Introduction
Senescence is defined as the decline of
physiological functions with aging and affects most living
organisms (1). Adverse changes at
cellular and molecular levels caused by senescence lead to
increased risks of several diseases, such as cancer, cardiovascular
diseases, diabetes, osteopenia and osteoporosis (2).
Bone is constantly remodelled throughout life by the
balanced activity of osteoblastic bone formation and osteoclastic
bone resorption; however, the balance between bone resorption and
formation is lost due to aging, which results in age-related bone
loss, including osteoporosis, and an increased risk of fracture
(3). Some aging mechanisms are
related to changes in the expression and signalling of local
factors in the bone microenvironment (4). There are three important cell types
for bone remodelling: Osteoblasts, osteocytes and osteoclasts (OCs)
(5). Osteocyte and osteoblast
senescence in the bone microenvironment has been investigated
(6), whereas OC and OC precursor
(OCP) senescence in the bone microenvironment has not been analysed
in detail. OCs differentiate from OCPs, which are precursor cells
derived from the monocyte/macrophage lineage (7). OCPs are recruited from the bloodstream
into bone by various factors released at sites undergoing
resorption in the bone microenvironment, and OCPs are subsequently
differentiated into OCs responsible for bone resorption (7). Cao et al (8) revealed that aging is accompanied by
increased receptor activator of nuclear factor (NF)-κB ligand
(RANKL) and macrophage colony-stimulating factor expression, which
can increase stromal/osteoblastic cell-induced osteoclastogenesis
and expansion of the OCP pool. If the number of OCPs that cannot
differentiate increases, the number of aging OCPs increases. Along
with stem cell aging, their ability to differentiate into various
cell types is also altered (9).
Senescence is an unexplained phenomenon, which
occurs in the microenvironment where bone remodelling occurs. In
recent years, it has become clear that senescent cells do not
simply cease growing but also secrete various proteins, such as
inflammatory cytokines and chemokines (10). This secretory phenomenon is known as
the senescence-associated secretory phenotype (SASP). SASP is
predominantly exhibited by senescent osteoprogenitors, myeloid
cells and osteocytes (11,12). Farr et al (12) suggested that senescent osteocytes
and their SASP contribute to age-related bone loss by assessing the
critical roles of osteocytes in orchestrating bone remodelling.
However, additional senescent cell types in the bone
microenvironment and their corresponding SASP may be uncovered. The
aim of the present study was to elucidate senescent OCPs and their
corresponding SASP.
Materials and methods
Cell culture
RAW264.7 cells for use as OCPs were obtained from
the RIKEN BioResource Center. Cells were cultured in α-minimum
essential medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% foetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin at 37°C in a humidified atmosphere containing 5%
CO2 and were passaged every 3–4 days. RAW264.7
cell-derived OCs appear to form better in response to RANKL
stimulation after passage (P)4, but cannot form after P18-20
(13). Therefore, it was assumed
that cells at P20 would be replicative senescent cells, and
RAW264.7 cells were grown until P5, P10 and P20.
Cell proliferation assay
RAW264.7 cells (P5, P10, and P20) were seeded on
96-well culture plates at 5×103 cells/well and were
cultured for 1–4 days. Subsequently, the WST-8 reagent in a Cell
Counting Kit-8 (Dojindo Molecular Technologies, Inc.) was added to
the cells (10 µl/well), followed by incubation for 2 h at 37°C. The
cell proliferation was determined by measuring the absorbance at
450 nm using a microplate reader (Benchmark Plus™ Microplate
Spectrophotometer; Bio-Rad Laboratories, Inc.) (14).
Senescence-associated β-galactosidase
(SA-β-gal) staining and flow cytometric analysis
SA-β-gal staining was performed using a Cellular
Senescence Detection Kit-SPiDER-βgal (Dojindo Molecular
Technologies, Inc.) in accordance with the manufacturer's
instructions. RAW264.7 cells (P5, P10 and P20) were seeded in a
35-mm culture dish at a density of 5×104 cells/dish and
incubated overnight. The cells were washed with 2 ml Hank's
balanced salt solution (HBSS) and bafilomycin A1 working solution
(1 ml) was added to the culture dish for 1 h. SPiDER-βgal working
solution (1 ml) and 1 mg/ml Hoechst 33342 (1 µl; Dojindo Molecular
Technologies, Inc.) were then added to the culture and the cells
were incubated for 30 min. After the supernatant was removed, the
cells were washed twice with HBSS (2 ml) and HBSS (2 ml) was added
to the culture dish. All images were obtained under a fluorescence
microscope (Ti-E; Nikon Corporation). The percentage of positive
cells was manually computed in five random fields per section at
×400 magnification (15).
Flow cytometry was performed using a protocol
similar to SA-β-gal staining. RAW264.7 cells (P5, P10, and P20)
were seeded in a 35-mm culture dish at a density of
1×105 cells/dish. Cells were treated in the same manner
as for SA-β-gal staining; however, they were not treated with
Hoechst 33342. Flow cytometric analysis was performed using a BD
FACSCalibur and BD FACSDiva (v8.0.1; BD Biosciences).
Quantification of telomere length
Genomic DNA was isolated from RAW264.7 cells (P5,
P10, and P20) by the phenol-chloroform extraction method using
phenol/chloroform/isoamyl alcohol (25:24:1; cat. no. 311-90151,
Nippon Gene Co., Ltd.). Total telomere length was determined by a
telomere hybridization protection assay using 0.2 µg denatured
genomic DNA as described previously (16).
Tartrate-resistant acid phosphatase
(TRAP) staining
RAW264.7 cells (P5, P10, and P20) were seeded on
12-well culture plates at 2.5×103 cells/well. The cells
were treated with RANKL (50 ng/ml; Oriental Yeast Co. Ltd.) for 7
days at 37°C. The culture medium supplemented with RANKL was
replaced every 3 days. Subsequently, the cells were stained using a
TRAP staining kit (Cosmo Bio Co., Ltd.) in accordance with the
manufacturer's instructions. TRAP-positive multinucleated cells
were identified as OCs under the light microscope (Ti-E; Nikon
Corporation) (14).
TRAP activity assay
RAW264.7 cells (P5, P10, and P20) were seeded on
96-well culture plates at 2×103 cells/well. The cells
were treated with RANKL (50 ng/ml) for 4 days at 37°C. The cells
were then fixed with ethanol/acetone as described previously
(17) and evaluated for TRAP
activity using a TRAP solution kit (Oriental Yeast Co. Ltd.) in
accordance with the manufacturer's instructions. Briefly, 150 µl 50
mM citrate buffer (pH 4.5) containing 5.5 mM p-nitrophenol
phosphate and 10 mM sodium tartrate was added to each well. After
incubation for 60 min at room temperature, 50 µl 0.1 N NaOH was
added and the absorbance at 405 nm was determined using a
microplate reader (14).
Measurement of interleukin (IL)-6,
tumour necrosis factor (TNF)-α and nitric oxide (NO)
production
RAW264.7 cells (P5, P10 and P20) at 1×106
cells/well in 24-well culture plates were cultured for 24 h in
serum-free medium. The culture supernatants were centrifuged at 4°C
for 10 min at 1,000 × g, harvested and used immediately. IL-6 was
measured via a quantitative sandwich enzyme immunoassay method
using a Mouse IL-6 Assay Kit (cat. no. 27768; Immuno-Biological
Laboratories Co., Ltd.) in accordance with the manufacturer's
instructions. Optical density (OD) values at 450 nm were measured
using a microplate reader. The levels of TNF-α in culture
supernatants were measured using a TNF-α Mouse ELISA kit Quantikine
(cat. no. MTA00B; R&D Systems, Inc.) in accordance with
manufacturer's instructions. OD values at 450 nm were measured
using a microplate reader. The levels of
NO2−/NO3− in culture
supernatants were measured using an
NO2−/NO3− Assay Kit-C
II consisting of Griess reagents (cat. no. NK05; Dojindo Molecular
Technologies, Inc.) according to the manufacturer's instructions.
Samples were reacted with Griess reagent and absorbance was
measured at 540 nm using a microplate reader. It is common practice
to quantitate total
NO2−/NO3− as a measure
of NO levels (18).
Isolation and quantification of
exosomes
Exosome isolation using an ExoQuick exosome
precipitation kit (System Biosciences) was performed in accordance
with the manufacturer's instructions. Briefly, 1×106
RAW264.7 cells (P5, P10 and P20) were cultured for 48 h in α-MEM
containing 10% exosome-depleted FBS (System Biosciences). The
culture supernatant was collected and centrifuged at 3,000 × g for
15 min at 4°C to remove intact cells and cell debris. ExoQuick
exosome precipitation solution (2 ml) was added to 10-ml culture
supernatant. The resulting solution was mixed by inverting the tube
and then incubated for 12 h at 4°C. This mixture was then
centrifuged at 1,500 × g for 30 min at room temperature. The
supernatant was discarded and the precipitate consisted of exosomes
(19). Exosome quantification and
western blot analysis of exosomes were performed using the exosome
pellets. For exosome quantification, the exosome pellets were
resuspended in Exosome Binding Buffer (a component of the CD63
ExoELISA kit). The total number of exosomes was determined using a
CD63 ExoELISA kit (cat. no. EXOEL-CD63A-1, System Bioscience)
following the manufacturers' instructions; this kit detects the
general exosome marker CD63 by ELISA (20). For western blot analysis of
exosomes, the exosome pellets were resuspended in
radioimmunoprecipitation buffer (RIPA; cat. no. sc-24948; Santa
Cruz Biotechnology, Inc.). Western blot analysis of exosomes was
performed using the resulting solution. Exosomes prepared from
RAW264.7 cells at P5, P10 and P20 were termed Exo-P5, Exo-P10 and
Exo-P20, respectively.
Nanosight particle tracking
analysis
The isolated exosome pellets were diluted at 1:5,000
with PBS and injected into the Nanosight LM10 system (Malvern
Panalytical Ltd.). Capture and analysis settings were manually set
in accordance with the manufacturer's instructions. Particles were
visualised by laser light scattering and their Brownian motion was
captured on digital video. Five separate runs were conducted for
each sample. The recorded videos were analysed using Nanoparticle
Tracking Analysis 2.3 software (Malvern Panalytical Ltd.) and the
size distribution of particles was determined (21).
Western blot analysis
RAW264.7 cells (P5, P10 and P20) were seeded in
60-mm culture plates and lysed using RIPA buffer (cat. no.
sc-24948; Santa Cruz Biotechnology, Inc.). The lysates and exosomal
proteins were analysed by western blotting as described previously
(22). The following primary
antibodies were used: Rabbit polyclonal antibodies against p53
(1:200; cat. no. sc-6243; Santa Cruz Biotechnology, Inc.),
transforming growth factor β1 (TGF-β1; 1:1,000; cat. no. 3711; Cell
Signaling Technology, Inc.), histone H2A.X (H2A.X; 1:1,000; cat.
no. 2595; Cell Signaling Technology, Inc.), inducible nitric oxide
synthase (iNOS; 1:2,000; cat. no. ab3523; Abcam), NF-κB (1:1,000;
cat. no. GTX102090; GeneTex, Inc.), CD63 (1:1,000; cat. no.
EXOAB-CD63A-1; System Biosciences) and mammalian target of
rapamycin (mTOR; 1:1,000; cat. no. 2972; Cell Signaling Technology,
Inc.), rabbit monoclonal antibodies against tumour susceptibility
gene 101 (TSG101; 1:1,000; cat. no. ab125011; Abcam) and
phosphorylated (p)-H2A.X (Ser139) (1:1,000; cat. no. 2577; Cell
Signaling Technology, Inc.), mouse monoclonal antibodies against
receptor activator of NF-κB (RANK; 1:500; cat. no. ab13918; Abcam),
hypoxia-inducible factor (HIF)-1α (1:1,000; cat. no. NB100-131;
Novus Biologicals LLC) and nuclear factor of activated T cells
cytoplasmic 1 (NFATc1; 1:200; cat. no. sc-7294; Santa Cruz
Biotechnology, Inc,), and a goat polyclonal antibody against
β-actin (1:1,000; cat. no. sc-1616; Santa Cruz Biotechnology,
Inc.). The secondary antibodies used were HRP-conjugated anti-goat
IgG (1:2,000; cat. no. P0449; Dako; Agilent Technologies, Inc.),
HRP-conjugated anti-mouse IgG (1:1,000; cat. no. 7076; Cell
Signaling Technology, Inc.) and HRP-conjugated anti-rabbit IgG
(1:10,000; cat. no. 458; MBL International Co.). Signals were
detected by chemiluminescence using a Pierce SuperSignal Western
Blotting Kit (Pierce; Thermo Fisher Scientific, Inc.). β-actin was
evaluated as an internal control to confirm equal amounts of total
protein.
Statistical analysis
All data are expressed as the mean ± standard
deviation of three independent experiments. Statistical analyses
were performed using one-way ANOVA and Tukey-Kramer test for
intergroup comparisons in each experiment. P<0.05 was considered
to indicate a statistically significant difference.
Results
Serial passaging leads to replicative
senescence
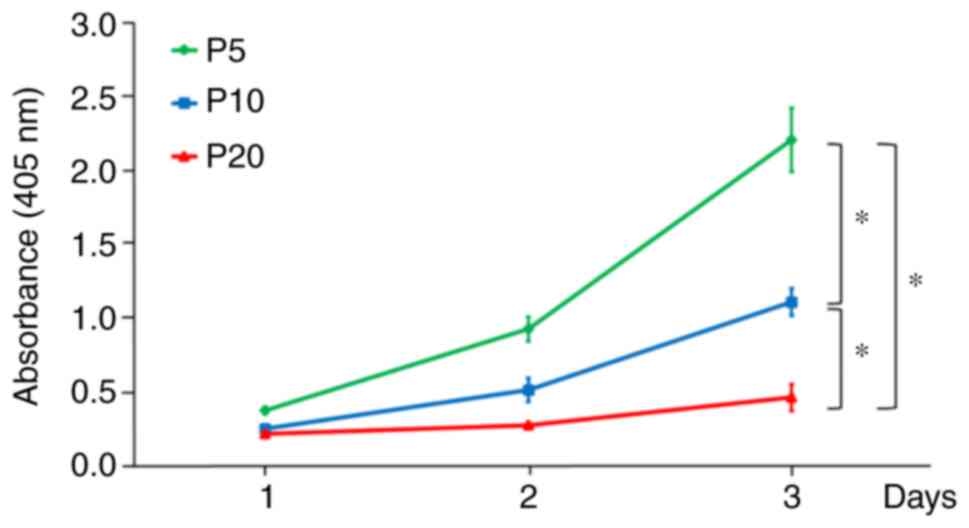
A decrease in cell proliferation was detected during
the progression of replicative senescence. The proliferation of
cells at P20 was significantly lower than that of cells at P5 and
P10 (Fig. 1). In addition, the
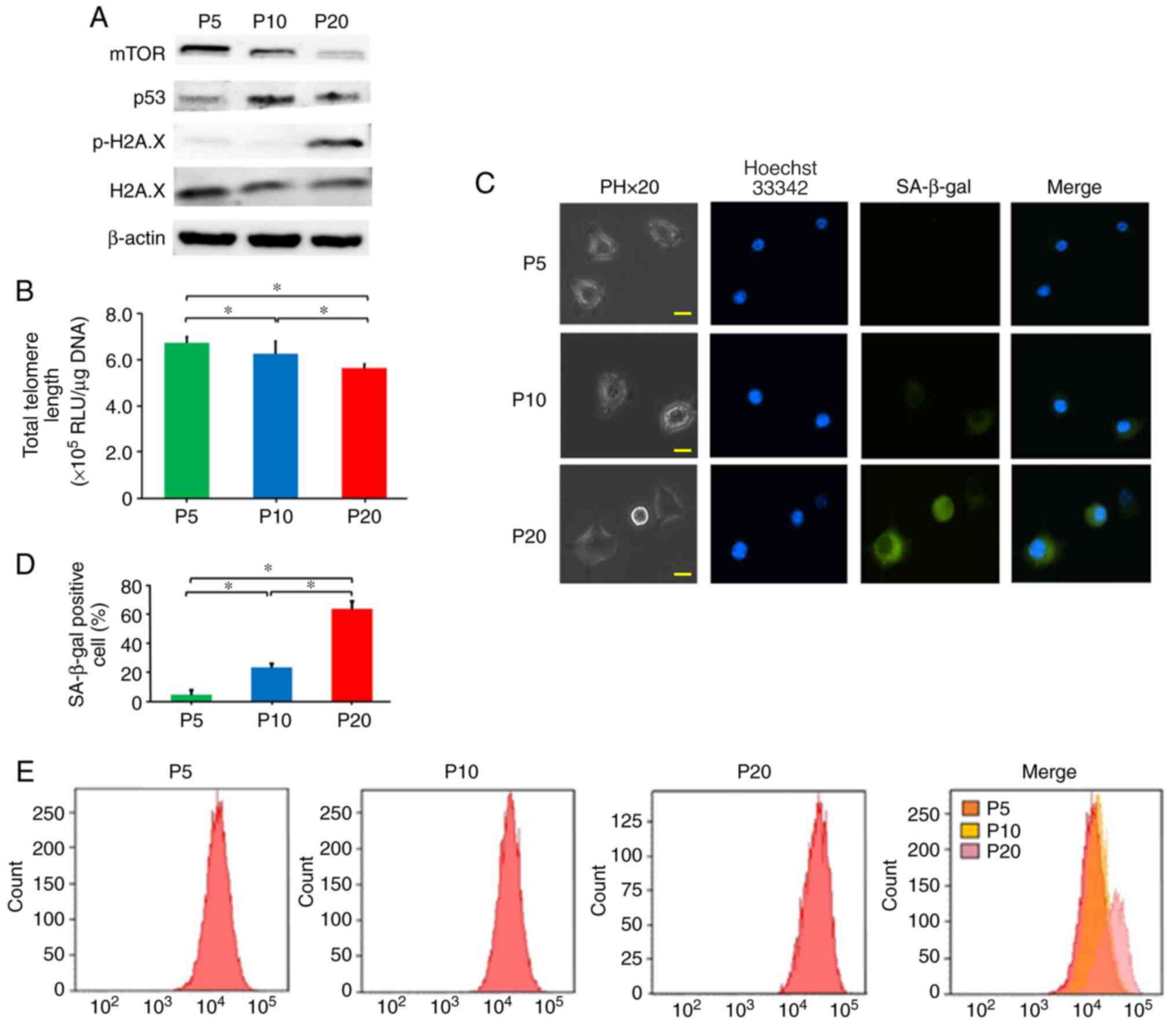
protein expression levels of mTOR in cells at P20 were decreased
compared with those at P5 and P10 (Fig.
2A). The protein expression levels of p-H2A.X in cells at P20
were increased compared with those at P5 and P10 (Fig. 2A). Telomere length at P20 was
decreased compared with that at P5 and P10 (Fig. 2B). The protein expression levels of
p53 in cells at P10 was increased compared with those at P5;
however, when cells reached P20 and exhibited reduced
proliferation, the protein expression levels of p53 were lower than
those at P10 (Fig. 2A).
RAW264.7 cells at P5, P10 and P20 were subjected to
SA-β-gal staining (Fig. 2C). The
senescent status of the cell cultures was measured by SA-β-gal
quantitation. SA-β-gal-positive cells at P20 were significantly
increased (63.2%) compared with those at P5 (4.5%) and P10 (23.2%)
(Fig. 2D). Furthermore, using flow
cytometry, the fluorescence intensity of SA-β-gal staining was
analysed (Fig. 2E). The
fluorescence intensity of cells at P20 was increased compared with
that at P5 and P10. Thus, it was confirmed that cells at P20 were
replicative senescent cells.
Differentiation of RAW264.7 cells
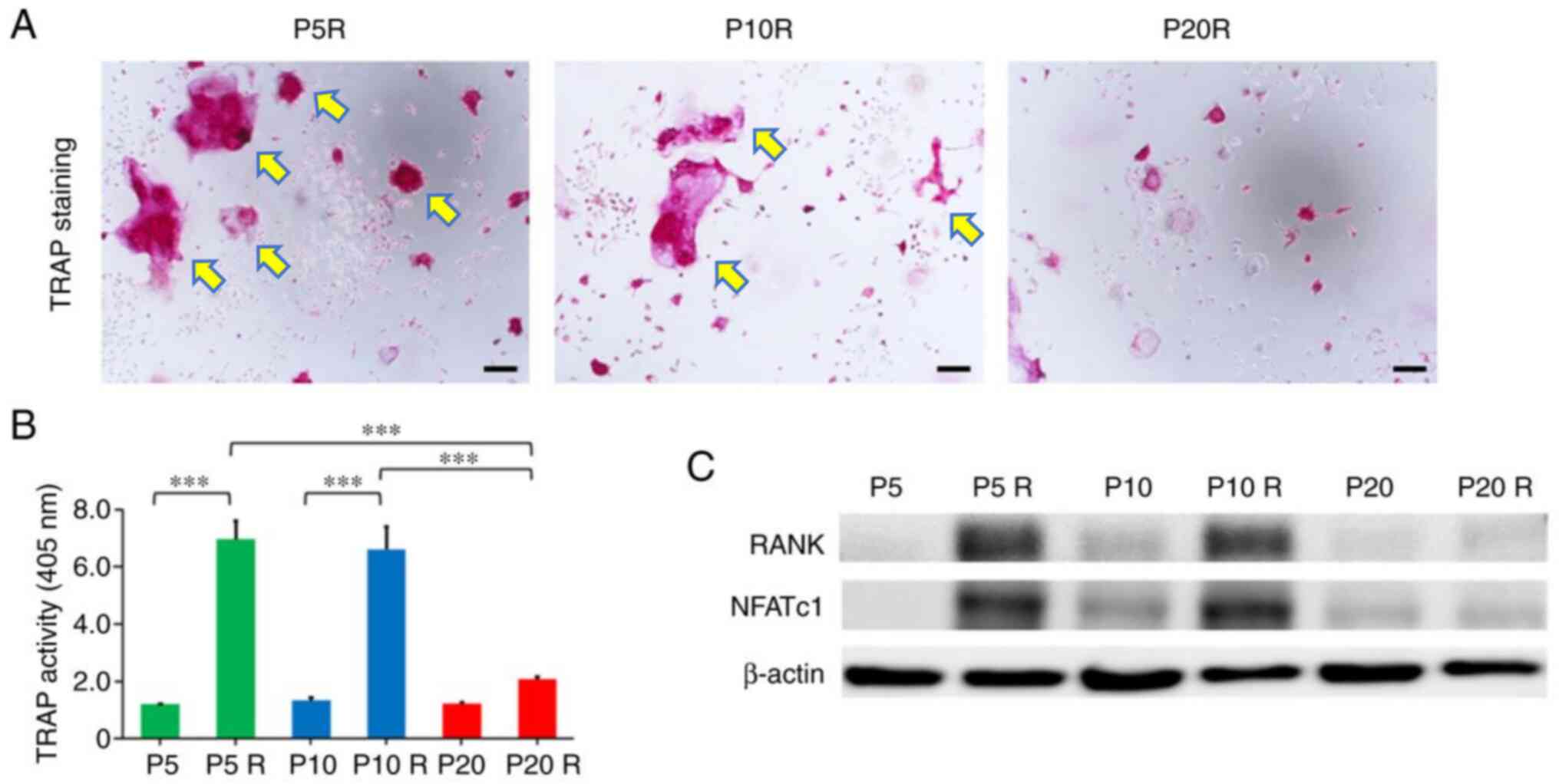
To investigate the effect of replicative senescence
on OC differentiation, TRAP staining was performed to determine
TRAP enzymatic activity. The level of TRAP enzymatic activity in
cultured cell lysates has been reported to be correlated with the
relative number of OCs observed by TRAP staining (23). On day 7, RANKL induced the formation
of large multinucleated (≥5 nuclei) OC-like cells at P5 and P10;
however, there were no TRAP-positive multinucleated cells detected
at P20 (Fig. 3A). To confirm the
potential effect of replicative senescence on RANKL-induced OC
differentiation, TRAP enzymatic activity was determined. The level
of TRAP activity in cells treated with RANKL at P20 was
significantly lower than that at P5 and P10 (Fig. 3B). Furthermore, the protein
expression levels of RANK and NFATc1 were increased in cells
treated with RANKL (50 ng/ml) at P5 and P10 compared with those at
P20 (Fig. 3C). These results
suggested that RAW264.7 cells cannot differentiate into OC-like
cells while aging.
Effect of senescence in RAW264.7 cells
on SASP factors
The expression levels of SASP factors TGF-β1, iNOS
and NF-κB in cells at P20 were increased compared with those at P5
and P10. In addition, HIF-1α protein expression was increased in
cells at P20 compared with that at P5 and P10 (Fig. 4A). Furthermore, the present study
detected secretion of SASP factors IL-6, TNF-α and NO from RAW264.7
cells. The levels of IL-6, TNF-α and NO in the culture supernatants
at P20 were significantly increased compared with those at P5 and
P10 (Fig. 4B).
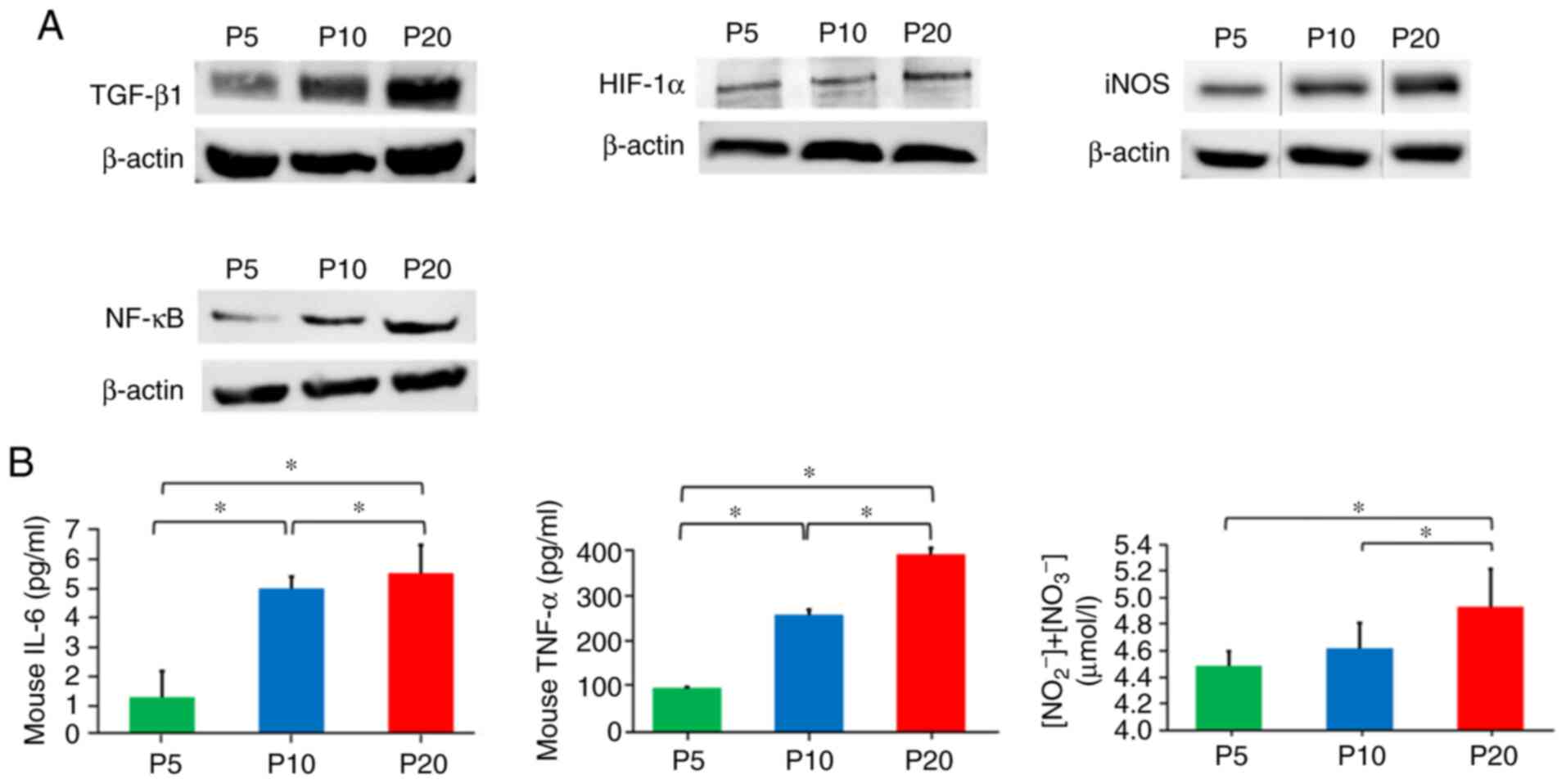 | Figure 4.Effect of senescence on
senescence-associated secretory phenotype factors in RAW264.7
cells. (A) Measurement of protein expression levels of TGF-β1,
HIF-1α, iNOS and NF-κB, as determined by western blot analysis. (B)
ELISA was used to determine
NO2−/NO3−, IL-6 and
TNF-α levels in the culture medium. Data are presented as the mean
± standard deviation. *P<0.05 (one-way ANOVA and Tukey-Kramer
test). HIF-1α, hypoxia-inducible factor-1α; IL-6, interleukin 6;
iNOS, inducible nitric oxide synthase; NF-κB, nuclear factor-κB;
NO, nitric oxide; P, passage; TGF-β1, transforming growth factor
β1; TNF-α, tumour necrosis factor-α. |
Effect of senescence in RAW264.7 cells
on isolation of exosomes
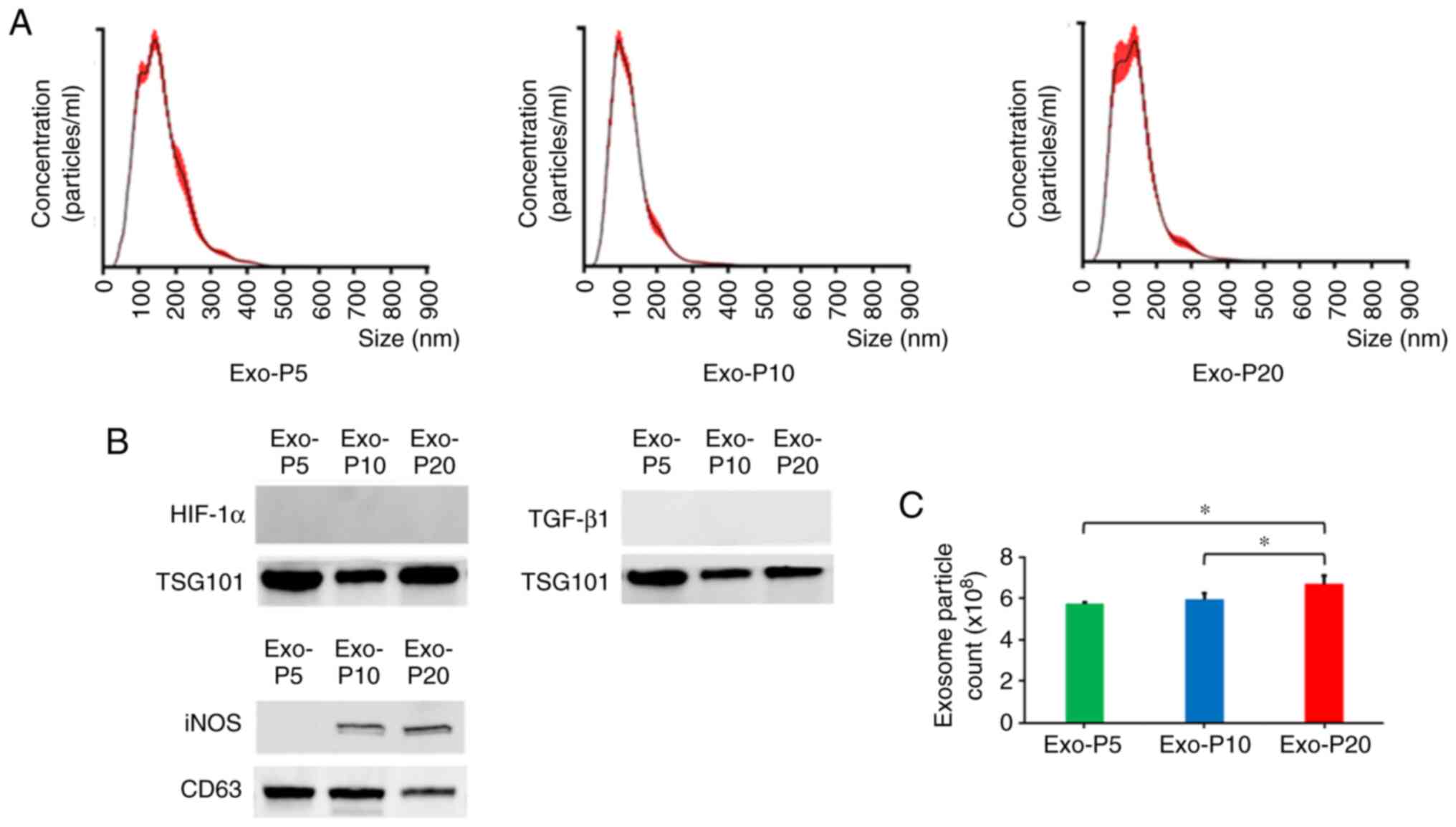
To characterise vesicles released from RAW264.7
cells, size detection was performed by Nanosight particle tracking
analysis (Fig. 5A). Vesicle size
peaks at P5, P10 and P20 were 128, 100 and 133 nm, respectively.
Exosomes are vesicles derived from the endosomal membrane with
diameters 40–150 nm (24,25). Subsequently, the expression of
exosome markers (CD63 and TSG101) were detected by western blotting
to confirm the identity of exosomes (Fig. 5B). The results indicated successful
isolation of exosomes from RAW264.7 cells. Expression of
tetraspanin protein CD63 was detected by ELISA to determine the
concentration of exosomes (Fig.
5C). The number of exosomes at P20 was increased compared with
that at P5 and P10. In addition, iNOS was detected in exosomes at
P10 and P20, but not at P5; however, TGF-β1 and HIF-1α were not
detected in exosomes at P5, P10 and P20 (Fig. 5B). In these SASP factors, it was
suggested that only iNOS may be contained in exosomes and
specifically released.
Discussion
Cellular senescence was initially described as the
finite proliferative capacity of cultured normal human fibroblasts
by Hayflick and Moorhead in 1961 (26). After its discovery, cellular
senescence was defined as a status characterised by irreversible
arrest of cell proliferation (27).
Growing evidence has suggested that replicative senescence can be
used as an in vitro surrogate for aging (6,28–30).
The present study established replicative senescent cells by serial
passaging of RAW264.7 cells. RAW264.7 cells are widely known as
OCPs and serve an important role in in vitro studies
(13,31,32).
Cellular senescence is defined by irreversible arrest of the cell
cycle, changes in cell morphology (large and flattened cells) and
expression of specific biomarkers (33). Among the specific biomarkers of
senescent cells, the lysosomal enzyme SA-β-gal is widely used
(34,35). Induction of replicative senescence
in RAW264.7 cells at P20 was confirmed by SA-β-gal staining. In
addition, induction of p53 causes cellular senescence by reversible
cell cycle arrest (36–38). It is known that p53 can inhibit the
mTOR pathway (39,40); mTOR signalling regulates cell growth
and metabolism (41). In the
present study, the protein expression levels of p53 in cells at P10
were increased compared with those at P5; however, the protein
expression levels of p53 in cells at P20 were comparatively lower
than those at P10. p53 functions decline with senescence and SASP
is more intense when cells lose the functions of p53 (10,42).
The growth curve of replicative senescent cells at P20 was almost
flat and mTOR protein expression in cells at P20 was decreased
compared with that at P5. Conversely, the protein expression levels
of p-H2A.X in cells at P20 were increased compared with those at
P5. Telomere length at P20 was also decreased compared with that at
P5. Therefore, the present study confirmed that cells at P20 were
replicative senescent cells.
The present study observed differences in the
ability of RAW264.7 cells to differentiate into OCs, which depended
on passage number. A previous study reported that RAW264.7 cells do
not differentiate into OCs in response to RANKL stimulation at
P18-20 (13). The present results
demonstrated that the level of TRAP activity in cells treated with
RANKL at P20 was significantly lower than that at P5 and P10.
Moreover, cellular senescence reduced RANKL-induced osteoclastic
differentiation of RAW264.7 cells by inhibiting RANK expression.
Cellular senescence also attenuated RANKL-induced TRAP activity and
the protein expression levels of NFATc1. These results suggested
that OCPs may gradually lose their self-renewal and regenerative
potentials, and cannot differentiate into OCs while aging. As a
result, the number of senescent OCPs in the bone microenvironment
may continue to increase because numerous OCPs are unable to
differentiate into OCs.
SASP is a phenomenon in which senescent cells
secrete proinflammatory cytokines, chemokines, growth factors and
proteases (43). A beneficial
function of SASP may be promotion of immune cell migration through
secretion of proinflammatory cytokines (44). SASP is characterised by high level
secretion of the cytokine IL-6, which is a crucial mediator of
inflammation (45). Circulation of
proinflammatory cytokines, such as IL-6, can induce not only
cancer, but also various chronic inflammatory diseases (46). There are >60 known SASP factors,
including ILs, chemokines and inflammatory molecules, such as
TGF-β1, IL-6, TNF-α and NO (47).
Accumulation of senescent cells can lead to tissue damage by
increased signalling of proinflammatory cytokines caused by
propagation of oxidative stress due to mitochondrial dysfunction in
neighbouring cells (48). Farr
et al (12,49) reported that multiple senescent cell
types, such as osteoprogenitors, myeloid cells, B cells, T cells
and osteocytes, accumulated in the bone microenvironment with aging
in vivo and suggested that senescent osteocytes may be
linked to aging-associated bone loss. The present study focused on
OCPs as senescent cells that affect the bone microenvironment. The
results of the present study detected increases in the protein
expression levels of TGF-β1, NF-κB and iNOS in senescent RAW264.7
cells, and increases in SASP factors, such as IL-6, TNF-α and NO,
in the culture supernatant of senescent RAW264.7 cells. These
results suggested high amounts of SASP factors (e.g., IL-6, TNF-α
and NO) in the bone microenvironment.
TNF-α is a proinflammatory cytokine and potent
inducer of bone resorption, which serves major roles in bone
metabolism and inflammatory bone diseases (50). TNF-α directly induces the formation
of TRAP-positive multinucleated OCs from OCPs in the absence of
RANKL-mediated activation of NF-κB signalling (51). In addition, TNF-α can induce RANK
expression in OCPs (52). However,
in the present study, RAW264.7 cells exhibited inhibition of
osteoclastogenesis after senescence, which were unaffected by
TNF-α. Although TNF-α expression was increased in P20 cells, it did
not induce RANK expression and formation of TRAP-positive
multinucleated cells.
IL-6 is believed to indirectly induce production of
RANKL by osteoblastic/stromal cells, which in turn stimulates OC
activity and the commitment of OCPs into mature OCs (53). In addition, IL-6 has a direct effect
on OC activity via differential regulation of RANKL-induced OC
formation by specifically modulating phosphorylation of NF-κB,
extracellular signal-regulated kinase and c-Jun N-terminal kinase
in a RANKL concentration-dependent manner; a stimulatory effect is
observed under the condition of low RANKL, whereas an inhibitory
effect is observed when the level of RANKL is markedly enhanced
(54). Yokota et al
(55) reported that the combination
of TNF-α and IL-6 differentiated TRAP-positive multinucleated cells
that resemble OCs, which had in vitro and in vivo
bone resorption activities. Although TNF-α and IL-6 drive
osteoclastogenesis, the present study revealed that RAW264.7 cells
had inhibited osteoclastogenesis after senescence and that
inhibition of osteoclastogenesis by NO exceeded acceleration of
osteoclastogenesis by TNF-α and IL-6 in senescent OCPs.
OC formation and bone resorption are inhibited by
elevated levels of the multifunctional signal molecule NO in
vivo and in vitro (56–58).
NO is a multifunctional gaseous molecule produced by NOS using
L-arginine and molecular oxygen as substrates. It is known that NOS
has three major isoforms: Neuronal NOS and endothelial NOS, which
are constitutively expressed in cells, and iNOS, the expression of
which is induced by inflammatory cytokines, endotoxins, hypoxia and
oxidative stress (59,60). iNOS continuously produces a high
concentration of NO (61). Zheng
et al (62) reported that
RANKL may induce iNOS expression in OCPs and revealed that high NO
concentrations produced by iNOS inhibited OC differentiation. NO
inhibited differentiation of OCPs after senescence in the bone
microenvironment, which increased senescent OCPs. The present
results demonstrated that the protein expression levels of HIF-1α
and iNOS at P20 were increased compared with those at P5 and P10.
HIF-1α induces NO production by induction of iNOS in mouse
macrophages (63). HIF signalling
is both necessary and sufficient to inhibit osteoclastogenesis
through modulation of osteoprotegerin (64). From this viewpoint of SASP factors,
the present results indicated that inhibition of osteoclastogenesis
by NO exceeded acceleration of osteoclastogenesis by TNF-α and IL-6
in senescent OCPs.
Exosomes are vesicles derived from the endosomal
membrane with diameters between 40 and 150 nm (24,25).
Pools of exosomes are packed in multivesicular endosomes (MVEs) and
released into the extracellular space after fusion of MVEs with the
plasma membrane (24,65). Various proteins, lipids and nucleic
acids in exosomes cause functional and physiological changes via
transmission to other cells, and exosomes have important roles in
intercellular communication (66,67).
The results of the present study demonstrated that the number of
exosomes at P20 was significantly higher than that at P5 and P10.
In addition to secretory proteins, senescent cells may exhibit
increased secretion of exosomes (68). In the present study, SASP factors in
exosomes were analysed by western blotting. The protein expression
of iNOS was detected in exosomes at P20 and P10, but not at P5.
Exosomes transmit carried signals to distant tissues and cells by
circulating in body fluids, the contents of exosomes can reflect
the physiological and pathological processes in cells (69). The present results suggested that
iNOS-containing senescent OCP-derived exosomes may affect cellular
communication not only locally in neighbouring cells, but also
distally and systemically. iNOS-containing exosomes may be involved
in inflammatory processes that serve pivotal roles in a large
number of pathological states, including bone-destructive diseases,
rheumatoid arthritis, inflammatory diseases and cancer.
In conclusion, cellular senescence reduced RANKL-
induced osteoclastic differentiation of RAW264.7 cells by
inhibiting RANK expression. In addition, senescent RAW264.7 cells
exhibited increased production of SASP factors (IL-6, TNF-α and NO)
and increased release of iNOS in exosomes. It is possible that
clarification of the association between cellular senescence and
SASP in OCPs may improve understanding of age-related bone changes
and bone disorders, and further studies are required in
vivo.
Acknowledgements
The authors would like to thank Ms. Shinobu Osawa
M.Sc. (Department of Oral and Maxillofacial Surgery, Hyogo College
of Medicine, Nishinomiya, Japan) for technical assistance.
Funding
The present study was supported by JSPS KAKENHI
(grant nos. 18K09825 and 21K10106 to KT, 18K17124 to JT and
19K19182 to MU) and by a Grant-in-Aid for Graduate Students, Hyogo
College of Medicine, 2020 (to HH).
Availability of data and materials
The datasets used and/or analysed during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
HH, KT and KN conceived and designed the study. HH,
KT, MU, MO and JT performed the experiments. HH, KT and MU analysed
the data. HH, KT, MU and HK interpretated the data. HH, KT and HK
wrote the manuscript. KT and HK confirm the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
López-Otín C, Blasco MA, Partridge L,
Serrano M and Kroemer G: The hallmarks of aging. Cell.
153:1194–1217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singh T and Newman AB: Inflammatory
markers in population studies of aging. Ageing Res Rev. 10:319–329.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roberts S, Colombier P, Sowman A, Mennan
C, Rolfing HD, Guicheux J and Edwards JR: Ageing in the
musculoskeletal system: Cellular function and dysfunction
throughout life. Acta Orthop. 87:15–25. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marie PJ: Bone cell senescence: Mechanisms
and perspectives. J Bone Miner Res. 29:1311–1321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katsimbri P: The biology of normal bone
remodelling. Eur J Cancer Care (Engl):. 26:e127402017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han BI, Hwang SH and Lee MA: Progressive
reduction in autophagic capacity contributes to induction of
replicative senescence in Hs68 cells. Int J Biochem Cell Biol.
92:18–25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boyce BF, Zuscik MJ and Xing L: Biology of
bone and cartilage. Genetics of Bone Biology and Skeletal Disease.
Thakker RV, Eisman J, Igarashi T and Whyte MP: Elsevier; London:
pp. 3–24. 2012
|
|
8
|
Cao JJ, Wronski TJ, Iwaniec U, Phleger L,
Kurimoto P, Boudignon B and Halloran BP: Aging increases
stromal/osteoblastic cell-induced osteoclastogenesis and alters the
osteoclast precursor pool in the mouse. J Bone Miner Res.
20:1659–1668. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ahmed AS, Sheng MH, Wasnik S, Baylink DJ
and Lau KW: Effect of aging on stem cells. World J Exp Med. 7:1–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Coppe JP, Patil CK, Rodier F, Sun Y, Munoz
DP, Goldstein J, Nelson PS, Desprez PY and Campisi J:
Senescence-associated secretory phenotypes reveal
cell-nonautonomous functions of oncogenic RAS and the p53 tumor
suppressor. PLoS Biol. 6:2853–2868. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim HN, Chang J, Shao L, Han L, Iyer S,
Manolagas SC, O'Brien CA, Jilka RL, Zhou D and Almeida M: DNA
damage and senescence in osteoprogenitors expressing Osx1 may cause
their decrease with age. Aging Cell. 16:693–703. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Farr JN, Fraser DG, Wang H, Jaehn K,
Ogrodnik MB, Weivoda MM, Drake MT, Tchkonia T, LeBrasseur NK,
Kirkland JL, et al: Identification of senescent cells in the bone
microenvironment. J Bone Miner Res. 31:1920–1929. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Collin-Osdoby P and Osdoby P:
RANKL-mediated osteoclast formation from murine RAW264.7 cells.
Methods Mol Biol. 816:187–202. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ueta M, Takaoka K, Yamamura M, Maeda H,
Tamaoka J, Nakano Y, Nouchi K and Kishimoto H: Effects of TGF-β1 on
the migration and morphology of RAW264.7 cells in vitro. Mol
Med Rep. 20:4331–4339. 2019.PubMed/NCBI
|
|
15
|
Wang Z, Gao J, Ohno Y, Liu H and Xu C:
Rosiglitazone ameliorates senescence and promotes apoptosis in
ovarian cancer induced by Olaparib. Cancer Chemother Pharmacol.
85:273–284. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nezu T, Hosomi N, Takahashi T, Anno K,
Aoki S, Shimamoto A, Maruyama H, Hayashi T, Matsumoto M and Tahara
H: Telomere G-tail length is a promising biomarker related to white
matter lesions and endothelial dysfunction in patients with
cardiovascular risk: A cross-sectional study. EBioMedicine.
30:960–967. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takahashi N, Yamana H, Yoshiki S, Roodman
GD, Mundy GR, Jones SJ, Boyde A and Suda T: Osteoclast-like cell
formation and its regulation by osteotropic hormones in mouse bone
marrow cultures. Endocrinology. 122:1373–1382. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guevara I, Iwanejko J, Dembinska-Kiec A,
Pankiewicz J, Wanat A, Anna P, Golabek I, Bartus S,
Malczewska-Malec M and Szczudlik A: Determination of
nitrite/nitrate in human biological material by the simple Griess
reaction. Clin Chim Acta. 274:177–188. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim JH, Lee CH and Lee SW: Exosomal
transmission of microRNA from HCV replicating cells stimulates
transdifferentiation in hepatic stellate cells. Mol Ther Nucleic
Acids. 14:483–497. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Yang Y, Xiong A, Wu X, Xie J, Han S
and Zhao S: Comparative gene expression analysis of lymphocytes
treated with exosomes derived from ovarian cancer and ovarian
cysts. Front Immunol. 8:6072017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wermuth PJ, Piera-Velazquez S and Jimenez
SA: Exosome isolated from serum of systemic sclerosis patients
display alterations in their content of profibrotic and
antifibrotic microRNA and induce a profibrotic phenotype in
cultured normal dermal fibroblasts. Clin Exp Rheumatol. 35:21–30.
2017.PubMed/NCBI
|
|
22
|
Hashitani S, Urade M, Nishimura N, Maeda
T, Takaoka K, Noguchi K and Sakurai K: Apoptosis induction and
enhancement of cytotoxicity of anticancer drugs by celecoxib, a
selective cyclooxygenase-2 inhibitor, in human head and neck
carcinoma cell lines. Int J Oncol. 23:665–672. 2003.PubMed/NCBI
|
|
23
|
Simonet WS, Lacey DL, Dunstan CR, Kelley
M, Chang MS, Luthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, et
al: Osteoprotegerin: A novel secreted protein involved in the
regulation of bone density. Cell. 89:309–319. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Harding CV, Heuser JE and Stahl PD:
Exosomes: Looking back three decades and into the future. J Cell
Biol. 200:367–371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hayflick L and Moorhead PS: The serial
cultivation of human diploid cell strains. Exp Cell Res.
25:585–621. 1961. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Campisi J and d'Adda di Fagagna F:
Cellular senescence: When bad things happen to good cells. Nat Rev
Mol Cell Biol. 8:729–740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zeng X: Human embryonic stem cells:
Mechanisms to escape replicative senescence? Stem Cell Rev.
3:270–279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Delfarah A, Parrish S, Junge JA, Yang J,
Seo F, Li S, Mac J, Wang P, Fraser SE and Graham NA: Inhibition of
nucleotide synthesis promotes replicative senescence of human
mammary epithelial cells. J Biol Chem. 294:10564–10578. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alique M, Bodega G, Giannarelli C,
Carracedo J and Ramírez R: MicroRNA-126 regulates hypoxia-inducible
factor-1α which inhibited migration, proliferation, and
angiogenesis in replicative endothelial senescence. Sci Rep.
9:73812019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gan K, Xu L, Feng X, Zhang Q, Wang F,
Zhang M and Tan W: Celastrol attenuates bone erosion in
collagen-induced arthritis mice and inhibits osteoclast
differentiation and function in RANKL-induced RAW264.7. Int
Immunopharmacol. 24:239–246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
GuezGuez A, Prod'Homme V, Mouska X, Baudot
A, Blin-Wakkach C, Rottapel R and Deckert M: 3BPs adapter protein
is required for receptor activator of NFκB ligand (RANKL)-induced
osteoclast differentiation of RAW264.7. J Biol Chem.
285:20952–20963. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Itahana K, Campisi J and Dimri GP: Methods
to detect biomarkers of cellular enescence: The
senescence-associated beta-galactosidase assay. Methods Mol Biol.
371:21–31. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dimri GP, Lee X, Basile G, Acosta M, Scott
G, Roskelley C, Medrano MM, Linskens M, Rubelj I, Pereira-Smith O,
et al: A biomarker that identifies senescent human cells in culture
and in aging skin in vivo. Proc Natl Acad Sci USA. 92:9363–9367.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Debacq-Chainiaux F, Erusalimsky JD,
Campisi J and Toussaint O: Protocols to detect
senescence-associated beta-galactosidase (SA-betagal) activity, a
biomarker of senescent cells in culture and in vivo. Nat Protoc.
4:1798–1806. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vogelstein B, Lane DP and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Levine AJ and Oren M: The first 30 years
of p53: Growing ever more complex. Nat Rev Cancer. 9:749–758. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vousden KH and Prives C: Blinded by the
light: The Growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feng Z, Zhang H, Levine AJ and Jin S: The
coordinate regulation of the p53 and mTOR pathways in cells. Proc
Natl Acad Sci USA. 102:8204–8209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Budanov AV and Karin M: p53 target genes
sestrin1 and sestrin2 connect genotoxic stress and mTOR signalling.
Cell. 134:451–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weichhart T: mTOR as regulator of
lifespan, aging, and cellular senescence: A mini-review.
Gerontology. 64:127–134. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Feng Z, Hu W, Teresky AK, Hernando E,
Cordon-Cardo C and Levine AJ: Declining p53 function in the aging
process: A possible mechanism for the increased tumor incidence in
older populations. Proc Natl Acad Sci USA. 104:16633–16638. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Campisi J: Aging, cellular senescence, and
cancer. Annu Rev Physiol. 75:685–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Soriani A, Zingoni A, Cerboni C, Lannitto
ML, Ricciardi RR, Gialleonardo VD, Cippitelli M, Fionda C, Petrucci
MT, Guarini A, et al: ATM-ATR-dependent up-regulation of DNAM-1 and
NKG2D ligands on multiple myeloma cells by therapeutic agents
results in enhanced NK-cell susceptibility and is associated with a
senescent phenotype. Blood. 113:3503–3511. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Takahashi S, Hakuta M, Aiba K, Ito Y,
Horikoshi N, Miura M, Hatake K and Ogata E: Elevation of
circulating plasma cytokines in cancer patients with high plasma
parathyroid hormone-related protein levels. Endocr Relat Cancer.
10:403–407. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mabrouk N, Ghione S, Laurens V, Plenchette
S, Bettaieb A and Paul C: Senescence and cancer: Role of nitric
oxide (NO) in SASP. Cancers (Basel). 12:11452020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Campisi J: Senescent cells, tumor
suppression, and organismal aging: Good citizens, bad neighbors.
Cell. 120:513–522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Farr JN, Kaur J, Doolittle ML and Khosla
S: Osteocyte cellular senescence. Curr Osteoporos Rep. 18:559–567.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Walsh MC, Kim N, Kadono Y, Rho J, Lee SY,
Lorenzo J and Choi Y: Osteoimmunology: Interplay between the immune
system and bone metabolism. Annu Rev Immunol. 24:33–63. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Azuma Y, Kaji K, Katogi R, Takeshita S and
Kudo A: Tumor necrosis factor-alpha induces differentiation of and
bone resorption by osteoclasts. J Biol Chem. 18:4858–4864. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Komine M, Kukita A, Kukita T, Ogata Y,
Hotokebuchi T and Kohashi O: Tumor necrosis factor-alpha cooperates
with receptor activator of nuclear factor kappaB ligand in
generation of osteoclasts in stromal cell-depleted rat bone marrow
cell culture. Bone. 28:474–483. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Palmqvist P, Persson E, Conaway HH and
Lerner UH: IL-6, leukemia inhibitory factor, and oncostatin M
stimulate bone resorption and regulate the expression of receptor
activator of NF-kappa B ligand, osteoprotegerin, and receptor
activator of NF-kappa B in mouse calvariae. J Immunol.
169:3353–3362. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Feng W, Liu H, Luo T, Liu D, Du J, Sun J,
Wang W, Han X, Yang K, Guo J, et al: Combination of IL-6 and sIL-6R
differentially regulate varying levels of RANKL-induced
osteoclastogenesis through NF-κB, ERK and JNK signaling pathways.
Sci Rep. 7:414112017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yokota K, Sato K, Miyazaki T, Kitaura H,
Kayama H, Miyoshi F, Araki Y, Akiyama Y, Takeda K and Mimura T:
Combination of tumor necrosis factor α and interleukin-6 induces
mouse osteoclast-like cells with bone resorption activity both in
vitro and in vivo. Arthritis Rheumatol. 66:121–129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
MacIntyre I, Zaidi M, Alam AS, Datta HK,
Moonga BS, Lidbury PS, Hecker M and Vane JR: Osteoclastic
inhibition: An action of nitric oxide not mediated by cyclic GMP.
Proc Natl Acad Sci USA. 88:2936–2940. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kasten TP, Collin-Osdoby P, Patel N,
Osdoby P, Krukuwski M, Misko TP, Settle SL, Currie MG and Nickols
GA: Potentiation of osteoclast bone-resorption activity by
inhibition of nitric oxide synthase. Proc Natl Acad Sci USA.
91:3569–3573. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Brandi M, Hukkanen M, Umeda T,
Moradi-Bidhendi N, Bianchi S, Gross SS, Polak JM and Maclntyre I:
Bidirectional regulation of osteoclast function by nitric oxide
synthase isoforms. Proc Natl Acad Sci USA. 92:2954–2958. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nathan C and Xie QW: Nitric oxide
synthases: Roles, tolls and controls. Cell. 78:915–918. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fukumura D, Kashiwagi S and Jain RK: The
role of nitric oxide in tumor progression. Nat Rev Cancer.
6:521–534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Beckman JS, Beckman YW, Chen J, Marshall
PA and Freeman BA: Apparent hydroxyl radical production by
peroxynitrite: Implications for endothelial injury from nitric and
superoxide. Proc Natl Acad Sci USA. 87:1620–1624. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zheng H, Yu X, Collin-Osdoby P and Osdoby
P: RANKL stimulates inducible nitric-oxide synthase expression and
nitric oxide production in developing osteoclasts. An autocrine
negative feedback mechanism triggered by RANKL-induced
interferon-beta via NF-kappaB that restrains osteoclastogenesis and
bone resorption. J Biol Chem. 281:15809–15820. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Takeda N, O'Dea EL, Doedens A, Kim JW,
Weidemann A, Stockmann C, Asagiri M, Simon MC, Hoffmann A and
Johnson RS: Differential activation and antagonistic function of
HIF-{alpha} isoforms in macrophages are essential for NO
homeostasis. Genes Dev. 24:491–501. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu C, Rankin EB, Castellini L, Alcudia JF,
LaGory EL, Andersen R, Rhodes SD, Wilson TL, Mohammad KS, Castillo
AB, et al: Oxygen-sensing PHDs regulate bone homeostasis through
the modulation of osteoprotegerin. Genes Dev. 29:817–831. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Schorey JS, Cheng Y, Singh PP and Smith
VL: Exosomes and other extracellular vesicles in host-pathogen
interactions. EMBO Rep. 16:24–43. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Valadi H, Ekstrom K, Bossios A, Sjostrand
M, Lee JJ and Lotvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lehmann BD, Paine MS, Brooks AM, McCubrey
JA, Renegar RH, Wang R and Terrian DM: Senescence-associated
exosome release from human prostate cancer cells. Cancer Res.
68:7864–7871. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yoshida M, Satoh A, Lin JB, Mills KF,
Sasaki Y, Rensing N, Wong M, Apte RS and Imai S: Extracellular
Vesicle-contained eNAMPT delays aging and extends lifespan in mice.
Cell Metab. 30:329–342. 2019. View Article : Google Scholar : PubMed/NCBI
|