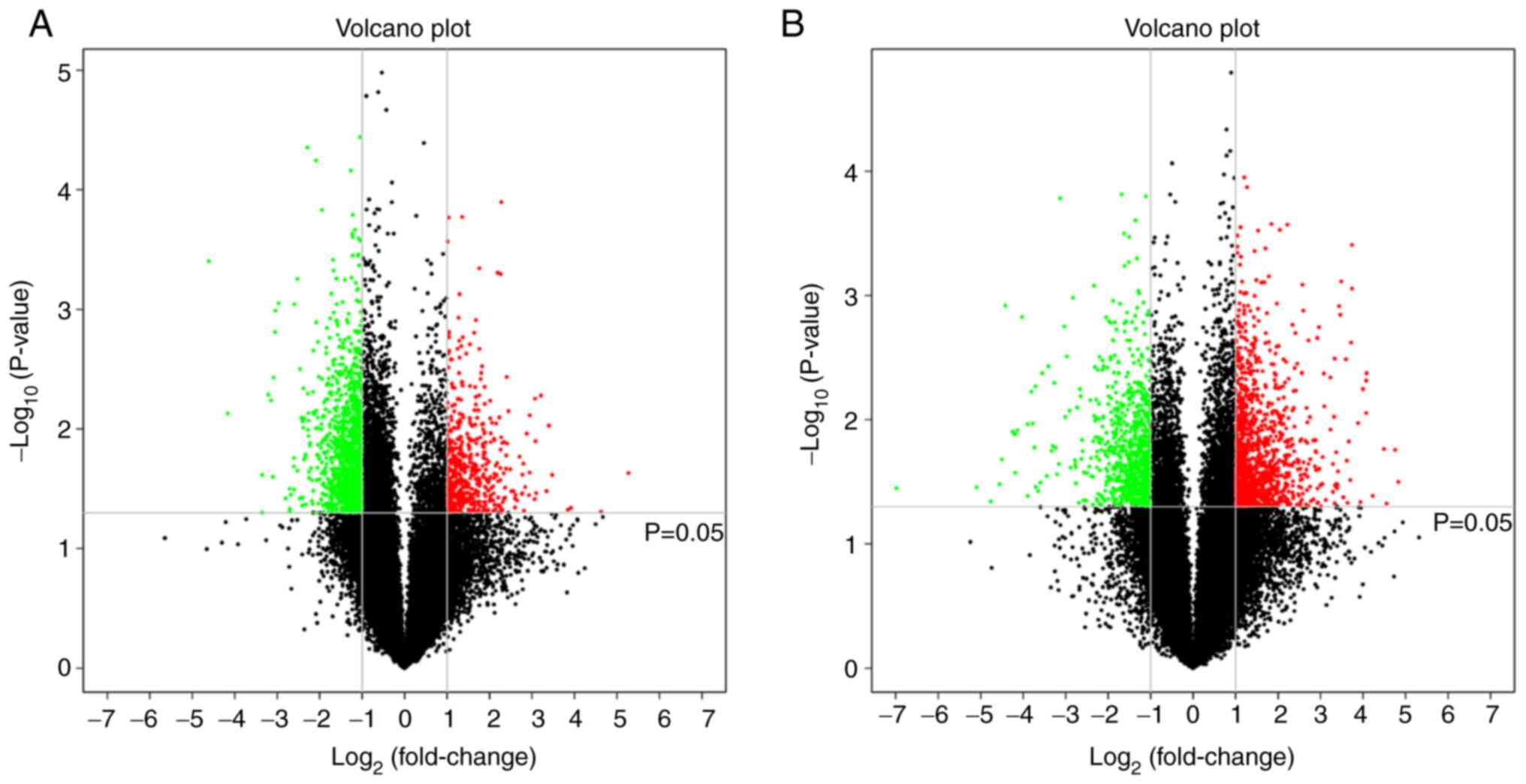
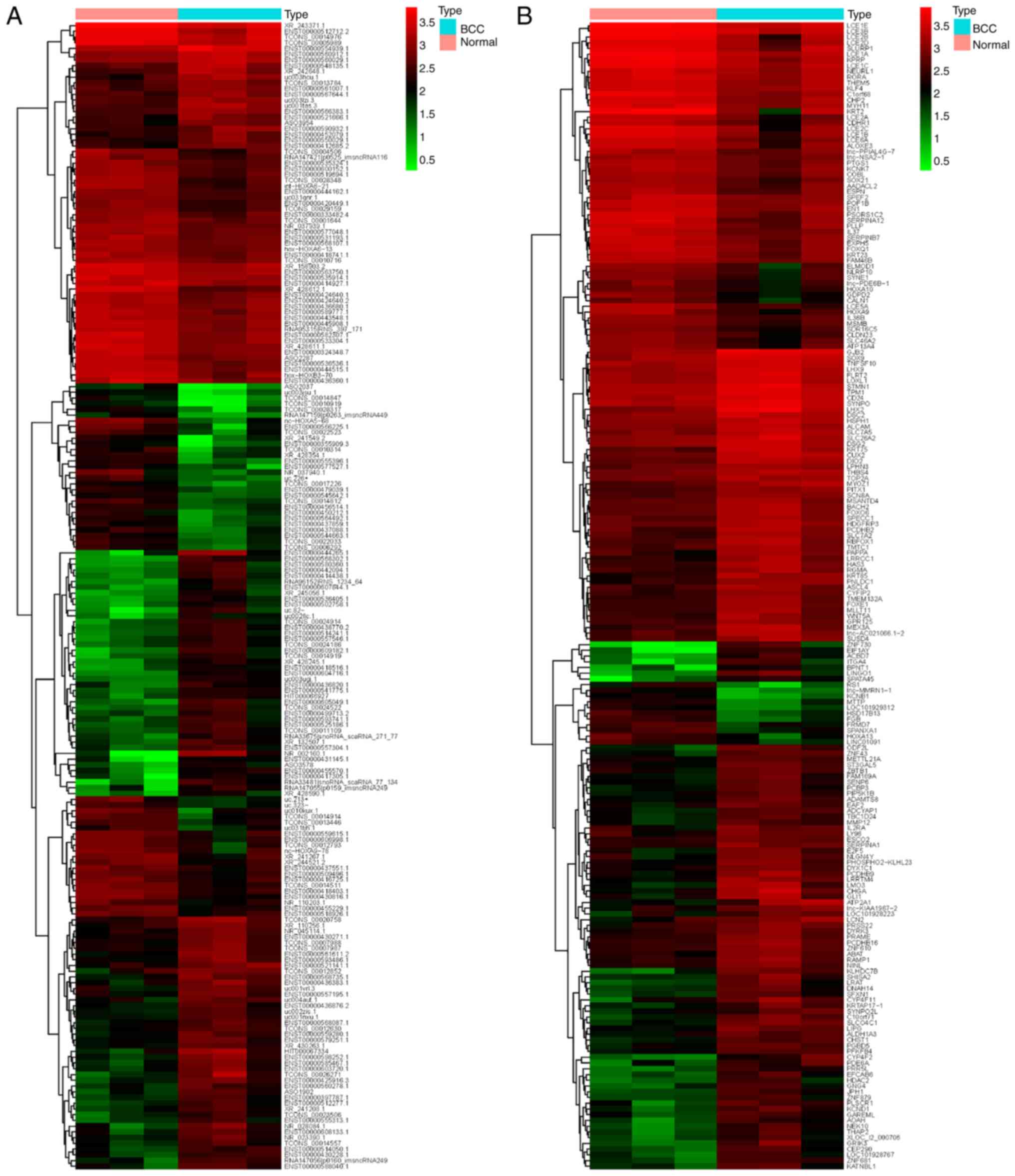
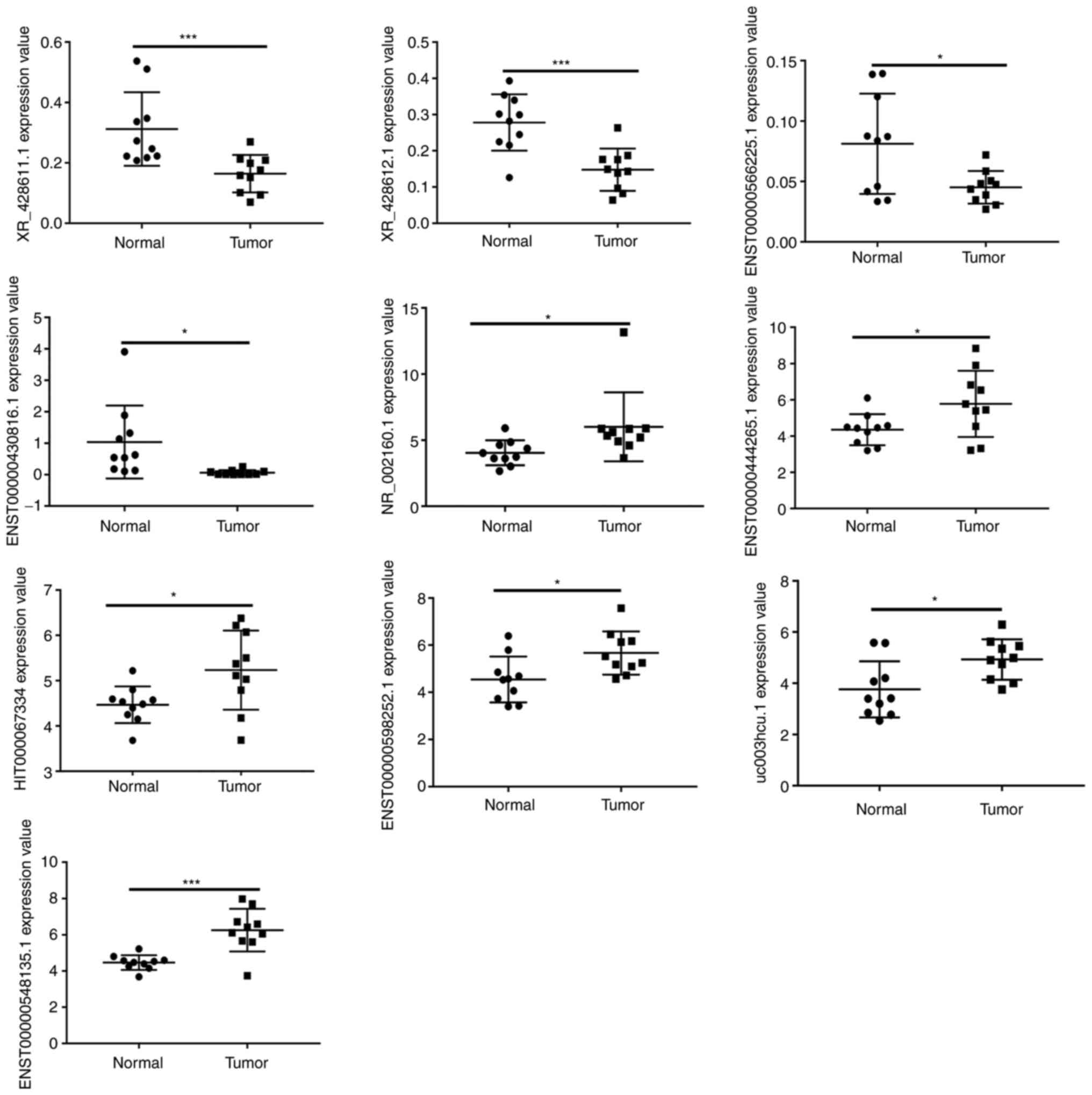
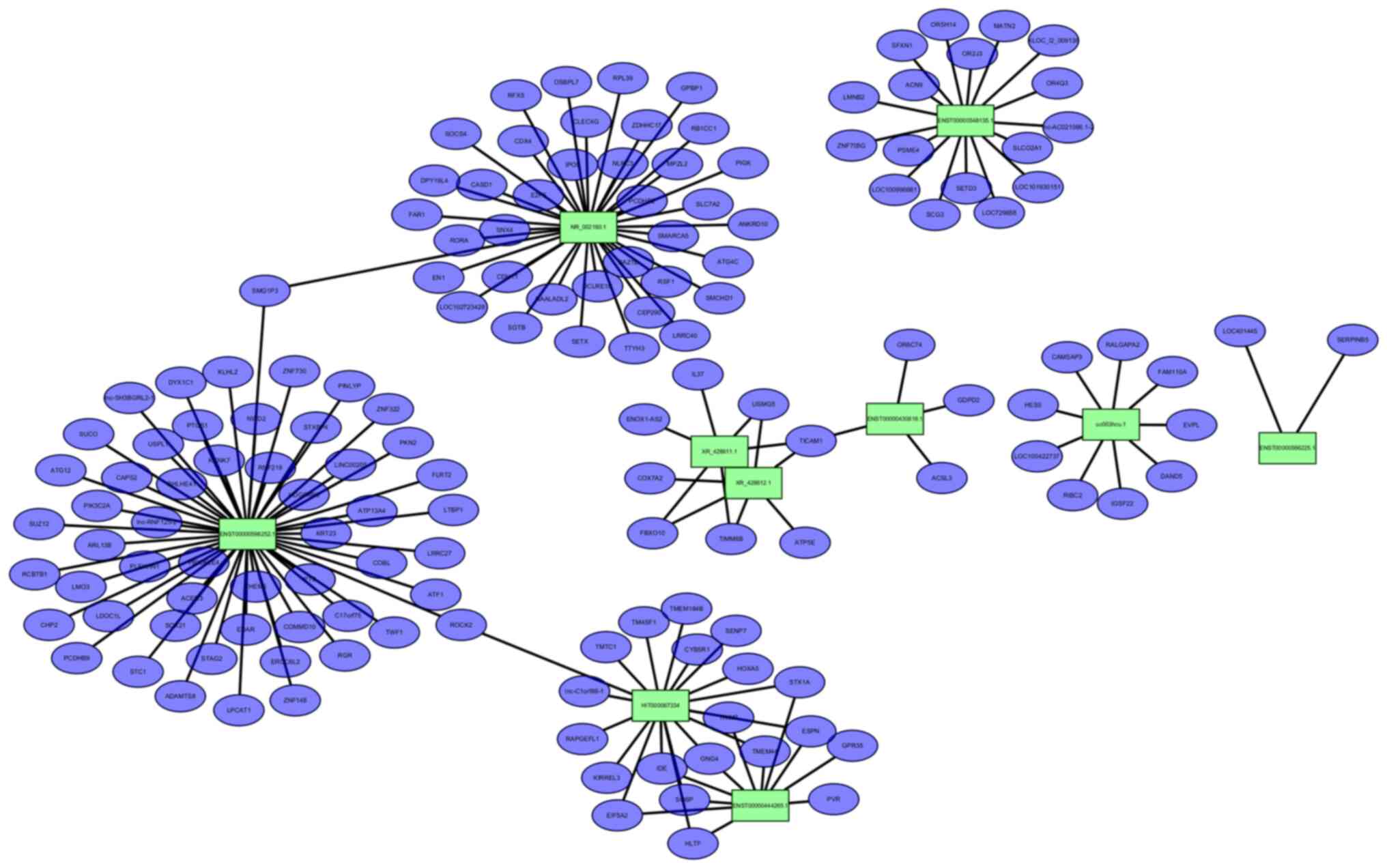
|
1
|
Puig S and Berrocal A: Management of
high-risk and advanced basal cell carcinoma. Clin Transl Oncol.
17:497–503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pellegrini C, Maturo MG, Di Nardo L,
Ciciarelli V, Gutierrez Garcia-Rodrigo C and Fargnoli MC:
Understanding the molecular genetics of basal cell carcinoma. Int J
Mol Sci. 18:24852017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ciążyńska M, Bednarski IA, Wódz K, Kolano
P, Narbutt J, Sobjanek M, Woźniacka A and Lesiak A: Proteins
involved in cutaneous basal cell carcinoma development. Oncol Lett.
16:4064–4072. 2018.
|
|
4
|
Sălan AI, Mărăşescu PC, Camen A, Ciucă EM,
Matei M, Florescu AM, Pădureanu V and Mărgăritescu C: The
prognostic value of CXCR4, α-SMA and WASL in upper lip basal cell
carcinomas. Rom J Morphol Embryol. 59:839–849. 2018.
|
|
5
|
Sun H and Jiang P: MicroRNA-451a acts as
tumor suppressor in cutaneous basal cell carcinoma. Mol Genet
Genomic Med. 6:1001–1009. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vand-Rajabpour F, Sadeghipour N, Saee-Rad
S, Fathi H, Noormohammadpour P, Yaseri M, Hesari KK, Bagherpour Z
and Tabrizi M: Differential BMI1, TWIST1, SNAI2 mRNA expression
pattern correlation with malignancy type in a spectrum of common
cutaneous malignancies: Basal cell carcinoma, squamous cell
carcinoma, and melanoma. Clin Transl Oncol. 19:489–497. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sand M, Hessam S, Amur S, Skrygan M,
Bromba M, Stockfleth E, Gambichler T and Bechara FG: Expression of
oncogenic miR-17-92 and tumor suppressive miR-143-145 clusters in
basal cell carcinoma and cutaneous squamous cell carcinoma. J
Dermatol Sci. 86:142–148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mohammad F, Pandey GK, Mondal T, Enroth S,
Redrup L, Gyllensten U and Kanduri C: Long noncoding RNA-mediated
maintenance of DNA methylation and transcriptional gene silencing.
Development. 139:2792–2803. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu X, Zheng H, Tse G, Chan MT and Wu WK:
Long non-coding RNAs in melanoma. Cell Prolif. 51:e124572018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McCarthy N: Epigenetics. Going places with
BANCR. Nat Rev Cancer. 12:4512012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L
and Sha N: Long non-coding RNA BANCR promotes proliferation in
malignant melanoma by regulating MAPK pathway activation. PLoS One.
9:e1008932014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Panda AC, Abdelmohsen K and Gorospe M:
SASP regulation by noncoding RNA. Mech Ageing Dev. 168:37–43. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koczan D, Fitzner B, Zettl UK and Hecker
M: Microarray data of transcriptome shifts in blood cell subsets
during S1P receptor modulator therapy. Sci Data. 5:1801452018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao L, Zhang P, Li J and Wu M: LAST, a
c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote
CCND1 mRNA stability in human cells. Elife. 6:e304332017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vila-Casadesús M, Gironella M and Lozano
JJ: MiRComb: An R package to analyse miRNA-mRNA interactions.
Examples across five digestive cancers. PLoS One. 11:e01511272016.
View Article : Google Scholar
|
|
19
|
Lan X, Zhang H, Wang Z, Dong W, Sun W,
Shao L, Zhang T and Zhang D: Genome-wide analysis of long noncoding
RNA expression profile in papillary thyroid carcinoma. Gene.
569:109–117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hulstaert E, Brochez L, Volders PJ,
Vandesompele J and Mestdagh P: Long non-coding RNAs in cutaneous
melanoma: Clinical perspectives. Oncotarget. 8:43470–43480. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Botchkareva NV: The molecular revolution
in cutaneous biology: Noncoding RNAs: New molecular players in
dermatology and cutaneous biology. J Invest Dermatol.
137:e105–e111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Biehs B, Dijkgraaf GJ, Piskol R, Alicke B,
Boumahdi S, Peale F, Gould SE and de Sauvage FJ: A cell identity
switch allows residual BCC to survive Hedgehog pathway inhibition.
Nature. 562:429–433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lobl M, Hass B, Clarey D, Higgins S,
Sutton A and Wysong A: Basal cell carcinoma gene mutations differ
between Asian, hispanic, and caucasian patients: A pilot study. J
Drugs Dermatol. 20:504–510. 2021.PubMed/NCBI
|
|
26
|
Stacey SN, Helgason H, Gudjonsson SA,
Thorleifsson G, Zink F, Sigurdsson A, Kehr B, Gudmundsson J, Sulem
P, Sigurgeirsson B, et al: New basal cell carcinoma susceptibility
loci. Nat Commun. 6:68252015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saijo S, Kuwano Y, Tange S, Rokutan K and
Nishida K: A novel long non-coding RNA from the HOXA6-HOXA5 locus
facilitates colon cancer cell growth. BMC Cancer. 19:5322019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jeannotte L, Gotti F and Landry-Truchon K:
Hoxa5: A key player in development and disease. J Dev Biol.
4:132016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aalijahan H and Ghorbian S: Long
non-coding RNAs and cervical cancer. Exp Mol Pathol. 106:7–16.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Abbastabar M, Sarfi M, Golestani A and
Khalili E: lncRNA involvement in hepatocellular carcinoma
metastasis and prognosis. EXCLI J. 17:900–913. 2018.PubMed/NCBI
|
|
31
|
Zhang J, Le TD, Liu L and Li J: Inferring
and analyzing module-specific lncRNA-mRNA causal regulatory
networks in human cancer. Brief Bioinform. 20:1403–1419. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du Y, Xia W, Zhang J, Wan D, Yang Z and Li
X: Comprehensive analysis of long noncoding RNA-mRNA co-expression
patterns in thyroid cancer. Mol Biosyst. 13:2107–2115. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lim LJ, Jin Y, Yang H, Chung AYF, Goh BKP,
Chow PKH, Chan CY, Blanks WK, Cheow PC, Lee SY, et al: Network of
clinically-relevant lncRNAs-mRNAs associated with prognosis of
hepatocellular carcinoma patients. Sci Rep. 10:111242020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guo Q, Cheng Y, Liang T, He Y, Ren C, Sun
L and Zhang G: Comprehensive analysis of lncRNA-mRNA co-expression
patterns identifies immune-associated lncRNA biomarkers in ovarian
cancer malignant progression. Sci Rep. 5:176832015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tao S, Zhu L, Lee P, Lee WM, Knox K, Chen
J, Di YP and Chen Y: Negative control of TLR3 signaling by TICAM1
down-regulation. Am J Respir Cell Mol Biol. 46:660–667. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun R, Zhang Y, Lv Q, Liu B, Jin M, Zhang
W, He Q, Deng M, Liu X, Li G, et al: Toll-like receptor 3 (TLR3)
induces apoptosis via death receptors and mitochondria by
up-regulating the transactivating p63 isoform alpha (TAP63alpha). J
Biol Chem. 286:15918–15928. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chiorazzi M, Rui L, Yang Y, Ceribelli M,
Tishbi N, Maurer CW, Ranuncolo SM, Zhao H, Xu W, Chan WC, et al:
Related F-box proteins control cell death in Caenorhabditis elegans
and human lymphoma. Proc Natl Acad Sci USA. 110:3943–3948. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Del Puerto-Nevado L, Santiago-Hernandez A,
Solanes-Casado S, Gonzalez N, Ricote M, Corton M, Prieto I, Mas S,
Sanz AB, Aguilera O, et al: Diabetes-mediated promotion of colon
mucosa carcinogenesis is associated with mitochondrial dysfunction.
Mol Oncol. 13:1887–1897. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kontro H, Cannino G, Rustin P, Dufour E
and Kainulainen H: DAPIT over-expression modulates glucose
metabolism and cell behaviour in HEK293T cells. PLoS One.
10:e01319902015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang YJ, Jan YH, Chang YC, Tsai HF, Wu
AT, Chen CL and Hsiao M: ATP synthase subunit epsilon
overexpression promotes metastasis by modulating AMPK signaling to
induce epithelial-to-mesenchymal transition and is a poor
prognostic marker in colorectal cancer patients. J Clin Med.
8:10702019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deng S, Li Y, Yi G, Lei B, Guo M, Xiang W,
Chen Z, Liu Y and Qi S: Overexpression of COX7A2 is associated with
a good prognosis in patients with glioma. J Neurooncol. 136:41–50.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Adamkov M, Halasova E, Rajcani J, Bencat
M, Vybohova D, Rybarova S and Galbavy S: Relation between
expression pattern of p53 and survivin in cutaneous basal cell
carcinomas. Med Sci Monit. 17:BR74–BR80. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chipuk JE, Kuwana T, Bouchier-Hayes L,
Droin NM, Newmeyer DD, Schuler M and Green DR: Direct activation of
Bax by p53 mediates mitochondrial membrane permeabilization and
apoptosis. Science. 303:1010–1014. 2004. View Article : Google Scholar : PubMed/NCBI
|