Introduction
Colorectal cancer (CRC) is a heterogeneous disease
of the intestinal epithelium, which is characterized by the
accumulation of mutations and immune response disorders (1). CRC is the third most common type of
cancer worldwide, with an incidence rate of 10.2% among all cancer
types and a mortality rate of 9.2% (2). Its prevalence has been gradually
increasing among individuals <50 years old (1), while the survival rate of patients
with metastasis is estimated to be only 18.5% in the United States
(3). Although early diagnosis via
fecal occult blood testing and colonoscopy can reduce the mortality
rate to a certain extent, the mortality rate of CRC remains high
(4). The currently available
conventional methods for treating CRC are surgery, chemotherapy and
radiation therapy. These therapeutic approaches may be used alone
or in combination, based on the progression of CRC during treatment
(5–7). In addition, for localized, easily
accessible CRC, total mesenteric rectal resection is considered as
an effective treatment approach (8–10). In
mesenteric rectal resection, the cancer tissue is removed via
laparoscopic and transanal surgery, followed by adjuvant
chemotherapy or radiotherapy (11).
However, these therapies are non-specifically cytotoxic to all
rapidly dividing cells, inevitably causing several side effects
(12). Following neoadjuvant
therapy, 54% of patients with CRC develop disease relapse (13). Therefore, developing alternative and
more effective therapeutic strategies for treating patients with
CRC is crucial.
It is estimated that there will be 2.2 million new
CRC cases and 1.1 million CRC-related deaths by 2030 worldwide
(14). Biomarkers are objective
indicators that can be isolated or measured in the human body and
used to evaluate the pathological process of cancer or predict
treatment effectiveness. In addition, biomarkers can be used as
tools for the early detection and personalized therapy for CRC
(15,16). More specifically, biomarkers provide
information in order to evaluate the progression or recurrence of
the disease at different stages and provide guidance for
individualized treatment. For asymptomatic patients, biomarkers can
be used for the diagnosis of early CRC and premalignant lesions, to
achieve favorable survival (17).
For patients with advanced disease, biomarkers can be used to
predict disease progression, including the risk of recurrence and
survival (16,18). Ongoing advances in the molecular
classification of CRC subtypes, DNA methylation and microRNA
biogenesis have resulted in the identification of numerous
biomarkers for CRC (19).
Therefore, further exploration and mechanistic research on
biomarkers in human cancer may prove to be of great diagnostic and
prognostic value.
It has been reported that salt-inducible kinase 2
(SIK2) is upregulated in breast (20), ovarian (21) and cervical cancer (22), and promotes cancer progression, thus
suggesting that SIK2 may act as an oncogene. To the best of our
knowledge, the role of SIK2 in CRC has not been previously
reported. Therefore, the present study aimed to reveal the effect
of SIK2 on CRC cell lines and further examine its underlying
mechanism of action. The results of the current study may provide a
novel approach to the targeted therapy and clinical diagnosis of
CRC.
Materials and methods
Cell lines and transfection
The normal colonic epithelial cell line, NCM460, and
the CRC cell lines, LoVo, SW480, SW620, CW2 and HCT116, were
purchased from the National Infrastructure of Cell Line Resource
(Beijing, China). Each cell line was cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 100
U/ml penicillin-streptomycin (all Thermo Fisher Scientific, Inc.)
at 37°C in a humidified incubator with 5% CO2.
To knock down SIK2 or overexpress tripartite motif
containing 28 (TRIM28), HCT116 cells were seeded into 6-well plates
at a density of 5×105 cells/well and transfected with
100 nM Silencer Select small interfering RNA (siRNA)-SIK2 (cat. no.
s23355) and Silencer Select Negative Control No. 1 siRNA (siRNA-NC;
cat. no. 4390843; both from Thermo Fisher Scientific, Inc.) or 4 µg
pCDNA3.1-TRIM28 and pCDNA3.1-NC [Yunzhou Biotechnology (Guangzhou)
Co., Ltd.], using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions, at 37°C for 5 h. Then, the cells were
incubated in DMEM supplemented with 10% FBS at 37°C for 48 h.
Following incubation for 48 h and the evaluation of transfection
efficacy, the cells were utilized for subsequent assays.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using the
MolPure® cell RNA kit (Shanghai Yeasen Biotechnology
Co., Ltd.) and cDNA was synthesized using a RT system kit (cat. no.
N8080234; Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Subsequently, for qPCR, the
QuantiNova SYBR Green PCR kit (Qiagen GmbH) was used according to
the manufacturer's instructions. The qPCR thermocycling conditions
were as follows: Initial denaturation at 95°C for 10 sec, followed
by 40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C
for 10 sec and extension at 72°C for 15 sec. GAPDH served as the
internal reference control. The differences in the expression of
the target genes were assessed using the 2−ΔΔCq method
(23). The primer sequences used
are listed in Table I.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR analysis |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR analysis
| Gene | Sequence |
|---|
| SIK2 | Forward:
5′-GGGTGGGGTTCTACGACATC-3′ |
|
| Reverse:
5′-TATTGCCACCTCCGTCTTGG-3′ |
| Glut1 | Forward:
5′-CCCCGTCCTGCTGCTATTG-3′ |
|
| Reverse:
5′-GCACCGTGAAGATGATGAAGAC-3′ |
| HK2 | Forward:
5′-CAAAGTGACAGTGGGTGTGG-3′ |
|
| Reverse:
5′-GCCAGGTCCTTCACTGTCTC-3′ |
| PKM2 | Forward:
5′-CTTGCAATTATTTGAGGAACTCCGC-3′ |
|
| Reverse:
5′-CACGGTACAGGTGGGCCTGAC-3′ |
| TRIM28 | Forward:
5′-AGCGGGTGAAAAACACCAAG-3′ |
|
| Reverse:
5′-ACCCAAAGCCATAGCCTTCC-3′ |
| GAPDH | Forward:
5′-TCCACCACCCTGTTGCTGTA-3′ |
|
| Reverse:
5′-ACCACAGTCCATGCCATCATC-3′ |
Western blot analysis
Total proteins were extracted from cells using a
pre-chilled RIPA lysis buffer (Beyotime Institute of Biotechnology)
for 10 min on ice and the protein concentration was determined
using a BCA kit (cat. no. P0012; Beyotime Institute of
Biotechnology). The protein samples (25 µg/lane) were separated via
12% SDS-PAGE. Subsequently, proteins were transferred onto PVDF
membranes. Following blocking with 5% skimmed milk for 2 h at room
temperature, the membranes were incubated with primary antibodies
against SIK2 (cat. no. PA1-41208; dilution, 1:500; Invitrogen;
Thermo Fisher Scientific, Inc.), MMP12 (cat. no. ab52897; dilution,
1:2,000), MMP9 (cat. no. ab76003; dilution, 1:1,000), glucose
transporter 1 (Glut1; cat. no. ab115730; dilution, 1:50,000),
hexokinase 2 (HK2; cat. no. ab209847; dilution, 1:1,000), pyruvate
kinase (PK) M2 (PKM2; cat. no. ab85555; dilution, 1:1,000), TRIM28
(cat. no. ab109287; dilution, 1:10,000) and GAPDH (cat. no. ab8245;
dilution, 1:1,000; all from Abcam) at 37°C overnight. The PVDF
membranes were then incubated with the corresponding HRP goat
anti-rabbit secondary antibody (cat. no. G-21234; dilution, 1:
50,000; Invitrogen; Thermo Fisher Scientific, Inc.) for 2 h at room
temperature, followed by washing with PBS. The chemiluminescent
reaction was performed using an ECL kit (iGene Biotechnology Co.,
Ltd.). GAPDH served as the internal reference and the results were
analyzed using ImageJ software (v. 1.6; National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
For CCK-8 assays, HCT116 cells were inoculated into
a 96-well plate at a density of 5×103 cells/well.
Following cell culture for 0, 24, 48 and 72 h at 37°C in a 5%
CO2 incubator, each well was supplemented with 10 µl
CCK-8 reagent (Beyotime Institute of Biotechnology), according to
the manufacturer's instructions, and cells were incubated for an
additional 1 h at 37°C in a 5% CO2 incubator. The
optical density was measured at a wavelength of 450 nm using a
microplate reader (Molecular Devices, LLC).
Wound healing assay
HCT116 cells were seeded into a 6-well plate at a
density of 5×105 cells/well until reaching 80%
confluence. A linear scratch was created with a 200-µl pipette tip
in the middle of wells and free cells were removed by washing with
PBS. Following culture in serum-free DMEM (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C for 24 h, images of the wound area were
captured in the same field at 0 and 24 h using a light microscope
(Nikon Corporation; magnification, ×100) and data were analyzed
using ImageJ (v. 1.8.0; National Institutes of Health).
Transwell assay
Transwell inserts for 24-well plates (8 µm; Corning,
Inc.) were coated with pre-diluted Matrigel (1:8; BD Biosciences)
at 37°C for 30 min. HCT116 cells, at a density of 5×105
cells in 200 µl DMEM, were seeded into the Matrigel-coated upper
chamber, while the lower chamber was filled with 400 µl DMEM
supplemented with 10% FBS. Following incubation at 37°C for 24 h,
the cells on the lower surface of the membrane were fixed with 10%
formaldehyde and stained with 0.1% crystal violet solution both for
15 min at room temperature. The number of invading cells was
counted under a light microscope (Nikon Corporation; magnification,
×100) and cells were analyzed using ImageJ (v. 1.8.0; National
Institutes of Health).
PK activity, and lactate and
glycolysis detection
PK activity and lactate production in HCT116 cells
were measured using a PK assay (cat. no. ab83432; Abcam) and a
lactic acid assay (cat. no. BC2235; Beijing Solarbio Science &
Technology Co., Ltd.) kits, respectively. Glycolysis was measured
using a glycolysis cell-based assay kit (cat. no. 600450-1; AmyJet
Scientific, Inc.). The analyses were conducted according to the
manufacturer's instructions.
Co-immunoprecipitation (co-IP)
assay
For co-IP experiments, cells were collected and
lysed with pre-cooled IP lysis buffer (cat. no. P0013) containing
protease inhibitors (cat. no. P1046; both from Beyotime Institute
of Biotechnology) for 10 min on ice. The supernatant was collected
after centrifugation at 13,000 x g for 10 min at 4°C. Subsequently,
the lysate (0.5 mg per IP reaction) was supplemented with 1 µg SIK
(cat. no. 6919; dilution, 1:50; Cell Signaling Technology, Inc.) or
TRIM28 antibody (cat. no. ab109545; dilution, 1:30; Abcam) and 10
µl protein A agarose beads (Beyotime Institute of Biotechnology)
followed by gentle rotation at 4°C for 4 h. Following
centrifugation at 1,000 x g for 3 min at 4°C, the supernatant was
carefully removed and the agarose beads in the bottom of the tube
were washed with lysis buffer. The pellets were then dissolved in
15 µl 2× SDS sample buffer and boiled for 5 min, followed by
western blot analysis.
Bioinformatics analyses
The Broad Institute Cancer Cell Line Encyclopedia
(CCLE; http://portals.broadinstitute.org/ccle) (24) incorporates genetic data from
numerous human cancer cell lines and has become one of the standard
databases for cancer genomics. Therefore, the mRNA expression
levels of SIK2 in CRC were downloaded from the CCLE database. The
Biological General Repository for Interaction Datasets (BioGRID;
http://thebiogrid.org) (25) includes data on protein, genetic and
drug interactions. In the present study, BioGRID was used to reveal
the interactions between SIK2 and other proteins, referring to
https://thebiogrid.org/116840/table/homo-sapiens/sik2.html.
Statistical analysis
All experiments were repeated at least three times.
Data are expressed as the mean ± SD of triplicate experiments. All
statistical analyses were carried out using GraphPad Prism 8.0
(GraphPad Software, Inc.). The differences between two groups were
compared using an unpaired Student's t-test, while those among
multiple groups with one-way ANOVA followed by Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of SIK2 in CRC cell
lines
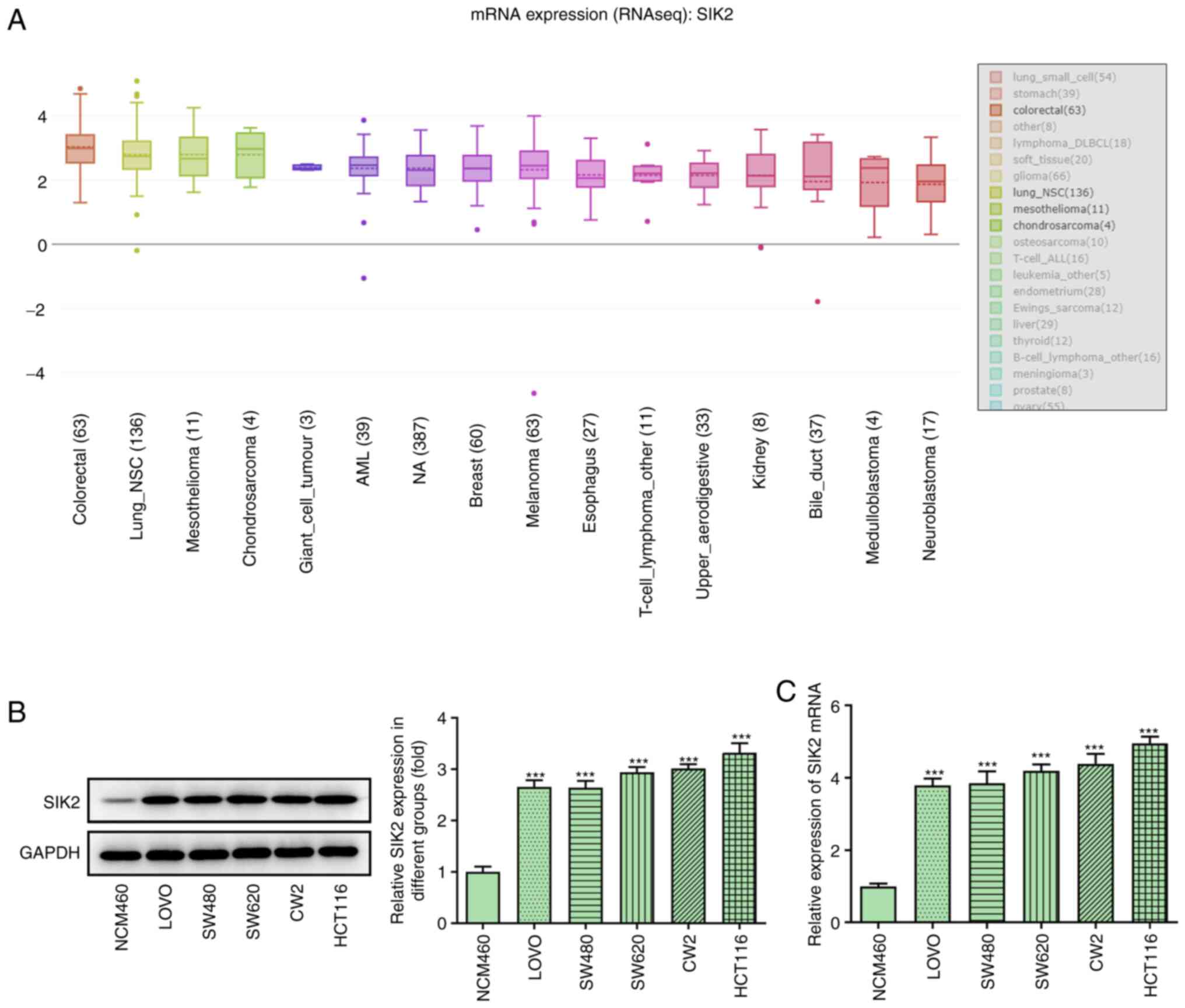
The mRNA expression levels of SIK2 were
significantly increased in all types of cancer in the CCLE
database, including CRC (Fig. 1A).
The mRNA and protein expression levels of SIK2 were determined in
the normal colonic epithelial cell line, NCM460, and different CRC
cell lines (LoVo, SW480, SW620, CW2 and HCT116) using RT-qPCR and
western blot analysis, respectively. The results demonstrated that
SIK2 was upregulated in all examined CRC cell lines compared with
NCM460 cells (Fig. 1B and C). The
expression of SIK2 was more potent in the HCT116 cell line;
therefore, HCT116 cells were used for the subsequent
experiments.
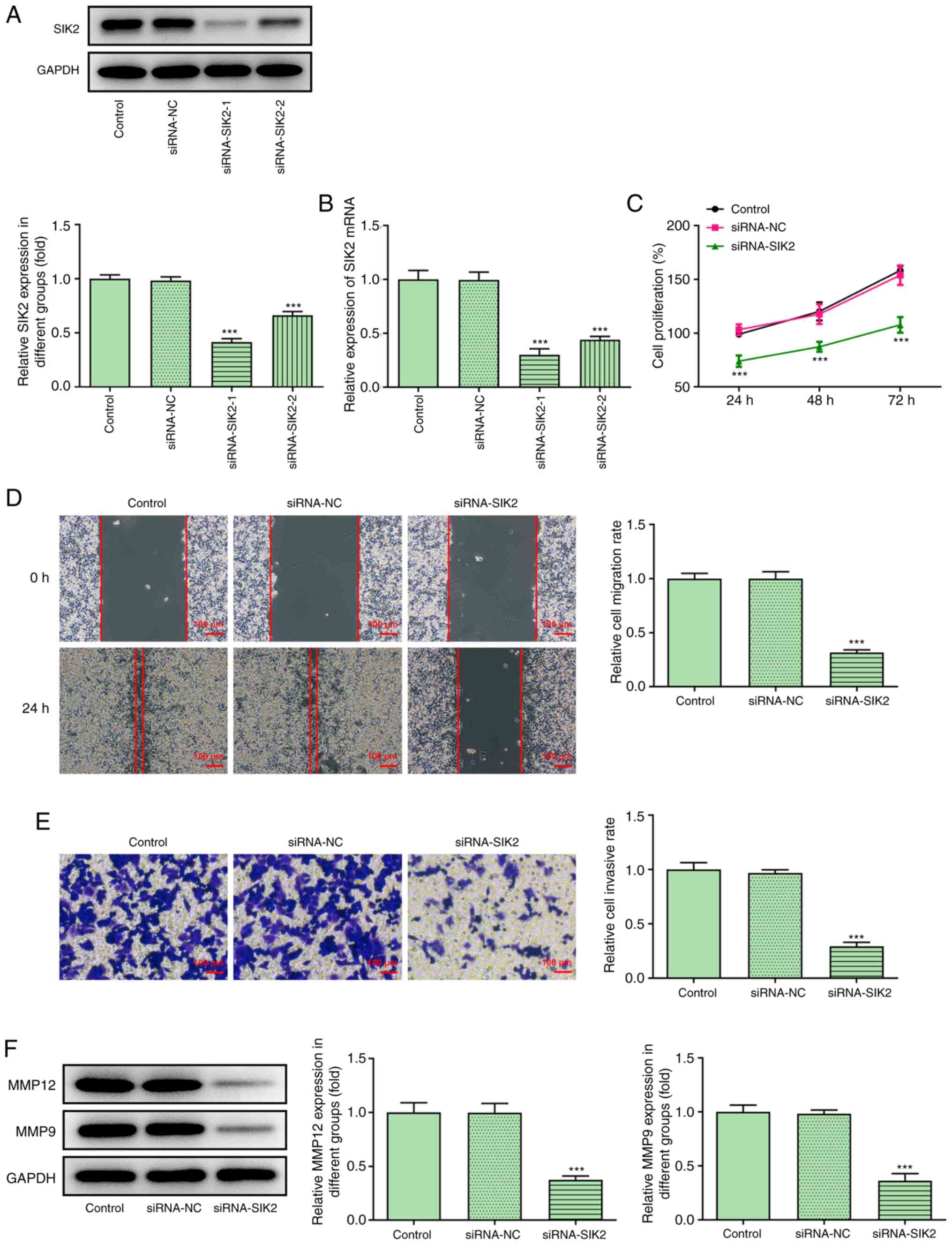
SIK2 silencing attenuates the
proliferation, migration and invasion of CRC cells
The silencing potency of siRNA-SIK2 on the
expression of SIK2 in HCT116 cells was evaluated using RT-qPCR and
western blot analysis. It was found that siRNA-SIK2-1 exerted the
strongest silencing effect on the expression of SIK2 compared with
siRNA-SIK2-2, and it was therefore used in the knockdown
experiments (Fig. 2A and B).
Subsequently, HCT116 cells were divided into the following three
groups: Control, siRNA-NC and siRNA-SIK2 groups. The proliferative
ability of HCT116 cells was determined using a CCK-8 assay. As
shown in Fig. 2C, the cell
proliferation rate in the siRNA-SIK2 group was notably lower
compared with the siRNA-NC group at 24 h following cell
transfection. This trend was maintained over time, suggesting that
SIK2 knockdown could attenuate HCT116 cell proliferation (Fig. 2C). In addition, the cell migratory
and invasive rates were measured in the same experimental groups.
The wound healing assays revealed that the wound was almost
completely closed in the siRNA-NC group at 24 h post-wounding.
However, the migratory ability of HCT116 cells in the siRNA-SIK2
group was significantly decreased (Fig.
2D). The Transwell assays also demonstrated that the invasive
ability of HCT116 cells was decreased in the siRNA-SIK2 group
(Fig. 2E). Furthermore, the protein
expression levels of MMP12 and MMP9, which are relevant to cell
movement (26), were measured via
western blot analysis. Consistent with the Transwell assay results,
both protein expression levels were downregulated in the siRNA-SIK2
group compared with the siRNA-NC group (Fig. 2F).
SIK2 knockdown attenuates glycolysis
in CRC cell lines
The activity of PK, lactate production and
glycolysis were evaluated using their corresponding kits. The
levels of all the aforementioned processes were significantly
attenuated following cell transfection with siRNA-SIK2 (Fig. 3A and B). Additionally, the
expression levels of the glycolysis-related genes Glut1, HK2 and
PKM2 were determined via RT-qPCR and western blot analyses. The
results demonstrated that the expression levels of Glut1, HK2 and
PKM2 were significantly downregulated in HCT116 cells transfected
with siRNA-SIK2 compared with the siRNA-NC group (Fig. 3C and D).
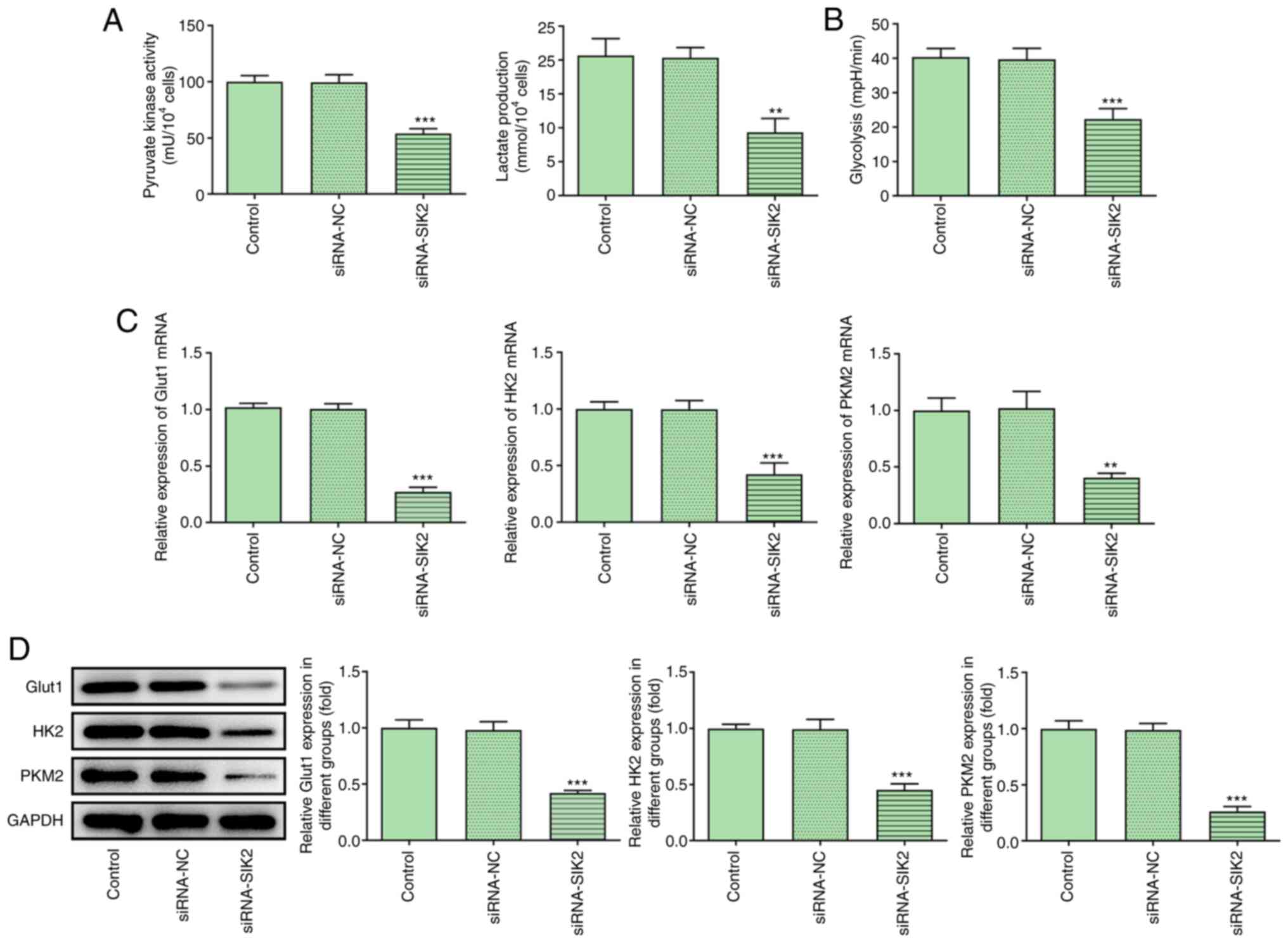 | Figure 3.SIK2 silencing attenuates glycolysis
in colorectal cancer cell lines. The levels of (A) pyruvate kinase
activity, lactate production and (B) glycolysis were detected using
their corresponding kits. The mRNA and protein expression levels of
the glycolysis-related genes, Glut1, HK2 and PKM2, were determined
via (C) reverse transcription-quantitative PCR and (D) western blot
analyses, respectively. **P<0.01, ***P<0.001 vs. siRNA-NC
group. PKM2, pyruvate kinase M2; Glut1, glucose transporter 1; HK2,
hexokinase 2; siRNA, small interfering RNA; NC, negative
control. |
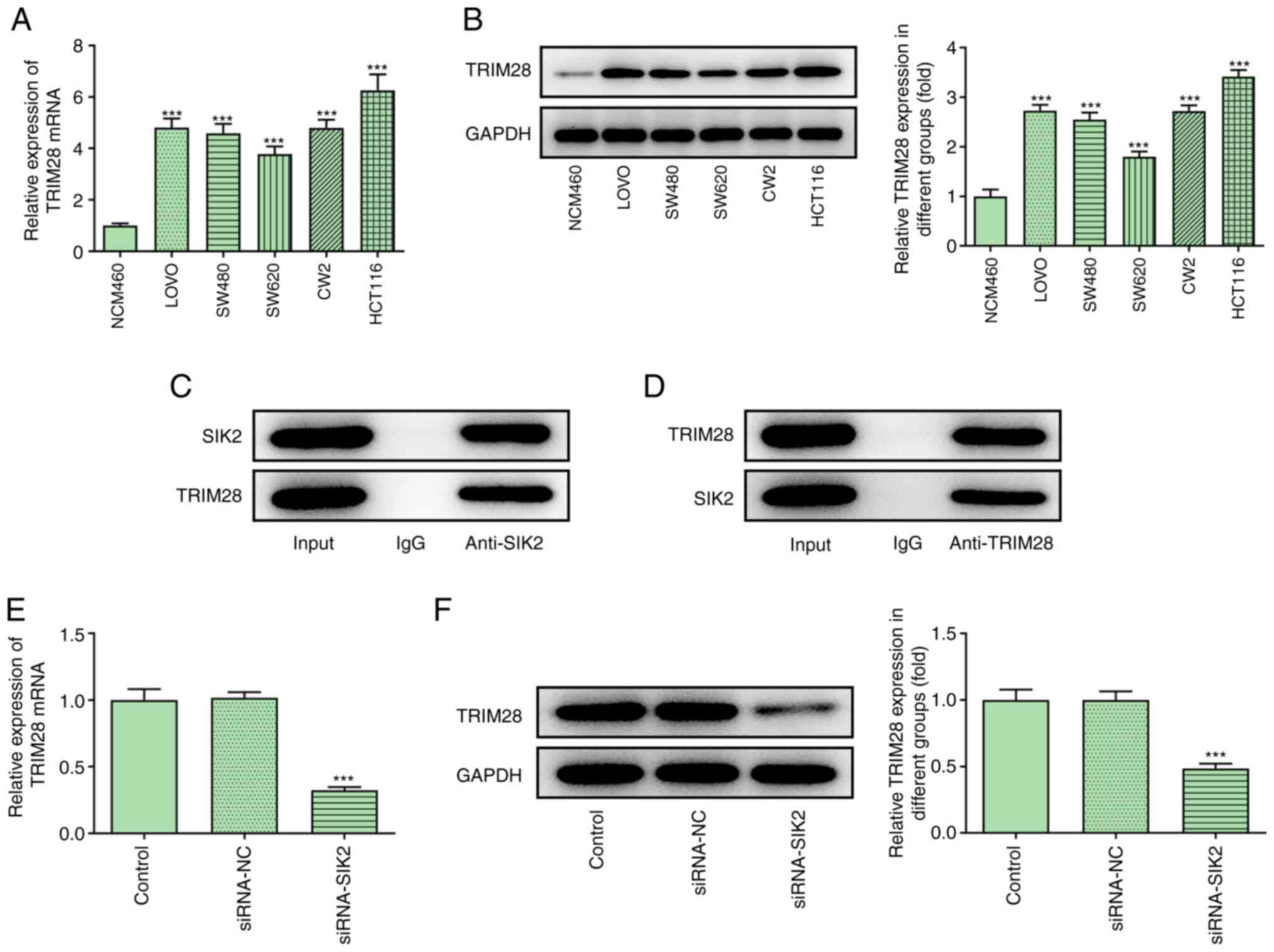
Association between SIK2 and
TRIM28
Bioinformatics analysis using the BioGRID database
predicted that TRIM28 could interact with SIK2. Therefore, the
expression levels of TRIM28 were determined in NCM460, LoVo, SW480,
SW620, CW2 and HCT116 cells via RT-qPCR and western blot analysis.
The results indicated that TRIM28 was upregulated in CRC cell lines
(Fig. 4A and B). In addition, co-IP
was conducted to verify the interaction between SIK2 and TRIM28,
and the results revealed that endogenous SIK2 was co-precipitated
with endogenous TRIM28 in HCT116 cells (Fig. 4C and D). Furthermore, TRIM28 mRNA
and protein expression was downregulated in the siRNA-SIK2 group
(Fig. 4E and F), supporting the
interaction between SIK2 and TRIM28.
SIK2 regulates the proliferation,
migration and invasion of CRC cells by regulating TRIM28
The transfection efficiency of HCT116 cells with
pCDNA3.1-TRIM28 was assessed via RT-qPCR and western blot analyses,
and the results demonstrated that TRIM28 was successfully
overexpressed (Fig. 5A and B).
Subsequently, HCT116 cells were divided into the following four
groups: Control, siRNA-SIK2, siRNA-SIK2 + overexpression vector
(ov)-NC and siRNA-SIK2 + ov-TRIM28 groups. The cell proliferation
rate in the four different groups was measured using a CCK-8 assay.
It was found that TRIM28 overexpression reversed the SIK2-silencing
mediated decreased in the proliferative ability of HCT116 cells
(Fig. 5C). Consistent with the
CCK-8 assay results, the cell migration and invasion assays
demonstrated that the reduced migratory and invasive abilities of
HCT116 cells with SIK2 knockdown were restored following TRIM28
overexpression (Fig. 5D and E).
Furthermore, MMP12 and MMP9 expression was upregulated in the
siRNA-SIK2 + ov-TRIM28 group compared with the siRNA-SIK2 + ov-NC
group (Fig. 5F). Collectively, the
aforementioned findings indicated that SIK2 could promote the
proliferation, migration and invasion of HCT116 cells by
upregulating TRIM28.
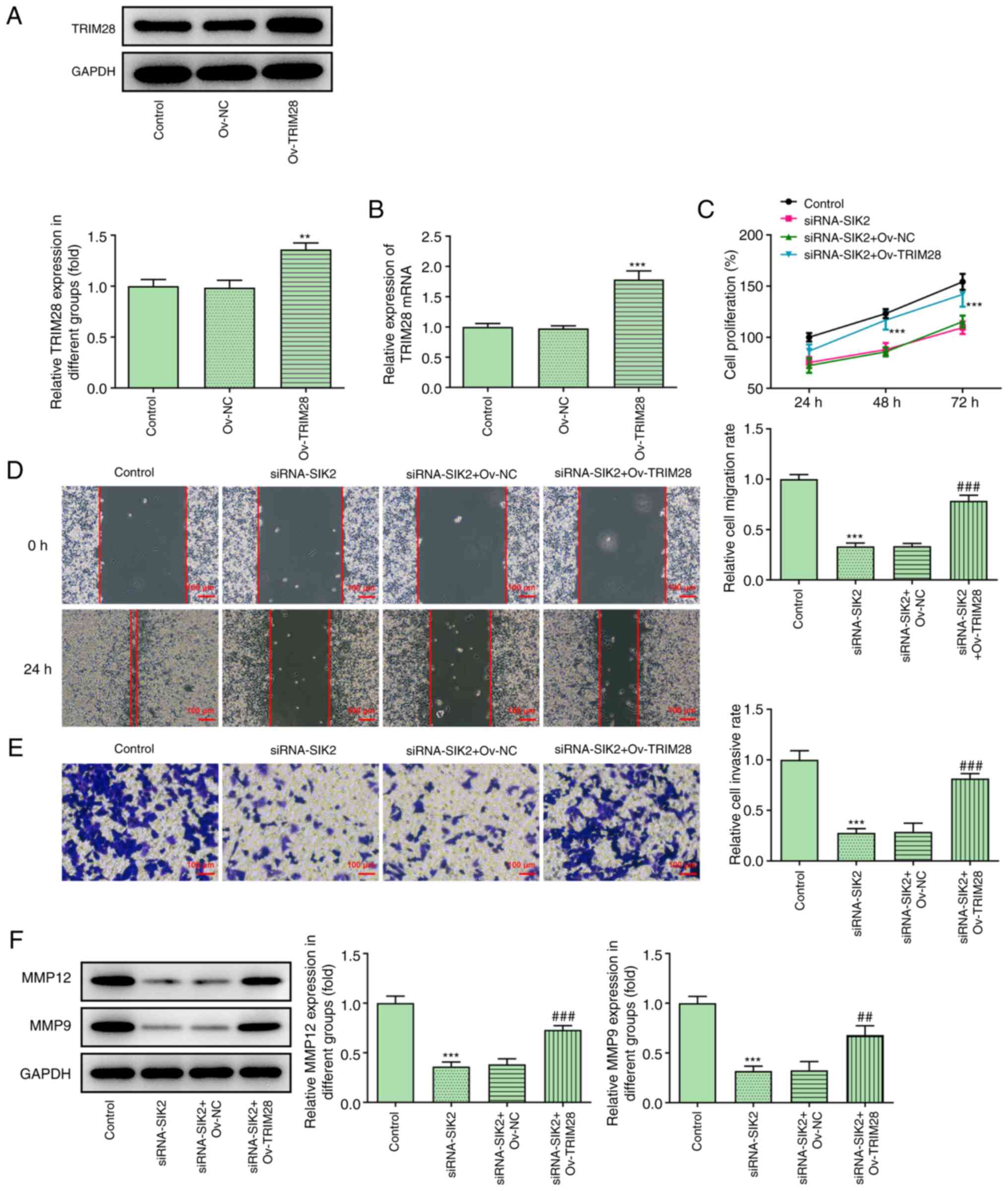 | Figure 5.SIK2 regulates the proliferation,
migration and invasion of colorectal cancer cell lines by
regulating TRIM28. The transfection efficiency of cells with
pCDNA3.1-TRIM28 was confirmed via (A) western blot and (B) reverse
transcription-quantitative PCR analyses. **P<0.01, ***P<0.001
vs. ov-NC group. (C) Cell proliferation ability in different groups
was evaluated using a Cell Counting Kit-8 assay. ***P<0.001 vs.
siRNA-SIK2 + ov-NC group. Cell migration and invasion were measured
using (D) wound healing and (E) Transwell assays, respectively.
Scale bar, 100 µm. (F) Expression levels of MMP12 and MMP9 were
determined via western blot analysis. ***P<0.001 vs. control
group; ##P<0.01, ###P<0.001 vs.
siRNA-SIK2 + ov-NC group. SIK2, salt-inducible kinase 2; TRIM28,
tripartite motif containing 28; ov, overexpression vector; NC,
negative control; siRNA, small interfering RNA. |
SIK2 modulates glycolysis in CRC cell
lines by regulating TRIM28
The effect of TRIM28 on glycolysis in HCT116 cells
was then evaluated following TRIM28 overexpression. The levels of
PK activity, lactate secretion and glycolysis were all restored in
the siRNA-SIK2 + ov-TRIM28 group compared with the siRNA-SIK2 +
ov-NC group (Fig. 6A and B).
Similarly, the RT-qPCR and western blotting results demonstrated
that the expression levels of Glut1, HK2 and PKM2 were reversed
following TRIM28 overexpression (Fig.
6C and D). Collectively, these results suggested that SIK2
could promote glycolysis in HCT116 cells by upregulating
TRIM28.
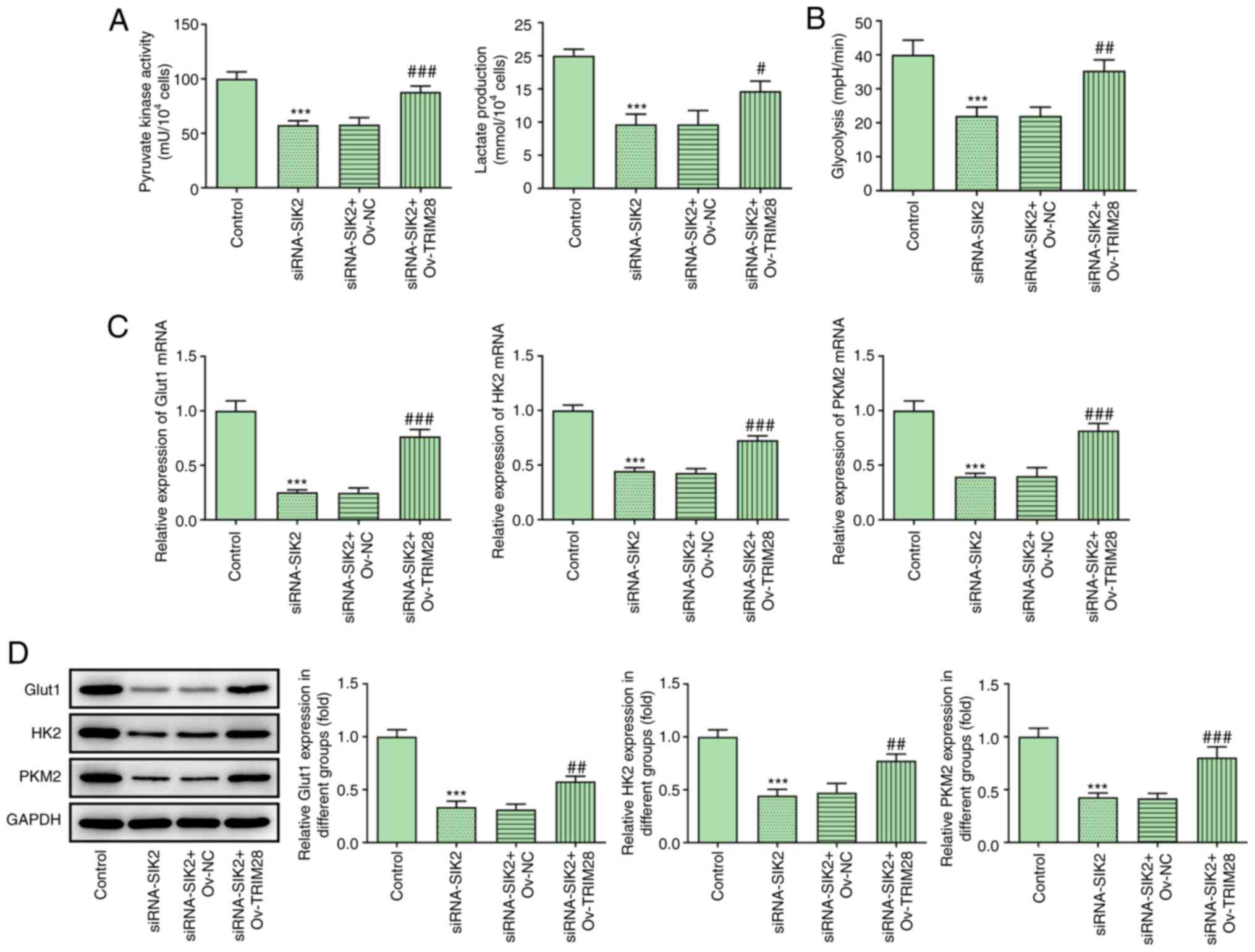 | Figure 6.SIK2 regulates glycolysis in
colorectal cancer cell lines by regulating TRIM28. The levels of
(A) pyruvate kinase activity, lactate production and (B) glycolysis
were measured using their corresponding kits. The mRNA and protein
expression levels of the glycolysis-related genes Glut1, HK2 and
PKM2 were determined via (C) reverse transcription-quantitative PCR
and (D) western blot analyses, respectively. ***P<0.001 vs.
control group; #P<0.05, ##P<0.01 and
###P<0.001 vs. siRNA-SIK2 + Ov-NC group. SIK2,
salt-inducible kinase 2; PKM2, pyruvate kinase M2; TRIM28,
tripartite motif containing 28; ov, overexpression vector; NC,
negative control; siRNA, small interfering RNA; Glut1, glucose
transporter 1; HK2, hexokinase 2. |
Discussion
CRC is a common malignant tumor, and its incidence
in the elderly population is decreasing with the wider application
of colonoscopy in the clinical practice. However, the morbidity and
mortality rates of CRC in young individuals are increasing,
particularly in developing countries (27). In addition, due to the lack of
typical clinical symptoms in the early stages of the disease,
numerous patients with CRC are already at an advanced stage at the
time of diagnosis. Therefore, the prognosis for these patients is
usually poor (28). In a study on
the association between CRC and geographic location and diet, Doll
and Peto (29) revealed that
>90% of intestinal tumors were associated with dietary habits,
suggesting that different foods could have a great influence on the
composition and diversity of the gut microbiome (30). Moreover, Liang et al
(31) showed that the structure of
the intestinal microbial community of patients with colorectal
adenoma was more susceptible to interference compared with that of
healthy subjects. Since the progression from detectable
precancerous lesions to CRC is relatively slow, patients diagnosed
early usually have a good prognosis and, therefore, CRC screening
has been shown to be effective in reducing mortality (32). Therefore, early diagnosis is
considered particularly important for disease outcome.
Cancer cells undergo a series of changes during the
process of carcinogenic transformation and the changes in cell
metabolism serve a key role for their new phenotypes (33). Compared with normal cells, cancer
cells exhibit enhanced proliferation and can rapidly adapt to
changes in metabolism, thereby meeting the requirements of cells
for metabolism and biosynthesis (34,35).
The most significant feature of metabolism is the ability of cancer
cells to increase the glycolysis rate in an aerobic environment,
which is referred to as aerobic glycolysis (36). Glycolysis is the degradation
reaction of glucose or glycogen into lactic acid or pyruvate that
resembles fermentation, thus resulting in reduced energy
consumption and ATP production from the cells (37). The glycolytic enzymes PKM2 and HK2
and the glucose transporter Glut1 are often dysregulated during
carcinogenesis, and they have been considered to be involved in the
crucial pathways of aerobic glycolysis in cancer cells (38). Previous studies reported that SIK2
was involved in the promotion of aerobic glycolysis in breast and
ovarian cancer cells (20,39). In addition to studying the effect of
SIK2 on the proliferation, migration and invasion of CRC cells, the
present study aimed to investigate the effect of SIK2 on
glycolysis. The results indicated that SIK2 could also promote
glycolysis in CRC cells, thus suggesting that SIK2 may be a
promising biomarker for CRC.
The current study also investigated the specific
mechanism underlying the effects of SIK2 on CRC. Bioinformatics
analysis using the BioGRID database revealed that TRIM28 could be a
target of SIK2. The association between the two proteins was
verified using a co-IP assay. It has been reported that TRIM28 is
involved in several types of cancer. For example, increased
expression of TRIM28 was found to be associated with cervical
cancer metastasis (40). In gastric
cancer, TRIM28 was upregulated in the peripheral blood of patients
and its expression was associated with poor prognosis (41,42).
In gliomas, TRIM28 was also highly expressed and was positively
associated with the degree of tumor malignancy, poor overall
survival and progression-free survival (43). Interestingly, the expression of
TRIM28 was elevated in the interstitial tissues of patients with
CRC, while its increased expression was associated with worse
prognosis (44,45). In the present study, TRIM28
overexpression reversed the effects of SIK2 silencing on cell
proliferation, migration, invasion and glycolysis, thus indicating
that the effects of SIK2 on the physiological behavior of CRC cells
may be mediated by regulating TRIM28. However, as only in
vitro experiments were performed, the results of the present
study must be further verified using in vivo experiments in
the future. Moreover, the mechanism underlying the downstream genes
of TRIM28 are worthy of a follow-up investigation.
In conclusion, the results of the present study
demonstrated that SIK2 was upregulated in CRC, while SIK2 knockdown
suppressed cell proliferation, migration, invasion and glycolysis,
suggesting that SIK2 may represent a promising biomarker for CRC.
Furthermore, TRIM28 overexpression could reverse these effects,
thus indicating that SIK2 may affect the malignant behavior and
glycolysis of CRC cells by regulating TRIM28. The findings of the
present study may provide a novel approach to the targeted therapy
and clinical diagnosis of CRC in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author upon
reasonable request.
Authors' contributions
XN designed and performed the experiments and made
substantial contributions to the manuscript writing. YF performed
the experiments and analyzed the data. XF made substantial
contributions to conception and design, guided the experiments and
revised the manuscript. All the authors have read and approved the
final manuscript. XN and XF confirm the authenticity of the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Janney A, Powrie F and Mann EH:
Host-microbiota maladaptation in colorectal cancer. Nature.
585:509–517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Johdi NA and Sukor NF: Colorectal cancer
immunotherapy: Options and strategies. Front Immunol. 11:16242020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmoll HJ, Van Cutsem E, Stein A,
Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde
CJ, Balmana J, Regula J, et al: ESMO consensus guidelines for
management of patients with colon and rectal cancer. A personalized
approach to clinical decision making. Ann Oncol. 23:2479–2516.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Cutsem E, Cervantes A, Nordlinger B
and Arnold D; ESMO Guidelines Working Group, : Metastatic
colorectal cancer: ESMO clinical practice guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 25 (Suppl 3):iii1–iii9. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yoshino T, Arnold D, Taniguchi H,
Pentheroudakis G, Yamazaki K, Xu RH, Kim TW, Ismail F, Tan IB, Yeh
KH, et al: Pan-Asian adapted ESMO consensus guidelines for the
management of patients with metastatic colorectal cancer: A
JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann
Oncol. 29:44–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Cutsem E, Cervantes A, Adam R, Sobrero
A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson
A, Bodoky G, et al: ESMO consensus guidelines for the management of
patients with metastatic colorectal cancer. Ann Oncol.
27:1386–1422. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bonjer HJ, Deijen CL, Abis GA, Cuesta MA,
van der Pas MH, de Lange-de Klerk ES, Lacy AM, Bemelman WA,
Andersson J, Angenete E, et al: A randomized trial of laparoscopic
versus open surgery for rectal cancer. N Engl J Med. 372:1324–1332.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
MacFarlane JK, Ryall RD and Heald RJ:
Mesorectal excision for rectal cancer. Lancet. 341:457–460. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park SC, Sohn DK, Kim MJ, Chang HJ, Han
KS, Hyun JH, Joo J and Oh JH: Phase II clinical trial to evaluate
the efficacy of transanal endoscopic total mesorectal excision for
rectal cancer. Dis Colon Rectum. 61:554–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garg MB, Lincz LF, Adler K, Scorgie FE,
Ackland SP and Sakoff JA: Predicting 5-fluorouracil toxicity in
colorectal cancer patients from peripheral blood cell telomere
length: A multivariate analysis. Br J Cancer. 107:1525–1533. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ogura A, Konishi T, Cunningham C,
Garcia-Aguilar J, Iversen H, Toda S, Lee IK, Lee HX, Uehara K, Lee
P, et al: Neoadjuvant (chemo) radiotherapy with total mesorectal
excision only is not sufficient to prevent lateral local recurrence
in enlarged nodes: Results of the multicenter lateral node study of
patients with low cT3/4 rectal cancer. J Clin Oncol. 37:33–43.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patel JN: Application of genotype-guided
cancer therapy in solid tumors. Pharmacogenomics. 15:79–93. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pellino G, Gallo G, Pallante P, Capasso R,
De Stefano A, Maretto I, Malapelle U, Qiu S, Nikolaou S, Barina A,
et al: Noninvasive biomarkers of colorectal cancer: Role in
diagnosis and personalised treatment perspectives. Gastroenterol.
2018:23978632018.PubMed/NCBI
|
|
17
|
Ogunwobi OO, Mahmood F and Akingboye A:
Biomarkers in colorectal cancer: Current research and future
prospects. Int J Mol Sci. 21:53112020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Patel JN, Fong MK and Jagosky M:
Colorectal cancer biomarkers in the era of personalized medicine. J
Pers Med. 9:32019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yiu AJ and Yiu CY: Biomarkers in
colorectal cancer. Anticancer Res. 36:1093–1102. 2016.PubMed/NCBI
|
|
20
|
Zong S, Dai W, Fang W, Guo X and Wang K:
SIK2 promotes cisplatin resistance induced by aerobic glycolysis in
breast cancer cells through PI3K/AKT/mTOR signaling pathway. Biosci
Rep. May 27–2020.(Epub ahead of Print).
|
|
21
|
Zhao J, Zhang X, Gao T, Wang S, Hou Y,
Yuan P, Yang Y, Yang T, Xing J, Li J and Liu S: SIK2 enhances
synthesis of fatty acid and cholesterol in ovarian cancer cells and
tumor growth through PI3K/Akt signaling pathway. Cell Death Dis.
11:252020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun Z, Niu S, Xu F, Zhao W, Ma R and Chen
M: CircAMOTL1 promotes tumorigenesis through miR-526b/SIK2 axis in
cervical cancer. Front Cell Dev Biol. 8:5681902020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oughtred R, Rust J, Chang C, Breitkreutz
BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, et
al: The BioGRID database: A comprehensive biomedical resource of
curated protein, genetic, and chemical interactions. Protein Sci.
30:187–200. 2021. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Xu X, Jiang Y, Hansbro NG, Hansbro
PM, Xu J and Liu G: Elastin is a key factor of tumor development in
colorectal cancer. BMC Cancer. 20:2172020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mauri G, Sartore-Bianchi A, Russo AG,
Marsoni S, Bardelli A and Siena S: Early-onset colorectal cancer in
young individuals. Mol Oncol. 13:109–131. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang X, Zhang H, Shen B and Sun XF:
Chromogranin-A expression as a novel biomarker for early diagnosis
of colon cancer patients. Int J Mol Sci. 20:29192019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doll R and Peto R: The causes of cancer:
Quantitative estimates of avoidable risks of cancer in the United
States today. J Natl Cancer Inst. 66:1191–1308. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rothschild D, Weissbrod O, Barkan E,
Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN,
Bar N, et al: Environment dominates over host genetics in shaping
human gut microbiota. Nature. 555:210–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang Q, Chiu J, Chen Y, Huang Y,
Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, et al:
Fecal bacteria act as novel biomarkers for noninvasive diagnosis of
colorectal cancer. Clin Cancer Res. 23:2061–2070. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ladabaum U, Dominitz JA, Kahi C and Schoen
RE: Strategies for colorectal cancer screening. Gastroenterology.
158:418–432. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cuezva JM, Ortega AD, Willers I,
Sánchez-Cenizo L, Aldea M and Sánchez-Aragó M: The tumor suppressor
function of mitochondria: Translation into the clinics. Biochim
Biophys Acta. 1792:1145–1158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vander Heiden MG, Lunt SY, Dayton TL,
Fiske BP, Israelsen WJ, Mattaini KR, Vokes NI, Stephanopoulos G,
Cantley LC, Metallo CM and Locasale JW: Metabolic pathway
alterations that support cell proliferation. Cold Spring Harb Symp
Quant Biol. 76:325–334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lunt SY and Vander Heiden MG: Aerobic
glycolysis: Meeting the metabolic requirements of cell
proliferation. Annu Rev Cell Dev Biol. 27:441–464. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Enzo E, Santinon G, Pocaterra A, Aragona
M, Bresolin S, Forcato M, Grifoni D, Pession A, Zanconato F, Guzzo
G, et al: Aerobic glycolysis tunes YAP/TAZ transcriptional
activity. EMBO J. 34:1349–1370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Samec M, Liskova A, Koklesova L, Samuel
SM, Zhai K, Buhrmann C, Varghese E, Abotaleb M, Qaradakhi T, Zulli
A, et al: Flavonoids against the warburg phenotype-concepts of
predictive, preventive and personalised medicine to cut the Gordian
knot of cancer cell metabolism. EPMA J. 11:377–398. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gao T, Zhang X, Zhao J, Zhou F, Wang Y,
Zhao Z, Xing J, Chen B, Li J and Liu S: SIK2 promotes reprogramming
of glucose metabolism through PI3K/AKT/HIF-1α pathway and
Drp1-mediated mitochondrial fission in ovarian cancer. Cancer
Letters. 469:89–101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin LF, Li CF, Wang WJ, Yang WM, Wang DD,
Chang WC, Lee WH and Wang JM: Loss of ZBRK1 contributes to the
increase of KAP1 and promotes KAP1-mediated metastasis and invasion
in cervical cancer. PLoS One. 8:e730332013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yokoe T, Toiyama Y, Okugawa Y, Tanaka K,
Ohi M, Inoue Y, Mohri Y, Miki C and Kusunoki M: KAP1 is associated
with peritoneal carcinomatosis in gastric cancer. Ann Surg Oncol.
17:821–828. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang YY, Li L, Zhao ZS and Wang HJ:
Clinical utility of measuring expression levels of KAP1, TIMP1 and
STC2 in peripheral blood of patients with gastric cancer. World J
Surg Oncol. 11:812013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qi ZX, Cai JJ, Chen LC, Yue Q, Gong Y, Yao
Y and Mao Y: TRIM28 as an independent prognostic marker plays
critical roles in glioma progression. J Neurooncol. 126:19–26.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fitzgerald S, Sheehan KM, O'Grady A, Kenny
D, O'Kennedy R, Kay EW and Kijanka GS: Relationship between
epithelial and stromal TRIM28 expression predicts survival in
colorectal cancer patients. J Gastroenterol Hepatol. 28:967–974.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fitzgerald S, Espina V, Liotta L, Sheehan
KM, O'Grady A, Cummins R, O'Kennedy R, Kay EW and Kijanka GS:
Stromal TRIM28-associated signaling pathway modulation within the
colorectal cancer microenvironment. J Transl Med. 16:892018.
View Article : Google Scholar : PubMed/NCBI
|