Introduction
The differentiation therapy of cancer cells is an
alternative therapeutic strategy used to treat leukemia regarding
its lower side effects in comparison with the traditional
chemotherapy (1,2). Natural agents have been used to
restart the process of differentiation which was stopped during
leukemic transformation of hematopoietic stem cells or progenitor
(3–5). In this context, previously in our
laboratory, Beneytout et al demonstrated that treating human
erythroleukemia (HEL) cells with 10 µM of a natural steroidal
saponin named diosgenin [(25R)-5-spirosten-3b-ol] induced
its megakaryocytic differentiation (6). Formerly, published results showed that
diosgenin-induced HEL megakaryocytic differentiation is
accomplished after 8 days. With time progression, the main features
of the megakaryocytic maturation began to stand out as progressive
cell enlargement and polyploidization e.g. the increase in the DNA
content up to 64N. Some cell receptors expression as CD41, a marker
of the megakaryocytic differentiation, appeared and increased
continuously until the end of the differentiation with the
regression of the erythroid marker glycophorin A (7,8). Many
teams in which our laboratory confirmed that the platelet formation
from mature megakaryocytes implicates apoptotic signaling pathways
and key effectors activation (9–11).
However, other researchers reported that the caspase-3, poly-ADP
ribose polymerase (PARP) cleavage are not only correlated to the
megakaryocyte's fragmentation driven by apoptosis (12,13).
In 2006, Leger et al demonstrated that, during the initial
differentiation stages, diosgenin-treated HEL cells undergo a
transient apoptosis-independent caspase-3 activation that could be
necessary to start the demarcation membrane system development
(9).
Autophagy is a housekeeping pathway that maintains
cell homeostasis against stress by recycling macromolecules and
organelles and plays an important role in cell development, death,
survival, and differentiation. There are different types of
autophagy: macroautophagy, microautophagy, and chaperone-mediated
autophagy. We were interested in macroautophagy in which
cytoplasmic materials are going to be enclosed in a double membrane
structure called autophagosomes to be recycled and degraded. This
autophagy canonical pathway is composed of three parts: the
initiation, the elongation, and the autophagosome maturation
(14,15). Autophagy is modulated by many
chemical molecules, among them 3-methyladenine (3-MA) and metformin
(Met). 3-MA is an autophagy inhibitor that will inhibit the PI3K
complex III during autophagy. Wu et al had demonstrated that
the 3-MA is not specific for the PI3K complex III, but it is a
selective inhibitor for the PI3K complex family. The principle
evidence on the autophagy flux blockage is the accumulation of the
form LC3A/B-II (Light chain 3 A/B-II) as evidence of the lysosomal
degradation interruption (16).
Furthermore, Met is an autophagy initiation activator that plays
essentially on the AMPK/LKB1 axis, an activator of the complex
ULK1/2 (autophagy initiation complex). On the other hand, Met can
abrogate the mTORC1 pathway that mainly regulates the autophagy
(17).
Interestingly, in the last few years, scientists
started to investigate the role of this second programmed cell
death pathway in megakaryocytic differentiation. In 2009, Colosetti
et al showed that the knockdown of the autophagy key
effectors, Beclin1, LC3 abrogated the K562 megakaryocytic
differentiation induced by phorbol myristate acetate (18). Cao et al confirmed that the
complete knockout Atg7 -/- mice abrogate the megakaryocytopoiesis
and the thrombopoiesis (19). In
2017, Wang et al demonstrated that the autophagy inhibition
or activation in fetal liver cells but not in mature megakaryocytes
altered significantly the megakaryocytopoiesis and thrombopoiesis
(20).
In our present study, we establish an experimental
model to investigate the autophagy involvement in the HEL
megakaryocytic differentiation induced by the diosgenin. We showed
that the autophagy effector's protein expression was changed during
the diosgenin-induced megakaryocytic differentiation. We proceed
with our investigation by modulating autophagy before and after the
differentiation induction to target the maximum of the critical
point that affect this procedure. At a late stage of the
differentiation, we showed that the autophagy inhibition by 3-MA
had a significant repression effect on the nuclear
(polyploidization) and membrane (Glycoprotein V (GpV) expression)
maturation. On the other hand, autophagy activation increased the
GpV genomic expression but did not changed the nuclear maturation
profile after HEL cells treatment with Met. Given together, this
study demonstrates that autophagy was implicated in the HEL
megakaryocytic differentiation induced by diosgenin.
Materials and methods
Materials
RPMI-1640 medium, fetal bovine serum (FBS),
L-glutamine and penicillin-streptomycin were purchased from Gibco
BRL. Diosgenin [(25R)-5-spirosten-3β-ol], 3-methyladenine
(3-MA), metformin (Met) were obtained from Sigma-Aldrich. The
monoclonal antibody against Beclin1, Atg7, Atg12-5, Atg3, LC3A/B
were acquired from Cell Signaling Technology-Ozyme
(Saint-Cyr-L′école, France). GAPDH antibody was purchased from
Santa Cruz Biotechnology (Santa Cruz Biotechnology-Clinisciences
(Nanterre, France). Goat anti-rabbit IgG H&L horseradish
peroxidase (HRP) secondary antibody was purchased from Abcam
(Paris, France). Rabbit anti-mouse IgG-IgM H&L HRP secondary
antibody and propidium iodide (PI) were obtained from
Invitrogen-Thermo Fisher Scientific, Inc. Immobilon Western
Chemiluminescent HRP Substrate was acquired from Merck.
Cell line, cell culture and
treatment
The HEL cell line was kindly provided by Professor
J.P. Cartron (INSERM U76) (6).
Cells were seeded at 105 cells/ml in tissue culture
flasks, grown in RPMI-1640 medium (Gibco BRL) supplemented with 10%
fetal calf serum (Gibco BRL), 1% sodium pyruvate, 1% HEPES (Gibco
BRL), 100 U/ml penicillin and 100 µg/ml streptomycin. Cultures were
maintained in a humified atmosphere with 5% CO2 at 37°C.
Diosgenin is widely used in many pharmacological applications.
Diosgenin is soluble in many organic solvents as propyl acetate,
acetone, isopropanol, methanol and ethanol. As the literature
suggest, diosgenin used on human cancer cells is dissolved in
absolute ethanol. With the exception of ethanol at a final
concentration of 0.1% in the culture medium, all the other solvents
mentioned above cannot be used in cell culture. At 1995, in our
laboratory, Beneytout et al confirmed that ethanol with a
concentration below 0.1% do not induce any differentiation effect
(6). In order to induce the
megakaryocytic differentiation, the cells were treated with
diosgenin 10 µM after 1 day of seeding and left for 10 days. The
same amount of vehicle (<0.1% ethanol) was added to control
cells. The autophagy modulation was done by 3-MA (2 mM) and Met
(0.25 mM). We pretreated the HEL cells with 3-MA or Met 2 h prior
to the differentiation induction by diosgenin (10 µM) or later at
day 2 or day 4 of the differentiation. Then, the cells were
harvested at day 1, 2, 4, 6 and 8 of the differentiation, washed
twice in phosphate buffered saline (PBS, pH 7.4), counted and cell
viability was determined by the trypan blue dye exclusion method.
Before starting our experiment, we tried to see whether 3-MA and
Met induce the megakaryocytic differentiation. There is no
difference between the control group (<0.1% ethanol) and the
treated group with different concentrations during 24 and 48 h
(data not shown). The photos do not reflect any morphological
changes comparing to the control group.
RNA extraction and semi-quantitative
RT-PCR analysis
Total RNA was isolated from HEL cells with the
RNeasy® Mini Kit following the RNA extraction and
semi-quantitative RT-PCR manufacturer's protocol (Qiagen). 2 µg
total RNA were retro-transcribed into cDNA using the first-strand
cDNA synthesis part of Omniscript RT Kit (Qiagen), oligo-dT (25 µg)
(Invitrogen, Cergy Pontoise, France) and RNase Out™ (40 U/ml)
(Invitrogen). 2 µl of reverse-transcribed cDNA was used for PCR
according to the Kit HotStar Taq DNA Polymerase (250 units)
protocol (Qiagen) with dNTP mix (100 mM) (Invitrogen) and 0.5 µM of
sense and antisense primers. The primers for PCR were chosen to
amplify human GpV and we used GAPDH as an internal control. PCR
resulting fragments were visualized as previously described
(21) by the GBOX (Syngene) and the
Genesnap version 7.09 software. Bands quantification was done by
Image J software.
Evaluation of nuclear ploidy
For DNA content analysis for all conditions,
106 cells were fixed and permeabilized in 70% ethanol in
PBS at −20°C overnight, washed in PBS, treated with RNase (40 U/µl,
Boehringer Mannheim) for 20 min at room temperature, and stained
with PI (50 µg/ml). Flow cytometric analyses
(fluorescence-activated cell sorter (FACS) were performed as
described previously (22) (Becton
Dickinson FACScalibur). The used software was CellQuest™ pro
version 6.0 (Becton Dickinson).
Protein expression analysis
For total protein extraction, control or treated
were washed in PBS, then, the total cell pool was centrifuged at
200 g for 5 min at 4°C and homogenized in RIPA lysis buffer (50 mM
HEPES, pH 7.5, 150 mM NaCl, 1% sodium deoxycholate, 1% NP-40, 0.1%
SDS, 20 mg/ml of aprotinin) containing protease inhibitors
(Complete™ Mini, Roche Diagnostics) according to the manufacturer's
instructions. Protein levels were determined using the Bradford
method. Proteins (40–70 µg) were separated by electrophoresis on
10–12% SDS-PAGE gels and transferred to polyvinylidene fluoride
(PVDF) membranes (Amersham Pharmacia Biotech). Western blotting was
performed on following proteins with respective antibodies against:
Beclin1, Atg7, Atg12-5, Atg3, LC3A/B: (1:1,000). After incubation
with the appropriate secondary antibodies, blots were developed
using the Immobilon Western Chemiluminescent HRP Substrate and
G:BOX system (Syngene) (23) and
Genesys version 1.6.1.0 software. The bands density are measured by
using Image J software. Membranes were then reblotted with human
anti-GAPDH (1:1,000) used as a loading control.
DNA fragmentation
HEL cells were seeded at 105 cells/ml in
75 cm2 tissue culture flasks and then treated as
described above. DNA fragmentation was quantified by ‘cell death’
enzyme-linked immunosorbent assay (ELISA) (Cell Death Detection
ELISAPLUS, Sigma-Aldrich, Saint-Quentin-Fallavier,
France). Cytosol extracts were obtained from treated or control
cells according to the manufacturer's protocol and DNA
fragmentation was measured as previously described (24) by Thermo Scientific™ Multiskan™ FC
Microplate Photometer (ThermoFisher SCIENTIFIC, Illkirch,
France).
Statistical analysis
All quantitative results are expressed as the mean ±
standard error of the mean (SEM) of separate experiments using
Excel (Microsoft Office, Version 98). Statistical significance was
evaluated by the two-tailed unpaired Student's t-test. A P-value of
<0.05 was considered to indicate significance.
Results
Autophagy flux is stimulated during
diosgenin-induced megakaryocytic differentiation of HEL cells
We first wanted to study the autophagy protein
expression during diosgenin-induced megakaryocytic differentiation
of HEL cells. We studied the protein expression of different key
autophagy mediators (Beclin1, Atg7, Atg12-5, Atg3, LC3A/B-I) and
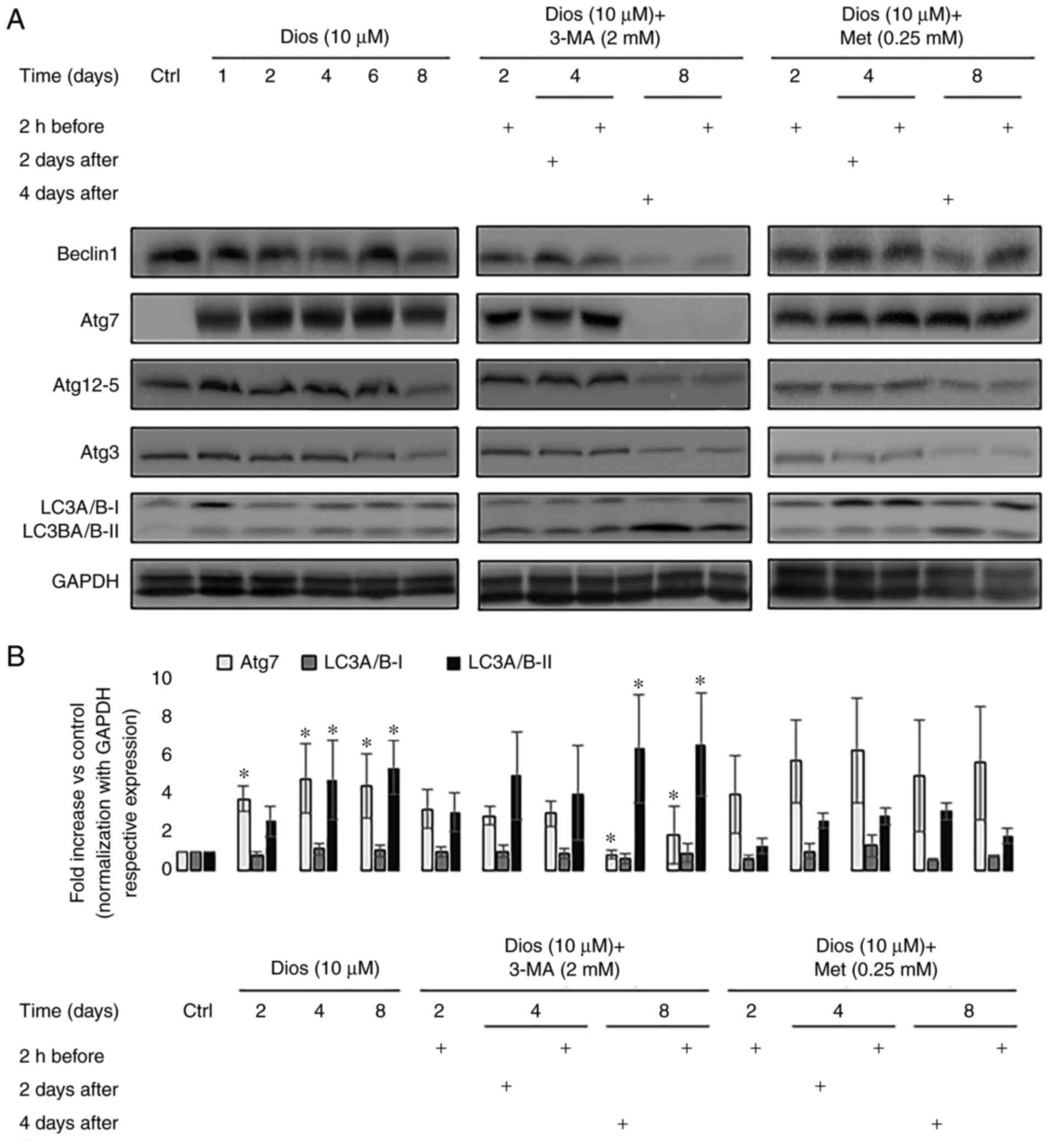
LC3A/B-II at days 1, 2, 4, 6 and 8 of the differentiation (Fig. 1A). As the Fig. 1A indicates, in comparison with
control cells, western blot results showed a significantly higher
expression of Atg7 starting from day 1. This pattern continued
until the end of diosgenin-induced megakaryocytic differentiation
of HEL cells (day 2, 4, 6 and 8, P<0.05). In parallel, we did
not see any significant changes in the expression of Beclin1,
Atg12-5 and Atg3 with the differentiation enhancement. Moreover, we
noted a significant increase in the conversion of the form LC3A/B-I
to LC3A/B-II at the last four days of the differentiation in
comparison with control cells (day 4: 4.7-fold and day 8: 5.3-fold,
P<0.05) (Fig. 1B). This result
showed that the autophagy flux was activated from day 1 after
treating HEL with diosgenin and this activation was characterized
by the continuously increased expression of autophagy initiation
protein Atg7 and the conversion of the form LC3A/B-I to LC3A/B-II,
an hallmark of autophagosome maturation.
Autophagy modulation by 3-MA but not
Met affects the expression of autophagy mediators at the end of the
diosgenin-induced megakaryocytic differentiation of HEL cells
In order to define precisely the autophagy role
during megakaryocytic differentiation, we modulated this pathway
before and after the HEL cells treatment with diosgenin. The
modulation was done by using two molecules that affect the
autophagy initiation: Met (activator) and 3-MA (inhibitor). First,
we decided to modulate autophagy 2 h before the differentiation
induction since we wanted to see if autophagy has any effect on the
differentiation beginning in a short term and the maturation in the
long term. In the second place, we wanted to modulate autophagy in
several time points after the differentiation launch: at day 2
(early stage of differentiation) and day 4 (cells reached a 32N DNA
content) (7). As the Fig. 1A illustrates, the autophagy
inhibition by 3-MA before or after the HEL treatment with diosgenin
inhibited the Atg7 protein expression but not Atg12-5 and Atg3. On
days 2 and 4, Atg7 expression reduction was not statistically
significant. On day 8, the protein expression of Atg7 was
significantly diminished whether the autophagy was inhibited 2 h
before the differentiation induction (2.5-fold) or after on day 4
(3.5-fold) in comparison with the cells treated only with diosgenin
(P<0.05) (Fig. 1B). This
abrogation was accompanied with an accumulation of the form
LC3A/B-II (P<0.05) validating the autophagy flux repression
(Fig. 1A and B). However, the
autophagy activation by Met did not have any significant effect on
the expression of Beclin1, Atg7, Atg12-5, Atg3 and the conversion
of LC3A/B-I to LC3A/B-II independently from the treatment time
(Fig. 1A and B). Taken together,
the autophagy activation did not have any effect on the autophagy
protein's expression whether at early or late-stage of the
megakaryocytes differentiation. On the other hand, the autophagy
inhibition by 3-MA had an impact exclusively at a late stage (day
8) by inhibiting ATG7 expression and accumulating LC3-A/B-II.
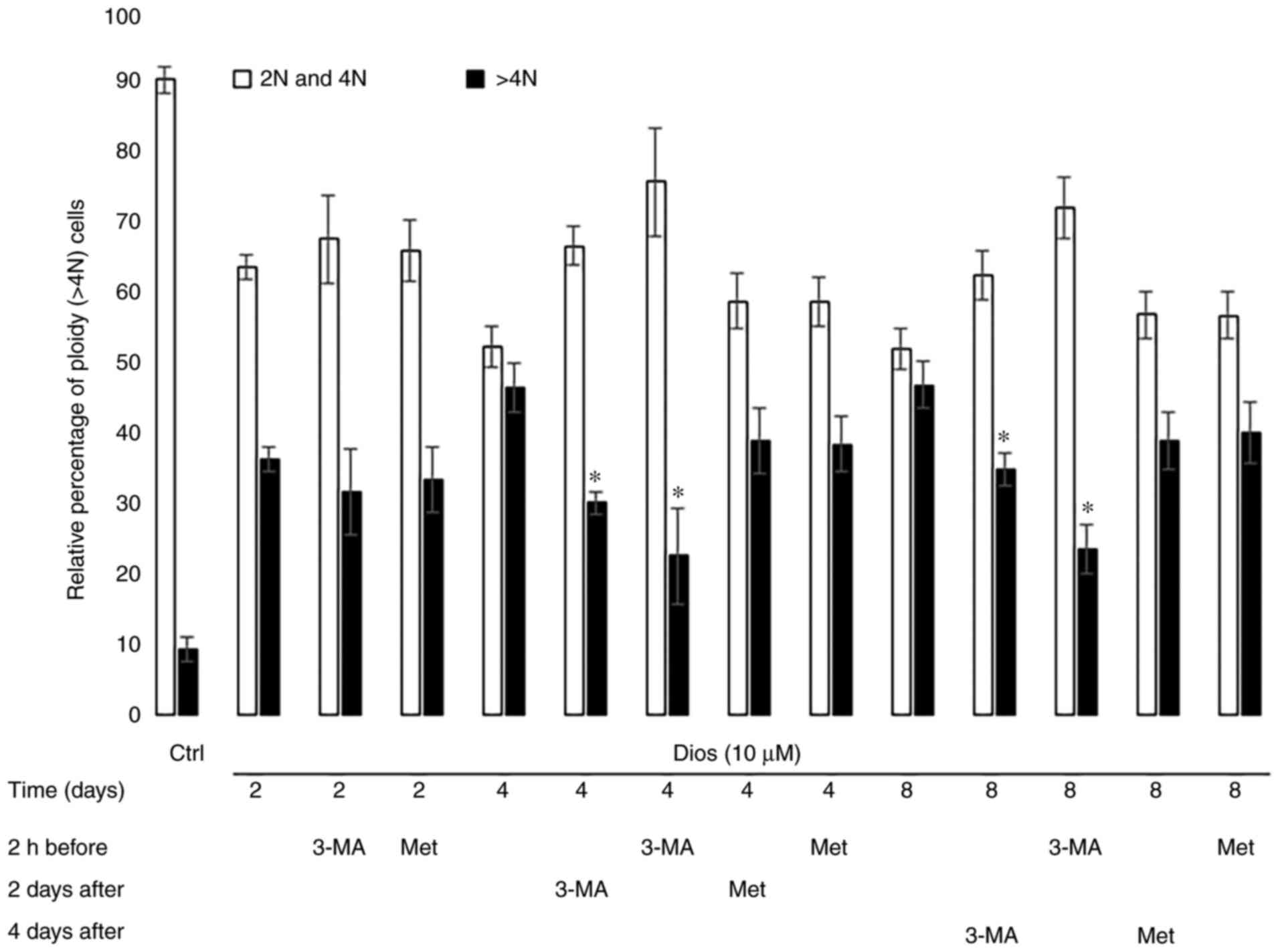
Autophagy inhibition by 3-MA blocked
polyploidization during diosgenin-induced megakaryocytic
differentiation where autophagy induction did not affect the
polyploidization process
Polyploidy is considered a major marker of the
megakaryocytic differentiation by increasing the DNA content up to
128N (25). Polyploidy was used by
Léger et al (7) to evaluate
the megakaryocytic differentiation of HEL cells after treatment
with diosgenin. Polyploidy in the diosgenin-differentiated cells
reach out 64N at the end of the differentiation (day 8) (7). Based on this, we evaluated the
polyploidization in the diosgenin-differentiated cells after
autophagy modulation by 3-MA or Met. The results in Fig. 2 show that the autophagy inhibition
by 3-MA reduced polyploidization at day 4 and 8 of the
differentiation but not at an early stage (day 2). Moreover, on day
4 and 8, we observed a strong inhibition of the polyploidy if the
HEL cells were 2 h pre-treated by 3-MA (55% and 45% of inhibition
respectively vs. the cells treated only with diosgenin on days 4
and 8). These inhibition rates were significantly higher in
comparison with the cells in which the autophagy was inhibited
after the differentiation induction (day 2 or day 4) by diosgenin
(33% and 22% of inhibition respectively vs. the cells treated only
with diosgenin at day 4 and 8). On the contrary, independently from
the treatment time, autophagy activation by Met did not had any
impact on the polyploidization (Fig.
2). Collectively, these results demonstrated a strong autophagy
inhibition effect on polyploidy on day 4 and 8, which is not the
case after autophagy activation.
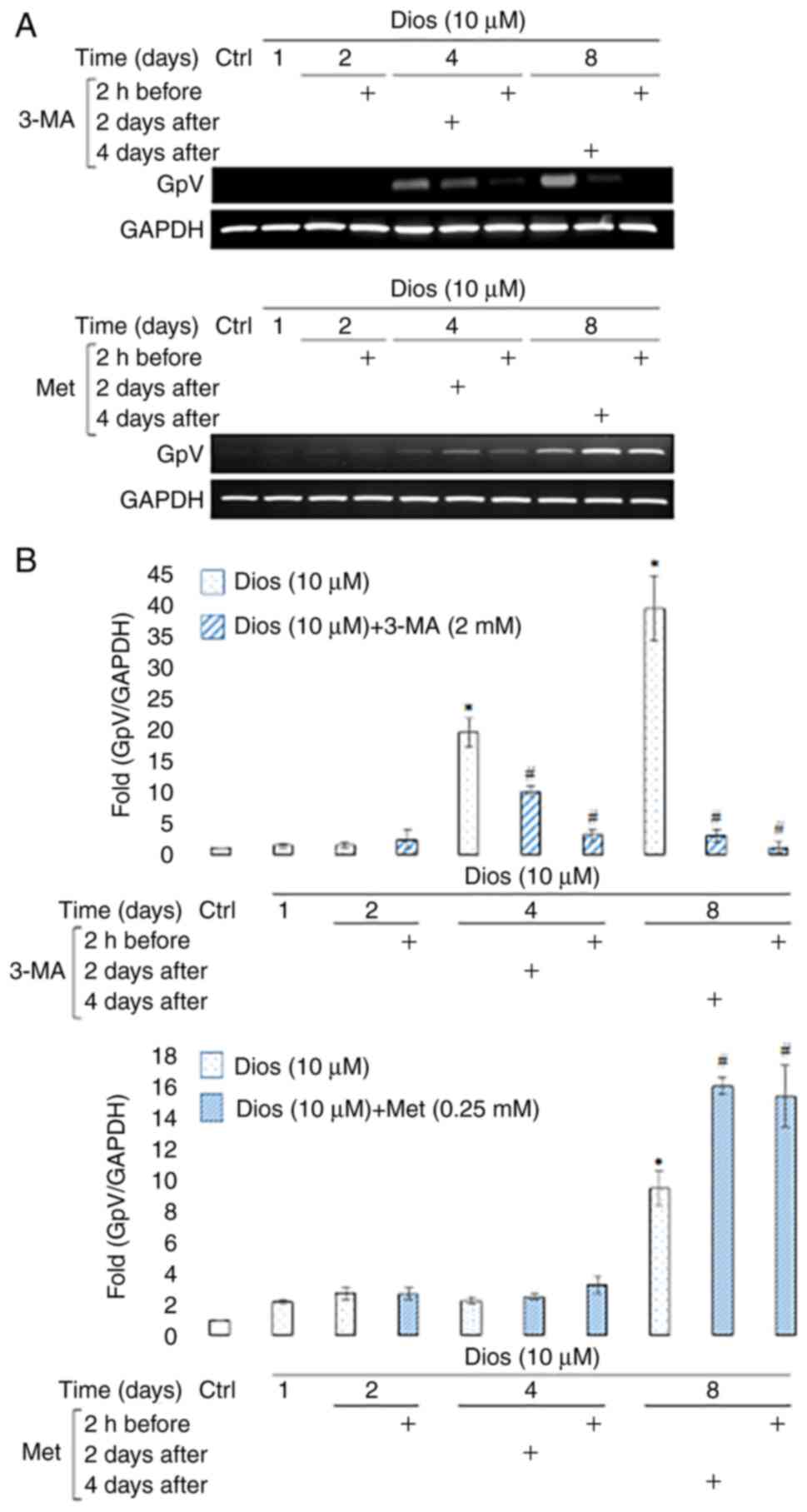
Autophagy inhibition blocked GpV
transcription where autophagy induction increased GpV transcription
during diosgenin-induced megakaryocytic differentiation
The next step was to see the effect of the autophagy
modulation on the megakaryocytes membrane receptor expression. GpV
was used to identify the maturation level after the induction of
HEL cells megakaryocytic differentiation by diosgenin (7). As the Fig.
3 demonstrates, 3-MA induced a dramatic decrease in GpV
transcription during diosgenin-induced megakaryocytic
differentiation (Fig. 3A). This
decrease occurred after 4 days of diosgenin-induced
differentiation, regardless of the treatment time at which 3-MA was
added (2-fold 2 days after and 9-fold 2 h before, P<0.05)
(Fig. 3B). This decrease was even
more drastic (>13-fold, P<0.05) at the end of differentiation
and was correlated with the decrease observed in polyploidization
process after 3-MA treatment (Figs.
2 and 3).
Concerning the effect of autophagy induction, we
observed that Met induced an increase in GpV transcription during
diosgenin-induced megakaryocytic differentiation. This increase
started after 4 days of diosgenin-induced differentiation and
lasted until the end of the treatment (day 8) regardless of the
treatment time at which Met was added (1.6-fold and 1.7-fold,
P<0.05) (Fig. 3A and B).
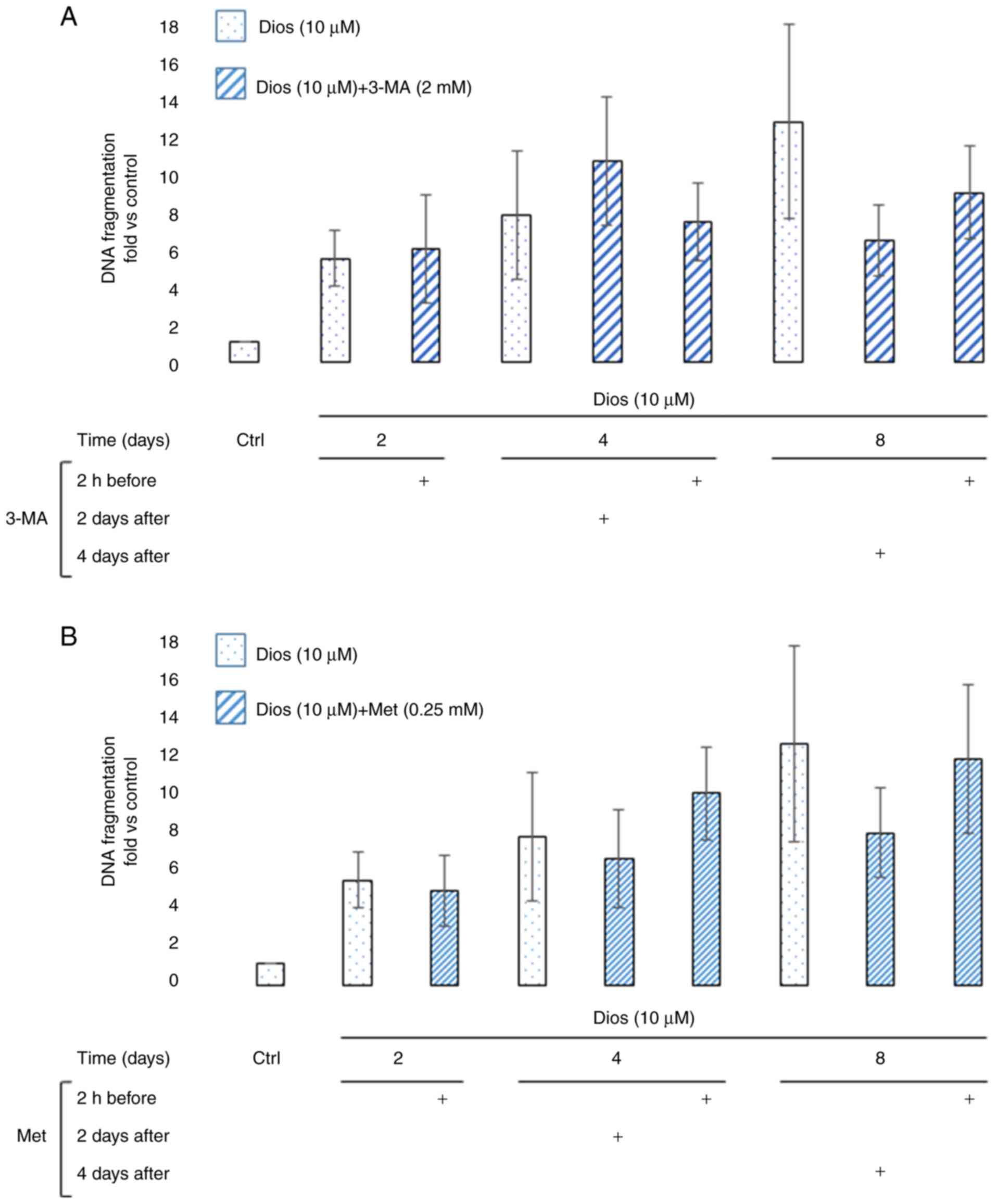
The autophagy flux modulation by 3-MA
or Met does not affect DNA fragmentation of HEL cells
In 2002, De Botton et al reported that during
the early stage of the megakaryocytic differentiation, caspase-3
activity was detected but not related to any DNA fragmentation
signal (12). Here, we investigated
the effect of autophagy modulation during the megakaryocytic
differentiation of HEL cells induced by diosgenin on DNA
fragmentation. As the Fig. 4
demonstrates, the DNA fragmentation fold increase compared to
control cells starting from day 2 to the end of the treatment. As
the Fig. 4A and B report, the
autophagy modulation 2 h before or on day 2 or day 4 after the HEL
cells treatment with diosgenin did not significantly changed DNA
fragmentation process.
Discussion
Most of the anti-cancer treatments aimed to induce
cancer cell cytotoxicity. Recently, cancer cell differentiation is
considered as an additional choice to eradicate cancer cells.
Talking about leukemia, differentiation therapy had been studied by
many teams. Several molecules are engaged in this context and many
of them are natural compounds such as retinoic acid (26). In our laboratory, the
diosgenin-induced megakaryocytic differentiation of HEL cells had
been studied since 1995 by Beneytout et al (6). Léger et al investigated the
molecular mechanisms induced after HEL cells treatment with
diosgenin. According to these works, apoptosis is indispensable for
mature megakaryocyte's fragmentation and pseudo-platelet release
in vitro (7–9). Recently, the implication of the
autophagy pathway during the megakaryocytic differentiation had
been discussed by the scientific community (18,19,27).
This assumption was built on the existence of common regulatory
pathways between those two procedures. A study conducted by Cao
et al demonstrated that Atg7 knockout in mice impaired the
megakaryocytic differentiation and altered the thrombopoiesis
(19). During the same year, an
additional study by Ouseph et al confirmed that the Atg7
knockdown in mature megakaryocytes does not have any effect on it
(27). Autophagy's precise role
during the megakaryocytic differentiation remain poorly understood
and needs more investigations to be clarified.
Autophagy is a homeostatic pathway aiming to recycle
the cytosolic content in cells (14). Since 1970, many pathologies linked
to megakaryocytes as ITP (immune thrombocytopenia) describe the
occurrence of autophagy vacuoles in those cells (28). We are interested in macro-autophagy.
This non-selective process implicates several Atgs that will make
two ubiquitin-like conjugation systems consisted of Atg7, Atg10,
Atg12, Atg5, Atg16L1 and Atg3. Atgs will conjugate LC3A/B-I to
phosphatidylethanolamine on the autophagosome membrane and induce
its enclosure (14). Our first step
was to see the key autophagy mediator's protein expressions. We
demonstrated that Atg7 expression is overexpressed since day 1
after the differentiation induction by diosgenin and was
accompanied by a significant elevation of the LC3-A/B-I conversion
to LC3-A/B-II. This first result demonstrates that the autophagy
flux is active during the diosgenin-induced megakaryocytic
differentiation of HEL cells. Given that the Atg3 and Atg12-5
protein expressions were unchanged during the diosgenin-induced
megakaryocytic differentiation, the autophagy non-canonical pathway
(a pathway independent from Beclin1, Atg12-5 and Atg3) could be
activated but this hypothesis remains under investigation. A
published study in 2009 by Colosetti et al mentioned that
the autophagy genes (Beclin1 and LC3) knockdown in K562 cells
impeded the induced megakaryocytic differentiation marker
CD41+ (Glycoprotein II-B) (18). Moreover, a study conducted by Huang
et al in 2011 reported that the autophagy inhibition by 3-MA
altered the lapatinib-induced megakaryocytic differentiation of
K562 cells (29). Recently, a
published article by Wang et al confirmed previous studies
by demonstrating that the autophagy modulation by bafilomycin A1
(autophagy inhibitor) and rapamycin (mTORC1 inhibitor) in fetal
liver cells abrogated the polyploidy as well as the expression of
the principle megakaryocytic maturation markers (20). However, autophagy modulation in
mature megakaryocytes does not affect these parameters (20). In our present study, we modulated
autophagy before or after the megakaryocytic differentiation
induction by using Met and 3-MA. We have mentioned that Met is
never been used in the megakaryocytic differentiation study field.
We noted that Met did not have any significant effect on the
autophagy flux in the diosgenin-differentiated cells independently
from the treatment strategy. This is normal to be reflected on the
GpV expression. There is no difference between the polyploidy and
the GpV genomic expression after autophagy activation at the second
and the fourth day of differentiation. Thus, the autophagy
activation by Met in our study did not induce a statistically
significant accumulation of the LC3A/B-II in HEL cells, indicating
the necessity to use another activator agent to decide if this is
the case in our cell line. Further, to explain the Met behavior in
our case, additional investigations should be done regarding the
detection of AMPK/LKB1 axis and phospho-Akt status.
However, 3-MA induced a significant accumulation of
the form LC3 A/B-II at day 8 of the differentiation and not before.
It is important to note that 3-MA is a selective PI3K complex
family inhibitor (16) therefore,
more investigations should be done regarding the PI3K/mTORC1/Akt
axis since mTORC1 is essential for the megakaryocytic
differentiation (30). In order to
see the effect of these observations on the megakaryocytic
differentiation, we examined the nuclear maturation (polyploidy) of
the diosgenin-differentiated megakaryocytes. In mature
megakaryocytes, the polyploidy rate could reach 128N and its
importance lies in the cell size enlargement and platelet formation
(25). In 2006, Drayer et al
reported that the autophagy activation by rapamycin in MO7e cells
and human megakaryocytic primary progenitors does not affect the
megakaryocytic polyploidization after the treatment with
thrombopoietin (31). Another
published work in 2020 by Sun et al reports that the
autophagy activation by rapamycin in TPA (12-O-Tetradecanoylphorbol
13-acetate)-differentiated megakaryocytes from Dami cells increased
significantly the polyploidy in a dose-dependent manner (32). Nevertheless, the autophagy
inhibition by bafilomycin A1 inhibits the polyploidy (32). In this paper, we demonstrated that
megakaryocytes are insensitive to the autophagy modulation on day 2
as the polyploidy rate did not change after modulating autophagy 2
h before the differentiation induction. On days 4 and 8, Met
continues to show no effect on the polyploidization independently
from the treatment time. Met did not affect the key autophagy
mediator's protein expression so this is might be reflected by the
absence of the effect on the polyploidy on days 2, 4 and 8. On the
other side, 3-MA inhibited the nuclear maturation of the
diosgenin-induced megakaryocytes at days 4 and 8 whatever is the
treatment time. This effect is stronger if the autophagy is
inhibited 2 h before the treatment of HEL cells with diosgenin,
which is compatible with the above-mentioned studies. According to
the literature, the autophagy depletion in the megakaryocytic
progenitors impedes the megakaryocytic differentiation. According
to Léger et al, after 5 min of diosgenin treatment, the cell
signaling is modulated toward the differentiation (7). Our results suggested that the
autophagy inhibition effect appears strongly at day 4 and not
before. The role of autophagy is to ensure that the cells are in a
good percentage of polyploidy. With a high polyploidy rate, the
cells are able to produce all the necessary materials to launch the
platelet formation. The role of autophagy in the platelet formation
during the diosgenin induced megakaryocytic differentiation needs
to be investigated as apoptosis is involved during this stage.
According to Léger et al on day 2 of differentiation, a
caspase-3 activity peak is observed as a sign of the platelet
formation initiation inside the differentiated megakaryocytes.
Furthermore, they showed an increase in the cleaved PARP expression
and a reduction in the phospho-ERK1/2 activity (7,9).
The majority of the studies focus on the expression
of CD41+ and CD61+ to characterize the
megakaryocytic differentiation. However, GpV is a specific marker
of the megakaryocytic lineages described since 1994 (33). According to this publication, the
GpV expression increase while the megakaryocytic differentiation
progression. In 2006, Léger et al presented GpV as a marker
of the diosgenin-induced megakaryocytic differentiation of HEL
cells (8). According to this study,
GpV will increase progressively with the differentiation
enhancement (8). Here in our study,
we measured the GpV genomic expression to evaluate the autophagy
modulation effect on the megakaryocytic differentiation. As claimed
by Sun et al, the activation of the autophagy flux by
rapamycin in TPA-differentiated megakaryocytes increased the % of
CD41+ megakaryocytes (MK) (32). Liu et al in 2011 demonstrated
that neonatal MKs (micro MKs) can achieve full maturation and a
mature membrane receptor profile with a low polyploidy level (4N)
(34). In our report, we
demonstrated that the autophagy activation by Met before and after
the megakaryocytic differentiation induce the genomic expression of
GpV despite of the ineffectiveness of the autophagy activation on
the diosgenin-differentiated cell's polyploidy and independently
from the treatment strategy. It is interesting to note that
autophagy inhibition with 3-MA reduced both membrane maturation
(GpV) and polyploidization and that autophagy induction with Met
promoted only membrane maturation (GpV) and not
polyploidization.
In 2000, Falcieri et al demonstrated that
megakaryocytes from late stages of differentiation did not show any
signal of DNA fragmentation (35).
Using electrophoresis analysis, they suggested that DNA
fragmentation was not internucleosomal like the majority of the
hematopoietic cell lines that undergo apoptosis thus the DNA
fragments did not appear on the agarose gel (35). In 2002, De Botton et al
reported an early caspase-3 activity localized in the cytosol and
independent from apoptosis as it did not lead to DNA fragmentation
(12). Otherwise, in later stages
of the megakaryocytic differentiation, the DNA fragmentation
increased with diffused caspase-3 inside the cells (12). In 2018, a TUNEL assay done on
histological cut belonged to immune thrombocytopenia patients
demonstrated the absence of DNA fragmentation signals in fully
mature megakaryocytes (36). Here
in this report, the DNA fragmentation assay showed no statistical
significant variation of the DNA fragmentation in the cells treated
only with diosgenin or the cells that underwent autophagy
modulation.
In conclusion, this study demonstrated for the first
time the autophagy implication in the diosgenin-induced
megakaryocytic differentiation of HEL cells. The autophagy
implication during the megakaryocytic differentiation was studied
after the modulation of this pathway by 3-MA and Met. We conclude
that the autophagy modulation induced an effect on the
diosgenin-induced megakaryocytic differentiation. It is interesting
to note that autophagy inhibition with 3-MA reduced both membrane
maturation and polyploidization and that autophagy induction with
Met promoted only membrane maturation and not polyploidization.
Acknowledgements
The authors would like to acknowledge Dr Ludovic
Bretin and Miss Lucie Paulus (University of Limoges, Limoges,
France) for their technical assistance during the experiments. The
authors would also like to thank Dr Jeanne Cook-Moreau (University
of Limoges, Limoges, France) for their help with manuscript
editing.
Funding
The present study was funded in part by the Ministry
of Higher Education, Research and Innovation by the New Aquitaine
Regional Council and by the Lebanese University of Beirut (Beirut,
Lebanon).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DYL participated in research design. DD, AP and CO
conducted the experiments. DD, BL and DYL performed data analysis.
DD, BL and DYL wrote or contributed to the writing of the
manuscript. RHS and MDA obtained doctoral funding and contributed
to the design of the research. DYL and BL confirm the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
FACS
|
fluorescence-activated cell sorter
|
|
FBS
|
fetal bovine serum
|
|
GpV
|
glycoprotein V
|
|
HEL
|
human erythroleukemia
|
|
HRP
|
horseradish peroxidase
|
|
LC3A/B
|
light chain 3 A/B
|
|
3-MA
|
3-methyladenine
|
|
Met
|
metformin
|
|
MK
|
megakaryocytes
|
|
PBS
|
phosphate buffered saline
|
|
PI
|
propidium iodide
|
|
PVDF
|
polyvinylidene fluoride
|
References
|
1
|
Sell S: Leukemia: Stem cells, maturation
arrest, and differentiation therapy. Stem Cell Rev. 1:197–205.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nowak D, Stewart D and Koeffler HP:
Differentiation therapy of leukemia: 3 decades of development.
Blood. 113:3655–3665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gocek E and Marcinkowska E:
Differentiation therapy of acute myeloid leukemia. Cancers (Basel).
3:2402–2420. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang J, Qiu J, Hu Y, Zhang Y, Chen L, Long
Q, Chen J, Song J, Rao Q, Li Y, et al: A natural small molecule
induces megakaryocytic differentiation and suppresses
leukemogenesis through activation of PKCδ/ERK1/2 signaling pathway
in erythroleukemia cells. Biomed Pharmacother. 118:1092652019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao X, Wu J, Zou W and Dai Y: Two ellagic
acids isolated from roots of Sanguisorba officinalis L. promote
hematopoietic progenitor cell proliferation and megakaryocyte
differentiation. Molecules. 19:5448–5458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beneytout JL, Nappez C, Leboutet MJ and
Malinvaud G: A plant steroid, diosgenin, a new megakaryocytic
differentiation inducer of HEL cells. Biochem Biophys Res Commun.
207:398–404. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Léger D, Battu S, Liagre B, Cardot P and
Beneytout JL: Sedimentation field flow fractionation to study human
erythroleukemia cell megakaryocytic differentiation after short
period diosgenin induction. J Chromatogr A. 1157:309–320. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Léger DY, Battu S, Liagre B, Beneytout JL
and Cardot PJP: Megakaryocyte cell sorting from
diosgenin-differentiated human erythroleukemia cells by
sedimentation field-flow fractionation. Anal Biochem. 355:19–28.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leger D, Liagre B and Beneytout JL: Role
of MAPKs and NF-kappaB in diosgenin-induced megakaryocytic
differentiation and subsequent apoptosis in HEL cells. Int J Oncol.
28:201–207. 2006.PubMed/NCBI
|
|
10
|
Luff SA, Kao CY and Papoutsakis ET: Role
of p53 and transcription-independent p53-induced apoptosis in
shear-stimulated megakaryocytic maturation, particle generation,
and platelet biogenesis. PLoS One. 13:e02039912018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kovuru N, Raghuwanshi S, Sharma DS,
Dahariya S, Pallepati A and Gutti RK: Endoplasmic reticulum stress
induced apoptosis and caspase activation is mediated through
mitochondria during megakaryocyte differentiation. Mitochondrion.
50:115–120. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Botton S, Sabri S, Daugas E, Zermati Y,
Guidotti JE, Hermine O, Kroemer G, Vainchenker W and Debili N:
Platelet formation is the consequence of caspase activation within
megakaryocytes. Blood. 100:1310–1317. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Solier S, Fontenay M, Vainchenker W, Droin
N and Solary E: Non-apoptotic functions of caspases in myeloid cell
differentiation. Cell Death Differ. 24:1337–1347. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Glick D, Barth S and Macleod KF:
Autophagy: Cellular and molecular mechanisms. J Pathol. 221:3–12.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wesselborg S and Stork B: Autophagy signal
transduction by ATG proteins: From hierarchies to networks. Cell
Mol Life Sci. 72:4721–4757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu YT, Tan HL, Shui G, Bauvy C, Huang Q,
Wenk MR, Ong CN, Codogno P and Shen HM: Dual role of
3-methyladenine in modulation of autophagy via different temporal
patterns of inhibition on class I and III phosphoinositide
3-kinase. J Biol Chem. 285:10850–10861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hawley SA, Gadalla AE, Olsen GS and Hardie
DG: The antidiabetic drug metformin activates the AMP-activated
protein kinase cascade via an adenine nucleotide-independent
mechanism. Diabetes. 51:2420–2425. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Colosetti P, Puissant A, Robert G, Luciano
F, Jacquel A, Gounon P, Cassuto JP and Auberger P: Autophagy is an
important event for megakaryocytic differentiation of the chronic
myelogenous leukemia K562 cell line. Autophagy. 5:1092–1098. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao Y, Cai J, Li X, Fang Y, Zhang S, Yuan
N, Mao X and Wang J: Loss of autophagy leads to megakaryocytes
differentiation failure and defective platelets function. Blood.
124:41482014. View Article : Google Scholar
|
|
20
|
Wang Q, You T, Fan H, Wang Y, Chu T, Poncz
M and Zhu L: Rapamycin and bafilomycin A1 alter autophagy and
megakaryopoiesis. Platelets. 28:82–89. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pinon A, Limami Y, Micallef L, Cook-Moreau
J, Liagre B, Delage C, Duval RE and Simon A: A novel form of
melanoma apoptosis resistance: Melanogenesis up-regulation in
apoptotic B16-F0 cells delays ursolic acid-triggered cell death.
Exp Cell Res. 317:1669–1676. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Léger D, Liagre B, Cardot P, Beneytout JL
and Battu S: Diosgenin dose-dependent apoptosis and differentiation
induction in human erythroleukemia cell line and sedimentation
field-flow fractionation monitoring. Anal Biochem. 335:267–278.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bretin L, Pinon A, Bouramtane S, Ouk C,
Richard L, Perrin ML, Chaunavel A, Carrion C, Bregier F, Sol V, et
al: Photodynamic therapy activity of new porphyrin-xylan-coated
silica nanoparticles in human colorectal cancer. Cancers (Basel).
11:14742019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bertrand J, Liagre B, Ghezali L, Beneytout
JL and Leger DY: Cyclooxygenase-2 positively regulates Akt
signalling and enhances survival of erythroleukemia cells exposed
to anticancer agents. Apoptosis. 18:836–850. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mazzi S, Lordier L, Debili N, Raslova H
and Vainchenker W: Megakaryocyte and polyploidization. Exp Hematol.
57:1–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Klobuch S, Steinberg T, Bruni E, Mirbeth
C, Heilmeier B, Ghibelli L, Herr W, Reichle A and Thomas S:
Biomodulatory treatment with azacitidine, all-trans retinoic acid
and pioglitazone induces differentiation of primary AML blasts into
neutrophil like cells capable of ROS production and phagocytosis.
Front Pharmacol. 9:13802018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ouseph MM, Huang Y, Banerjee M, Joshi S,
MacDonald L, Zhong Y, Liu H, Li X, Xiang B, Zhang G, et al:
Autophagy is induced upon platelet activation and is essential for
hemostasis and thrombosis. Blood. 126:1224–1233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ortega Aramburu JJ: Idiopathic
thrombocytopenic purpura-autoimmune thrombocytopenia in children.
An Esp Pediatr. 23:145–151. 1985.(In Spanish). PubMed/NCBI
|
|
29
|
Huang HL, Chen YC, Huang YC, Yang KC, Pan
HY, Shih SP and Chen YJ: Lapatinib induces autophagy, apoptosis and
megakaryocytic differentiation in chronic myelogenous leukemia K562
cells. PLoS One. 6:e290142011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Raslova H, Baccini V, Loussaief L, Comba
B, Larghero J, Debili N and Vainchenker W: Mammalian target of
rapamycin (mTOR) regulates both proliferation of megakaryocyte
progenitors and late stages of megakaryocyte differentiation.
Blood. 107:2303–2310. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Drayer AL, Olthof SG and Vellenga E:
Mammalian target of rapamycin is required for
thrombopoietin-induced proliferation of megakaryocyte progenitors.
Stem Cells. 24:105–114. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun RJ, Yuan D, Liu SY, Zhu JJ and Shan
NN: Rapamycin induces megakaryocytic differentiation through
increasing autophagy in Dami cells. Blood Coagul Fibrinolysis.
31:310–316. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takafuta T, Fujimura K, Kawano H, Noda M,
Fujimoto T, Oda K, Shimomura T and Kuramoto A: Expression of
platelet membrane glycoprotein V in human megakaryocytes and
megakaryocytic cell lines: A study using a novel monoclonal
antibody against GPV. Thromb Haemost. 72:762–769. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu ZJ, Italiano J Jr, Ferrer-Marin F,
Gutti R, Bailey M, Poterjoy B, Rimsza L and Sola-Visner M:
Developmental differences in megakaryocytopoiesis are associated
with up-regulated TPO signaling through mTOR and elevated GATA-1
levels in neonatal megakaryocytes. Blood. 117:4106–4117. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Falcieri E, Bassini A, Pierpaoli S,
Luchetti F, Zamai L, Vitale M, Guidotti L and Zauli G:
Ultrastructural characterization of maturation, platelet release,
and senescence of human cultured megakaryocytes. Anat Rec.
258:90–99. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vrbensky JR, Nazy I, Toltl LJ, Ross C,
Ivetic N, Smith JW, Kelton JG and Arnold DM: Megakaryocyte
apoptosis in immune thrombocytopenia. Platelets. 29:729–732. 2018.
View Article : Google Scholar : PubMed/NCBI
|