Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor,
which is rare in most parts of the world, but relatively common in
Southeast Asia, North Africa and Southern China (1,2). In
2012, there were ~86,700 new cases and 50,800 deaths associated
with NPC worldwide (3). A variety
of risk factors, such as environmental factors, genetic variation
and Epstein-Barr virus (EBV) infection, are associated with the
occurrence of NPC (4). In recent
years, due to the limitations of surgery, radiotherapy and
chemotherapy remain the most promising and effective treatments for
early-stage NPC (5). Although
improvements in treatment methods have increased survival, some
patients with advanced NPC develop distant metastasis with poor
prognosis (6). Therefore, further
understanding of the molecular mechanisms involved in the
progression of NPC may provide a new direction for the therapeutic
efficiency of NPC.
Forkhead-box gene 1 (FOXG1), a member of the
forkhead box family of transcription factors, is often specifically
expressed in human brain tissue and is associated with the
developmental lesions of the nervous system (7–9).
Previous studies have reported that FOXG1 expression was
upregulated in several cancer types, such as hepatoblastoma
(10), ovarian cancer (11) and glioblastoma (12), and upregulation of FOXG1 was
positively correlated with high tumor grade, suggesting that FOXG1
may be an oncogene (13). However,
there is little knowledge regarding the role of FOXG1 in NPC, which
requires further study.
Uncontrolled cell energetics are a feature of
malignant cancer cells (14,15).
Mitochondria are the main cellular sites of energy production
(16). Changes in the structure and
function of mitochondria in tumor cells lead to an increase in the
absorption and utilization of glutamine, thereby meeting the
bioenergy requirements of tumor cells, which is known as tumor
mitochondrial metabolic reprogramming (17). Accumulating evidence has revealed
that mitochondria serve an important role in cancer metabolism,
proliferation, apoptosis and metastasis (18,19).
However, the specific regulatory relationship between FOXG1 and NPC
progression remains unknown.
The present study investigated the effects of FOXG1
on NPC progression and further examined its role in cell
proliferation, apoptosis, migration, invasion and mitochondrial
function. The results of the current study may provide a foundation
for the treatment of NPC.
Materials and methods
Bioinformatics analysis
The mRNA expression profiles were obtained from Gene
Expression Omnibus (GEO) dataset website (ncbi.nlm.nih.gov/geo/). GSE12452 was used to analyze
the mRNA expression of FOXG1 in NPC tissues and normal tissues.
FOXG1 mRNA expression data was processed using the R software
(3.4.0 version, r-project.org/) and limma R package (20).
Tissue samples
A total of 70 NPC tumor tissues and matching
para-carcinoma tissues were obtained from the Department of
Otolaryngology-Head and Neck Surgery, The Second Affiliated
Hospital of Kunming Medical University between January 2018 and
March 2020. The inclusion criteria were as follows: i) Patients had
never received radiotherapy or chemotherapy before surgery; ii)
patients had no medical history of other malignant tumors; and iii)
the diagnosis of all samples was confirmed by histopathology of
NPC. The exclusion criteria were: i) Patients had incomplete
clinicopathological data; and ii) patients were unwilling to
cooperate with treatment. The collected samples were immediately
frozen in liquid nitrogen after surgical resection and stored at
−80°C until use in subsequent assays. The study was approved by the
Ethics Committee of The Second Affiliated Hospital of Kunming
Medical University (approval no. KYDE201801012), and all patients
(20 females and 50 males; age, 28–75 years) signed informed consent
forms.
Immunohistochemistry (IHC)
First, 10% neutral buffered formalin was used to fix
the tissue samples for 24 h at room temperature, which were then
dehydrated and embedded in paraffin wax. Next, the
paraffin-embedded samples were cut into 4-µm thick sections, and
the sections were dewaxed with xylene. They were then dehydrated
with gradient ethanol, and endogenous enzymes were removed with 3%
H2O2 for 10 min at room temperature.
Subsequently, the sections were treated with 10 mM Tris-EDTA
(Thermo Fisher Scientific, Inc.) at 125°C in a pressure cooker for
antigenic retrieval and then incubated with primary antibody
(FOXG1; 1:200; cat. no. ab196868; Abcam) overnight at 4°C, followed
by incubation with goat anti-rabbit secondary antibody (1:250; cat.
no. ab150081; Abcam) for 30 min at room temperature. Finally, the
sections were stained with 3, 3′-diaminobenzidine (Thermo Fisher
Scientific, Inc.) for 5 min at room temperature, counterstained
with hematoxylin for 3 min at room temperature and mounted with
neutral gum. Images were captured using an Olympus FV1000 laser
scanning confocal microscope (Olympus Corporation; magnification,
×100 and ×400).
Cell culture
Human immortalized nasopharyngeal epithelial cells
(NP69; BeNa Culture Collection; Beijing Beina Chunglian Institute
of Biotechnology) were cultured in a keratinocyte/serum-free medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 0.2 ng/ml
human recombinant epidermal growth factor, 2% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and 1% streptomycin and penicillin (Gibco;
Thermo Fisher Scientific, Inc.). The NPC cells (C666-1 and SUNE-1;
The Cell Bank of Type Culture Collection of Chinese Academy of
Sciences) were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 5% FBS and 1% streptomycin and
penicillin. Both cells were grown in an incubator with 5%
CO2 at 37°C.
Cell transfection
SUNE-1 cells were divided into four groups: Control
(without treatment), small interfering (si)RNA-negative control
(siNC; cells transfected with 5 µg non-targeting siRNAs), si1-FOXG1
(cells transfected with 5 µg si1-FOXG1;
5′-GCCCTTCAGTTCAGGTACAAT-3′) and si2-FOXG1 (cells transfected with
5 µg si2-FOXG1; 5′-GGCTGTTTACCCACAATGAAA-3′). C666-1 cells were
divided into three groups: Control (without treatment), pcDNA3.1-NC
(cells transfected with 4 µg pcDNA3.1-NC vector) and pcDNA3.1-FOXG1
(cells transfected with 4 µg pcDNA3.1-FOXG1 vector). Cells
(1×105 cells/well) were seeded into 12-well plates and
transfected using Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C. The
pcDNA3.1-FOXG1, siRNA-FOXG1 and NCs were purchased from Guangzhou
RiboBio Biotech Co., Ltd. At 48 h post-transfection, subsequent
experiments were performed.
Reverse transcription-quantitative PCR
(RT-qPCR)
The relative mRNA expression level of FOXG1 and
mitochondrial DNA (mtDNA) copy number was measured via RT-qPCR.
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to extract total RNA from tissues and cells, and a
Prime-Script™ reverse transcription kit (Takara Bio, Inc.) was used
to synthesize cDNA according to the manufacturer's instructions.
The SYBR-Green PCR kit (Takara Bio, Inc.) was used to perform
RT-qPCR on a 7500 Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The RT-qPCR procedure was as
follows: Initial denaturation at 94°C for 2 min, followed by 40
cycles of denaturation at 94°C for 30 sec, annealing at 47°C for 1
min (FOXG1) or 47°C for 30 sec (D-Loop), elongation at 72°C for 30
sec and final extension at 72°C for 5 min. The relative mRNA
expression level of FOXG1 was measured using the 2−ΔΔCq
method (21). GAPDH was used as
internal controls for FOXG1. The mtDNA copy number was calculated
as the ratio of mitochondrial D-Loop to 18S rRNA (22). The primer sequences (Invitrogen;
Thermo Fisher Scientific, Inc.) for RT-qPCR are shown in Table I.
 | Table I.Primers sequences for reverse
transcription-quantitative PCR. |
Table I.
Primers sequences for reverse
transcription-quantitative PCR.
| Primers | Sequences (5–3) |
|---|
| FOXG1-F |
GGAATTCAACATITCCTCCAAGGACACA |
| FOXG1-R |
CGGGATCCGGGTFGCTAGAGCCTGGTAAT |
| GAPDH-F |
CGGGAAACTGTGGCGTGAT |
| GAPDH-R |
AGTGGGTGTCGCTGTFGAAGT |
| D-Loop-F |
GATTTGGGTACCACCCAAGTATTG |
| D-Loop-R |
AATATTCATGGTGGCTGGCATGTA |
| 18S rRNA-F |
TCTCCTACTTGGATAACTGTGG |
| 18S rRNA-R |
GGCGACTACCATCGAAAGTTG |
Western blot analysis
RIPA buffer (Cell Signaling Technology, Inc.) was
used to obtain total protein from tissues or cells. A BCA assay kit
(Pierce; Thermo Fisher Scientific, Inc.) was used to determine the
protein concentration. Protein samples (30 µg) were separated via
10% SDS-PAGE and then transferred onto PVDF membranes. The PVDF
membranes were blocked with 5% non-fat milk in 0.1% TBS-Tween-20
for 1 h at room temperature. Next, the PVDF membranes were
incubated with primary antibodies [FOXG1, cat. no. ab196868;
N-cadherin, cat. no. ab18203; Snail, cat. no. ab229701; caspase-9,
cat. no. ab219590; heat shock protein (HSP) 60, cat. no. ab190828;
1:1,000, all from Abcam; Bax, cat. no. 5023; Bcl-2, cat. no. 3498;
cleaved poly(ADP-ribose) polymerase 1 (PARP), cat. no. 9185;
E-cadherin, cat. no. 3195; caspase-3, cat. no. 14220; caspase-8,
cat. no. 4790; succinate dehydrogenase complex flavoprotein subunit
A (SDHA), cat. no. 11998; pyruvate dehydrogenase (PDH), cat. no.
3205; cleaved caspase-3, cat. no. 9654; cleaved caspase-8, cat. no.
9496; cleaved caspase-9, cat. no. 20750; 1:1,000, all from Cell
Signaling Technology, Inc.; PARP, cat. no. SAB4500487; 1:1,000,
Sigma-Aldrich; Merck KGaA) overnight at 4°C. The PVDF membranes
were then incubated with horseradish peroxidase-conjugated goat
polyclonal anti-rabbit IgG secondary antibodies (cat. no. ab150077;
1:2,000; Abcam) for 1 h at room temperature. Finally, the protein
bands were visualized using an ECL reagent (Pierce; Thermo Fisher
Scientific, Inc.). The intensity of protein bands were quantified
using the Image Lab Software (V3.0; Bio-Rad Laboratories,
Inc.).
MTT assay
SUNE-1 and C666-1 cells (1×105
cells/well) were seeded in 96-well plates. Next, the cells were
incubated in fresh RPMI-1640 medium for 24, 48, 72 or 96 h at 37°C.
Then, MTT solution (10 µl) was added to each well, and the cells
were cultured for 4 h in the incubator at 37°C. Subsequently, the
cells were incubated with DMSO (150 µl) for 15 min at 37°C. A
microplate reader (Bio-Rad Laboratories, Inc.) was used to measure
the optical density value at 450 nm.
5-Ethynyl-20-deoxyuridine (EdU)
assays
An EdU labelling/detection kit (Guangzhou RiboBio
Co., Ltd.) was used to assess cell proliferation. Briefly, after
transfection for 48 h, the cells were incubated with 50 µM EdU
labelling medium for 2 h at 37°C. Next, the cells were fixed with
4% paraformaldehyde for 25 min at room temperature and treated with
0.5% Triton X-100 for 15 min at room temperature. Then, the cells
were incubated with 1X Apollo reaction reagents (15 µl) for 25 min
at room temperature and stained with DAPI solution for 10 min at
room temperature. Finally, the number of EdU-positive cells was
examined under a fluorescent microscope at a magnification of ×100,
and the percentage of EdU-positive cells was counted from five
random fields.
Wound healing assay
After transfection for 48 h, the cells (1×106
cells/well) were seeded in 6-well plates and cultured in RPMI-1640
medium containing 10% FBS to reach 90% confluence. A sterile 200-µl
pipette tip was used to create wounded monolayers, and the cells
were cultured in serum-free medium for 48 h at 37°C. Images of the
monolayer wound were imaged at 0 and 48 h under an Olympus CKX53
microscope (Olympus Corporation) at a magnification of ×200, and
the migratory ability was analyzed using ImageJ software
(V1.8.0.112; National Institutes of Health) from three randomly
chosen fields.
Transwell invasion assay
The cell invasive ability was detected using 24-well
Transwell chambers (pre-coated with Matrigel for 1 h at 37°C;
Corning, Inc.). In brief, after transfection for 48 h, cells
(2×104 cells/well) were resuspended in 200 µl serum-free
medium and then added into the upper chambers. Next, 500 µl
RPMI-1640 medium containing 20% FBS was added into the lower
chambers. After incubation for 48 h at 37°C, cells on the upper
surface were wiped using a cotton swab, and the cells were fixed
with 4% paraformaldehyde for 10 min at room temperature and stained
with 0.1% crystal violet for 15 min at room temperature. Finally,
the number of invasive cells was counted from three random fields
under an inverted light microscope at a magnification of ×200.
Cell apoptosis assay
After transfection for 48 h, the Annexin V-FITC
Apoptosis Detection kit (Sigma-Aldrich; Merck KGaA) was used to
detect cell apoptosis. Cells were harvested with 0.25% trypsin and
resuspended in binding buffer. Subsequently, the cells were
incubated with PI (5 µl) and Annexin V-FITC (2.5 µl) for 20 min at
room temperature in the dark. The cells were washed with cold PBS,
and then flow cytometry (FACScan™; BD Biosciences) equipped with
CellQuest software (V6.0; BD Biosciences) was conducted to
calculate the proportion of apoptotic cells.
Measurement of ATP/ADP ratio
The ATP/ADP ratio was measured using the ADP/ATP
ratio assay kit (Abcam). After transfection for 48 h, the cells
(5×104) were incubated with nucleotide-releasing buffer
(200 µl) for 10 min at room temperature. Next, the cells were
treated with ATP monitoring enzyme (10 µl) at room temperature for
10 min and ATP levels were measured immediately (A). ADP levels
were measured (B), then cells were treated with ADP converting
enzyme (10 µl) at room temperature for 5 min and ADP levels were
measured (C). Finally, the ATP/ADP ratio was calculated as follows:
A/(C-B).
Detection of mitochondrial membrane
potential (MMP)
After transfection for 48 h, the cells
(5×105) were grown in a medium containing 10 µg/ml
5,5′,6,6
-Tetrachloro-1,1′,3,3′-tetraethyl-benzimidazolylcarbocyanine iodide
(Thermo Fisher Scientific, Inc.). After incubation for 30 min at
37°C in the dark, the cells were washed using PBS and flow
cytometry (FACScan™; BD Biosciences) equipped with CellQuest
software (V6.0; BD Biosciences) was used to detect changes in
MMP.
Statistical analysis
All experiments were repeated ≥3 times, and the data
are expressed as the mean ± SD. GraphPad Prism 7.0 (GraphPad
Software, Inc.) was used to perform statistical analyses.
χ2 test was used to analyze the association between
FOXG1 expression and clinic characteristics in patients with NPC.
An unpaired Student's t-test was used for comparison between two
groups, and one-way ANOVA with Tukey's post-hoc test was used for
comparison among ≥3 groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
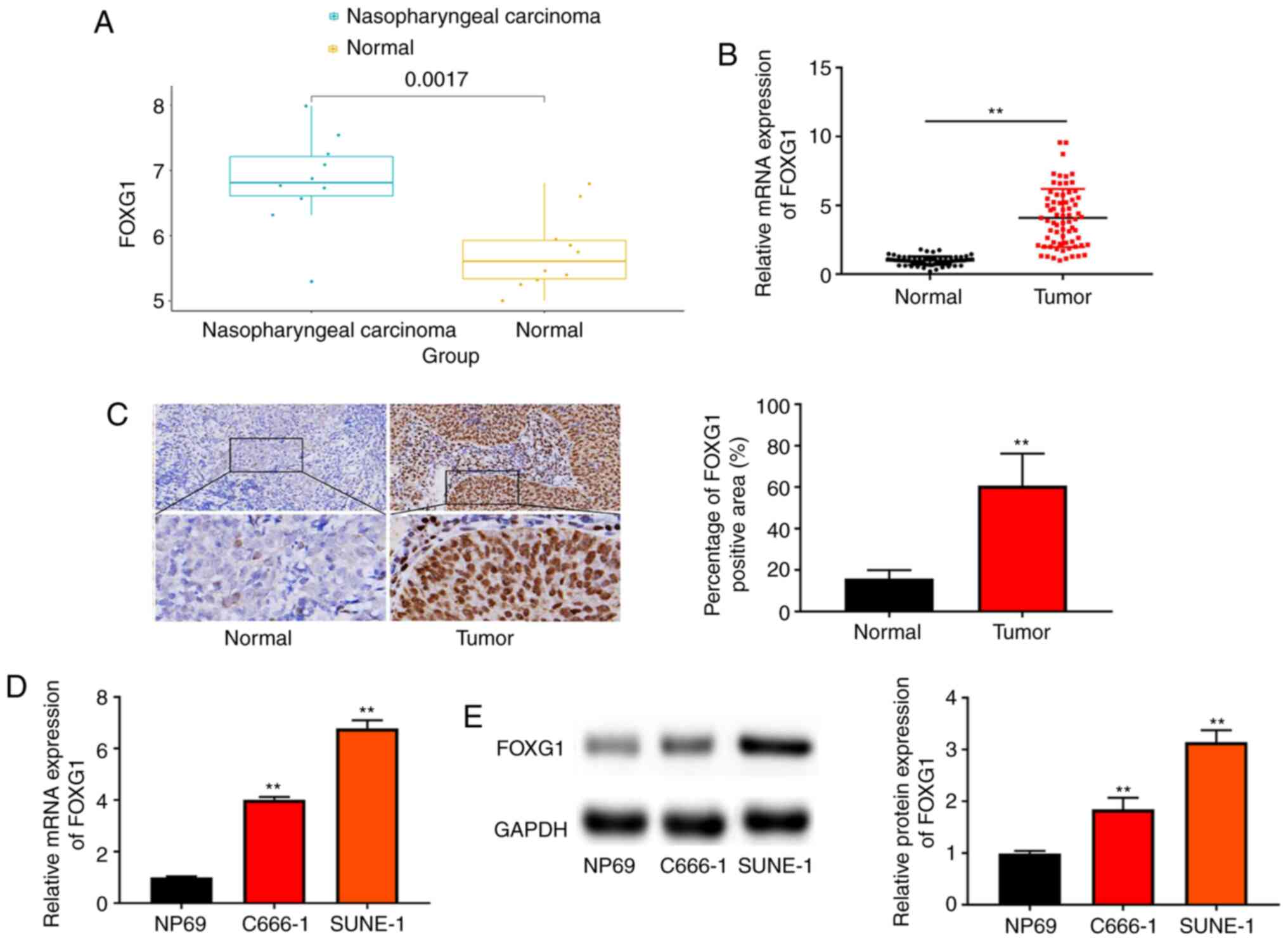
FOXG1 expression is upregulated in NPC
tissues and cells
Analysis of GEO dataset (GSE12452) revealed that
FOXG1 expression in NPC tissues was upregulated compared with that
in normal tissues (Fig. 1A). To
examine the function of FOXG1 in NPC progression, FOXG1 expression
was first measured in NPC tissues and para-carcinoma tissues
(Normal) via RT-qPCR and IHC. As shown in Fig. 1B and C, FOXG1 expression in NPC
tissues was higher compared with that in normal tissues. According
to the median mRNA expression of FOXG1, patients with NPC were
divided into low and high expression groups. It was found that
FOXG1 expression was associated with distant metastasis and TNM
stage, while there was no significant association with sex,
smoking, EBV infection and age (Table
II).
 | Table II.Association between FOXG1 expression
and clinicopathological characteristics of patients with
nasopharyngeal cancer. |
Table II.
Association between FOXG1 expression
and clinicopathological characteristics of patients with
nasopharyngeal cancer.
|
|
| FOXG1
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Number of
cases | High | Low | P-value |
|---|
| Sex |
|
|
| 0.112 |
|
Male | 50 | 28 | 22 |
|
|
Female | 20 | 7 | 13 |
|
| Age, years |
|
|
| 0.632 |
|
<50 | 34 | 18 | 16 |
|
|
≥50 | 36 | 17 | 19 |
|
| Smoking |
|
|
| 0.212 |
|
Yes | 45 | 25 | 20 |
|
| No | 25 | 10 | 15 |
|
| EBV infection |
|
|
| 0.147 |
|
Negative | 40 | 23 | 17 |
|
|
Positive | 30 | 12 | 18 |
|
| Distant
metastasis |
|
|
| 0.005a |
|
Yes | 23 | 17 | 6 |
|
| No | 47 | 18 | 29 |
|
| TNM stage |
|
|
| 0.003a |
|
I–II | 24 | 6 | 18 |
|
|
III–IV | 46 | 29 | 17 |
|
Next, FOXG1 expression in NPC cells (C666-1 and
SUNE-1) and NP69 cells was evaluated using RT-qPCR and western
blotting. As presented in Fig. 1D and
E, FOXG1 expression was upregulated in NPC cells compared with
that in NP69 cells.
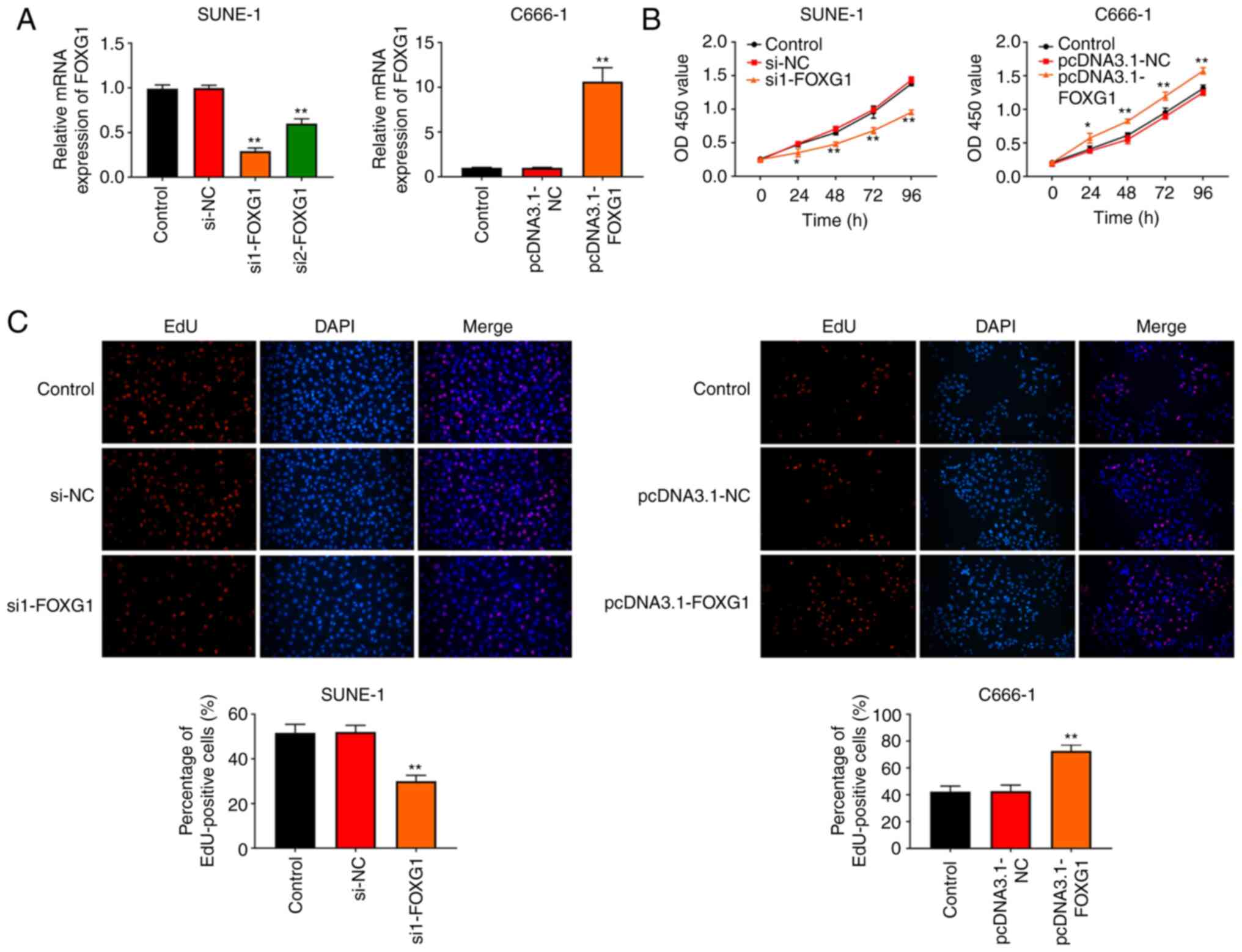
FOXG1 promotes the proliferation of
NPC cells
To investigate the effects of FOXG1 in NPC, SUNE-1
and C666-1 cells were transfected with siRNA-FOXG1 and
pcDNA3.1-FOXG1, respectively. Transfection efficiency was assessed
using RT-qPCR. As presented in Fig.
2A, FOXG1 expression in the si1-FOXG1 and si2-FOXG1groups was
lower compared with that in the control and si-NC groups, and FOXG1
expression in the pcDNA3.1-FOXG1 group was higher compared with
that in the control and pcDNA3.1-NC groups. These results indicated
that transfection had been successful. Subsequently, cell
proliferation was detected using MTT and EdU assays. It was found
that knockdown of FOXG1 inhibited the proliferation of SUNE-1 cells
compared with the control and si-NC groups, while overexpression of
FOXG1 promoted the proliferation of C666-1 cells compared with the
control and pcDNA3.1-NC groups (Fig. 2B
and C).
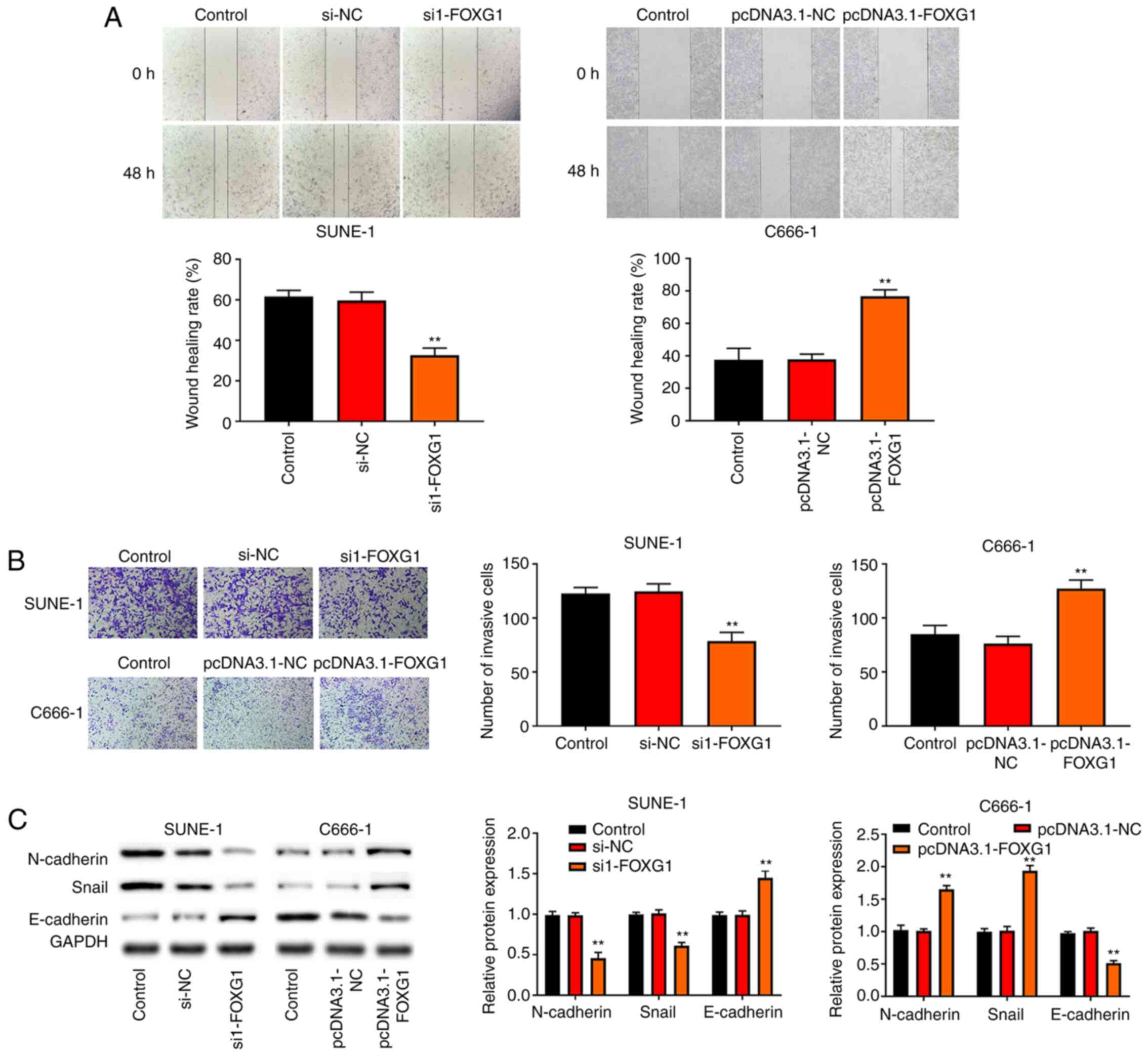
FOXG1 promotes the migration, invasion
and epithelial-mesenchymal transition (EMT) of NPC cells
To investigate the role of FOXG1 in migration and
invasion of NPC cells, cells were detected via wound healing and
Transwell assays. The results demonstrated that knockdown of FOXG1
inhibited the migration and invasion of SUNE-1 cells, whereas the
overexpression of FOXG1 promoted the migration and invasion of
C666-1 cells (Fig. 3A and B).
Subsequently, western blotting was conducted to
detect the expression levels of EMT-related proteins (N-cadherin,
Snail and E-cadherin) to further determine the molecular mechanism
mediating the aggressive effect of FOXG1. It was identified that
knockdown of FOXG1 decreased the protein expression levels of
N-cadherin and Snail in SUNE-1 cells and increased the protein
expression level of E-cadherin (Fig.
3C). Conversely, overexpression of FOXG1 increased the protein
expression levels of N-cadherin and Snail in C666-1 cells and
decreased E-cadherin protein expression.
FOXG1 inhibits the apoptosis of NPC
cells
To observe the effect of FOXG1 on cell apoptosis,
flow cytometry analysis was performed on C666-1 and SUNE-1 cells.
The results (Fig. 4A) demonstrated
that knockdown of FOXG1 promoted the apoptosis of SUNE-1 cells,
whereas overexpression of FOXG1 inhibited the apoptosis of C666-1
cells. Next, western blotting was performed to detect the
expression levels of apoptosis-related proteins (Bax, Bcl-2, PARP,
cleaved PARP, cleaved caspase-3, cleaved caspase-8, cleaved
caspase-9, caspase-3, caspase-8 and caspase-9) in C666-1 and SUNE-1
cells. As shown in Fig. 4B,
knockdown of FOXG1 increased the expression levels of Bax/Bcl-2,
PARP, cleaved PARP, cleaved caspase-3, cleaved caspase-8, cleaved
caspase-9, caspase-3, caspase-8 and caspase-9 in SUNE-1 cells.
Notably, the opposite results were observed in the C666-1 cells
with FOXG1 overexpression.
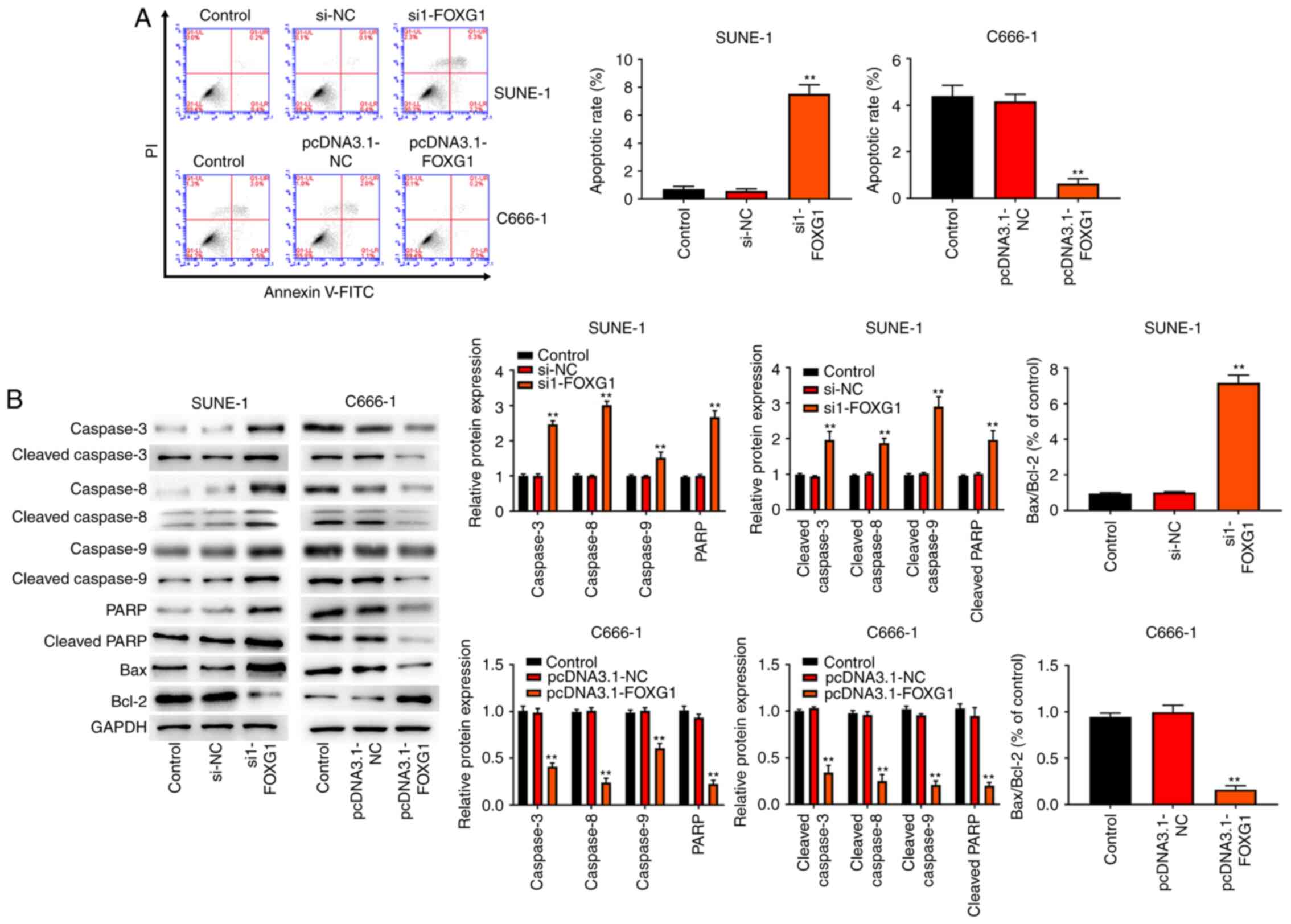 | Figure 4.FOXG1 inhibits the apoptosis of NPC
cells. (A) Flow cytometry analysis was used to detect the apoptosis
of C666-1 and SUNE-1 cells. (B) Western blot analysis was conducted
to detect the expression levels of apoptosis-related proteins (Bax,
Bcl-2, PARP, cleaved PARP, caspase-3, cleaved caspase-3, caspase-8,
cleaved caspase-8, cleaved caspase-9 and caspase-9) in C666-1 and
SUNE-1 cells. **P<0.01 vs. Control, si-NC or pcDNA3.1-NC group.
NC, negative control; si, small interfering RNA; FOXG1,
forkhead-box gene 1; PARP, poly(ADP-ribose) polymerase 1. |
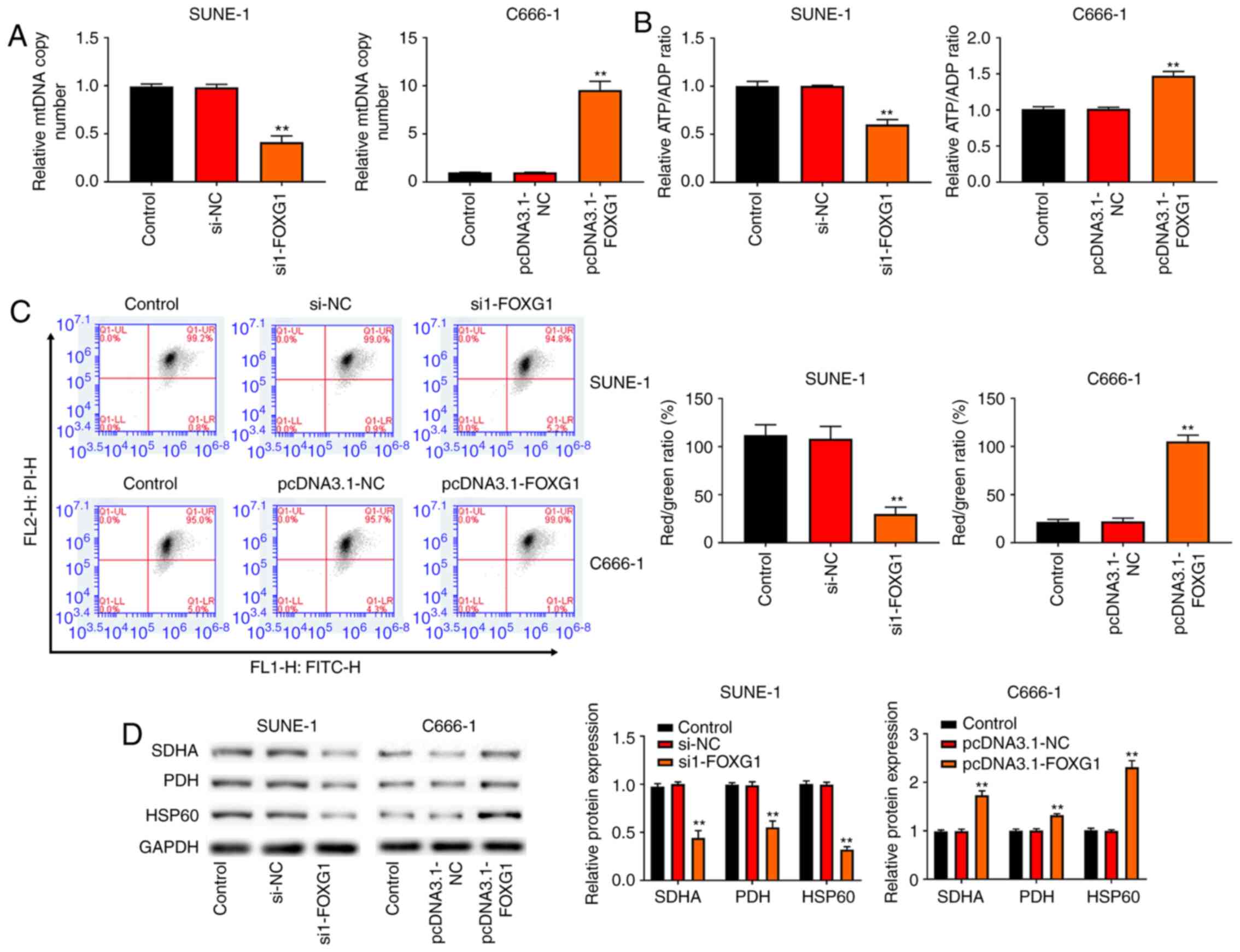
FOXG1 improves mitochondrial function
in NPC cells
Mitochondrial dysfunction is reported to be
associated with cancer cell death (19). To investigate whether FOXG1 was
involved in the regulation of NPC mitochondrial function, the copy
number of mtDNA was detected via RT-qPCR. As shown in Fig. 5A, knockdown of FOXG1 reduced the
mtDNA copy number, whereas overexpression of FOXG1 increased the
mtDNA copy number. Next, the ATP/ADP ratio was measured, and it was
found that knockdown of FOXG1 resulted in a decreased ATP/ADP
ratio, but overexpression of FOXG1 resulted in an increased ATP/ADP
ratio (Fig. 5B). Moreover,
knockdown of FOXG1 decreased the Red/Green ratio (indicative of
MMP), while overexpression of FOXG1 increased the Red/Green ratio
(Fig. 5C). Notably, the western
blotting results revealed that knockdown of FOXG1 decreased the
expression levels of mitochondrial markers (SDHA, HSP60 and PDH) in
SUNE-1 cells, while the opposite results were observed in C666-1
cells with FOXG1 overexpression (Fig.
5D).
Discussion
It is well known that NPC is one of the most common
malignant head and neck cancer types and lead to ~50,800 deaths
worldwide in 2012 (3,23). Previous studies have reported that
FOXG1 exerted antitumor or carcinogenic effects in the progression
of various cancer types (12,24,25).
In the present study, FOXG1 expression was upregulated in NPC
tissues and cells, and FOXG1 expression was associated with distant
metastasis and TNM stage. Moreover, knockdown of FOXG1 inhibited
the proliferation, migration, invasion, EMT and mitochondrial
function of SUNE-1 cells, as well as promoted cell apoptosis.
Notably, the opposite results were observed in the C666-1 cells
with FOXG1 overexpression.
FOXG1 participates in the proliferation and
differentiation of various cells, and its dysregulated expression
can cause the occurrence and development of different diseases
(26–29). It has been reported that FOXG1 can
induce the carcinogenic transformation of chicken embryo
fibroblasts, indicating that FOXG1 may act as an oncogene in cancer
(30,31). Adesina et al (10) revealed that FOXG1 expression was
upregulated in hepatoblastoma, while Chan et al (11) reported that FOXG1 was upregulated in
ovarian cancer, and its expression level was positively correlated
with ovarian cancer stage. In addition, a study by Chen et
al (32) indicated that FOXG1
expression was elevated in glioma tissues and its expression was
associated with glioma grade. The present study also demonstrated
that FOXG1 expression was upregulated in NPC tissues and cells, and
was associated with distant metastasis and TNM stage, which was
consistent with a previous study (32). Taken together, these results
indicate that FOXG1 may serve a role in the carcinogenesis of
NPC.
In recent years, previous studies have reported that
the members of the FOX transcription factor family serve an
important role in the development of tumors and participate in the
regulation of various malignant biological phenotypes of tumors
(33,34). The overexpression of FOXG1 has been
shown to suppress the TGF-β/Smad pathway
induced-p21WAF1/CIP1 expression and promote the
proliferation of ovarian cancer cells (11). Verginelli et al (9) also observed that knockdown of FOXG1
inhibited the proliferation of brain tumor-initiating cells.
Moreover, Chen et al (32)
reported that knockdown of FOXG1 inhibited cell proliferation and
promoted glioma cell apoptosis. In the present study, knockdown of
FOXG1 inhibited proliferation and promoted apoptosis of SUNE-1
cells, while overexpression of FOXG1 promoted proliferation and
inhibited apoptosis of C666-1 cells.
Apoptosis is regarded as a promising treatment for
cancer, and the activation of caspases serves important parts in
the process (35). The death
receptor-mediated caspase-8 and the mitochondria-dependent
caspase-9 pathways are the main caspase-dependent pathways in
apoptosis (36). Furthermore, the
activation of caspase-9 and caspase-8 results in the activation of
caspase-3 (37,38). PARP, a downstream substrate of
caspase-3, can directly affect cell apoptosis (39). Bax, a member of the Bcl-2 family, is
a pro-apoptotic protein (40),
while it has been shown that a high expression of Bcl-2 (an
anti-apoptotic protein) could prevent cell apoptosis in cancer
(41). Therefore, cell apoptosis is
affected by the ratio of Bax/Bcl-2 (42). Zhang et al (43) confirmed that the overexpression of
forkhead box protein O1 (FOXO1; a downstream target of FOXG1)
induced cell-cycle arrest and apoptosis, as well as the
upregulation of caspases-3 and caspases-9 expression in cervical
cancer. The present study demonstrated that knockdown of FOXG1
increased the expression levels of Bax/Bcl-2, PARP, cleaved PARP,
cleaved caspase-3, cleaved caspase-8, cleaved caspase-9, caspase-3,
caspase-8 and caspase-9 in SUNE-1 cells, while the opposite results
were observed in C666-1 cells. In summary, these findings indicate
that FOXG1 promotes cell proliferation and inhibits apoptosis in
NPC cells.
Distant metastases, rather than primary tumors,
cause the majority of deaths associated with NPC (44). Metastasis is a multi-factor and
multi-step dynamic process that involves a variety of gene
regulatory cascade reactions (45,46).
The FOX gene family include types of transcription factors with
diverse biological functions (33,47).
The current results suggested that knockdown of FOXG1 inhibited the
migration and invasion of SUNE-1 cells, whereas overexpression of
FOXG1 promoted the migration and invasion of C666-1 cells. EMT is
defined as the loss of epithelial morphology and acquisition of a
mesenchymal phenotype, and it is usually considered as an important
factor for promoting cell invasion and migration in malignant
diseases (48,49). In the present study, knockdown of
FOXG1 decreased the protein expression levels of N-cadherin and
Snail in SUNE-1 cells and increased the protein expression level of
E-cadherin. By contrast, overexpression of FOXG1 increased the
protein expression levels of N-cadherin and Snail in C666-1 cells
and decreased the protein expression level of E-cadherin. These
findings suggest that FOXG1 promotes NPC metastasis by inducing
EMT.
Mitochondria serve an important role in maintaining
cellular energy homeostasis (50).
These are the main consumers of oxygen and glucose and can produce
sufficient ATP, which is necessary for cancer behaviors (51,52).
However, mitochondrial damage can impair the metabolism of cancer
and activate the activity of mitochondria-related apoptosis
(53,54). In addition, injured mitochondria
cannot produce sufficient energy, which is associated with the
failure of cancer cells to adhere and invade (55). The change of mtDNA copy number is
considered as an indicator of mitochondrial damage (56). Previous studies have reported that
the decrease of mtDNA copy number is the result of the decrease of
biogenesis (57, 58). Furthermore, MMP is a marker for evaluating
the biological function of mitochondria, and a decrease of MMP
indicates mitochondrial biological dysfunction (59–61).
Chen et al (62) revealed
that the mitochondrial dysfunction, including reactive oxygen
species production and reduction of MMP, could be caused by the
overexpression of FOXO1. In the present study, it was found that
knockdown of FOXG1 reduced the mtDNA copy number, ATP/ADP ratio and
MMP, while overexpression of FOXG1 increased the mtDNA copy number,
ATP/ADP ratio and MMP.
SDHA, a subunit of succinate dehydrogenase,
participates in the tricarboxylic acid cycle and oxidative
phosphorylation, and serves an important role in the process of
cell energy metabolism (63,64).
PDH can transform pyruvate into acetyl-CoA and regulate energy
metabolism of cells (65), while
HSP60, a mitochondrial protein, serves an important role in
maintaining mitochondrial integrity and ATP generation (66). In the present study, knockdown of
FOXG1 decreased the expression levels of mitochondrial markers
(SDHA, HSP60 and PDH) in SUNE-1 cells, while the opposite results
were obtained in C666-1 cells. These data suggest that FOXG1
improves mitochondrial function in NPC cells.
The present study had certain limitations. Firstly,
the prognosis of FOXG1 was not assessed and the detailed mechanism
of FOXG1 in NPC progression requires further investigated. In
vivo assays are necessary to verify the present
conclusions.
In conclusion, the present study demonstrated that
FOXG1 expression was upregulated in NPC tissues and cells, and it
was associated with distant metastasis and TNM stage. In addition,
FOXG1 enhanced cell proliferation, migration and invasion, induced
EMT and improved mitochondrial function in NPC cells. These
findings may provide further insights into the interactive
mechanism between FOXG1 and NPC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CL designed the study. HX and ZH performed the
research and analyzed the data. HX and ZH confirmed the
authenticity of the raw data. HX wrote the paper. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The protocol of this research has been approved by
the Ethics Committee of Affiliated Hospital of The Second
Affiliated Hospital of Kunming Medical University (approval no.
KYDE201801012). All patients have signed written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FOXG1
|
forkhead-box gene 1
|
|
NPC
|
nasopharyngeal cancer
|
|
IHC
|
immunohistochemistry
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
MMP
|
mitochondrial membrane potential
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Tang LL, Chen WQ, Xue WQ, He YQ, Zheng RS,
Zeng YX and Jia WH: Global trends in incidence and mortality of
nasopharyngeal carcinoma. Cancer Lett. 374:22–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu MC and Yuan JM: Epidemiology of
nasopharyngeal carcinoma. Semin Cancer Biol. 12:421–429. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng H, Chen L, Chen Y-P, Li WF, Tang LL,
Lin AH, Sun Y and Ma J: The current status of clinical trials
focusing on nasopharyngeal carcinoma: A comprehensive analysis of
ClinicalTrials.gov database. PLoS One. 13:e01967302018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chan JYW, To VSH, Wong STS and Wei WI:
Radiation-induced squamous cell carcinoma of the nasopharynx after
radiotherapy for nasopharyngeal carcinoma. Head Neck. 36:772–775.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
8
|
Brancaccio M, Pivetta C, Granzotto M,
Filippis C and Mallamaci A: Emx2 and Foxg1 inhibit gliogenesis and
promote neuronogenesis. Stem Cells. 28:1206–1218. 2010.PubMed/NCBI
|
|
9
|
Verginelli F, Perin A, Dali R, Fung KH, Lo
R, Longatti P, Guiot MC, Del Maestro RF, Rossi S, di Porzio U, et
al: Transcription factors FOXG1 and Groucho/TLE promote
glioblastoma growth. Nat Commun. 4:29562013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Adesina AM, Nguyen Y, Guanaratne P,
Pulliam J, Lopez-Terrada D, Margolin J and Finegold M: FOXG1 is
overexpressed in hepatoblastoma. Hum Pathol. 38:400–409. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan DW, Liu VW, To RM, Chiu PM, Lee WY,
Yao KM, Cheung AN and Ngan HY: Overexpression of FOXG1 contributes
to TGF-β resistance through inhibition of p21WAF1/CIP1
expression in ovarian cancer. Br J Cancer. 101:1433–1443. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seoane J, Le HV, Shen L, Anderson SA and
Massagué J: Integration of Smad and forkhead pathways in the
control of neuroepithelial and glioblastoma cell proliferation.
Cell. 117:211–223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirschey MD, DeBerardinis RJ, Diehl AME,
Drew JE, Frezza C, Green MF, Jones LW, Ko YH, Le A, Lea MA, et al
Target Validation Team, : Dysregulated metabolism contributes to
oncogenesis. Semin Cancer Biol. 35 (Suppl 1):S129–S150. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ru P, Williams TM, Chakravarti A and Guo
D: Tumor metabolism of malignant gliomas. Cancers (Basel).
5:1469–1484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Attardi G and Schatz G: Biogenesis of
mitochondria. Annu Rev Cell Biol. 4:289–333. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li HJ, Sun XM, Li ZK, Yin QW, Pang H, Pan
JJ, Li X and Chen W: LncRNA UCA1 promotes mitochondrial function of
bladder cancer via the MiR-195/ARL2 signaling pathway. Cell Physiol
Biochem. 43:2548–2561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song IS, Jeong YJ, Jeong SH, Kim JE, Han
J, Kim TH and Jang SW: Modulation of mitochondrial ERβ expression
inhibits triple-negative breast cancer tumor progression by
activating mitochondrial function. Cell Physiol Biochem.
52:468–485. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Li F, Cui Y, Liu S and Sun H:
Mst1 overexpression combined with Yap knockdown augments thyroid
carcinoma apoptosis via promoting MIEF1-related mitochondrial
fission and activating the JNK pathway. Cancer Cell Int.
19:1432019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo C, Li Y, Wang H, Feng Z, Li Y, Long J
and Liu J: Mitochondrial accumulation under oxidative stress is due
to defects in autophagy. J Cell Biochem. 114:212–219. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong Z, Zhang S, Zeng Z, Wu H, Yang Q,
Xiong F, Shi L, Yang J, Zhang W, Zhou Y, et al: LOC401317, a
p53-regulated long non-coding RNA, inhibits cell proliferation and
induces apoptosis in the nasopharyngeal carcinoma cell line HNE2.
PLoS One. 9:e1106742014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zeng F, Xue M, Xiao T, Li Y, Xiao S, Jiang
B and Ren C: MiR-200b promotes the cell proliferation and
metastasis of cervical cancer by inhibiting FOXG1. Biomed
Pharmacother. 79:294–301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li JV, Chien CD, Garee JP, Xu J, Wellstein
A and Riegel AT: Transcriptional repression of AIB1 by FoxG1 leads
to apoptosis in breast cancer cells. Mol Endocrinol. 27:1113–1127.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pancrazi L, Di Benedetto G, Colombaioni L,
Della Sala G, Testa G, Olimpico F, Reyes A, Zeviani M, Pozzan T and
Costa M: Foxg1 localizes to mitochondria and coordinates cell
differentiation and bioenergetics. Proc Natl Acad Sci USA.
112:13910–13915. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Manoranjan B, Venugopal C, McFarlane N,
Doble BW, Dunn SE, Scheinemann K and Singh SK: Medulloblastoma stem
cells: Modeling tumor heterogeneity. Cancer Lett. 338:23–31. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Filippis R, Pancrazi L, Bjørgo K,
Rosseto A, Kleefstra T, Grillo E, Panighini A, Cardarelli F, Meloni
I, Ariani F, et al: Expanding the phenotype associated with FOXG1
mutations and in vivo FoxG1 chromatin-binding dynamics. Clin Genet.
82:395–403. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mariani J, Coppola G, Zhang P, Abyzov A,
Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M,
et al: FOXG1-dependent dysregulation of GABA/glutamate neuron
differentiation in autism spectrum disorders. Cell. 162:375–390.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang HW, Li J and Vogt PK: Domains of the
qin protein required for oncogenic transformation. Oncogene.
13:441–444. 1996.PubMed/NCBI
|
|
31
|
Li J, Thurm H, Chang HW, Iacovoni JS and
Vogt PK: Oncogenic transformation induced by the Qin protein is
correlated with transcriptional repression. Proc Natl Acad Sci USA.
94:10885–10888. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen J, Wu X, Xing Z, Ma C, Xiong W, Zhu X
and He X: FOXG1 expression is elevated in glioma and inhibits
glioma cell apoptosis. J Cancer. 9:778–783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Laissue P: The forkhead-box family of
transcription factors: Key molecular players in colorectal cancer
pathogenesis. Mol Cancer. 18:52019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Wang HQ, Wang AC, Li YX, Ding SS, An
XJ and Shi HY: Overexpression of Forkhead box Q1 correlates with
poor prognosis in papillary thyroid carcinoma. Clin Endocrinol
(Oxf). 90:334–342. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Qu Z, Jing S, Li X, Zhao C, Man S,
Wang Y and Gao W: Dioscin-6-O-acetate inhibits lung cancer cell
proliferation via inducing cell cycle arrest and caspase-dependent
apoptosis. Phytomedicine. 53:124–133. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang SH, Wu LW, Huang AC, Yu CC, Lien JC,
Huang YP, Yang JS, Yang JH, Hsiao YP, Wood WG, et al: Benzyl
isothiocyanate (BITC) induces G2/M phase arrest and apoptosis in
human melanoma A375.S2 cells through reactive oxygen species (ROS)
and both mitochondria-dependent and death receptor-mediated
multiple signaling pathways. J Agric Food Chem. 60:665–675. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Won YS and Seo KI: Lupiwighteone induces
caspase-dependent and -independent apoptosis on human breast cancer
cells via inhibiting PI3K/Akt/mTOR pathway. Food Chem Toxicol.
135:1108632020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lkhagvasuren K and Kim JK: Ziyuglycoside
II induces caspases-dependent and caspases-independent apoptosis in
human colon cancer cells. Toxicol In Vitro. 59:255–262. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zeng Y, Zhang Z, Wang W, You L, Dong X,
Yin X, Qu C and Ni J: Underlying mechanisms of apoptosis in HepG2
cells induced by polyphyllin I through Fas death and mitochondrial
pathways. Toxicol Mech Methods. 30:397–406. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nechushtan A, Smith CL, Hsu YT and Youle
RJ: Conformation of the Bax C-terminus regulates subcellular
location and cell death. EMBO J. 18:2330–2341. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Del Gaizo Moore V, Schlis KD, Sallan SE,
Armstrong SA and Letai A: BCL-2 dependence and ABT-737 sensitivity
in acute lymphoblastic leukemia. Blood. 111:2300–2309. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bivik CA, Larsson PK, Kågedal KM, Rosdahl
IK and Ollinger KM: UVA/B-induced apoptosis in human melanocytes
involves translocation of cathepsins and Bcl-2 family members. J
Invest Dermatol. 126:1119–1127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang B, Gui LS, Zhao XL, Zhu LL and Li
QW: FOXO1 is a tumor suppressor in cervical cancer. Genet Mol Res.
14:6605–6616. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pencheva N and Tavazoie SF: Control of
metastatic progression by microRNA regulatory networks. Nat Cell
Biol. 15:546–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li Y, Zhou X, Liu J, Gao N, Yang R, Wang
Q, Ji J, Ma L and He Q: Dihydroartemisinin inhibits the
tumorigenesis and metastasis of breast cancer via downregulating
CIZ1 expression associated with TGF-β1 signaling. Life Sci.
248:1174542020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jin Y, Liang Z and Lou H: The emerging
roles of fox family transcription factors in chromosome
replication, organization, and genome stability. Cells. 9:2582020.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xia YY, Yin L, Tian H, Guo WJ, Jiang N,
Jiang XS, Wu J, Chen M, Wu JZ and He X: HMGA2 is associated with
epithelial-mesenchymal transition and can predict poor prognosis in
nasopharyngeal carcinoma. OncoTargets Ther. 8:169–176. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Thiery JP, Acloque H, Huang RYJ and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
1390-890.
|
|
50
|
Bai M, Chen H, Ding D, Song R, Lin J,
Zhang Y, Guo Y, Chen S, Ding G, Zhang Y, et al: MicroRNA-214
promotes chronic kidney disease by disrupting mitochondrial
oxidative phosphorylation. Kidney Int. 95:1389–1404. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Biernacki M, Ambrożewicz E, Gęgotek A,
Toczek M, Bielawska K and Skrzydlewska E: Redox system and
phospholipid metabolism in the kidney of hypertensive rats after
FAAH inhibitor URB597 administration. Redox Biol. 15:41–50. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Davidson SM, Arjun S, Basalay MV, et al:
The 10th Biennial Hatter Cardiovascular Institute workshop:
cellular protection - evaluating new directions in the setting of
myocardial infarction, ischaemic stroke, and cardio-oncology. Basic
Res Cardiol. 113:432018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gonzalez NR, Liou R, Kurth F, Jiang H and
Saver J: Antiangiogenesis and medical therapy failure in
intracranial atherosclerosis. Angiogenesis. 21:23–35. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
DeLeon-Pennell KY, Mouton AJ, Ero OK, Ma
Y, Padmanabhan Iyer R, Flynn ER, Espinoza I, Musani SK, Vasan RS,
Hall ME, et al: LXR/RXR signaling and neutrophil phenotype
following myocardial infarction classify sex differences in
remodeling. Basic Res Cardiol. 113:402018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chandra M, Escalante-Alcalde D, Bhuiyan
MS, Orr AW, Kevil C, Morris AJ, Nam H, Dominic P, McCarthy KJ,
Miriyala S, et al: Cardiac-specific inactivation of LPP3 in mice
leads to myocardial dysfunction and heart failure. Redox Biol.
14:261–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sanyal T and Bhattacharjee P,
Bhattacharjee S and Bhattacharjee P: Hypomethylation of
mitochondrial D-loop and ND6 with increased mitochondrial DNA copy
number in the arsenic-exposed population. Toxicology. 408:54–61.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rice AC, Keeney PM, Algarzae NK, Ladd AC,
Thomas RR and Bennett JP Jr: Mitochondrial DNA copy numbers in
pyramidal neurons are decreased and mitochondrial biogenesis
transcriptome signaling is disrupted in Alzheimer's disease
hippocampi. J Alzheimers Dis. 40:319–330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wen SL, Zhang F and Feng S: Decreased copy
number of mitochondrial DNA: A potential diagnostic criterion for
gastric cancer. Oncol Lett. 6:1098–1102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Weber LW, Boll M and Stampfl A:
Hepatotoxicity and mechanism of action of haloalkanes: Carbon
tetrachloride as a toxicological model. Crit Rev Toxicol.
33:105–136. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Quarato G, Piccoli C, Scrima R and
Capitanio N: Functional imaging of membrane potential at the single
mitochondrion level: Possible application for diagnosis of human
diseases. Mitochondrion. 11:764–773. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lugli E, Troiano L and Cossarizza A:
Polychromatic analysis of mitochondrial membrane potential using
JC-1. Curr Protoc Cytom Chapter. 7:322007.PubMed/NCBI
|
|
62
|
Chen C, Luo Y, Su Y and Teng L: The
vitamin D receptor (VDR) protects pancreatic beta cells against
Forkhead box class O1 (FOXO1)-induced mitochondrial dysfunction and
cell apoptosis. Biomed Pharmacother. 117:1091702019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D,
Bartlam M and Rao Z: Crystal structure of mitochondrial respiratory
membrane protein complex II. Cell. 121:1043–1057. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liu X, Zhou Z, Wang Z, Li X, Lu G and Tong
J: SDHA-mediated Warburg effect in malignantly transformed human
bronchial epithelial cells following long-term exposure to radon.
Environ Toxicol. 35:861–866. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Eguchi K and Nakayama K: Prolonged hypoxia
decreases nuclear pyruvate dehydrogenase complex and regulates the
gene expression. Biochem Biophys Res Commun. 520:128–135. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Song E, Tang S, Xu J, Yin B, Bao E and
Hartung J: Lenti-siRNA Hsp60 promote bax in mitochondria and
induces apoptosis during heat stress. Biochem Biophys Res Commun.
481:125–131. 2016. View Article : Google Scholar : PubMed/NCBI
|