Introduction
Thoracic radiotherapy is an important cancer
treatment for patients diagnosed with malignant tumors, such as
breast cancer, lung cancer, Hodgkin's lymphoma and esophageal
cancer (1–4). Radiotherapy improves overall survival
(OS) in patients with thoracic cancer, but it can result in some
inevitable complications, particularly radiation-induced heart
damage (RIHD), which has gradually become an increasing concern of
oncologists and cardiologists (5,6). The
incidence of RIHD has increased due to the prolonged OS of patients
with cancer, and RIHD usually occurs 5–10 years after radiation
(7,8). Radiation influences various aspects of
the heart physiology, including the myocardium, pericardium,
valves, coronary arteries and conduction system (9–11).
Ionizing radiation exposure accelerates endothelial injury and
facilitates the development of atherosclerotic plaques in coronary
arteries clinically (10). The
rates of major coronary events increase by 7.4% per gray as the
mean heart dose (MHD) is increased, and these coronary events
usually occur 5–20 years after radiotherapy amongst breast cancer
survivors (12). It has been
reported that the risk of coronary heart disease was increased
2.5-fold in a 20 Gy MHD group compared with patients with Hodgkin's
lymphoma treated without radiation (2). Myocardial injury is another common
complication of radiation. In a previous study, the prevalence of
heart failure was nearly 24% at 20 years after receiving thoracic
ionizing radiation (13). In
preclinical studies, mice exposed to high doses of radiation often
exhibited a decrease in left ventricular ejection fraction (LVEF)
and an increase in expression of myocardial injury markers
(14,15). In our previous study, soluble
suppression of tumorigenicity-2 was identified as a cardiac
biomarker that was shown to be increased amongst patients with
cancer following thoracic radiotherapy, and it was positively
associated with the radiation parameters V5,
V10, V20 of heart and MHD (16).
High-dose exposure (>5 Gy) radiation, which is
mostly observed in patients receiving radiation therapy, leads to
cardiac injury (17). Low dose
exposure (<0.5 Gy) radiation has also been investigated with
cardiovascular effects, which is more frequently observed in
Japanese atomic bomb survivors, Mayak workers or other nuclear
workers (18). To further evaluate
the heart damage of patients who received radiotherapy, the
relevant literature was examined (19,20),
and consequently a high dose of 16 Gy was selected in the current
study. In previous studies, RIHD progression was considered to be
associated with energy deposition and the generation of reactive
oxygen species (ROS), followed by molecular changes, damage to DNA,
lipids and proteins, as well as the activation of early response
transcription factors, cytokines, endothelial cell injury and
signaling transduction pathways (21–25).
This radiation stimulation to heart tissue of mice may result in
increasing epicardial thickness and infiltrating inflammatory
factors in the epicardium, as well as interstitial collagen
deposition (20). To investigate
RIHD in-depth, high-throughput proteomics analysis was used in the
present study.
The latest advances in the high-throughput
technologies of multiple ‘combinatorial data’ analyses may allow
for a more direct and precise understanding of the molecular
mechanisms underlying diseases (26,27).
Notably, proteomics are used to provide functional context in
interpretation of genomic abnormalities, particularly in cancer
biology (28,29). Proteomics has gradually become an
important technique in the fields of disease diagnosis, drug
research and drug development, whilst also serving a predominant
role in the study of the molecular basis of diseases and the
understanding of various biological processes at the protein level
(30,31). Proteomics analysis of radiation
injury may provide novel directions for the study of the mechanism
underlying RIHD. Thus, in the present study, proteomics analysis
was used to identify the changes that occurred in protein
expression levels in cells or tissues.
The aim of the present study was to use a novel
high-throughput proteomics technology to evaluate the effects of
radiation on expression of proteins in the heart by establishing a
RIHD mouse model to further understand the potential molecular
mechanisms underlying the development of RIHD.
Materials and methods
Animal model of RIHD and local cardiac
irradiation
In total, 18 C57BL/6 male mice (weight, 20–25 g;
age, 8 weeks) were purchased from the Shanghai Institute of
Biochemistry and Cell Biology. The mice were irradiated at the age
of 8–9 weeks. All mice were maintained with controlled humidity
(40–60%) and temperature (22–24°C) and a 12-h light/dark cycle,
with ad libitum access to food and water. The irradiated
mice were sacrificed after 1, 3 or 5 months, and the mice in the
control group mice were sacrificed after 5 months. Mice were
sacrificed by dislocating the cervical spine under intraperitoneal
injection of 75 mg/kg pentobarbital sodium. Each group included 3–6
mice. Ethical approval for the animal experiments was obtained from
the Institutional Review Board of The Second Affiliated Hospital of
Nanchang University.
The whole heart was locally irradiated with a dose
of 16 Gy using a precise small-animal radiation research platform
(XStrahl Medical and Life Sciences) in the Zhejiang Key Radiation
Laboratory. Mice in the control group were sham-irradiated and
heart tissue was harvested after 5 months. All mice were
anesthetized by intraperitoneal injection of 75 mg/kg pentobarbital
sodium and placed in the supine position in the irradiation area of
the small animal X-ray radiometer. The laser system was used to
establish a 3D coordinate system. Irradiated mice underwent whole
heart radiation. Control mice received sham irradiation (0 Gy).
Cone-beam computed tomography, using 50 kV and 0.8 mA photons
filtered with aluminum (1 mm), was performed for each mouse to
visualize the tomographic scanning of the thorax. Heart, lungs and
the spinal cord were drawn by the same physicist during the
tomographic scanning of the thorax, then the physicists designed
and evaluated the radiotherapy plan, and limited the irradiated
volume of the lung tissue and spinal cord in the mice as much as
possible. A dose-volume histogram of the heart, lungs and spine was
obtained (Fig. S1B). For arc
radiation, a 0.15-mm copper filter was fitted with a 10×10
mm2 collimator (−90° to 90°) at a source-skin distance
of 342.5 mm, energized at 220 kV and 13.0 mA a single X-ray beam
with a dose rate of 2.32 Gy/min (Fig.
S1A, C-E).
Cardiac echocardiogram
Transthoracic echocardiography was performed using
the Vevo 2100 ultrasound system (Visualsonics, Inc.) as described
in our previous study (32). The
mice were anesthetized by isoflurane (induction and maintenance
doses of isoflurane were 3 and 1.5%, respectively). Mice were
placed in the supine position on an electrical heating pad at 37°C.
At the level of the largest left ventricle (LV), 2D guided M-mode
echoes were obtained. The left ventricular posterior wall at the
end of diastole was measured from the M-mode image in short axis.
The LVEF and fractional shortening were calculated from the
measured ventricle dimensions. Each group contained ≥3 mice.
Measurement of serum brain natriuretic
peptide (BNP) levels
Mice were anesthetized by intraperitoneal injection
of 75 mg/kg pentobarbital sodium, and blood was collected from the
vena orbital sinus of the 1-, 3- and 5-month irradiated mice group
and the control group. Then, mice were sacrificed quickly via
cervical dislocation. Blood samples (~1 ml per mouse) were
collected in tubes with EDTA and the serum was separated via
centrifugation at 4°C at 600 × g for 10 min. BNP was determined
using a high sensitivity ELISA kit [Presage BNP assay; USCNK,
https://www.uscnk.cn/uscn/ELISA-Kit-for-Brain-Natriuretic-Peptide-(BNP)-49004.htm]
according to the manufacturer's protocol. BNP levels were evaluated
after determining the optical density of the samples at 450 nm
(Thermo Fisher Scientific, Inc.). Each group contained ≥3 mice.
H&E, Masson staining and
immunohistochemistry (IHC)
Half of the apical part of mouse hearts in control
(n=3) and experiment (n=3) group was quickly excised, immersed in
10% paraformaldehyde at room temperature for 24 h and embedded in
paraffin. The heart tissue was cut into 5-µm thick sections, and
the slides were stained with the H&E staining kit (Beijing
Solarbio Science & Technology Co., Ltd.; cat. no. G1120).
Masson staining of heart tissue was conducted using a Trichrome
stain (Masson) kit (Beijing Solarbio Science & Technology Co.,
Ltd.; cat. no. G1340) according to manufacturer's instructions.
IHC was performed as described previously (33). The 5-μM sections were baked at 65°C
for 30 min, deparaffinized with xylene and rehydrated with a
decreasing series of ethanol concentrations (100, 95, 90, 80 and
70%). Then, antigen retrieval was performed using EDTA, after which
tissues were treated with 3% hydrogen peroxide in methanol to
quench endogenous peroxidase activity for 30 min at room
temperature, and blocked with 5% goat serum (Solarbio, Inc.) at
room temperature for 1 h. Primary antibodies against mouse
anti-collagen type 1 α 1 chain (Col1a1; 1:500; ProteinTech Group,
Inc.; cat. no. BA0325) or mouse anti-collagen type III α 1 chain
(Col3a1; 1:500; ProteinTech Group, Inc.; cat. no. M00788) were
added, and samples were incubated at 4°C overnight, following which
they were incubated with biotinylated secondary antibody
(anti-rabbit; 1:5,000; OriGene Technologies, Inc.; cat. no.
TA130017) for 1 h at room temperature and stained with DAB for 15
min at room temperature. After fully washing under tap water,
sections were stained with 10% hematoxylin for 3 min at room
temperature and sealed with neutral gum. Sections were examined
using an Olympus CX33 light microscope (Olympus Corporation)
Proteomics analysis
Proteomics analysis was conducted and analyzed by
Jingjie PTM Biolab Co., Ltd. The 5-month and control groups
contained three mice each. The primary processes included protein
extraction of heart apex with lysis buffer-8 M urea (Sigma-Aldrich;
Merck KGaA), 1% Protease Inhibitor (Calbiochem; Merck KGaA), 3 µM
TSA (Sigma-Aldrich; Merck KGaA), 50 mM NAM (Sigma-Aldrich; Merck
KGaA), 2 mM EDTA (Sigma-Aldrich; Merck KGaA), trypsin digestion and
tandem mass tag (TMT) labeling, followed by high performance liquid
chromatography (LC) fractionation and LC-mass spectrometry (MS)/MS
analysis. MS/MS data were analyzed using the Maxquant search engine
(https://www.maxquant.org/,version
1.5.2.8), and the tandem mass spectra were analyzed using the
SwissProt Mouse database (version 201808,16992 sequences,
http://www.uniprot.org/), along with the reverse
decoy database to calculate the false positive rate caused by
random matching. Detailed descriptions can be found in the Data S1.
Bioinformatics analysis
Gene Ontology (GO) analysis
A GO annotation proteome was derived from the
UniProt-GOA database (version Goa_uniprot_gcrp.gaf.163;
ebi.ac.uk/GOA/).
Subcellular localization analysis
Wolfpsort (version 0.2; genscript.com/psort/wolf_psort.html), a subcellular
localization predication program, was used to predict the
subcellular localization of the proteins.
Cluster of Orthologous
Groups/Eukaryotic Orthologous Groups (COG/KOG) function
classification
The KOG database (version 2003; http://ftp.ncbi.nih.gov/pub/COG/KOG/)
was uses to classified DEPs for function annotations.
Functional enrichment
Enrichment of GO analysis
GO annotations can be divided into three categories:
Biological Process (BP), Cellular Component (CC) and Molecular
Function (MF), which categorize the biological functions of
proteins based on different features. A two-tailed Fisher's exact
test was used to assess the enrichment of the differentially
expressed proteins (DEPs) against all identified proteins.
Enrichment of pathway analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG)
database (version 2.5; http://www.kegg.jp/kegg/pathway) was used to study the
enriched pathways using a two-tailed Fisher's exact test.
Enrichment of the protein domain analysis. InterPro
(version 5.14–53.0; ebi.ac.uk/interpro/), a database providing
functional analysis of protein sequences and predicting the
presence of domains and important sites, was used and a two-tailed
Fisher's exact test was used to assess the enrichment of the
DEPs.
Protein-protein interaction (PPI)
network mapping
All DEP database accession nos. or sequences were
searched against the Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database version 11.0 (https://www.string-db.org/) to determine the PPI
network. Only interactions between the proteins belonging to the
searched dataset were selected, thereby excluding external
candidates. STRING defines a metric termed ‘confidence score’ to
describe the interaction confidence; all interactions that had a
confidence score >0.7 (high confidence) were fetched.
Interaction networks from STRING were visualized in Cytoscape 3.7.2
(34). Multi Contrast Delayed
Enhancement (MCODE) version 1.5.1 was used to identify significant
modules (MCODE score ≥10) (35).
Additionally, cytoHubba (version 1.6) was used to study essential
nodes in the network using 11 methods [Density of maximum
neighborhood component (DMNC) exhibits a satisfied comparative
performance], and was completed to examine the hub genes (36).
Transmission electron microscopy
Parts of fresh apical heart portions of the mice
after 5 months were quickly sectioned into 1-mm3 cubes
and fixed in paraformaldehyde (Beijing Solarbio Science &
Technology Co., Ltd.) for 2 h at 4°C. Sections were fixed in 1%
osmium tetroxide for 2 h at room temperature, followed by stepwise
dehydration in graded acetone, after which, tissues were
infiltrated, embedded (1:1 acetone/812 embedding agent for 3 h at
37°C, 1:2 acetone/812 embedding agent overnight at 37°C and 812
embedding agent for 6 h at 37°C) and polymerized. The pieces were
sectioned with an ultramicrotome into 1–2 µm pieces, and then
stained with a toluidine blue dye solution for 30 sec at room
temperature. The myocardial ultrastructure of the heart was
observed on a HITACHI-7700 electron microscope (Hitachi, Ltd.).
Each group contained three mice.
ATP and lactic acid assays
Samples of the apex of the heart were mixed with
lytic fluid, homogenized with a homogenizer and then centrifuged at
4°C at 12,000 × g for 5 min. ATP production was detected using an
ATP Assay kit (Beyotime Institute of Biotechnology) according to
the manufacturer's instructions. Relative light unit values were
collected using with the Luminometer mode using a multimode
Varioskan LUX microplate reader (Thermo Fisher Scientific,
Inc.).
Lactic acid was detected using a lactic acid kit
(Nanjing Jiancheng Bioengineering Institute) according to the
manufacturer's protocol. The optical density value was measured
using a multimode microplate reader at 530 nm.
The protein concentration was measured using a BCA
assay. Absolute ATP and lactate levels were calculated from the
corresponding standard curve and normalized to the total protein
concentration. Each group contained three mice.
Western blotting
Western blotting was performed using a standard
protocol, as described previously (37). Samples of the apex of the heart from
all mice were briefly homogenized using a homogenizer (PRO
Scientific), and lysed in lysis buffer [50 mmol/l Tris-HCl (pH
8.0), 150 mmol/l NaCl, 0.5% NP-40, 0.1% SDS and 5 mmol/l EDTA (pH
8.0)] containing aprotinin phosphatase inhibitor (Beyotime
Institute of Biotechnology) and protease inhibitor (Beyotime
Institute of Biotechnology). Protein concentrations were quantified
using a BCA Protein assay kit (Beyotime Institute of
Biotechnology), and 10 µg protein was loaded per a lane on an 8 or
12% SDS-gel. Proteins were resolved using SDS-PAGE and transferred
to PVDF membranes (MilliporeSigma). The membranes were blocked
using 5% fat-free milk for 1 h at room temperature and incubated
with primary antibodies at 4°C overnight, followed by incubation
with secondary antibodies (anti-rabbit; 1:5,000; Wuhan Boster
Biological Technology, Ltd.) for 1 h at room temperature. Signals
were visualized using chemiluminescence reagent (Bio-Rad
Laboratories, Inc.) and semi-quantified using ImageJ 1.49 (National
Institutes of Health). The primary antibodies used in the present
study were: Anti-Col1a1 polyclonal antibody (1:1,000; Wuhan Boster
Biological Technology, Ltd.; cat. no. BA0325), anti-Col3a1
monoclonal antibody (1:1,000; Wuhan Boster Biological Technology,
Ltd.; cat. no. M00788), anti-CCCTC-binding factor (CTGF) polyclonal
antibody (1:1,000; Wuhan Boster Biological Technology, Ltd.; cat.
no. PB0570), anti-vimentin (VIM) monoclonal antibody (1:1,000;
Wuhan Boster Biological Technology, Ltd.; cat. no. BM0135),
anti-fatty acid synthase (Fasn) monoclonal antibody (1:1,000; Wuhan
Boster Biological Technology, Ltd.; cat. no. BM4865), anti-solute
carrier family 25 member 1 (Slc25a1) polyclonal antibody (1:1,000;
Wuhan Boster Biological Technology, Ltd.; cat. no. A05995-2) and
anti-α-tubulin monoclonal antibody (1:1,000; Wuhan Boster
Biological Technology, Ltd.; cat. no. M03989-2). The secondary
antibodies used were anti-rabbit (1:10,000; cat. no. SA00001-2) and
anti-mouse IgG (1:10,000; cat. no. SA00001-1), both of which were
obtained from ProteinTech Group, Inc.
Statistical analysis
All data are presented as the mean ± SD of ≥3
repeats. Multiple comparisons difference involving ≥3 groups were
analyzed using one-way ANOVA with a Tukey's post hoc test and the
differences between two groups were analyzed using unpaired
Student's t-test. All analyses were performed using SPSS version 20
(IBM Corp.). P<0.05 was considered to indicate a statistically
significant difference.
Results
High-dose ionizing radiation causes
cardiac structural remodeling and functional injury to the hearts
of mice
To investigate the effects of high-energy X-rays on
mouse heart tissue, an RIHD animal model was first established
using local 16 Gy heart irradiation. Cardiac echocardiography,
serum myocardial biomarkers, and H&E and Masson staining were
used to verify cardiac injury at the different time points. The
results showed that LVEF and LV systolic posterior wall thickness
were decreased, while LV systolic internal dimension was increased
in the 5-month group compared with the 3-month group (Fig. 1A-G). Moreover, pericardial effusion
was not observed in any of the groups (Fig. 1A-D). In addition, the levels of
serum BNP, a cardiac biomarker, were significantly increased 5
months after exposure to ionizing radiation (Fig. 1H).
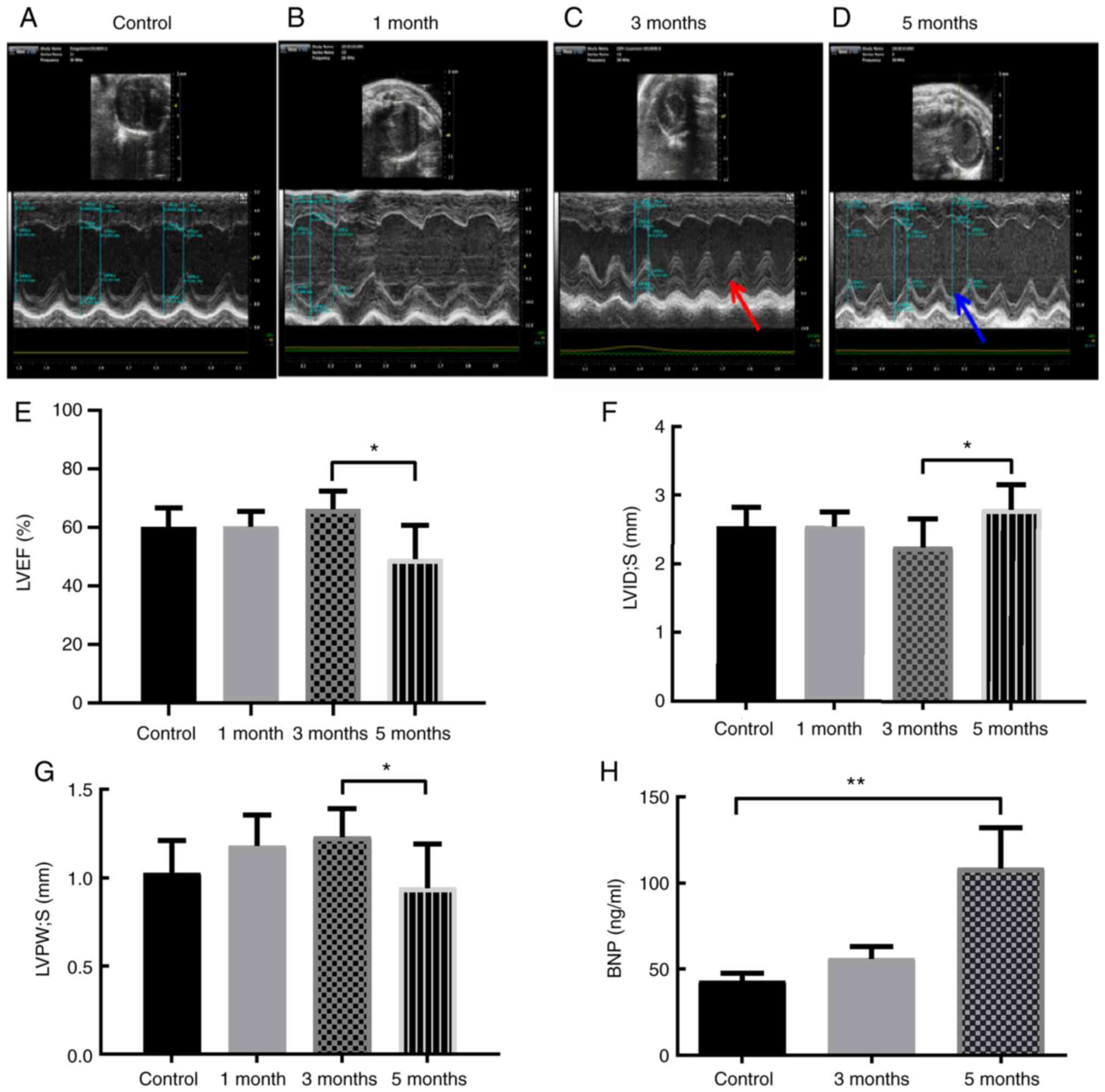 | Figure 1.Local heart irradiation leads to RIHD
after 5 months in mice. Representative images of mice
echocardiographs in the (A) control mice, and mice (B) 1, (C) 3 and
(D) 5 months after local heart irradiation with a dose of 16 Gy.
Red arrow, systolic thickness of the LV posterior wall 3 months
after ionizing irradiation. Blue arrow, systolic thickness of the
LV posterior wall 5 months after ionizing irradiation. (E) Cardiac
parameter: LVEF (%) in the control mice, and mice 1, 3 and 5 months
after local heart irradiation with a dose of 16 Gy. (F) Cardiac
parameter: LVID; s (mm) in the control mice, and mice 1, 3 and 5
months after local heart irradiation with a dose of 16 Gy. (G)
Cardiac parameter: LVPW; s (mm) in the control mice, and mice 1, 3
and 5 months after local heart irradiation with a dose of 16 Gy.
(H) Serum BNP (ng/ml). *P<0.05, **P<0.01; n=3-6 per group.
LV, left ventricular; RIHD, radiation-induced heart damage; BNP,
brain natriuretic peptide; LVID;s, Left ventricle internal
dimension in systole; LVPW; s, LV posterior wall thickness in
systole; LVEF, left ventricular ejection fraction. |
H&E staining of sections indicated that the
cardiomyocytes had degenerated and the myofilament boundary was
blurred after radiation, and these changes became more obvious as
the time of follow-up increased (Fig.
2A). In addition, compared with the control mice, collagen
fibers in the 5-month irradiated heart tissues were notably
increased (Fig. 2B). Collectively,
cardiomyocyte degeneration, fibrosis deposition, ventricular wall
remodeling, BNP elevation and LV systolic dysfunction were evident
in the 5-month irradiated mice. This indicated that myocardial
damage and cardiac remodeling manifested in the mice in the 5
months after local heart radiation exposure.
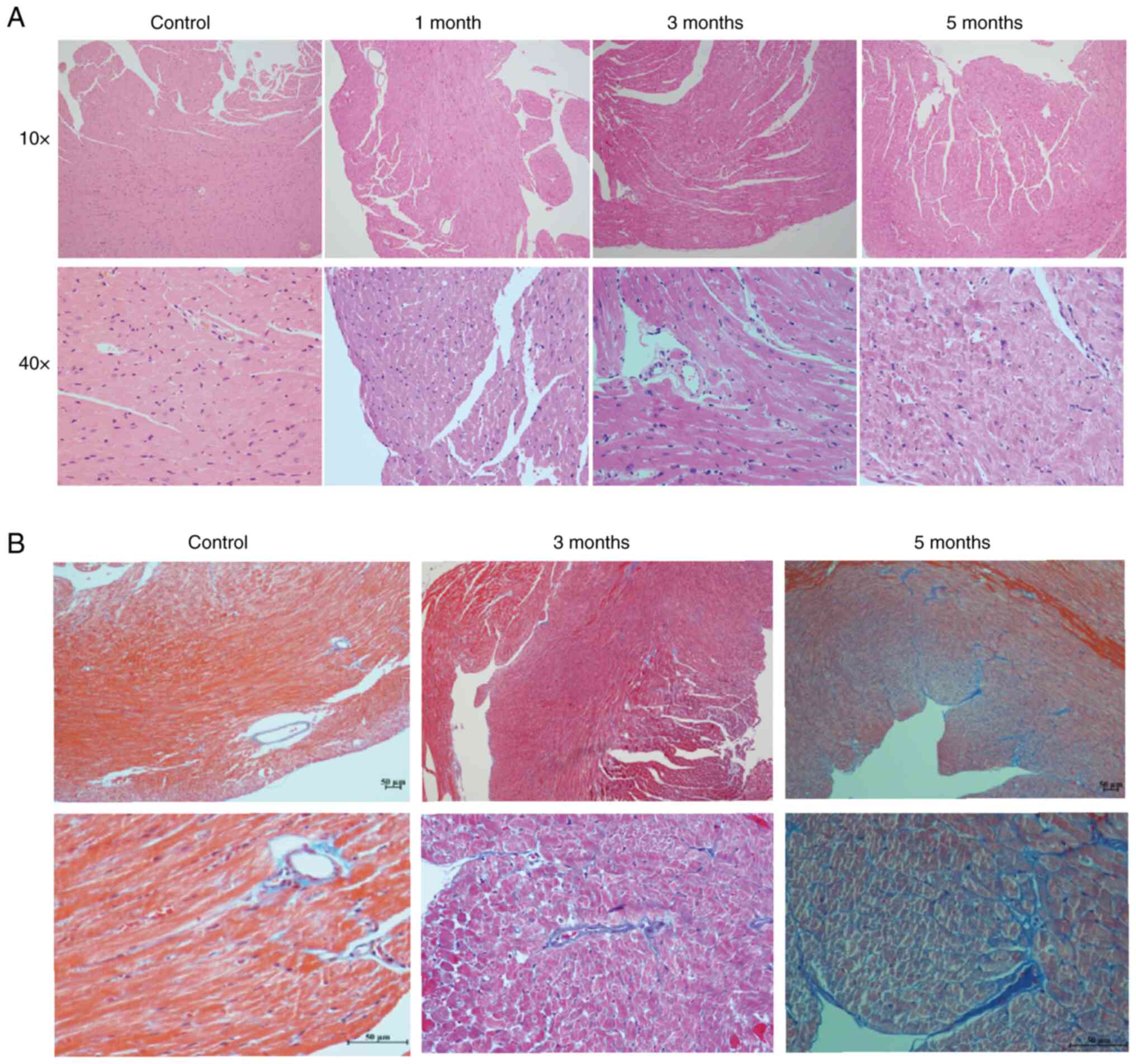 | Figure 2.H&E and Masson staining of
locally irradiated and control mouse heart tissues. (A) H&E
staining of heart tissues from the control mice, and 1, 3 and
5-month groups. Upper panel, light microscope at magnification,
10×10; lower panel, light microscope at magnification, 10×40. (B)
Masson's staining of heart tissues from the control mice, and 1, 3
and 5-month groups. Upper panel, light microscope at magnification,
10×10; lower panel, light microscope at magnification, 10×40. |
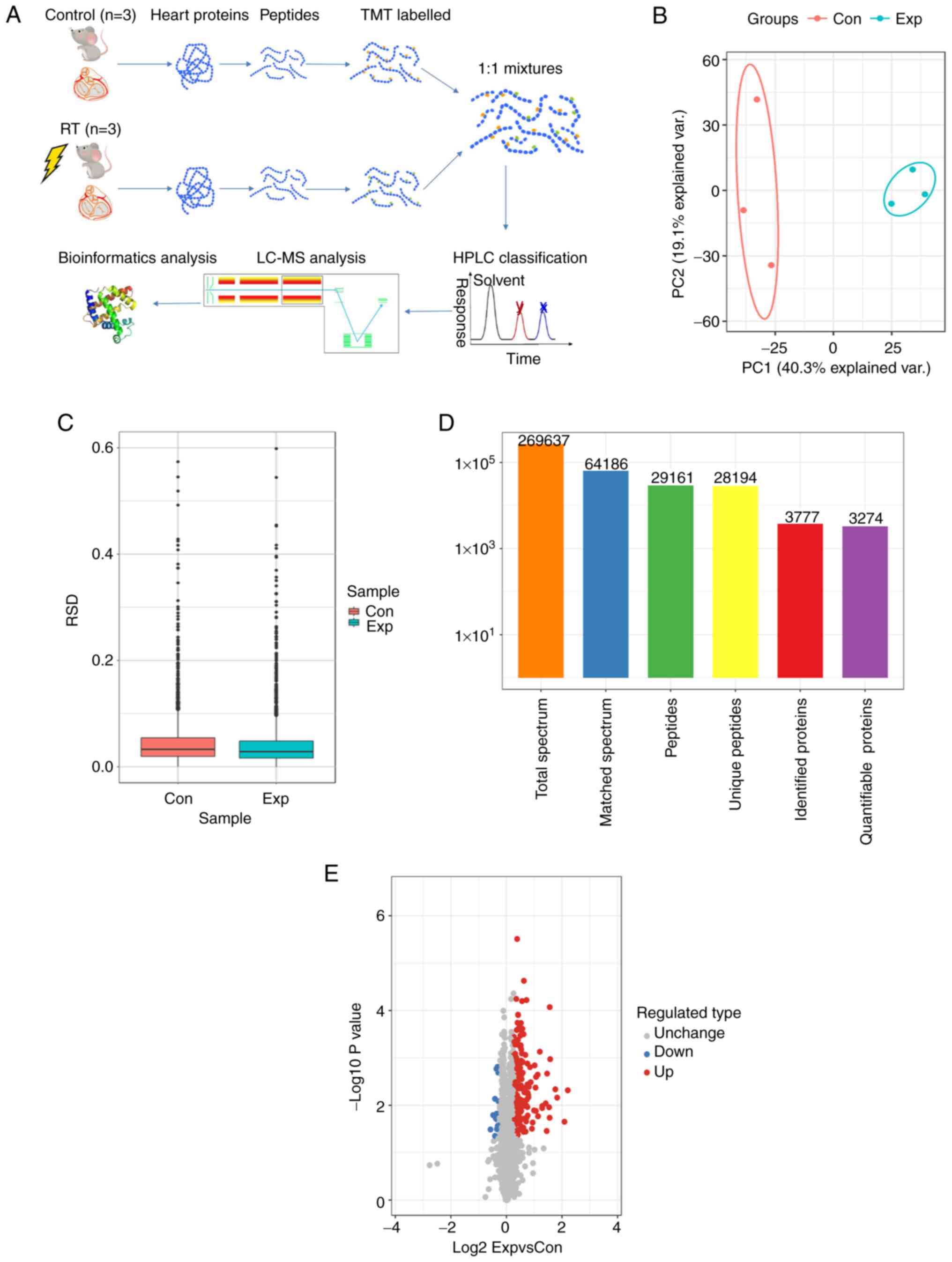
Proteomics analysis
Protein quantification
In the aforementioned results, it was shown that
mice manifested a RIHD phenotype 5 months after administration of
16 Gy ionizing radiation. Therefore, proteomics was performed using
heart tissues from the control and 5-month irradiated mice
(Fig. 3A). Principal component
analysis and relative standard deviation were used to evaluate the
quantitative repeatability of the proteins (Fig. 3B and C). A total of 269,637
secondary spectra were obtained by MS. After the secondary spectrum
of the MS was analyzed using the protein theory database, the
available number of spectra was 64,186 and the utilization rate of
the spectrum was 23.8%. A total of 29,161 peptides were identified
by the spectral diagrams, amongst which, there were 28,194 unique
peptides. From these peptides, 3,777 proteins were identified, and
3,274 were quantifiable (Fig. 3D).
High-throughput proteomics analysis identified 234 proteins in
total, and 219 proteins were found to be significantly increased
[log2 fold change (FC)>1.2], whereas only 15 proteins
were considered downregulated (log2FC<1:1.2) in the
irradiated mouse hearts (Fig.
3E).
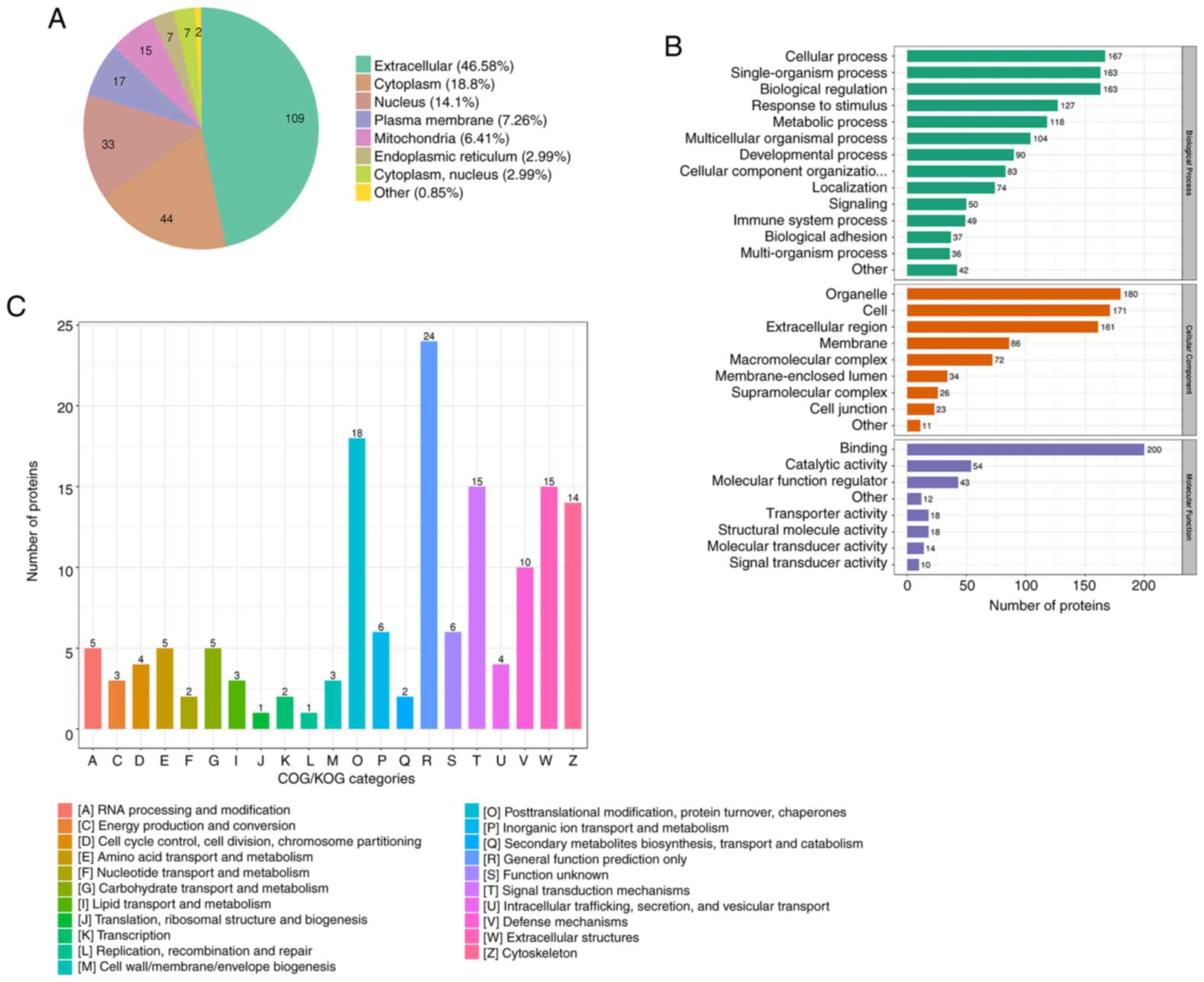
Subcellular localization and
functional classification analysis
The 234 DEPs were searched in the WolfPsort database
for prediction of their subcellular localization. The subcellular
localization analysis found that the distributions of the DEPs were
extracellular (46.58%), cytoplasmic (18.8%), nuclear (14.1%),
plasma membrane-bound (7.26%), mitochondrial (6.41%), endoplasmic
reticulum (2.99%) and cytoplasm, nucleus (2.99%), with the
remainder of proteins considered to be in other locations (0.85%)
(Fig. 4A).
According to the GO category results, the proteins
related to BP were primarily ‘cellular process’, ‘single-organism
process’, ‘biological regulation’, ‘response to stimulus’ and
‘metabolic process’, while CC was primarily composed of
‘organelle’, ‘cell’ and ‘extracellular region’ components.
‘Binding’ and ‘catalytic activity’ proteins were the primary MF
(Fig. 4B).
The DEPs in the heart tissue induced by radiation
were classified using COG/KOG (Fig.
4C). The results of the COG/KOG functional classification
showed that the DEPs were primarily distributed in the following
functions: (W) Extracellular structure, (O) Posttranslational
modification, protein turnover, chaperones. (T) Signal transduction
mechanisms and (V) Defense mechanism. Moreover, there were also
significant DEPs in substance metabolism after radiation, such as
(C) energy production and conversion, (E) amino acid transport and
metabolism, (F) nucleotide transport and metabolism, (G)
carbohydrate transport and metabolism and (I) lipid transport and
metabolism (Fig. 4C). Thus,
metabolism-related proteins were significantly altered according to
their subcellular localization and functional classification
analysis.
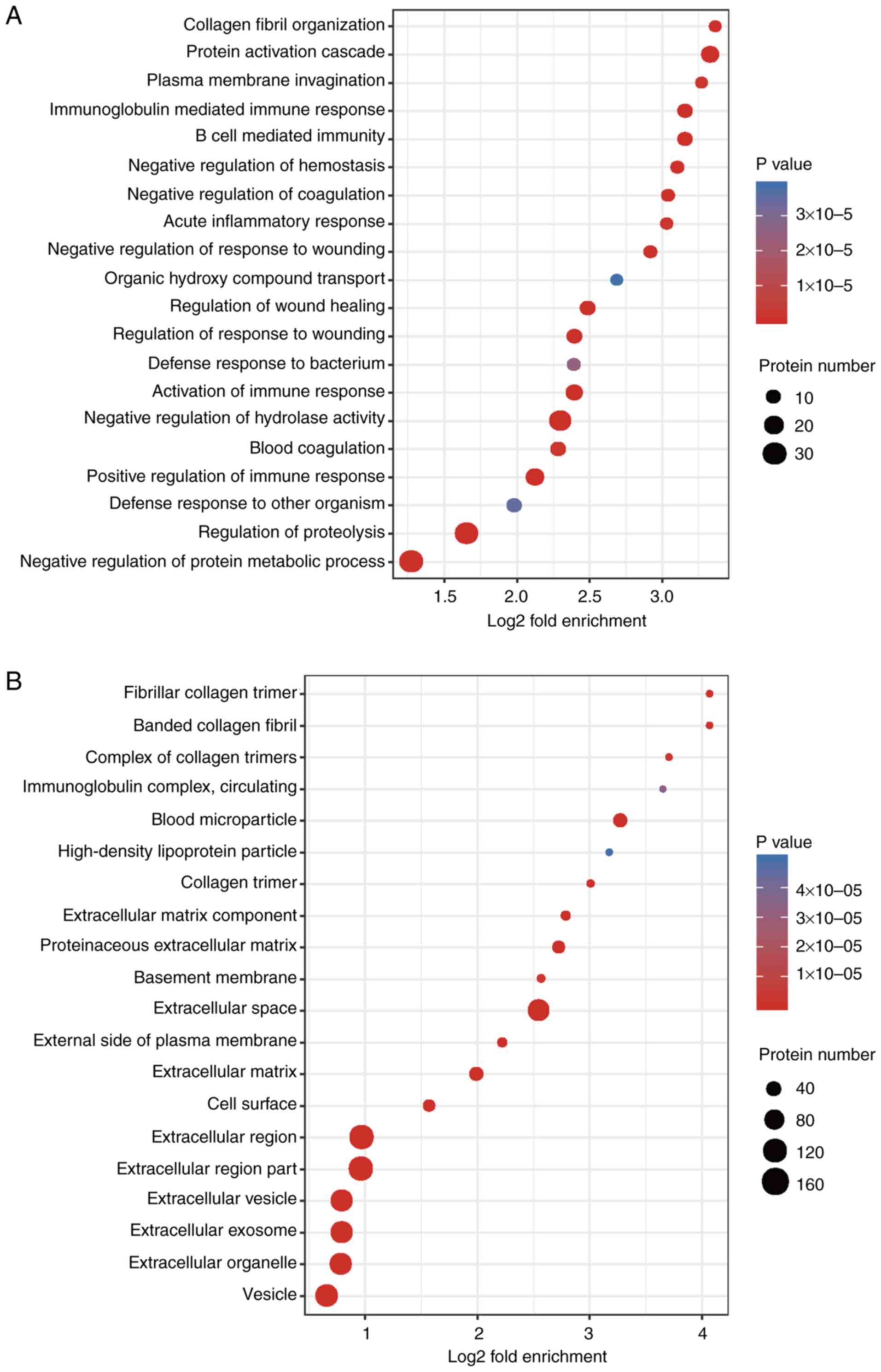
Functional enrichment analysis
To determine the significant enrichment trend of the
DEPs amongst the functional types, the enrichment of the DEPs in
the irradiated group compared with the control group were analyzed
through three aspects: GO term enrichment, KEGG pathway enrichment
and protein domain enrichment.
The BP analysis results of the GO secondary
classification showed that ‘collagen fibril organization’, ‘protein
activation cascade’ and ‘plasma membrane invagination’ were amongst
the most apparent differences. Abundant proteins were mainly
enriched in ‘negative regulation of protein metabolic process’ and
‘regulation of proteolysis’ (Fig.
5A). CC enrichment analysis demonstrated that the proteins
related to ‘fibrillar collagen trimer’, ‘banded collagen fibril’,
‘extracellular matrix components’, ‘extracellular vesicles’,
‘extracellular region’ or the parts of extracellular structures
were increased significantly after irradiation (Fig. 5B). MFs, such as ‘platelet-derived
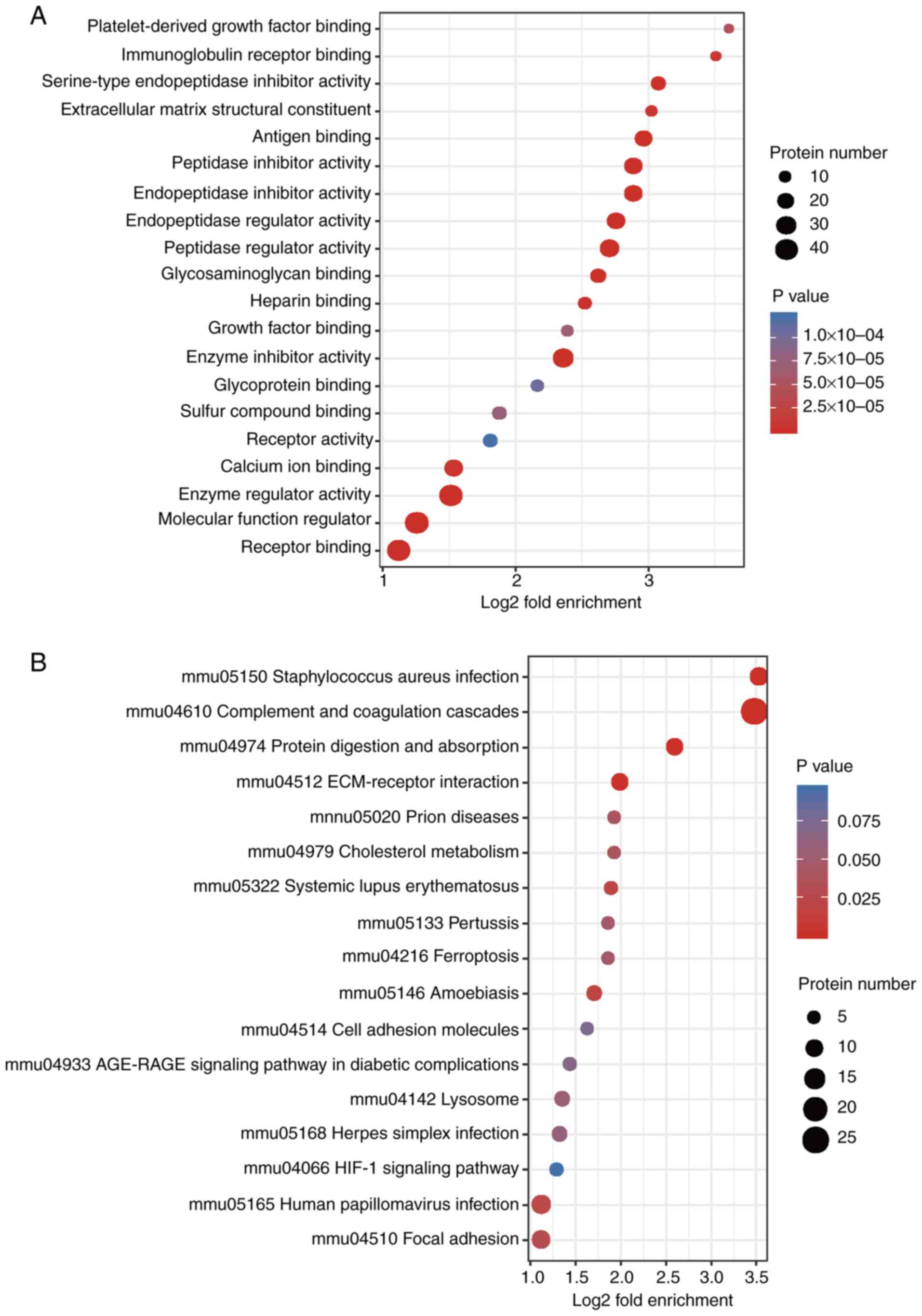
growth factor binding’, ‘immunoglobulin receptor binding’,
‘serine-type endopeptidase inhibitor activity’, ‘extracellular
matrix structural constituent’, ‘peptidase regulator activity’ and
‘enzyme inhibitor activity’ were also significantly increased after
irradiation (Fig. 6A).
KEGG enrichment analysis found that the
radiation-induced altered proteins were primarily enriched in
‘Staphylococcus aureus infection’, ‘complement and collagen
cascade’, ‘protein digestion and absorption’, ‘extracellular matrix
(ECM) interactions’ and ‘folding and adhesion related signaling
pathways’ (Fig. 6B). The domains of
these differentially expressed proteins were primarily enriched in
‘immunoglobulin subtypes’, ‘immunoglobulin folding’, ‘EGF-like
domains’ and ‘immunoglobulin-like domains’, amongst others
(Fig. S2). From the three
functional enrichment analyses, fibrosis was considered to be
increased following irradiation.
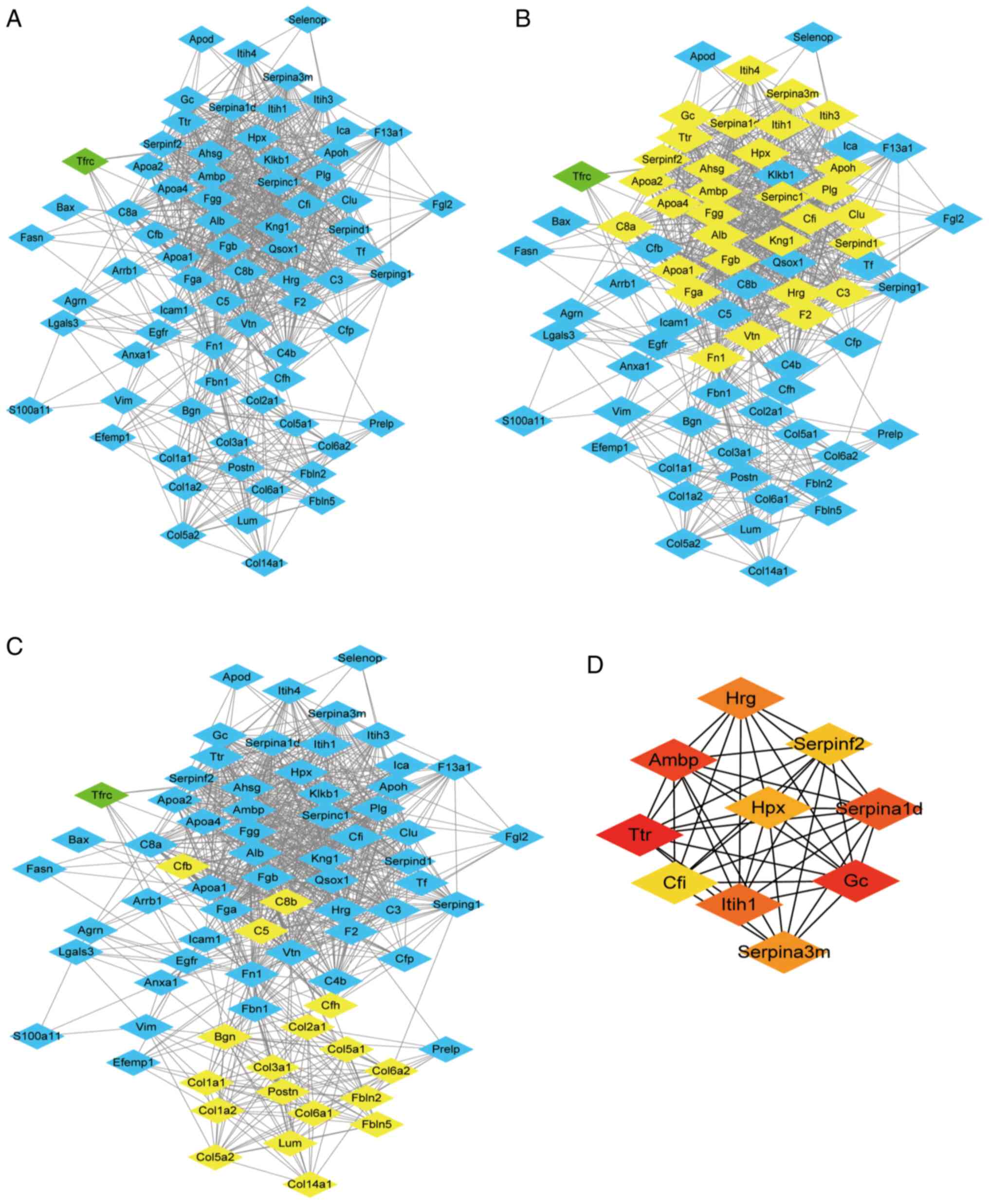
PPI network analysis
In order to identify the significant clusters
(confidence score >0.7), 74 proteins were filtered into the DEP
PPI network complex using STRING, and the resultant PPI network
contained 74 nodes and 870 edges (Fig.
7A). MCODE was used to identify the most significant module
that was comprised of two clusters (Fig. 7B and C) that consisted of
upregulated DEPs (MCODE score >10). The top 10 hub genes
selected using the DMNC method (score ≥5,000) and node degree
(score ≥10) in the cytoHubba plug-in included transthyretin, GC
vitamin D binding protein, α-1-microglobulin/bikunin precursor,
α-1-antitrypsin 1–4, inter-α-trpsin inhibitor heavy chain 1,
histidine rich glycoprotein, serine protease inhibitor A3M,
hemopexin (Hpx), serpin family F member 2 and complement factor I
(Fig. 7D). The results of MCODE and
cytoHubba analysis indicated that core DEPs were associated with
abnormal lipid metabolism, negative regulation of fibrinolysis and
ECM (Table SI,Table SII,Table SIII,Table SIV,Table SV,Table SVI).
Irradiation induces ECM and mitochondrial metabolism
damage. According to the proteomics analysis results, it was found
that irradiation led to the upregulation of ECM proteins, such as
Col14a1, perisostin (Postn), galectin 3 (Lgals3), Hpx, Tgfβi,
Col2a1, Col5a2, Col3a1, Col1a2, Col1a1, SPARC like 1 (Sparcl1),
Col5a1, vitronectin (Vtn), Col6a1 and laminin subunit α 5 (Lama5)
(Table I). Col1a1 and Col3a1 in the
irradiated heart were shown to be upregulated, as determine by IHC
and western blotting experiments (Fig.
8A-C). Additionally, other fibrosis-associated proteins (Vim
and CTGF) were also upregulated (Fig.
8B and C). The results of the proteomics analysis indicated
that metabolism-related proteins associated with carbohydrate
transport and metabolism, energy production and conversion, amino
acid transport and metabolism, nucleotide transport metabolism and
lipid transport metabolism were increased (Table II). Metabolism-related proteins,
such as Fasn and Slc25a1 were confirmed to be overexpressed by
western blotting (Fig. 8D and
E).
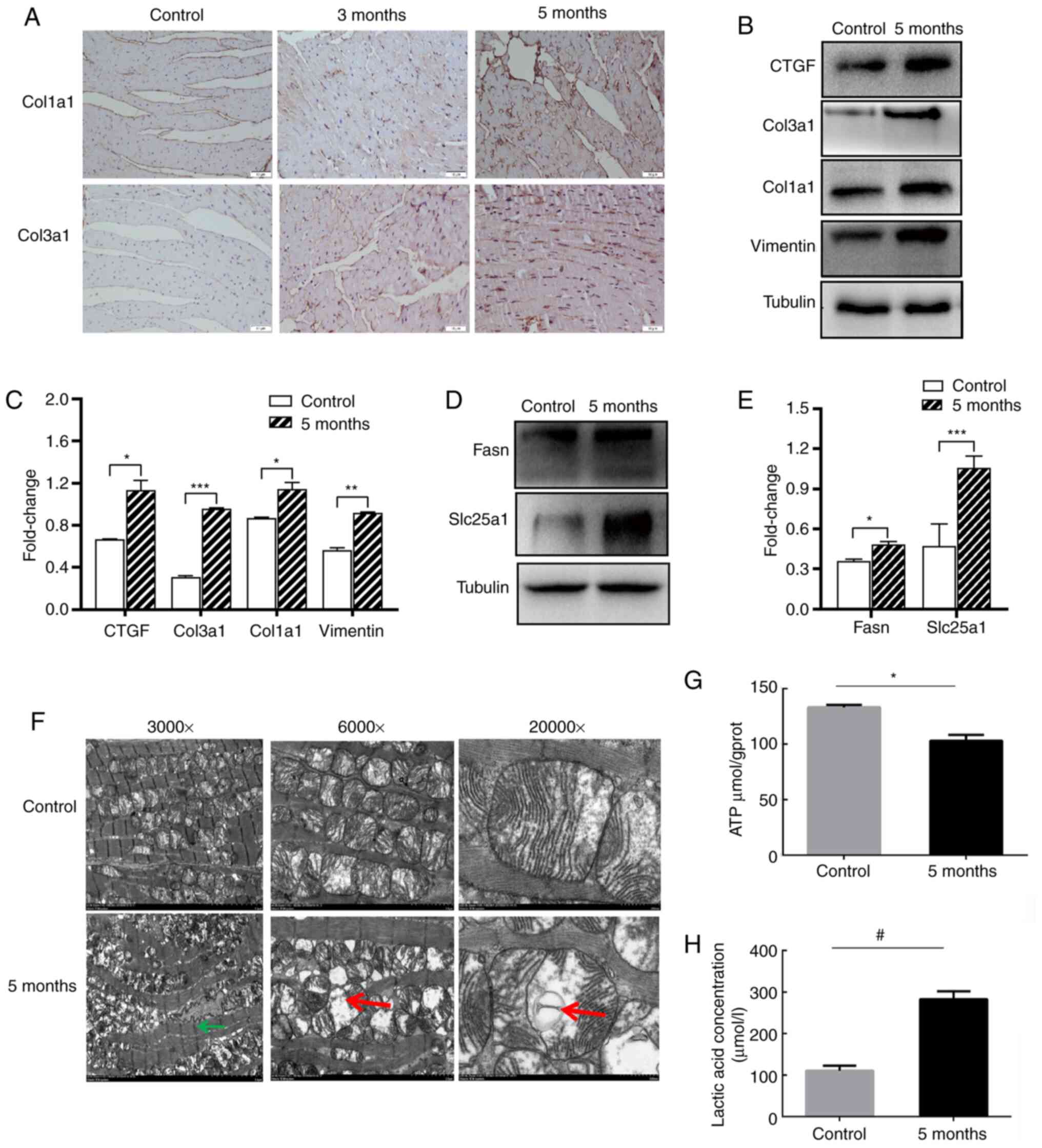 | Figure 8.Irradiation induces alterations to
the extracellular matrix and mitochondrial metabolism. (A)
Immunohistochemistry staining analysis of Col1a1 and Col3a1 in the
heart tissues. Magnification, ×400. (B) Western blotting and (C)
semi-quantitative analysis of Col1a1, Col3a1, Vimentin and CTGF.
(D) Western blotting and (E) semi-quantitative analysis of
metabolism related genes Fasn and Slc25al. Relative protein levels
of Col1a1, Col3a1, Vimentin, CTGF, Fasn and Slc25al were normalized
to Tubulin. (F) Electron micrographs of the cardiac mitochondria
from the mice in sham-irradiated and 5-month groups after 16 Gy
radiation. Left magnification, ×3,000; Middle magnification,
×6,000; Right magnification, ×20,000. Green arrows show the
myofilaments with fuzzy boundaries. Red arrows show swollen
mitochondria with cavitation. Measurement of (G) ATP levels and (H)
lactic acid concentration in the sham-irradiated and 5-month mice
heart tissues. *P<0.05, **P<0.01, ***P<0.001;
#P<0.05; n=3 per group. Col1a1, collagen type 1 α 1
chain; Col3a1, collagen type III α 1 chain; CTGF, CCCTC-binding
factor; Fasn, fatty acid synthase; Slc25a1, solute carrier family
25 member 1. |
 | Table I.Upregulation of extracellular matrix
proteins in mouse hearts 5 months after exposure to ionizing
radiation. |
Table I.
Upregulation of extracellular matrix
proteins in mouse hearts 5 months after exposure to ionizing
radiation.
| Category | Protein name | Ratio | Regulated type | P-value |
|---|
| Extracellular
structures | Col14a1 | 1.832 | Up | 0.0032566 |
|
| Postn | 1.643 | Up | 0.0184358 |
|
| Lgals3 | 1.637 | Up | 0.0100236 |
|
| Hpx | 1.531 | Up | 0.0092418 |
|
| Tgfbi | 1.468 | Up | 0.0008027 |
|
| Col2a1 | 1.419 | Up | 0.0005821 |
|
| Col5a2 | 1.406 | Up | 0.0023836 |
|
| Col3a1 | 1.342 | Up | 0.0002423 |
|
| Col1a2 | 1.334 | Up | 0.0001224 |
|
| Col1a1 | 1.331 | Up | 0.0004185 |
|
| Sparcl1 | 1.314 | Up | 0.043479 |
|
| Col5a1 | 1.299 | Up | 0.0041625 |
|
| Vtn | 1.287 | Up | 0.0076994 |
|
| Col6a1 | 1.24 | Up | 0.0013383 |
|
| Lama5 | 1.219 | Up | 0.0004591 |
 | Table II.Upregulation of metabolism-related
proteins in the mouse heart 5 months after exposure to ionizing
radiation. |
Table II.
Upregulation of metabolism-related
proteins in the mouse heart 5 months after exposure to ionizing
radiation.
| Category | Protein name | Ratio | Regulated type | P-value |
|---|
| [C] Energy
production and conversion | Slc25a1 | 1.228 | Up | 0.0136395 |
|
| Aldh1a1 | 1.268 | Up | 0.0105642 |
|
| Aldh1b1 | 1.257 | Up | 0.013897 |
| [G] Carbohydrate
transport and metabolism | Hexb | 1.245 | Up | 0.0017016 |
|
| Manba | 1.311 | Up | 0.038884 |
|
| Slc2a1 | 1.309 | Up | 0.0061217 |
|
| Tkt | 1.217 | Up | 0.0160025 |
| [E] Amino acid
transport and metabolism | Cfi | 1.318 | Up | 0.0001813 |
|
| Abat | 1.335 | Up | 0.0140643 |
|
| Cfb | 1.429 | Up | 0.0005422 |
|
| Klkb1 | 1.605 | Up | 0.0008644 |
|
| Lta4h | 1.204 | Up | 0.0122628 |
| [F] Nucleotide
transport and metabolism | Dpysl3 | 1.247 | Up | 0.0009192 |
|
| Entpd1 | 1.232 | Up | 0.022776 |
| [I] Lipid transport
and metabolism | Fasn | 1.617 | Up | 0.0052028 |
|
| Lta4h | 1.204 | Up | 0.0122628 |
|
| Ttr | 1.444 | Up | 0.0011445 |
Subsequently, the electron micrograph results of the
5-month irradiated heart mitochondria were found to have
myofilaments with fuzzy boundaries, swelling and cavitation of the
mitochondria were observed, and fewer mitochondria were present
compared with control group (Fig.
8F). The 3-month irradiated heart mitochondria electron
micrograph results are shown in Fig.
S3. It was also found that ATP production was decreased
following irradiation, from 133.2 µmol/gprot in the control to
103.0 µmol/gprot in the irradiated mice (Fig. 8G). However, lactate production in
the irradiated heart tissue was notably increased from 110.7 µmol/l
in the control to 269.4 µmol/l in the irradiated mice (Fig. 8H).
Discussion
The aim of the present study was to elucidate the
potential mechanisms involved in RIHD in mice using a novel
high-throughput proteomics technology. Using this technology,
29,161 peptides were identified, including 28,194 specific
peptides, and 3,274 quantitative proteins amongst the 3,777
identified proteins. Of these, 219 proteins were significantly
upregulated and 15 proteins were downregulated. By contrast,
Azimzadeh et al (38)
investigated the proteomics of 16-week-old irradiated mouse heart
(16 Gy) and found that there were 662 myocardial proteins
identified and 371 quantified proteins. Moreover, Subramanian et
al (39) studied the proteomics
of the mouse hearts at 40 weeks after 16 Gy radiation, and
identified 1,038 proteins, of which 940 proteins were quantified.
In the present study, 3,777 proteins were screened and 3,274
proteins were quantified using TMT labeling, which may be the
highest flux and labeling rate in the historical data of RIHD, and
this increased quantity of data may further show potential
mechanisms associated with RIHD.
The majority of these altered proteins were
extracellular proteins (46.58%). Fibrosis is the abnormal
deposition of ECM, which can lead to organ dysfunction, morbidity
and necrosis (40–42). According to COG/KOG functional
classification, 15 upregulated proteins associated with the ECM or
fibrosis were increased; these included Col14a1, Postn, Lgals3,
Hpx, Tgfbi, Col2a1, Col5a2, Col3a1, Col1a2, Col1a1, Sparcl1,
Col5a1, Vtn, Col6a1 and Lama5. According to GO enrichment analysis,
the results of BP and CC classifications indicated that there were
significant changes in fibrotic proteins. Additionally, protein
domain enrichment, KEGG pathway enrichment and PPI analysis showed
that radiated injury was associated with the ECM and fibrillar
collagen. Intriguingly, the most significantly correlated enriched
pathway based on KEGG pathway analysis was ‘Staphylococcus
aureus’ infection. Stoddard et al (43) used molecular techniques to
investigate the microbial flora in chronic sinusitis caused by
head-and neck radiotherapy, and found that Staphylococcus
aureus was the most common microorganism present following
radiation. Moreover, the microbiota induces alterations of
metabolism and inflammatory responses following radiotherapy
(44). The change in the flora of
microbiota may lead to alterations in metabolism and the
inflammatory response of irradiated heart tissues, although this
requires further analysis.
It has been reported that the mechanism via which
RIHD occurs may be associated with endothelial cell injury,
inflammatory reactions and ROS (9,21,25).
Irradiation leads to endothelial cell injury via pro-inflammatory
cytokines and damage of DNA or proteins mediated by ROS.
Endothelial dysfunction influences vascular intimal collagen
deposition, which can cause vessel wall thickening and luminal
stenosis (45). A series of
pathological effects, such as ischemia and hypoxia of the
irradiated heart, may aggravate the inflammatory reaction (25). With the continuous release of
inflammatory cytokines, such as TNF, IL-1, IL-6 and IL-8, cardiac
parenchyma cell necrosis and excessive deposition of ECM is
observed in irradiated heart tissue (9). Clinically, the rate of chronic
infections and fibrosis are increased following thoracic radiation
therapy and Staphylococcus aureus infections (46). These clinical results are consistent
with the proteomic data in the present study, in that high-energy
radiation can indeed cause inflammation, ECM and fibrosis.
In recent years, metabolism-associated disorders
have become increasingly recognized as important pathogenic
processes of several diseases, including neurological disorders,
type 2 diabetes mellitus, cancer and organ fibrosis, amongst others
(47,48). In the present study, COG/KOG
functional classification of high-throughput proteomics indicated
that proteins associated with various metabolic processes were
increased. Mitochondria are best known as the sites of production
of respiratory ATP, and are essential for metabolic process
(49,50). Electron micrographs of the
mitochondria were obtained in the present study, and ATP levels and
lactate production of the irradiated heart tissue were measured.
High-dose ionizing radiation resulted in increased structural
alterations of cardiac mitochondria. Consistent with the
histological results, changes in the inner membrane of the
mitochondria led to alterations in expression of metabolism-related
enzymes.
Several ECM- and metabolism-related proteins have
been reported to be upregulated in irradiated heart tissue.
Previous studies have highlighted the metabolic processes promoting
factors, including increasing glycine, ATP synthesis and
mitochondria respiration, involved in the pathogenesis of fibrosis
(51–53). Glucose metabolism can provide energy
for anabolic processes and collagen production. It is well
established that not only glucose metabolism, but also protein and
lipid metabolism are closely correlated with mitochondrial function
(54–56). Indeed, mitochondria are considered
to be the major suppliers of cellular energy in the form of ATP,
which is produced by oxidative phosphorylation, and is the primary
means of energy production under baseline conditions (57,58).
Vincent et al (59)
investigated keloid scars with excessive amounts of collagen
production, and concluded that keloid-associated fibroblasts
consume unusually large amounts of glucose and produce more lactate
to fulfill their ATP needs, which means the secretion of collagen
relies on ATP produced by glycolysis. However, in normal heart
tissue, fatty acids are the main source of energy metabolism,
accounting for 40–80% (60,61).
Metabolic changes are regarded as important
pathogenic processes of fibrosis in various organs, and metabolic
targeted therapy may become an important strategy to reduce
fibrosis (41,49,62).
Disturbance of myocardial energy metabolism is an important cause
of myocardial heart disease, and mitochondria are the primary
organelles involved in energy metabolism (50,63,64).
Similarly, mitochondrial dysfunction may also be an important cause
of radiation-induced heart disease. It has been shown that the
respiratory capacity of myocardial mitochondria among C57BL/6 mice
was significantly decreased 40 weeks after local 2 Gy radiation,
but not in the 0.2 Gy group (12).
Sridharan et al (65) used
Sprague Dawley rats to establish a radiation and tyrosine kinase
inhibitors-related heart injury model and showed enhanced effects
on mitochondrial morphology and mitochondrial permeability
transition pore opening. Another study confirmed that late
application of a palladium lipoate complex improved myocardial
mitochondrial function by reducing the presence of a cellular
inflammatory microenvironment, although it had no effect on
repairing the damaged mitochondrial structures (66). In 2019, Dai et al (67) also reported that radiation caused
damage to heart function, and this was mediated by ROS produced by
the mitochondria. The simple ganoderma lucidum spore oil system
these authors developed targeted the myocardial mitochondria and
slowed down the production of ROS, thus exhibiting a preventative
and therapeutic effect on RIHD. Additionally, the differences in
expression levels of mitochondrial genes caused by genetic variants
may lead to differences in sensitivity to ionizing radiation
(68). Collectively, the
aforementioned studies revealed that mitochondrial damage was an
important target in RIHD, and drugs or drug-loaded materials for
the repair of mitochondrial damage can reduce radiation-induced
heart injury.
Radiation induces oxidative changes and persistent
inflammation last days and months after the initial exposure,
possibly due to the continuous generation of ROS and nitrogen
species (69). Defects in
mitochondrial DNA, protein import, proteins and metabolic enzymes
resulting from exposure to ionizing radiation may lead to late
health effects, including degenerative diseases and metabolic
disorders (23). In the present
study, via electron microscopic analysis of mitochondria it was
indicated that myocardial mitochondria damage persisted after high
dose radiation. Therefore, the mitochondrial damage caused by
ionizing radiation may be a significant cause of the pathogenic
mechanisms underlying RIHD.
The present study has some limitations. The heart
tissue samples for proteomics were obtained from the heart apex,
including the left ventricle and right ventricle. Thus, it is hard
to distinguish the LV alternations alone. Additionally, the study
was performed in male mice only, and differences in sex should be
considered.
In the present study, the DEPs between irradiated
and control mice heart tissue, as well as the alterations enriched
in fibrosis and energy metabolism, were identified. Ionizing
radiation indeed led to cardiac fibrosis and metabolic disorders,
and these were validated by subsequent experiments. No direct
mechanism was identified in the present study; however, further
investigations regarding the mechanisms between ATP alterations and
radiation induced cardiac fibrosis are ongoing.
In conclusion, the results of the present study
demonstrated that ionizing radiation caused structural remodeling,
functional injury and fibrotic alterations in the heart.
Radiation-induced mitochondrial damage and metabolic alterations in
cardiac tissue may underlie the pathogenic mechanisms of RIHD.
Supplementary Material
Supporting Data
Acknowledgements
We thank physicist Dr Peng Zhang (Zhejiang Key
Laboratory of Radiation Oncology) for supporting our work of
establishing the animal model.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81760566 and 82060577),
Science and Technology Innovation Platform of Jiangxi Province
(grant no. 20171BCD40022), Development Project of Jiangxi province
(grant no. 20171ACB20034), Project in The Second Affiliated
Hospital of Nanchang University (grant no. Y578#) and Postgraduates
Special Funds Innovation of Jiangxi Province (grant no.
YC2020-B048).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AL and ZZ designed the study. PX, YY, YL and ZL
performed the experiments. YX and JC analyzed the bioinformatics
data. PX and ZZ prepared the manuscript. PX and ZZ confimed the
authenticity of raw data. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Animal ethical approval was obtained from the
Institutional Review Board of The Second Affiliated Hospital of
Nanchang University (approval no. A801).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RIHD
|
radiation-induced heart damage
|
|
BNP
|
brain natriuretic peptide
|
|
IHC
|
immunohistochemistry
|
|
MHD
|
mean heart dose
|
|
DEPs
|
differentially expressed proteins
|
|
PPI
|
protein-protein interactions
|
|
ROS
|
reactive oxygen species
|
|
GO
|
Gene Ontology
|
|
FC
|
fold change
|
|
LVEF
|
left ventricle ejection fraction
|
References
|
1
|
Atkins KM, Rawal B, Chaunzwa TL, Lamba N,
Bitterman DS, Williams CL, Kozono DE, Baldini EH, Chen AB, Nguyen
PL, et al: Cardiac radiation dose, cardiac disease, and mortality
in patients with lung cancer. J Am Coll Cardiol. 73:2976–2987.
2019. View Article : Google Scholar
|
|
2
|
van Nimwegen FA, Schaapveld M, Cutter DJ,
Janus CP, Krol AD, Hauptmann M, Kooijman K, Roesink J, van der
Maazen R, Darby SC, et al: Radiation dose-response relationship for
risk of coronary heart disease in survivors of hodgkin lymphoma. J
Clin Oncol. 34:235–243. 2016. View Article : Google Scholar
|
|
3
|
Witt JS, Jagodinsky JC, Liu Y, Yadav P,
Kuczmarska-Haas A, Yu M, Maloney JD, Ritter MA, Bassetti MF and
Baschnagel AM: Cardiac toxicity in operable esophageal cancer
patients treated with or without chemoradiation. Am J Clin Oncol.
42:662–667. 2019. View Article : Google Scholar
|
|
4
|
Wang H, Mu X, He H and Zhang XD: Cancer
radiosensitizers. Trends Pharmacol Sci. 39:24–48. 2018. View Article : Google Scholar
|
|
5
|
Zamorano JL, Lancellotti P, Rodriguez
Muñoz D, Aboyans V, Asteggiano R, Galderisi M, Habib G, Lenihan DJ,
Lip GY, Lyon AR, et al ESC Scientific Document Group, : 2016 ESC
Position Paper on cancer treatments and cardiovascular toxicity
developed under the auspices of the ESC Committee for Practice
Guidelines: The Task Force for cancer treatments and cardiovascular
toxicity of the European Society of Cardiology (ESC). Eur Heart J.
37:2768–2801. 2016. View Article : Google Scholar
|
|
6
|
Schaue D and McBride WH: Opportunities and
challenges of radiotherapy for treating cancer. Nat Rev Clin Oncol.
12:527–540. 2015. View Article : Google Scholar
|
|
7
|
Boerma M, Sridharan V, Mao XW, Nelson GA,
Cheema AK, Koturbash I, Singh SP, Tackett AJ and Hauer-Jensen M:
Effects of ionizing radiation on the heart. Mutat Res Rev Mutat
Res. 770:319–327. 2016. View Article : Google Scholar
|
|
8
|
Bhattacharya S and Asaithamby A: Ionizing
radiation and heart risks. Semin Cell Dev Biol. 58:14–25. 2016.
View Article : Google Scholar
|
|
9
|
Wang H, Wei J, Zheng Q, Meng L, Xin Y, Yin
X and Jiang X: Radiation-induced heart disease: A review of
classification, mechanism and prevention. Int J Biol Sci.
15:2128–2138. 2019. View Article : Google Scholar
|
|
10
|
Raghunathan D, Khilji MI, Hassan SA and
Yusuf SW: Radiation-induced cardiovascular disease. Curr
Atheroscler Rep. 19:222017. View Article : Google Scholar
|
|
11
|
Yusuf SW, Venkatesulu BP, Mahadevan LS and
Krishnan S: Radiation-induced cardiovascular disease: A clinical
perspective. Front Cardiovasc Med. 4:662017. View Article : Google Scholar
|
|
12
|
Barjaktarovic Z, Shyla A, Azimzadeh O,
Schulz S, Haagen J, Dörr W, Sarioglu H, Atkinson MJ, Zischka H and
Tapio S: Ionising radiation induces persistent alterations in the
cardiac mitochondrial function of C57BL/6 mice 40 weeks after local
heart exposure. Radiother Oncol. 106:404–410. 2013. View Article : Google Scholar
|
|
13
|
Guldner L, Haddy N, Pein F, Diallo I,
Shamsaldin A, Dahan M, Lebidois J, Merlet P, Villain E, Sidi D, et
al: Radiation dose and long term risk of cardiac pathology
following radiotherapy and anthracyclin for a childhood cancer.
Radiother Oncol. 81:47–56. 2006. View Article : Google Scholar
|
|
14
|
Mezzaroma E, Di X, Graves P, Toldo S, Van
Tassell BW, Kannan H, Baumgarten C, Voelkel N, Gewirtz DA and
Abbate A: A mouse model of radiation-induced cardiomyopathy. Int J
Cardiol. 156:231–233. 2012. View Article : Google Scholar
|
|
15
|
Dreyfuss AD, Goia D, Shoniyozov K, Shewale
SV, Velalopoulou A, Mazzoni S, Avgousti H, Metzler SD, Bravo PE,
Feigenberg SJ, et al: A novel mouse model of radiation-induced
cardiac injury reveals biological and radiological biomarkers of
cardiac dysfunction with potential clinical relevance. Clin Cancer
Res. 27:2266–2276. 2021. View Article : Google Scholar
|
|
16
|
Zeng ZM, Xu P, Zhou S, Du HY, Jiang XL,
Cai J, Huang L and Liu AW: Positive association between heart
dosimetry parameters and a novel cardiac biomarker, solubleST-2, in
thoracic cancer chest radiation. J Clin Lab Anal. 34:e231502020.
View Article : Google Scholar
|
|
17
|
Adams MJ, Hardenbergh PH, Constine LS and
Lipshultz SE: Radiation-associated cardiovascular disease. Crit Rev
Oncol Hematol. 45:55–75. 2003. View Article : Google Scholar
|
|
18
|
Kreuzer M, Auvinen A, Cardis E, Hall J,
Jourdain JR, Laurier D, Little MP, Peters A, Raj K, Russell NS, et
al: Low-dose ionising radiation and cardiovascular diseases -
Strategies for molecular epidemiological studies in Europe. Mutat
Res Rev Mutat Res. 764:90–100. 2015. View Article : Google Scholar
|
|
19
|
Sayler E, Dolney D, Avery S and Koch C:
Shielding considerations for the small animal radiation research
platform (SARRP). Health Phys. 104:471–480. 2013. View Article : Google Scholar
|
|
20
|
Hoving S, Seemann I, Visser NL, te Poele
JA and Stewart FA: Thalidomide is not able to inhibit
radiation-induced heart disease. Int J Radiat Biol. 89:685–691.
2013. View Article : Google Scholar
|
|
21
|
Wang B, Wang H, Zhang M, Ji R, Wei J, Xin
Y and Jiang X: Radiation-induced myocardial fibrosis: Mechanisms
underlying its pathogenesis and therapeutic strategies. J Cell Mol
Med. 24:7717–7729. 2020. View Article : Google Scholar
|
|
22
|
Slezak J, Kura B, Babal P, Barancik M,
Ferko M, Frimmel K, Kalocayova B, Kukreja RC, Lazou A, Mezesova L,
et al: Potential markers and metabolic processes involved in the
mechanism of radiation-induced heart injury. Can J Physiol
Pharmacol. 95:1190–1203. 2017. View Article : Google Scholar
|
|
23
|
Azzam EI, Jay-Gerin JP and Pain D:
Ionizing radiation-induced metabolic oxidative stress and prolonged
cell injury. Cancer Lett. 327:48–60. 2012. View Article : Google Scholar
|
|
24
|
Zhao W and Robbins ME: Inflammation and
chronic oxidative stress in radiation-induced late normal tissue
injury: Therapeutic implications. Curr Med Chem. 16:130–143. 2009.
View Article : Google Scholar
|
|
25
|
Baselet B, Sonveaux P, Baatout S and Aerts
A: Pathological effects of ionizing radiation: Endothelial
activation and dysfunction. Cell Mol Life Sci. 76:699–728. 2019.
View Article : Google Scholar
|
|
26
|
Zhang Z, Wu S, Stenoien DL and Paša-Tolić
L: High-throughput proteomics. Annu Rev Anal Chem (Palo Alto,
Calif). 7:427–454. 2014. View Article : Google Scholar
|
|
27
|
Sharon D, Tilgner H, Grubert F and Snyder
M: A single-molecule long-read survey of the human transcriptome.
Nat Biotechnol. 31:1009–1014. 2013. View Article : Google Scholar
|
|
28
|
Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi
Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al NCI CPTAC, :
Proteogenomic characterization of human colon and rectal cancer.
Nature. 513:382–387. 2014. View Article : Google Scholar
|
|
29
|
Mun DG, Bhin J, Kim S, Kim H, Jung JH,
Jung Y, Jang YE, Park JM, Kim H, Jung Y, et al: Proteogenomic
characterization of human early-onset gastric cancer. Cancer Cell.
35:111–124.e10. 2019. View Article : Google Scholar
|
|
30
|
Li X, Wang W and Chen J: Recent progress
in mass spectrometry proteomics for biomedical research. Sci China
Life Sci. 60:1093–1113. 2017. View Article : Google Scholar
|
|
31
|
Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C,
Quan S, Zhang F, Sun R, Qian L, et al: Proteomic and Metabolomic
Characterization of COVID-19 Patient Sera. Cell. 182:59–72.e15.
2020. View Article : Google Scholar
|
|
32
|
Zeng ZM, Du HY, Xiong L, Zeng XL, Zhang P,
Cai J, Huang L and Liu AW: BRCA1 protects cardiac microvascular
endothelial cells against irradiation by regulating p21-mediated
cell cycle arrest. Life Sci. 244:1173422020. View Article : Google Scholar
|
|
33
|
Li J, Zhang N, Song LB, Liao WT, Jiang LL,
Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS, et al: Astrocyte elevated
gene-1 is a novel prognostic marker for breast cancer progression
and overall patient survival. Clin Cancer Res. 14:3319–3326. 2008.
View Article : Google Scholar
|
|
34
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar
|
|
35
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast Delayed Enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar
|
|
36
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar
|
|
37
|
Wang Y, Cai J, Zeng X, Chen Y, Yan W,
Ouyang Y, Xiao D, Zeng Z, Huang L and Liu A: Downregulation of
toll-like receptor 4 induces suppressive effects on hepatitis B
virus-related hepatocellular carcinoma via ERK1/2 signaling. BMC
Cancer. 15:8212015. View Article : Google Scholar
|
|
38
|
Azimzadeh O, Sievert W, Sarioglu H,
Yentrapalli R, Barjaktarovic Z, Sriharshan A, Ueffing M, Janik D,
Aichler M, Atkinson MJ, et al: PPAR alpha: A novel radiation target
in locally exposed Mus musculus heart revealed by quantitative
proteomics. J Proteome Res. 12:2700–2714. 2013. View Article : Google Scholar
|
|
39
|
Subramanian V, Seemann I, Merl-Pham J,
Hauck SM, Stewart FA, Atkinson MJ, Tapio S and Azimzadeh O: Role of
TGF beta and PPAR alpha signaling pathways in radiation response of
locally exposed heart: Integrated global transcriptomics and
proteomics analysis. J Proteome Res. 16:307–318. 2017. View Article : Google Scholar
|
|
40
|
Tsuchida T and Friedman SL: Mechanisms of
hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol.
14:397–411. 2017. View Article : Google Scholar
|
|
41
|
Zhao X, Kwan JY, Yip K, Liu PP and Liu FF:
Targeting metabolic dysregulation for fibrosis therapy. Nat Rev
Drug Discov. 19:57–75. 2020. View Article : Google Scholar
|
|
42
|
Meziani L, Mondini M, Petit B, Boissonnas
A, Thomas de Montpreville V, Mercier O, Vozenin MC and Deutsch E:
CSF1R inhibition prevents radiation pulmonary fibrosis by depletion
of interstitial macrophages. Eur Respir J. 51:512018. View Article : Google Scholar
|
|
43
|
Stoddard TJ, Varadarajan VV, Dziegielewski
PT, Boyce BJ and Justice JM: Detection of microbiota in post
radiation sinusitis. Ann Otol Rhinol Laryngol. 128:1116–1121. 2019.
View Article : Google Scholar
|
|
44
|
Roy S and Trinchieri G: Microbiota: A key
orchestrator of cancer therapy. Nat Rev Cancer. 17:271–285. 2017.
View Article : Google Scholar
|
|
45
|
Liu LK, Ouyang W, Zhao X, Su ShF, Yang Y,
Ding WJ, Luo X, He ZX and Lu B: Pathogenesis and prevention of
radiation-induced myocardial fibrosis. Asian Pac J Cancer Prev.
18:583–587. 2017.
|
|
46
|
Soh J, Sugimoto S, Namba K, Miura A,
Shiotani T, Yamamoto H, Suzawa K, Shien K, Yamamoto H, Okazaki M,
et al: Chronic lung injury after trimodality therapy for locally
advanced non-small cell lung cancer. Ann Thorac Surg. 112:279–288.
2020. View Article : Google Scholar
|
|
47
|
Annesley SJ and Fisher PR: Mitochondria in
health and disease. Cells. 8:82019. View Article : Google Scholar
|
|
48
|
Khomich O, Ivanov AV and Bartosch B:
Metabolic hallmarks of hepatic stellate cells in liver fibrosis.
Cells. 9:92019. View Article : Google Scholar
|
|
49
|
Sabbah HN: Targeting the mitochondria in
heart failure: A translational perspective. JACC Basic Transl Sci.
5:88–106. 2020. View Article : Google Scholar
|
|
50
|
Gollmer J, Zirlik A and Bugger H:
Mitochondrial mechanisms in diabetic cardiomyopathy. Diabetes Metab
J. 44:33–53. 2020. View Article : Google Scholar
|
|
51
|
de Paz-Lugo P, Lupiáñez JA and
Meléndez-Hevia E: High glycine concentration increases collagen
synthesis by articular chondrocytes in vitro: Acute glycine
deficiency could be an important cause of osteoarthritis. Amino
Acids. 50:1357–1365. 2018. View Article : Google Scholar
|
|
52
|
Nishikawa T, Bellance N, Damm A, Bing H,
Zhu Z, Handa K, Yovchev MI, Sehgal V, Moss TJ, Oertel M, et al: A
switch in the source of ATP production and a loss in capacity to
perform glycolysis are hallmarks of hepatocyte failure in advance
liver disease. J Hepatol. 60:1203–1211. 2014. View Article : Google Scholar
|
|
53
|
Zhao YD, Yin L, Archer S, Lu C, Zhao G,
Yao Y, Wu L, Hsin M, Waddell TK, Keshavjee S, et al: Metabolic
heterogeneity of idiopathic pulmonary fibrosis: A metabolomic
study. BMJ Open Respir Res. 4:e0001832017. View Article : Google Scholar
|
|
54
|
Im MJ, Freshwater MF and Hoopes JE: Enzyme
activities in granulation tissue: Energy for collagen synthesis. J
Surg Res. 20:121–125. 1976. View Article : Google Scholar
|
|
55
|
Wang SP, Yang H, Wu JW, Gauthier N, Fukao
T and Mitchell GA: Metabolism as a tool for understanding human
brain evolution: Lipid energy metabolism as an example. J Hum Evol.
77:41–49. 2014. View Article : Google Scholar
|
|
56
|
Vidali S, Aminzadeh S, Lambert B,
Rutherford T, Sperl W, Kofler B and Feichtinger RG: Mitochondria:
The ketogenic diet - A metabolism-based therapy. Int J Biochem Cell
Biol. 63:55–59. 2015. View Article : Google Scholar
|
|
57
|
Brookes PS, Yoon Y, Robotham JL, Anders MW
and Sheu SS: Calcium, ATP, and ROS: A mitochondrial love-hate
triangle. Am J Physiol Cell Physiol. 287:C817–C833. 2004.
View Article : Google Scholar
|
|
58
|
du Plessis SS, Agarwal A, Mohanty G and
van der Linde M: Oxidative phosphorylation versus glycolysis: What
fuel do spermatozoa use? Asian J Androl. 17:230–235. 2015.
View Article : Google Scholar
|
|
59
|
Vincent AS, Phan TT, Mukhopadhyay A, Lim
HY, Halliwell B and Wong KP: Human skin keloid fibroblasts display
bioenergetics of cancer cells. J Invest Dermatol. 128:702–709.
2008. View Article : Google Scholar
|
|
60
|
Liu Y, Li Y, Zhang T, Zhao H, Fan S, Cai
X, Liu Y, Li Z, Gao S, Li Y, et al: Analysis of biomarkers and
metabolic pathways in patients with unstable angina based on ultra
high performance liquid chromatography quadrupole time of flight
mass spectrometry. Mol Med Rep. 22:3862–3872. 2020.
|
|
61
|
Michel M, Dubowy KO, Zlamy M, Karall D,
Adam MG, Entenmann A, Keller MA, Koch J, Odri Komazec I, Geiger R,
et al: Targeted metabolomic analysis of serum phospholipid and
acylcarnitine in the adult Fontan patient with a dominant left
ventricle. Ther Adv Chronic Dis. Apr 27–2020.(Epub ahead of print).
doi: 10.1177/2040622320916031. View Article : Google Scholar
|
|
62
|
Ma CX, Zhao XK and Li YD: New therapeutic
insights into radiation-induced myocardial fibrosis. Ther Adv
Chronic Dis. Aug 8–2019.(Epub ahead of print). doi:
10.1177/2040622319868383. View Article : Google Scholar
|
|
63
|
Del Re DP, Amgalan D, Linkermann A, Liu Q
and Kitsis RN: Fundamental mechanisms of regulated cell death and
implications for heart disease. Physiol Rev. 99:1765–1817. 2019.
View Article : Google Scholar
|
|
64
|
Vásquez-Trincado C, García-Carvajal I,
Pennanen C, Parra V, Hill JA, Rothermel BA and Lavandero S:
Mitochondrial dynamics, mitophagy and cardiovascular disease. J
Physiol. 594:509–525. 2016. View Article : Google Scholar
|
|
65
|
Sridharan V, Thomas CJ, Cao M, Melnyk SB,
Pavliv O, Joseph J, Singh SP, Sharma S, Moros EG and Boerma M:
Effects of local irradiation combined with sunitinib on early
remodeling, mitochondria, and oxidative stress in the rat heart.
Radiother Oncol. 119:259–264. 2016. View Article : Google Scholar
|
|
66
|
Sridharan V, Seawright JW, Antonawich FJ,
Garnett M, Cao M, Singh P and Boerma M: Late Administration of a
Palladium Lipoic Acid Complex (POLY-MVA) modifies cardiac
mitochondria but not functional or structural manifestations of
radiation-induced heart disease in a rat model. Radiat Res.
187:361–366. 2017. View Article : Google Scholar
|
|
67
|
Dai C, He L, Ma B and Chen T: Facile
nanolization strategy for therapeutic ganoderma lucidum spore oil
to achieve enhanced protection against radiation-induced heart
disease. Small. 15:e19026422019. View Article : Google Scholar
|
|
68
|
Schlaak RA, Frei A, SenthilKumar G, Tsaih
SW, Wells C, Mishra J, Flister MJ, Camara AKS and Bergom C:
Differences in expression of mitochondrial complexes due to genetic
variants may alter sensitivity to radiation-induced cardiac
dysfunction. Front Cardiovasc Med. 7:232020. View Article : Google Scholar
|
|
69
|
Ping Z, Peng Y, Lang H, Xinyong C, Zhiyi
Z, Xiaocheng W, Hong Z and Liang S: Oxidative stress
radiation-induced cardiotoxicity. Oxid Med Cell Longev. Mar
1–2020.(Epub ahead of print). doi: 10.1155/2020/3579143. View Article : Google Scholar
|