. Future Sci OA. 4:FSO2822018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun Y, Zhang L, Zhao L, Wu X and Gu J:
Association of leukocyte telomere length in peripheral blood
leukocytes with endometrial cancer risk in Caucasian Americans.
Carcinogenesis. 36:1327–1332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Benati M, Montagnana M, Danese E, Mazzon
M, Paviati E, Garzon S, Laganà AS, Casarin J, Giudici S, Raffaelli
R, et al: Aberrant telomere length in circulating cell-free DNA as
possible blood biomarker with high diagnostic performance in
endometrial cancer. Pathol Oncol Res. 26:2281–2289. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Blasco MA, Rizen M, Greider CW and Hanahan
D: Differential regulation of telomerase activity and telomerase
RNA during multi-stage tumorigenesis. Nat Genet. 12:200–204. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yashima K, Milchgrub S, Gollahon LS,
Maitra A, Saboorian MH, Shay JW and Gazdar AF: Telomerase enzyme
activity and RNA expression during the multistage pathogenesis of
breast carcinoma. Clin Cancer Res. 4:229–234. 1998.PubMed/NCBI
|
|
45
|
Yi X, Tesmer VM, Savre-Train I, Shay JW
and Wright WE: Both transcriptional and posttranscriptional
mechanisms regulate human telomerase template RNA levels. Mol Cell
Biol. 19:3989–3997. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gazzaniga FS and Blackburn EH: An
antiapoptotic role for telomerase RNA in human immune cells
independent of telomere integrity or telomerase enzymatic activity.
Blood. 124:3675–3684. 2014. View Article : Google Scholar : PubMed/NCBI
|