Introduction
Lung cancer is the most common malignancy worldwide
(1). Globally, >1.6 million new
cases of lung cancer, which account for 12.7% of all new cancer
cases, are reported each year, making this cancer type the most
lethal malignancy (2). In total,
>1.4 million patients with lung cancer succumb to the disease
each year, and this number accounts for 19% of all cancer-related
deaths annually (3). Although the
treatment of lung cancer has been improved in recent years, the
5-year survival rate of patients with lung cancer, especially those
with advanced stages, remains extremely low (4). Local recurrence, lymph node metastasis
and distant metastasis are the main causes of death (5–7).
Long non-coding RNAs (lncRNAs) refer to functional
RNA molecules with transcriptional lengths of >200 nucleotides
that are not translated into proteins (8,9).
lncRNA is closely related to human health and disease (10–12).
lncRNA plays an important role in the occurrence and development of
tumors, including lung cancer (13,14).
The lncRNA hedgehog-interacting protein antisense RNA 1 (HHIP-AS1)
inhibits hepatocellular carcinoma progression by stabilizing
hedgehog-interacting protein mRNA (15). However, the mechanism of HHIP-AS1 in
lung adenoma has yet to be clarified. Thus, in the present study,
the focus was on the role of HHIP-AS1 in the occurrence and
development of lung cancer.
MicroRNAs (miRNAs/miRs) are highly conserved
non-coding RNAs (16). miRNAs are
involved in cell proliferation, apoptosis, cell differentiation and
other biological behaviors by regulating the expression of
corresponding genes. miRNAs can also be involved in the invasion
and migration of malignant tumors, the regulation of the tumor
microenvironment and the regulation of tumor stem cells (TSCs)
(17,18). miR-153-3p is closely associated with
tumor detection (19–21). However, the mechanism of miR-153-3p
in the development and progression of lung cancer is unclear.
Protocadherin (PCDH), as a member of the cadherin
family, may play an important role in the establishment and
function of specific cell-cell connections in the brain (22). The extracellular coding sequence of
classical cadherin is interrupted by numerous introns (23). In comparison, the coding sequence of
the corresponding part of PCDH does not contain introns (24). High levels of PCDH are produced in
the brain in specific forms, but their function is unknown. At
least one homophobic binding tendency has been observed in PCDH.
This binding tendency is similar to that in classical cadherin
(25). Little information is
available on the relationship between PCDHs and tumorigenesis or
nuclear signaling (26).
Protocadherinγ subfamily A9 (PCDHGA9) is a potential new biomarker
in gastric cancer and is closely associated with the outcome of
patients with gastric cancer (27,28).
However, to the best of our knowledge, the role and function of
PCDHGA9 in lung cancer have not been reported.
The aim of the current study was to investigate the
expression level of HHIP-AS1 in lung cancer tissues and adjacent
tissues. HHIP-AS1 was overexpressed in A549 and NCI-H1299 cells.
Changes in cell proliferation, invasion, epithelial-mesenchymal
transition (EMT) and cellular stemness were then investigated. The
associations between HHIP-AS1 and miR-153-3p and PCDHGA9 were
further explored.
Materials and methods
Lung cancer tissue collection
A total of 20 patients, including 12 males and 8
females, aged between 54–82 years, (average age, 58.7±11.8 years)
who were clinically and pathologically diagnosed with lung cancer
and treated surgically between March and October 2020, were
selected as the study subjects. Specimens were collected in the
Department of Tumor Surgery of The Second Hospital of Tianjin
Medical University. All patients met the following inclusion
criteria: i) The selected specimens were pathologically confirmed
as NSCLC and paracancer tissue; ii) All patients were not treated
with radiotherapy or chemotherapy; and iii) Complete follow-up data
of patients. Exclusion criteria included: Presence of other tumors
or systemic diseases that threaten life and health. Cancer tissues
and adjacent tissues (5 cm away from the cancer tissue) were
collected. Patients with a preoperative history of chemotherapy
were excluded.
All patients provided written informed
consent
The present study was reviewed and approved by the
Ethics Review Committee of the Second Hospital of Tianjin Medical
University (Tianjin, China).
Cell culture
A549 and NCI-H1299 cell lines were obtained from the
Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The
cells were cultured in DMEM containing 10% FBS (both Gibco; Thermo
Fisher Scientific, Inc.). The cells were placed in an incubator at
37°C with 5% CO2. Logarithmic growth cells were
collected. Cell passage was performed every 2 days.
Cell transfection
Cells in the logarithmic phase and in good growth
condition were inoculated into a six-well plate a day before
transfection. The cell density was 70–80% after 12 h of adherence
to the cell-culture dish. Prior to transfection, the cells were
replaced with a medium without penicillin-streptomycin mixed
solution. Lipofectamine® 3000 (Thermo Fisher Scientific,
Inc.) transfection reagent was added according to the instructions.
The negative control (NC; empty vector, pcDNA3.1),
pcDNA3.1-HHIP-AS1, pcDNA3.1-PCDHGA9, mimic-NC
(5′-UUGUACUACACAAAAGUACUG-3′), miR-153-3p mimic
(5′-TTGCATAGTCACAAAAGTGATC-3′), inhibitor-NC
(5′-CAGUACUUUUGUGUAGUACAA-3′) and miR-153-5p inhibitor
(5′-GATCACTTTTGTGACTATGCAA-3′) were purchased from Shanghai
GeneChem Co., Ltd., and separately transfected into the cells. The
culture medium was replaced with DMEM (10% FBS) after 8 h. Cells
were transfected with 2 µg HHIP-AS1 vector (1 µg/ml), 40 nM
miR-153-3p mimic and 20 nM miR-153-3p inhibitor. After the cells
had been incubated at 37°C for 48 h, further experimentation was
performed.
Reverse transcription-quantitative
(RT-q)PCR experiments
The cancer tissues and adjacent tissues were washed
with 0.9% sodium chloride solution, placed into the RNA sample
storage solution, and stored in a refrigerator at −80°C. The total
RNA of lung cancer tissue and cells was extracted with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The integrity of the total RNA was determined by
electrophoresis. cDNA was reverse transcribed using the Prime
Script™ 1st Strand cDNA Synthesis kit (Takara Bio, Inc.). The
primer sequences are shown in Table
I. The reaction system (20 µl) comprised 10 µl, 0.6 µl of
upstream primer (10 µmol/l), 0.6 µl of downstream primer (10
µmol/l), 1 µl of cDNA and 7.8 µl of double distilled water. The
reaction conditions were as follows: 95°C for 10 min, 95°C for 15
sec and 60°C for 1 min (40 cycles). The coefficients of
determination of the standard curves generated by the software were
all >0.99, and the amplification efficiency was 90–110%. The
2−ΔΔCq method was used to calculate the difference in
gene expression (29).
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Forward primer
(5–3) | Forward primer
(5–3) |
|---|
| HHIP-AS1 |
GGCTGAAGAAGCAGAGGATAG |
TTCACCACTCTGTCGGTTTAG |
| miR-153-3p |
GGGTTGCATAGTCACAAAAG |
TTTGGCACTAGCACATT |
| PCDHGA9 |
GCTCATTTCGGTGGAAGAT |
CACTGGGCTAAACAGAGAT |
| N-cadherin |
GGTGGAGGAGAAGAAGACCAG |
GGCATCAGGCTCCACAGTG |
| Vimentin |
GAGAACTTTGCCGTTGAAGC |
GCTTCCTGTAGGTGGCAATC |
| Snail |
CCTCCCTGTCAGATGAGGAC |
CCAGGCTGAGGTATTCCTTG |
| Slug |
GGGGAGAAGCCTTTTTCTTG |
TCCTCATGTTTGTGCAGGAG |
| E-cadherin |
TGCCCAGAAAATGAAAAAGG |
GTGTATGTGGCAATGCGTTC |
| CD44 |
TTGCAGTCAACAGTCGAAGAAG |
CCTTGTTCACCAAATGCACCA |
| Oct4 |
CTTGCTGCAGAAGTGGGTGGAGGAA |
CTGCAGTGTGGGTTTCGGGCA |
| CD133 |
TGGATGCAGAACTTGACAACGT |
ATACCTGCTACGACAGTCGTGGT |
| SOX2 |
GCCGATGTGAAACTTTTGTCG |
GGCAGCGTGACTTATCCTTCT |
| GAPDH |
TGCACCACCAACTGCTTAGC |
GGCATGGACTGTGGTCATGAG |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
Cell proliferation assay
The cells were inoculated into a 96-well plate at a
density of 5×103 cells/well. Fresh culture medium (90
µl) containing 10% FBS and 10 µl Cell Counting Kit-8 (CCK-8)
solution (Dojindo Molecular Technologies, Inc.) was placed into
each well. The CCK-8 assay was used according to the manufacturer's
protocols. The reaction was then placed in an incubator at 37°C
with 5% CO2 for 4 h. The optical density of each well at
450 nm was determined using an immunoplate reader. The average
value of each group was used to reflect the proliferation ability
of the cells.
Transwell invasion assay
All reagents and equipment were pre-cooled on ice.
The Transwell chamber was placed in a 24-well plate, and 50 µl (0.2
µg/µl) of Matrigel (BD Biosciences) was evenly spread on the
membrane of the Transwell chamber (MilliporeSigma). The precoating
occurred at 37°C and was left for 15 min to solidify. The cells
were digested using 0.125% trypsin (37°C, 5 min), centrifuged at
200 × g/min for 5 min at 4°C, counted, and then diluted in
serum-free medium to 2.5×104 cells/ml to prepare a cell
suspension. The cell suspension was added to the upper Transwell
chamber at 200 µl/well, and 500 µl medium with 10% FBS was added to
the lower Transwell chamber. The Transwell chamber was placed in a
37°C incubator for culturing (48 h). The cells were fixed with 4%
formaldehyde for 15 min at room temperature. Then, stained with
0.1% crystal violet for 15 min at room temperature. Next, the cells
on the inner membrane were gently wiped with a cotton swab. The
number of cells that had passed through the filter membrane was
counted under a fluorescence microscope (Nikon Corporation) in four
high-power fields (magnification, ×40). The experiment was repeated
three times.
Clonogenic analysis
Cells at a density of 200 cells/well were inoculated
into a 12-well plate after 24 h of transfection. The cells were
cultured in an incubator with 5% CO2 at 37°C. The liquid
was changed every 2 days. After 15 days, the medium was discarded
and 10% precooled methanol (4°C) was used for fixation for 20 min.
A total of 400 µl 1% crystal violet (Solarbio) was added to each
well. The cells were stained at room temperature for 10 min and
rinsed with water for 5 min. Cell clones were counted under a
fluorescence microscope (Nikon Corporation).
Western blotting analysis
The transfected cells were collected and lysed with
RIPA buffer (Beyotime Institute of Biotechnology), and total
protein was extracted. Protein concentration was determined by the
BCA method. Loading buffer was added to denatured protein. 12%
SDS-PAGE was performed on 30 µg protein samples of each group. The
cells were then transferred to PVDF membranes. Cells were then
blocked using 5% skimmed milk powder (1X PBS) for 2 h at room
temperature. Primary antibodies for E-cadherin (cat. no. ab40772,
1:1,000), Vimentin (cat. no. ab92547, 1:1,000), N-cadherin (cat.
no. ab76011, 1:1,000), Snail1 (cat. no. ab216347, 1:1,000), Slug
(cat. no. ab51772, 1:1,000), Twist1 (cat. no. ab50887, 1:1,000),
Oct4 (cat. no. ab200834, 1:1,000), SOX2 (cat. no. ab171380,
1:1,000), CD44 (cat. no. ab189524, 1:1,000) and CD133 (cat. no.
ab222782, 1:1,000) were added and incubated overnight at 4°C. All
antibodies were purchased from Abcam. HRP-labeled secondary
antibodies (1:10,000, Thermo Fisher Scientific, Inc.; cat. nos.
A16072 and A16104) were added and incubated at room temperature for
2 h. After washing in TBST (0.1% Tween-20; Thermo Fisher
Scientific, Inc.), cells were developed in ECL solution
(MilliporeSigma; cat. no. MA01821). The images were exposed and
analyzed using a Bio-Rad gel imaging system.
Online database analysis
In this study, two database websites, StarBase
(version: v2.0; http://starbase.sysu.edu.cn/) (30) and TargetScan (version: 7.1;
http://www.targetscan.org/vert_71/)
(31), were used simultaneously.
The binding sites of HHIP-AS1 and miR-153-3p were analyzed through
StarBase. The binding sites of miR-153-3p and PCDHGA9 were analyzed
by TargetScan. The Cancer Genome Atlas data were used to analyze
the expression of PCDHGA9 in lung adenocarcinoma (LUAD) tissues and
normal tissues. Data were obtained from the UALCAN website
(http://ualcan.path.uab.edu/analysis.html) (32). Immunohistochemical images of PCDHGA9
were downloaded from The Human Protein Atlas (https://www.proteinatlas.org/) website (33).
Double luciferase reporter gene
detection experiment
The pmirGLO dual-luciferase reporter gene detection
system from Promega Corporation was used. The 3-untranslated
regions of HHIP-AS1 and PCDHGA9 and the sequence fragments of the
binding site of miR-153-3p were chemically synthesized. The pmirGLO
luciferase expression vector (Promega Corporation) was inserted to
obtain the wild-type vectors, pmirGLO/HHIP-AS1-wt and
pmirGLO/PCDHGA9-wt. The mutant sequence was synthesized in the same
way to obtain the mutant vectors pmirGLO/HHIP-AS1-mt and
pmirGLO/PCDHGA9-mt. pmirGLO/HHIP-AS1-wt, pmirGLO/PCDHGA9-wt,
pmirGLO/HHIP-AS1-mt, pmirGLO/PCDHGA9-mt, miR-153-3p mimic and the
mimics-NC were separately mixed with Lipofectamine 3000 for cell
transfection. The cells were collected after 48 h of transfection,
as per the instructions of the dual luciferase reporter gene kit,
and Renilla was used as the internal reference. A total of
150 µl 1X lysis buffer PLB was added to each well of 24-well plate
to cover cells, following which they were completely lysed. Then,
50 µl LAR II was added to 1.5 ml EP tubes. Next, 10 µl cell lysate
was added to the 1.5 ml EP tube containing LAR II, the solution was
then mixed and placed in detector for analysis. After the
measurement, the EP tube was removed and 50 µl Stop & GloR
reagent was added and mixed, following which it was placed in the
detector again (Promega Corporation). The ratio of the fluorescence
value of the luciferase to the fluorescence value of the
Renilla was calculated, and the relative viability of the
reporter gene in the cell was evaluated.
Statistical analysis
SPSS19.0 software (IBM Corp.) was used for
statistical processing. Each set of experiments was repeated three
times. Measurement data are expressed as the mean ± SD. The paired
Student's t-test was used in Figs.
1A and 6A. The other data with
two groups were compared using unpaired Student's t-test. One-way
ANOVA followed by Tukey's multiple comparisons test was used for
the comparisons among groups. Spearman's correlation analysis was
used for assessing the correlation between expression levels.
P<0.05 was considered to indicate a statistically significant
difference.
Results
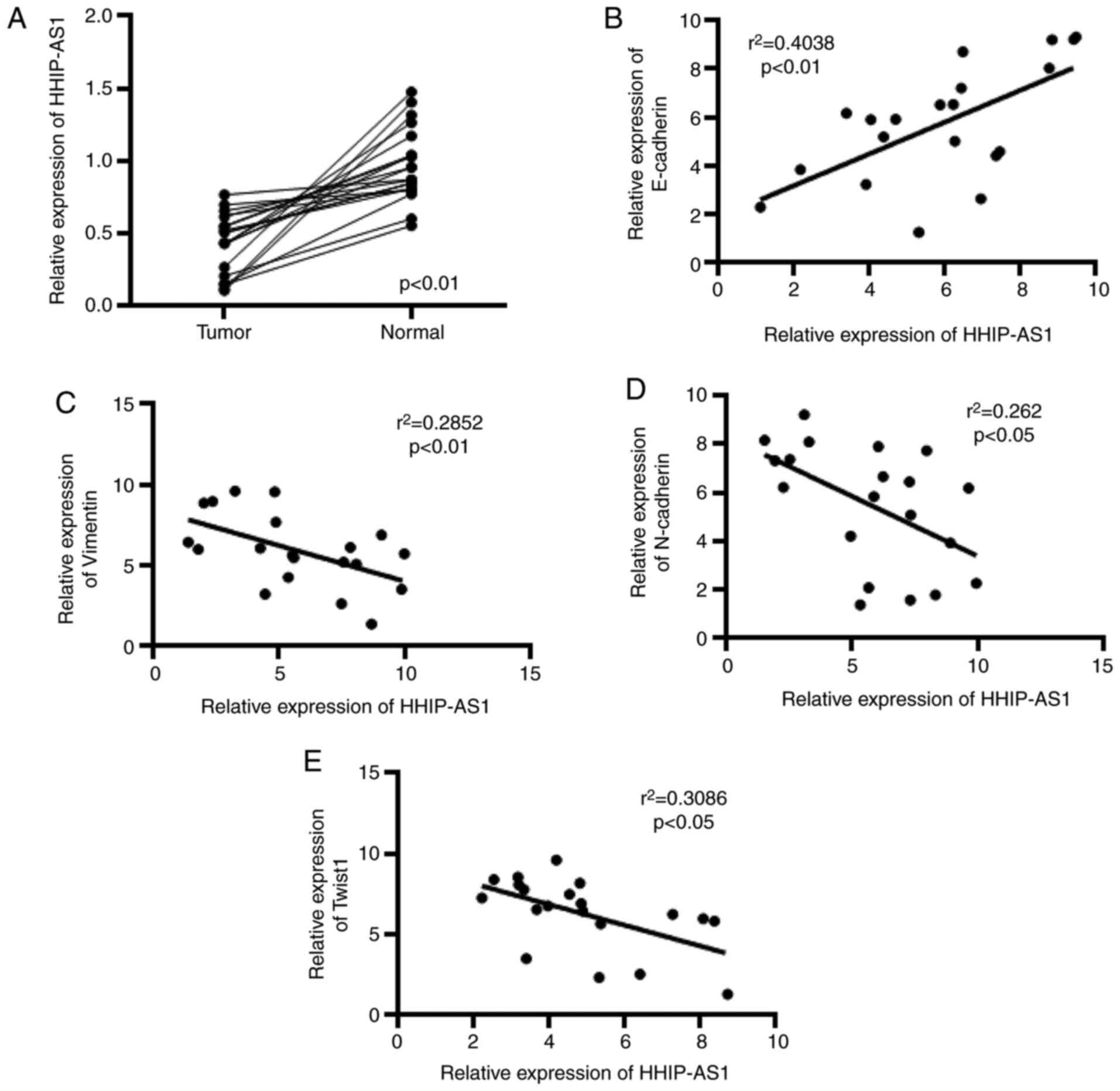
HHIP-AS1 is poorly expressed in lung
cancer
In this study, the expression of HHIP-AS1 in lung
cancer tissues and adjacent tissues was first measured. The
detection results showed that the expression level of HHIP-AS1 in
lung cancer tissues was decreased compared with that in adjacent
tissues (Fig. 1A). The correlation
of HHIP-AS1 with E-cadherin, Vimentin, N-cadherin and Twist1 was
also analyzed. The experimental results showed that the expression
of HHIP-AS1 and E-cadherin was positively correlated (Fig. 1B), whereas HHIP-AS1 was negatively
correlated with Vimentin, N-Cadherin and Twist1 (Fig. 1C-E).
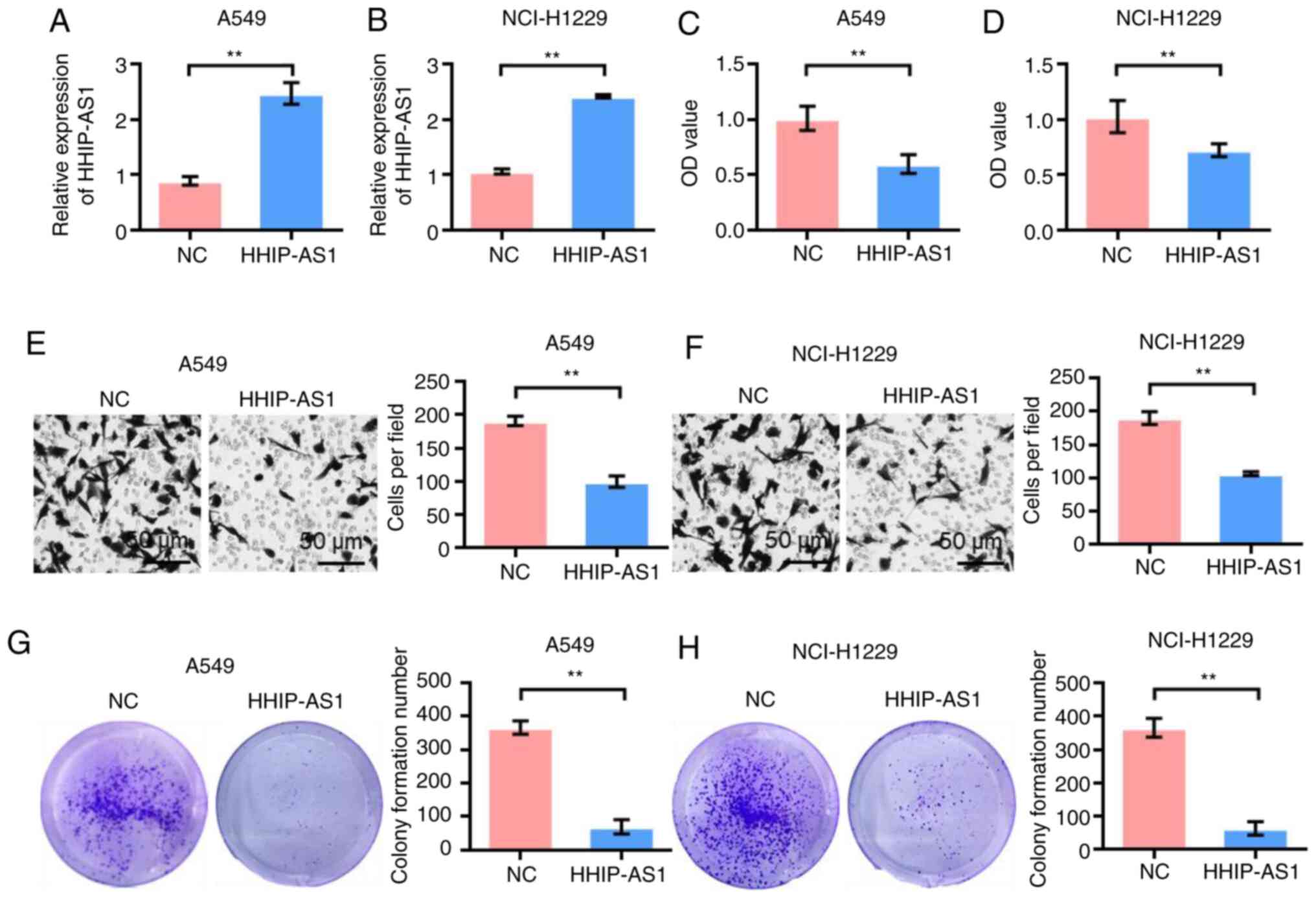
HHIP-AS1 overexpression inhibits the
proliferation, invasion and clonal formation of lung cancer
cells
The effect of HHIP-AS1 overexpression on lung cancer
cell lines was studied to investigate its biological role. First,
the transfection efficiency of overexpressing HHIP-AS1 in A549 and
NCI-H1299 cells was detected. The experimental results showed that
the expression of HHIP-AS1 was higher in the HHIP-AS1 transfection
group than that in the NC group (pcDNA3.1-NC) after
pcDNA3.1-HHIP-NC was transfected into the A549 and NCI-H1299 cell
lines (P<0.01; Fig. 2A and B).
The CCK-8 results showed that the proliferation of the A549 and
NCI-H1299 cells was significantly lower after HHIP-AS1
overexpression compared with that of the control group (P<0.01;
Fig. 2C and D). The Transwell test
results showed that the invasion ability of the A549 and NCI-H1299
cells was significantly lower after HHIP-AS1 overexpression than
that in the control group (P<0.01; Fig. 2E and F). The results of the clonal
formation experiment showed that the number of colony formations of
A549 and NCI-H1299 cell lines overexpressing HHIP-AS1 was
significantly lower than that of the control group (P<0.01;
Fig. 2G and H).
HHIP-AS1 overexpression inhibits the
EMT and cell stemness of lung cancer cells
The effect of HHIP-AS1 overexpression on EMT and
stem cell markers in lung cancer cells was examined. RT-qPCR
analysis confirmed that the overexpression of the HHIP-AS1 gene
significantly increased the expression of the epithelial marker,
E-cadherin (P<0.01; Fig. 3A) and
decreased the expression of mesenchymal markers (Fig. 3B-F). RT-qPCR analysis showed that
the upregulation of HHIP-AS1 significantly reduced the expression
of stemness-related genes Oct4 and SOX2 (Fig. 3G and H) and CD44- and CD133-related
surface antigens in cancer stem cells (P<0.01; Fig. 3I and J). In order to further verify
the role of lncRNA HHIP-AS1, changes in the expression levels of
E-cadherin, Vimentin, N-cadherin, Snail1, Slug, Twist1, Oct4, SOX2,
CD44 and CD133 after the overexpression of HHIP-AS1 were identified
using western blot analysis. The results showed that overexpression
of HHIP-AS1 increased the expression of the epithelial marker
E-cadherin. However, overexpression of HHIP-AS1 inhibited the
expression of the mesenchymal marker Vimentin and N-cadherin. In
addition, after overexpression of HHIP-AS1, the expression levels
of Snail1, Slug, Twist1, Oct4, SOX2, CD44 and CD133 were also
downregulated (Fig. 3K-L).
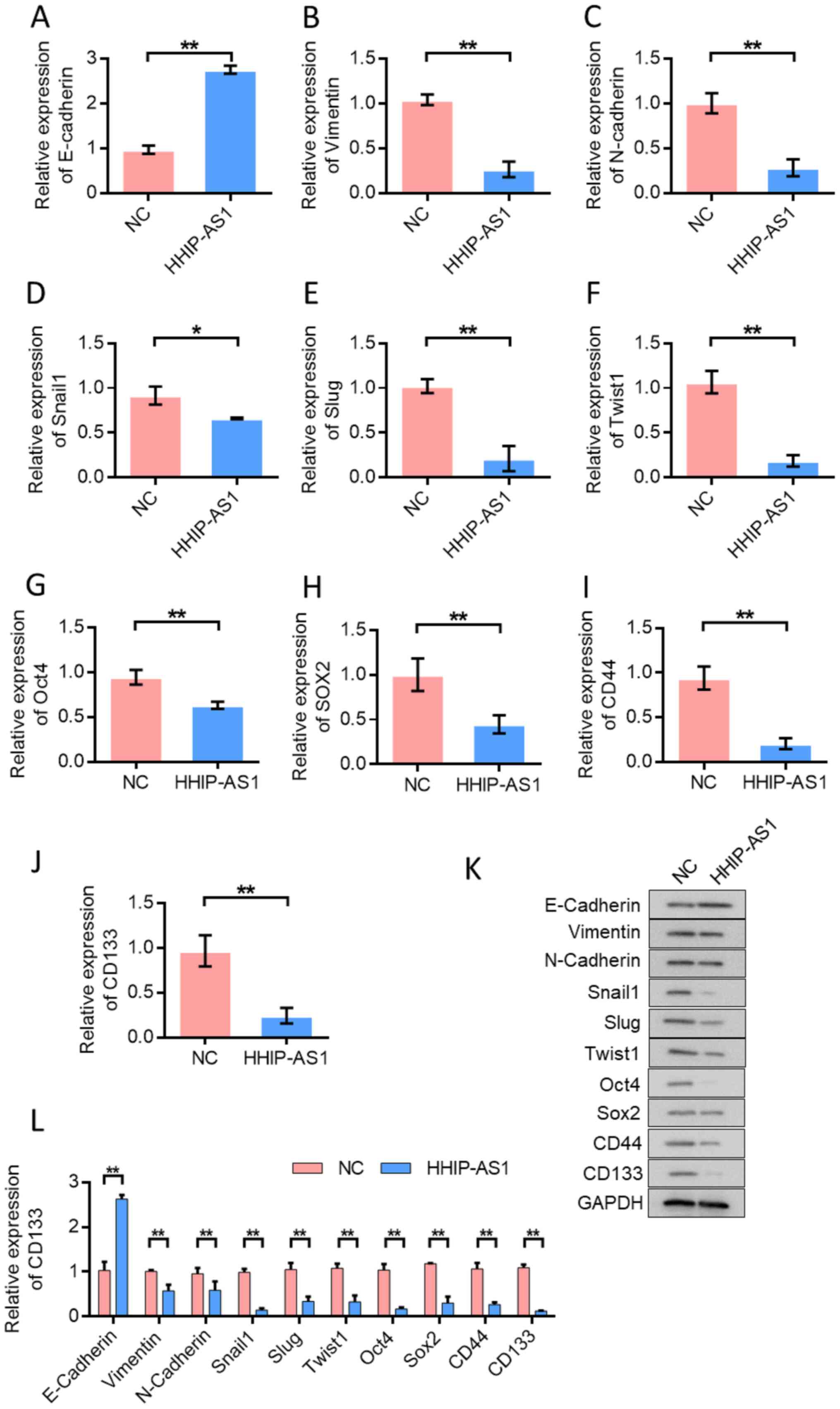 | Figure 3.Overexpression of HHIP-AS1 inhibits
EMT and cell stemness of lung cancer cells. (A) Detection of
E-cadherin expression in lung cancer cells. Overexpression of
HHIP-AS1 upregulated the expression of E-cadherin. Detection of (B)
Vimentin, (C) N-cadherin, (D) Snail1, (E) Slug, (F) Twist1, (G)
Oct4, (H) SOX2, (I) CD44and (J) CD133 expression in lung cancer
cells. (K) Western blotting detected the changes in the expression
levels of E-cadherin, Vimentin, N-cadherin, Snail1, Slug, Twist1,
Oct4, SOX2, CD44 and CD133 after the overexpression of HHIP-AS1.
(L) The semi-quantification for the western blotting results. Error
bars represent the mean ± SD values. *P<0.05, **P<0.01.
HHIP-AS1, hedgehog-interacting protein antisense RNA 1; NC,
negative control. |
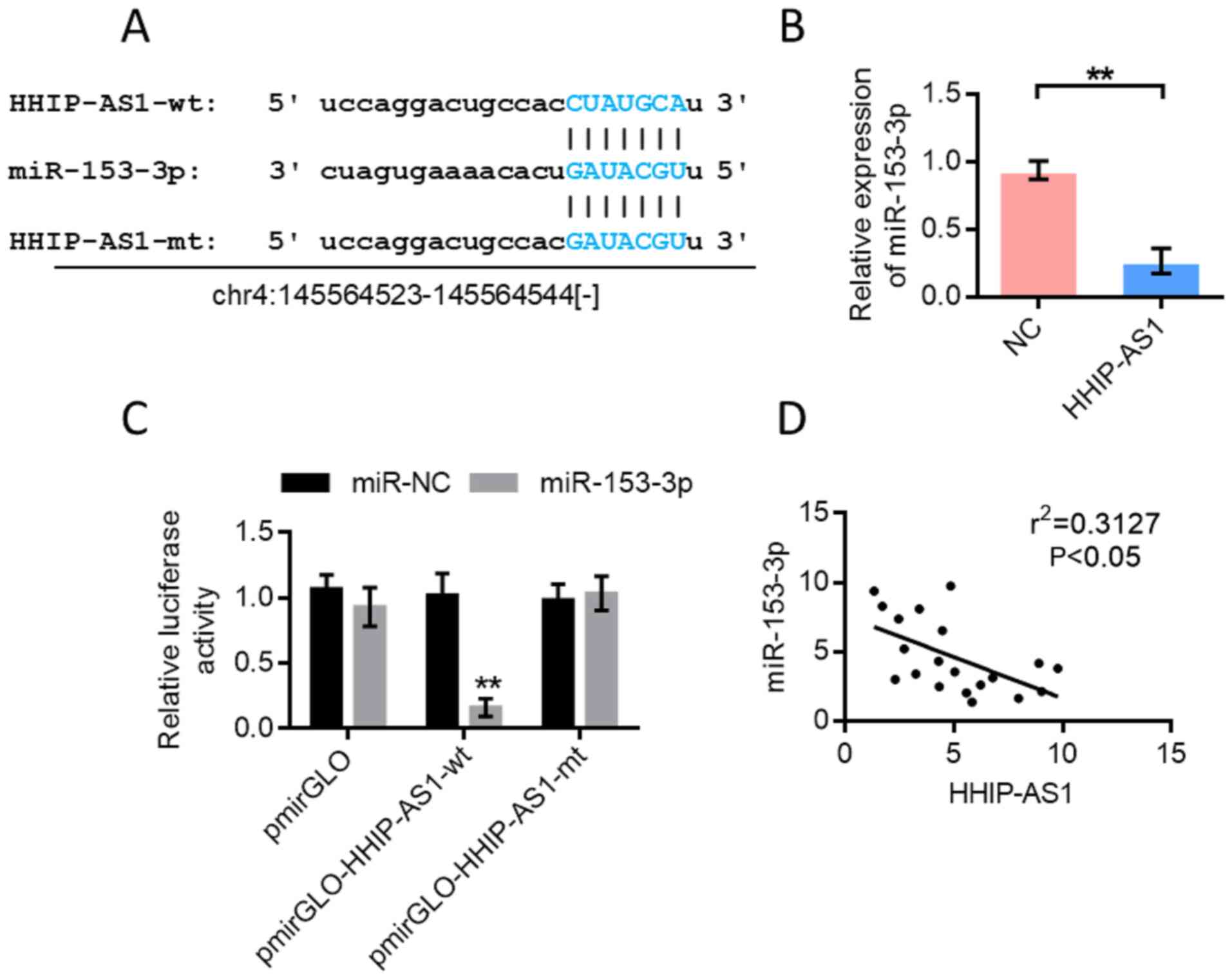
miR-153-3p is the target gene of
HHIP-AS1
Bioinformatics analysis was performed using the
StarBase database (http://starbase.sysu.edu.cn/) to predict the binding
sites of miR-153-3p and HHIP-AS1 (Fig.
4A). Further experiments showed that HHIP-AS1 overexpression
inhibited the expression of miR-153-3p (Fig. 4B). The pmirGLOdual-luciferase
reporter system was used for further verification. The results
showed that miR-153-3p could inhibit the luciferase expression of
wild-type HHIP-AS1 (P<0.01). However, the inhibitory effect
disappeared when the HHIP-AS1 binding site was mutated (Fig. 4C). The double luciferase reporter
gene assay showed that HHIP-AS1 could adsorb miR-153-3p. The
correlation test results of miR-153-3p and HHIP-AS1 expression
showed a negative correlation (Fig.
4D).
miR-153-3p directly targets PCDHGA9 in
lung cancer
TargetScan (http://www.targetscan.org/vert_72/) predicted that
miR-153-3p could bind to PCDHGA9. Fig.
5A shows the schematic diagram of the binding site of
miR-153-3p with PCDHGA9. The RT-qPCR results showed that
overexpression of miR-153-3p inhibited the expression of PCDHGA9
(Fig. 5B and C). Subsequently, the
pmirGLOdual-luciferase reporter system was used for further
verification. The results showed that miR-153-3p inhibited the
luciferase expression of wild-type PCDHGA9 (P<0.01). However,
the inhibitory effect disappeared when the binding site of PCDHGA9
was mutated (Fig. 5D). The
correlation test results of miR-153-3p and PCDHGA9 expression
showed a negative correlation (Fig.
5E). The expression of PCDHGA9 in 515 patients with lung cancer
was lower and the difference was statistically significant
(Fig. 5F), based on data from The
Cancer Genome Atlas (http://ualcan.path.uab.edu/analysis.html).Transfection
experiments using PCDHGA9 expression vector and PCDHGA9 +
miR-153-3p mimic were performed. CCK-8 was used to detect the
change in cell proliferation ability after different treatments.
The transfection efficiency results for the overexpression of
PCDHGA9 are provided in Fig. 5G.
The results showed that the proliferation ability of A549 and
NCI-H1299 cells was decreased after PCDHGA9 overexpression compared
with the control group. However, cell proliferation ability was
enhanced by the simultaneous transfection of PCDHGA9 overexpression
plasmid and miR-153-3p mimic compared with the PCDHGA9
overexpression group (Fig.
5H-K).
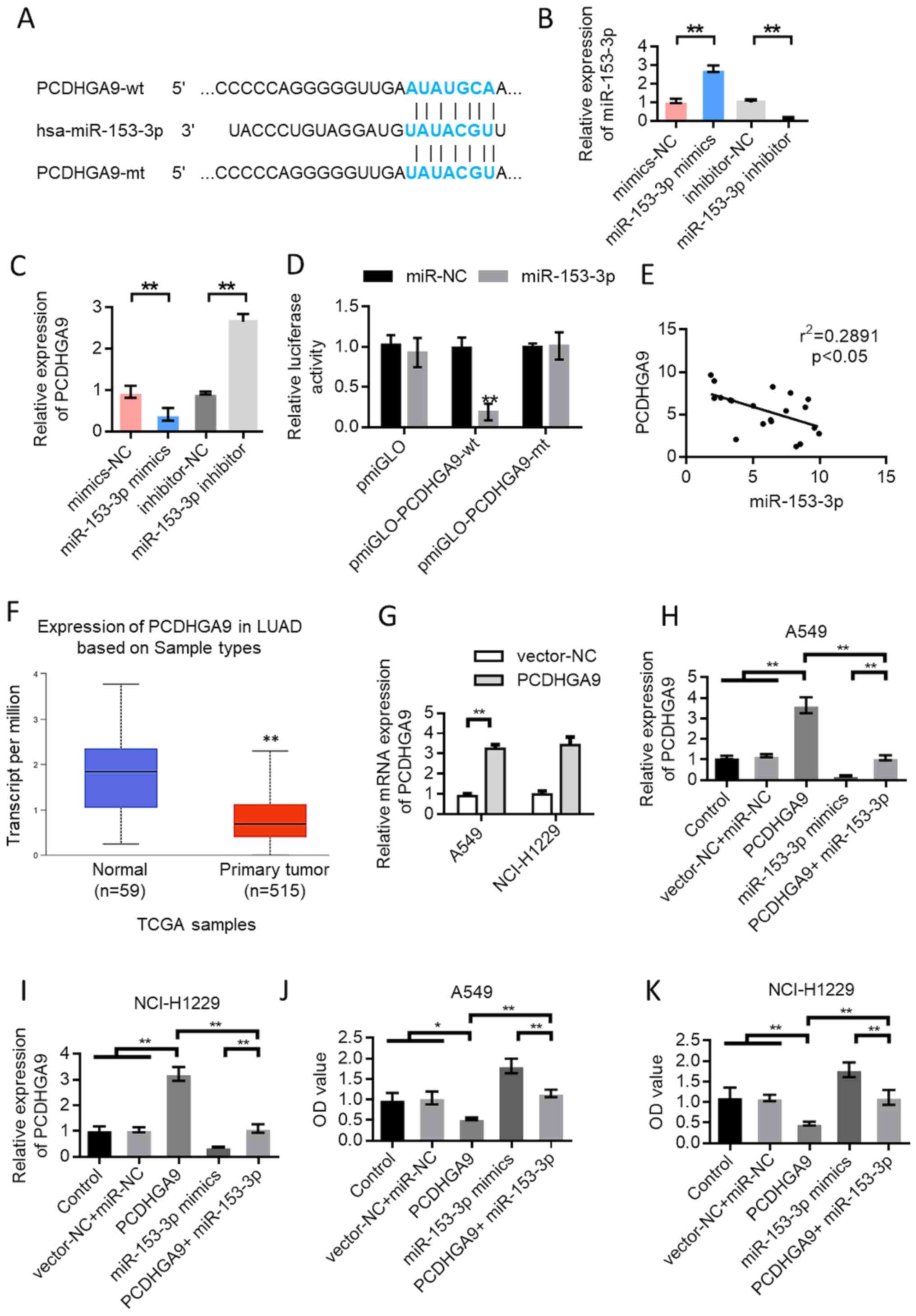 | Figure 5.In lung cancer, miR-153-3p directly
targets PCDHGA9. (A) Schematic diagram of the binding site of
miR-153-3p and PCDHGA9. (B) Verification of the transfection
efficiency of miR-153-3p mimics. (C) Overexpression of miR-153-3p
inhibited the expression of PCDHGA9. (D) The dual luciferase
reporter gene detection experiment verified the binding of
miR-153-3p to PCDHGA9. (E) The expression of miR-153-3p and PCDHGA9
was negatively correlated. (F) TCGA data were used to analyze the
expression of PCDHGA9 in LUAD tissues and normal tissues. Data from
UALCAN website (http://ualcan.path.uab.edu/analysis.html). (G) The
transfection efficiency results for the overexpression of PCDHGA9.
After transfection with PCDHGA9 expression vector and
PCDHGA9+miR-153-3p mimics, the expression level of PCDHGA9 in (H)
A549 and (I) NCI-H1299 cells was detected. After transfection with
PCDHGA9 expression vector and PCDHGA9+miR-153-3p mimics, the cell
proliferation ability of (J) A549 and (K) NCI-H1299 cells was
detected. Error bars represent the mean ± SD values. *P<0.05,
**P<0.01. HHIP-AS1, hedgehog-interacting protein antisense RNA
1; NC, negative control; miR, microRNA; wt, wild-type; mt, mutant;
PCDHGA9, protocadherinγ subfamily A9; LUAD, lung
adenocarcinoma. |
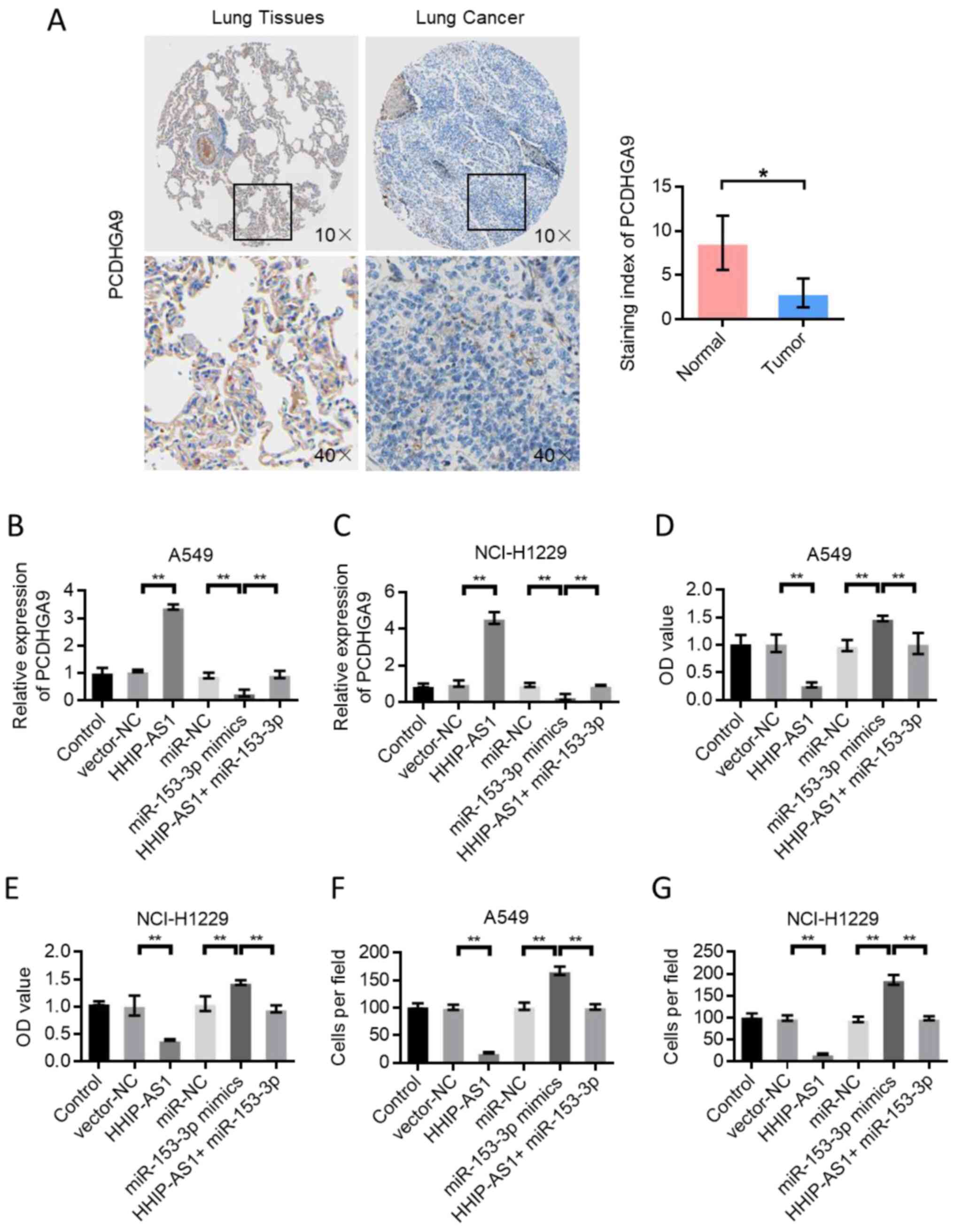
PCDHGA9 expression mediates the
biological effect of HHIP-AS1
The Human Proteins Atlas database (http://www.proteinatlas.org/) in data analysis was
employed and the results showed that PCDHGA9 was decreased in LUAD
tissue compared with normal lung tissue (Fig. 6A) (33). Immunohistochemical images of PCDHGA9
were downloaded from The Human Protein Atlas (https://www.proteinatlas.org/ENSG00000261934-PCDHGA9/pathology/lung+cancer)
website. Subsequently, PCDHGA9 expression was detected in A549 and
NCI-H1299 cells after different transfections. The results showed
that HHIP-AS1 overexpression upregulated the expression of PCDHGA9.
However, miR-153-3p overexpression inhibited the expression of
PCDHGA9. A549 and NCI-H1299 cells were simultaneously transfected
with HHIP-AS1 overexpression vector and miR-153-3p, and the result
showed that miR-153-3p could reverse the effect of HHIP-AS1
(Fig. 6B and C). Cell proliferation
and invasion experiments showed that HHIP-AS1 overexpression could
inhibit the proliferation and invasion of A549 and NCI-H1299 cells.
miR-153-3p overexpression promoted cell proliferation and invasion.
A549 and NCI-H1299 cells were simultaneously transfected with
HHIP-AS1 overexpression vector and miR-153-3p, and the results
showed that miR-153-3p could reverse the anticancer effect of
HHIP-AS1 (Fig. 6D-G).
Discussion
Tumor invasion and metastasis are the main causes of
death in patients with lung cancer (34). The manner in which to inhibit the
growth of lung cancer cells and block the invasion and metastasis
of cancer cells remains an urgent issue to address (35). EMT is a process in which epithelial
cells are transformed into mesenchymal cells and gain mesenchymal
properties (36). EMT can enable
epithelial tumor cells to acquire a mesenchymal phenotype and
further enhance the invasion and metastatic abilities of tumor
cells to obtain self-renewal ability and other stem-like
characteristics. EMT promotes the ability of tumor cells to acquire
stem cell properties; thus, making tumor therapy more difficult
(37–41).
Evidence shows that lncRNAs are involved in the
self-renewal of embryonic and pluripotent stem cells, but whether
lncRNAs drive the stem cell transformation process and their role
in maintaining stem cell stemness are still unknown (42). Therefore, a better understanding of
the molecular regulatory mechanisms and pathways of lncRNA in
tumorigenesis provides a guarantee for the establishment of new
treatment strategies and more effective cancer treatment methods
(43). HHIP-AS1 is widely involved
in tumor development. HHIP-AS1 inhibits the progression of
hepatocellular carcinoma by stabilizing HHIP mRNA (15). However, it has also been reported
that in Hedgehog (Hh)-driven human brain tumors, HHIP-AS1 promotes
tumor survival by stabilizing the dynein complex 1 (15). In addition, HHIP-AS1 plays an
important role in oncogenesis driven by the Sonic Hh pathway, an
attractive therapeutic target (15). Thus, HHIP-AS1 can regulate cell
proliferation and metastasis in a variety of tumors. Exploring the
mechanism of HHIP-AS1 in inhibiting EMT and stem cell function in
lung cancer is of great importance.
The mechanism of action of lncRNA is complex. One of
its important mechanisms is to bind miRNA as a molecular sponge to
inhibit its silencing effect on the corresponding target mRNA
downstream (44). A retrieval of
TargetScan online bioinformatics indicated that miR-153-3p may be
one of the binding molecules of HHIP-AS1 (45). Thus, we speculated that the
anti-lung cancer cell effect of HHIP-AS1 may be realized through
the adsorption of miR-153-3p. This hypothesis was verified through
the overexpression of HHIP-AS1 in A549 cells. RT-qPCR detection
showed that HHIP-AS1 overexpression markedly inhibited the
expression level of miR-153-3p in A549 cells. TargetScan analysis
indicated that PCDHGA9 was the downstream inhibition target
of miR-153-3p. The present study preliminarily indicated that the
mechanism of action of HHIP-AS1 is the adsorption of miR-153-3p and
the release of the inhibitory effect on PCDHGA9. The
dual-luciferase reporter system was used to further verify the
targeted relationship between miR-153-3p and PCDHGA9. lncRNA can
regulate the expression of target genes through competing
endogenous RNAs (ceRNAs). In the present study, it was found that
HHIP-AS1 could target miR-153-3p and regulate the expression of
PCDHGA9. HHIP-AS1/miR-153-3p/PCDHGA9 constituted the regulatory
mechanism of ceRNA. PCDHGA9, as a tumor suppressor gene, can
regulate the expression of E-cadherin. In the present study, the
overexpression of PCDHGA9 upregulated the expression of
E-cadherin.
EMT is essential for the maintenance of stem cell
identity (46). Multipotent
transcription factors, such as Oct4, Sox2, Fascin1 and Nanog, are
highly expressed in TSCs (CD133+ side population cells)
(47). Transcription factors
inhibits the expression of E-cadherin by acting on EMT-related
regulatory factors and increases the expression levels of
N-cadherin, E-box-binding homeobox 2 and Vimentin (48). These regulatory factors promote the
occurrence of EMT and thus promote the invasion, metastasis and
self-renewal of stem cells. The results of the present study showed
that HHIP-AS1 inhibited the expression of stem cell markers.
HHIP-AS1 can target Oct4, Sox2 and Nanog to decrease the sphere
formation efficiency and colony formation of lung cancer cells. In
addition, HHIP-AS1 upregulated the expression of epithelial markers
while inhibiting the expression of mesenchymal markers. Thus,
HHIP-AS1 can inhibit the EMT of lung cancer.
PCDHGA9 can induce autophagy, apoptosis and cell
cycle arrest in gastric cancer cells, and inhibit EMT through the
TGF-β/Smad2/3 pathway (27). In
addition, PCDHGA9 overexpression in gastric cancer cells can
enhance the expression of E-cadherin and decrease the expression of
N-cadherin, Vimentin, Slug and Twist. PCDHGA9 knockdown has the
opposite effect (28). Slug and
Twist are key transcription factors in EMT. In the present study,
HHIP-AS1 expression inhibited the expression of Slug and Twist by
regulating the expression of PCDHGA9. HHIP-AS1 upregulated the
expression of E-cadherin. These two factors are important for EMT
in lung cancer progression. Cadherin downregulation leads to the
disruption of cell-cell junctions, which is a key step in EMT that
promotes tumor metastasis. PCDHGA9 is a member of the cadherin
family, and PCDHGA9 overexpression can upregulate the expression of
E-cadherin. The expression of Vimentin decreased after the
overexpression of PCDHGA9. PCDHGA9 interacts directly with
β-catenin and weakens the EMT-induced effect of the TGF-β and
Wnt/β-catenin pathways (28).
Therefore, PCDHGA9 inhibits EMT. This effect may also be the
downstream molecular mechanism of PCDHGA9 regulated by HHIP-AS1 in
targeting miR-153-3p.
In summary, HHIP-AS1 expression was decreased in
lung cancer tissues. Mechanism of action studies have shown that
HHIP-AS1 exerts an antitumor effect by inhibiting the EMT and
cellular stemness of lung cancer cells through the adsorption of
miR-153-3p and the inhibition of PCDHGA9 expression. As a new
molecule closely related to lung cancer, HHIP-AS1 is a potential
target for lung cancer treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ and HT designed the experiments and were
responsible for confirming the authenticity of the raw data. ZZ and
YL performed the experiments and data analysis. ZZ, SC, QZ and HT
performed data analysis of cell experiments and wrote the
manuscript, with contributions from all authors. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Clinical sample collection was approved by the
Ethics Committee of The Second Hospital of Tianjin Medical
University, and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small-cell lung cancer
|
|
PCDHGA9
|
protocadherinγ subfamily A9
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ZEB2
|
E-box binding homeobox 2
|
|
lncRNA
|
long non-coding RNA
|
|
HHIP-AS1
|
hedgehog-interacting protein antisense
RNA 1
|
|
LUAD
|
lung adenocarcinoma
|
References
|
1
|
Howlader N, Forjaz G, Mooradian MJ, Meza
R, Kong CY, Cronin KA, Mariotto AB, Lowy DR and Feuer EJ: The
effect of advances in lung-cancer treatment on population
mortality. N Engl J Med. 383:640–649. 2020. View Article : Google Scholar
|
|
2
|
de Groot PM, Wu CC, Carter BW and Munden
RF: The epidemiology of lung cancer. Transl Lung Cancer Res.
7:220–233. 2018. View Article : Google Scholar
|
|
3
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar
|
|
4
|
Oudkerk M, Devaraj A, Vliegenthart R,
Henzler T, Prosch H, Heussel CP, Bastarrika G, Sverzellati N,
Mascalchi M, Delorme S, et al: European position statement on lung
cancer screening. Lancet Oncol. 18:e754–e766. 2017. View Article : Google Scholar
|
|
5
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar
|
|
6
|
Xi X, Liu N, Wang Q, Chu Y, Yin Z, Ding Y
and Lu Y: ACT001, a novel PAI-1 inhibitor, exerts synergistic
effects in combination with cisplatin by inhibiting PI3K/AKT
pathway in glioma. Cell Death Dis. 10:7572019. View Article : Google Scholar
|
|
7
|
Zhong W, Hou H, Liu T, Su S, Xi X, Liao Y,
Xie R, Jin G, Liu X, Zhu L, et al: Cartilage oligomeric matrix
protein promotes epithelial-mesenchymal transition by interacting
with transgelin in colorectal cancer. Theranostics. 10:8790–8806.
2020. View Article : Google Scholar
|
|
8
|
Sanli O, Dobruch J, Knowles MA, Burger M,
Alemozaffar M, Nielsen ME and Lotan Y: Bladder cancer. Nat Rev Dis
Primers. 3:170222017. View Article : Google Scholar
|
|
9
|
Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y,
Zhou JK, Guo C, Xia Z, Liu L, et al: Long non-coding RNA linc00460
promotes epithelial-mesenchymal transition and cell migration in
lung cancer cells. Cancer Lett. 420:80–90. 2018. View Article : Google Scholar
|
|
10
|
Liao J, Wang J, Liu Y, Li J and Duan L:
Transcriptome sequencing of lncRNA, miRNA, mRNA and interaction
network constructing in coronary heart disease. BMC Med Genomics.
12:1242019. View Article : Google Scholar
|
|
11
|
Li Y, Zhu G, Ma Y and Qu H: LncRNA CCAT1
contributes to the growth and invasion of gastric cancer via
targeting miR-219-1. J Cell Biochem. 120:19457–19468. 2019.
View Article : Google Scholar
|
|
12
|
Li P, Zhang N, Ping F, Gao Y and Cao L:
lncRNA SCAL1 inhibits inducible nitric oxide synthase in lung cells
under high-glucose conditions. Exp Ther Med. 18:1831–1836.
2019.
|
|
13
|
Wang S, Ke H, Zhang H, Ma Y, Ao L, Zou L,
Yang Q, Zhu H, Nie J, Wu C, et al: LncRNA MIR100HG promotes cell
proliferation in triple-negative breast cancer through triplex
formation with p27 loci. Cell Death Dis. 9:8052018. View Article : Google Scholar
|
|
14
|
Tian T, Wang M, Lin S, Guo Y, Dai Z, Liu
K, Yang P, Dai C, Zhu Y, Zheng Y, et al: The impact of lncRNA
dysregulation on clinicopathology and survival of breast cancer: A
systematic review and meta-analysis. Mol Ther Nucleic Acids.
12:359–369. 2018. View Article : Google Scholar
|
|
15
|
Bo C, Li X, He L, Zhang S, Li N and An Y:
A novel long noncoding RNA HHIP-AS1 suppresses hepatocellular
carcinoma progression through stabilizing HHIP mRNA. Biochem
Biophys Res Commun. 520:333–340. 2019. View Article : Google Scholar
|
|
16
|
Xi X, Chu Y, Liu N, Wang Q, Yin Z, Lu Y
and Chen Y: Joint bioinformatics analysis of underlying potential
functions of hsa-let-7b-5p and core genes in human glioma. J Transl
Med. 17:1292019. View Article : Google Scholar
|
|
17
|
Pipan V, Zorc M and Kunej T: MicroRNA
polymorphisms in cancer: A literature analysis. Cancers (Basel).
7:1806–1814. 2015. View Article : Google Scholar
|
|
18
|
Wong HA, Fatimy RE, Onodera C, Wei Z, Yi
M, Mohan A, Gowrisankaran S, Karmali P, Marcusson E, Wakimoto H, et
al: The Cancer Genome Atlas analysis predicts microRNA for
targeting cancer growth and vascularization in glioblastoma. Mol
Ther. 23:1234–1247. 2015. View Article : Google Scholar
|
|
19
|
Li L, Wang M, Mei Z, Cao W, Yang Y, Wang Y
and Wen A: lncRNAs HIF1A-AS2 facilitates the up-regulation of
HIF-1α by sponging to miR-153-3p, whereby promoting angiogenesis in
HUVECs in hypoxia. Biomed Pharmacother. 96:165–172. 2017.
View Article : Google Scholar
|
|
20
|
Li C, Zhang Y, Zhao W, Cui S and Song Y:
miR-153-3p regulates progression of ovarian carcinoma in vitro and
in vivo by targeting MCL1 gene. J Cell Biochem. 120:19147–19158.
2019. View Article : Google Scholar
|
|
21
|
Wang T, Zhai M, Xu S, Ponnusamy M, Huang
Y, Liu CY, Wang M, Shan C, Shan PP, Gao XQ, et al: NFATc3-dependent
expression of miR-153-3p promotes mitochondrial fragmentation in
cardiac hypertrophy by impairing mitofusin-1 expression.
Theranostics. 10:553–566. 2020. View Article : Google Scholar
|
|
22
|
Canzio D and Maniatis T: The generation of
a protocadherin cell-surface recognition code for neural circuit
assembly. Curr Opin Neurobiol. 59:213–220. 2019. View Article : Google Scholar
|
|
23
|
Goodman KM, Rubinstein R, Dan H, Bahna F,
Mannepalli S, Ahlsén G, Aye Thu C, Sampogna RV, Maniatis T, Honig
B, et al: Protocadherin cis-dimer architecture and recognition unit
diversity. Proc Natl Acad Sci USA. 114:E9829–E9837. 2017.
View Article : Google Scholar
|
|
24
|
Strehl S, Glatt K, Liu QM, Glatt H and
Lalande M: Characterization of two novel protocadherins (PCDH8 and
PCDH9) localized on human chromosome 13 and mouse chromosome 14.
Genomics. 53:81–89. 1998. View Article : Google Scholar
|
|
25
|
Frank M, Ebert M, Shan W, Phillips GR,
Arndt K, Colman DR and Kemler R: Differential expression of
individual gamma-protocadherins during mouse brain development. Mol
Cell Neurosci. 29:603–616. 2005. View Article : Google Scholar
|
|
26
|
Hirayama T and Yagi T: Regulation of
clustered protocadherin genes in individual neurons. Semin Cell Dev
Biol. 69:122–130. 2017. View Article : Google Scholar
|
|
27
|
Weng J, Xiao J, Mi Y, Fang X, Sun Y, Li S,
Qin Z, Li X, Liu T, Zhao S, et al: PCDHGA9 acts as a tumor
suppressor to induce tumor cell apoptosis and autophagy and inhibit
the EMT process in human gastric cancer. Cell Death Dis. 9:272018.
View Article : Google Scholar
|
|
28
|
Weng J, Li S, Lin H, Mei H, Liu Y, Xiao C,
Zhu Z, Cai W, Ding X, Mi Y, et al: PCDHGA9 represses
epithelial-mesenchymal transition and metastatic potential in
gastric cancer cells by reducing β-catenin transcriptional
activity. Cell Death Dis. 11:2062020. View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. µethods. 25:402–408. 2001.
|
|
30
|
Chiang HR, Schoenfeld LW, Ruby JG, Auyeung
VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al:
Mammalian microRNAs: Experimental evaluation of novel and
previously annotated genes. Genes Dev. 24:992–1009. 2010.
View Article : Google Scholar
|
|
31
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar
|
|
32
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar
|
|
33
|
Pontén F, Jirström K and Uhlen M: The
Human Protein Atlas - a tool for pathology. J Pathol. 216:387–393.
2008. View Article : Google Scholar
|
|
34
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TB, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al TRACERx Consortium, : Tracking the evolution of
non-small-cell lung cancer. N Engl J Med. 376:2109–2121. 2017.
View Article : Google Scholar
|
|
35
|
Blandin Knight S, Crosbie PA, Balata H,
Chudziak J, Hussell T and Dive C: Progress and prospects of early
detection in lung cancer. Open Biol. 7:1700702017. View Article : Google Scholar
|
|
36
|
Tulchinsky E, Demidov O, Kriajevska M,
Barlev NA and Imyanitov E: EMT: A mechanism for escape from
EGFR-targeted therapy in lung cancer. Biochim Biophys Acta Rev
Cancer. 1871:29–39. 2019. View Article : Google Scholar
|
|
37
|
Otsuki Y, Saya H and Arima Y: Prospects
for new lung cancer treatments that target EMT signaling. Dev Dyn.
247:462–472. 2018. View Article : Google Scholar
|
|
38
|
Zhang X, Sai B, Wang F, Wang L, Wang Y,
Zheng L, Li G, Tang J and Xiang J: Hypoxic BMSC-derived exosomal
miRNAs promote metastasis of lung cancer cells via STAT3-induced
EMT. Mol Cancer. 18:402019. View Article : Google Scholar
|
|
39
|
Zhong W, Yang W, Qin Y, Gu W, Xue Y, Tang
Y, Xu H, Wang H, Zhang C, Wang C, et al: 6-Gingerol stabilized the
p-VEGFR2/VE-cadherin/β-catenin/actin complex promotes microvessel
normalization and suppresses tumor progression. J Exp Clin Cancer
Res. 38:2852019. View Article : Google Scholar
|
|
40
|
Xiao T, Zhong W, Zhao J, Qian B, Liu H,
Chen S, Qiao K, Lei Y, Zong S, Wang H, et al: Polyphyllin I
suppresses the formation of vasculogenic mimicry via
Twist1/VE-cadherin pathway. Cell Death Dis. 9:9062018. View Article : Google Scholar
|
|
41
|
Zhang Q, Li X, Li X, Li X and Chen Z:
LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by
targeting miR-484 in human lung cancer cells. J Cell Biochem.
119:4447–4457. 2018. View Article : Google Scholar
|
|
42
|
Gong W, Su Y, Liu Y, Sun P and Wang X:
Long non-coding RNA Linc00662 promotes cell invasion and
contributes to cancer stem cell-like phenotypes in lung cancer
cells. J Biochem. 164:461–469. 2018. View Article : Google Scholar
|
|
43
|
Bartl J, Forget A, Zanini M, Picard D, Qin
N, Borkhardt A, Reifenberger G, Ayrault O and Remke M: SIG-03.
HHIP-AS1 promotes tumor survival through stabilizing dynein Complex
1 in hedgehog driven human brain tumors. Neuro Oncol. 21 (Suppl
2):ii113–ii114. 2019. View Article : Google Scholar
|
|
44
|
Ma R, Wang C, Wang J, Wang D and Xu J:
miRNA-mRNA interaction network in non-small cell lung cancer.
Interdiscip Sci. 8:209–219. 2016. View Article : Google Scholar
|
|
45
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
46
|
Bartl J, Picard D, Borkhardt A,
Reifenberger G and Remke M: Targeting the long non-coding RNA
HHIP-AS1 in sonic hedgehog driven brain tumors. Klin Padiatr.
228:A52016. View Article : Google Scholar
|
|
47
|
Wang H, Zhong W, Zhao J, Zhang H, Zhang Q,
Liang Y, Chen S, Liu H, Zong S, Tian Y, et al: Oleanolic acid
inhibits epithelial-mesenchymal transition of hepatocellular
carcinoma by promoting iNOS dimerization. Mol Cancer Ther.
18:62–74. 2019. View Article : Google Scholar
|
|
48
|
Chiou G-Y, Cherng J-Y, Hsu H-S, Wang ML,
Tsai CM, Lu KH, Chien Y, Hung SC, Chen YW, Wong CI, et al: Cationic
polyurethanes-short branch PEI-mediated delivery of Mir145
inhibited epithelial-mesenchymal transdifferentiation and cancer
stem-like properties and in lung adenocarcinoma. J Control Release.
159:240–250. 2012. View Article : Google Scholar
|