Introduction
Glioma is a primary cerebral neoplasm that
originates from glial tissue and spreads to the central nervous
system. Glioma accounts for 31% of the central nervous system
tumors in adults and is characterized by diverse histopathological
changes, poor clinical outcomes and high aggressiveness (1,2). The
clinical heterogeneity, rapid cell dissemination, invasion and
frequent recurrence of glioma results in high incidence and
mortality rates, with the 5-year survival rate of patients with
glioma being ~5% (3,4). Studies show that every year, ~100,000
people worldwide are diagnosed as having diffuse glioma (5), and the proportion of patient death
within 1 month and 3 months after the diagnosis was 9.24 and 19.15%
respectively for all glioma patients (6). Moreover, patients with glioma often
suffer from complications, including epilepsy, extensive
neurodegeneration and cognitive impairment, due to aberrant
glutamate secretion in the glioma microenvironment (7). Glioma grading is beneficial for the
prevention, control and prognosis of glioma (8). Multiple chemical and biomedical
management strategies, including ultrasound, receptor-targeted
methods, and local therapeutic delivery by interstitial spray and
polymeric hydrogels, are all beneficial for patients with glioma
(9). Despite these available
treatments, the prognosis and survival rate of patients with glioma
remain poor, and the development of new effective treatments for
glioma is challenging (10).
Therefore, the identification of novel therapeutic targets for
glioma is desperately needed.
Aerobic glycolysis, the metabolism of glucose to
lactate, is commonly enhanced in cancer and is a malignant
adaptation that allows for continued cell proliferation in diverse
brain tumor microenvironments (11). Lactic acid, the final product of the
glycolysis pathway, can only accumulate in cells or be transported
out of cells by specific transporters. Intracellular lactic acid
content can therefore act as one of the principal biochemical
markers of the intracellular glycolysis rate (12). Glucose transporter (GLUT) is
required for the transport of glucose into cells and hexokinase
(HK) is required for glucose phosphorylation. Next, the
phosphorylated glucose is isomerized into fructose, whereas
phosphofructokinase (PFK) is responsible for the phosphorylation of
fructose. Following a series of reactions, pyruvate kinase (PK)
catalyzes ADP to generate ATP (13–15).
Compared with healthy cells, the altered metabolism of cancer cells
means they depend heavily on aerobic glycolysis for proliferation.
Interference with tumor-specific glycolysis-related enzymes is a
potential and promising method for the treatment of glioma
(16). The present study therefore
aimed to explore the mechanism of aerobic glycolysis in human
glioma cell proliferation.
Long noncoding RNAs (lncRNAs) have been identified
as promising biomarkers that serve as oncogenes or tumor
suppressors, with lncRNA dysregulation being a necessary step in
tumor growth and metastasis (17).
Furthermore, lncRNAs are involved in the progression of a wide
range of glioma neoplasms, regulating cell cycle progression,
mediating apoptotic pathways and changing cancer cell behavior
(18). Long intergenic non-protein
coding RNA (LINC) 01138 is a known oncogene, and is highly
expressed in hepatocellular cancer, prostate cancer and clear cell
renal cell carcinoma (ccRCC). LINC01138 expression is also closely
correlated with poor overall survival rates (19). However, the expression patterns and
function of LINC01138 during glioma progression remain largely
unknown. Therefore, the present study attempted to determine the
role and underlying mechanisms of LINC01138 in glioma.
Salmena et al (20) demonstrated that lncRNAs may act as
competing endogenous RNAs (ceRNAs), interacting with microRNAs
(miRs) in human cancer. A recent study reported that LINC01138 may
facilitate gastric cancer progression by sponging miR-1273e
(21). However, to the best of our
knowledge, the ceRNA network of LINC01138 has not been reported.
Furthermore, miRs have been demonstrated to be promotors or
inhibitors of different types of cancer, including glioma (22). Evidence has shown that miR-375
expression is often downregulated in various types of cancer
(23–25). Notably, the ceRNA interaction
between miR-375 and lncRNAs could modulate the pathogenesis and
development of colon adenocarcinoma and triple-negative breast
cancer (26,27). Several studies have demonstrated
that the lncRNA/miR/mRNA axis serves a role in colorectal cancer,
ovarian cancer and thyroid carcinoma and thus affects cancer
progression (28–30). Furthermore, specificity protein 1
(SP1) upregulation has been reported to enhance glioma self-renewal
and to predict a poor prognosis (31). As a downstream gene in the
lncRNA/miR/mRNA axis, SP1 has been determined to be regulated by
lncRNA-miR crosstalk in various types of cancer, including
non-small cell lung cancer and lung adenocarcinoma (32,33).
SP1 is involved in the occurrence and malignancy of multiple
neoplasms; however, how it functions in glioma remains unclear
(34). It was therefore
hypothesized that there may be crosstalk between LINC01138, miR-375
and SP1 in glioma. The present study aimed to explore the effect of
LINC01138, miR-375 and SP1 in glioma.
Materials and methods
Ethical standards
The present study was approved and supervised by the
Ethics Committee of The First Affiliated Hospital of Soochow
University (Suzhou, China; approval no. SDU-MED-2017-085), and was
in accordance with the Declaration of Helsinki. Animal experiments
were approved by the Animal Ethics Committee of The First
Affiliated Hospital of Soochow University (approval no.
S20200616018) and followed the guidelines of Animal Research:
Reporting of In Vivo Experiments 2.0 (35).
Tissue sample collection
Between January 2018 and January 2019, 108 pairs (54
males and 54 females) of glioma tissues and adjacent normal brain
tissues were collected at The First Affiliated Hospital of Soochow
University. According to the World Health Organization (WHO) tumor
grade criteria (36), 62 tissue
samples were grade I + II tumors, and 46 tissue samples were grade
III + IV tumors. No patient received any treatment prior to sample
collection. The extracted tissues were immediately frozen and
stored in liquid nitrogen.
Cell culture
Immortalized normal human astrocytes (NHAs)
HASTR/ci35, human glioma cell lines (A172, U251MG, TJ899 and TJ905
cells) and 293T cells (all purchased from The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences), were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS;
Thermo Fisher Scientific, Inc.) and penicillin (100
µg/ml)-streptomycin (100 U/ml) at 37°C in 5% CO2. Cells
were detached using 0.25% trypsin and subcultured at a ratio of
1:3. Cells were then seeded into 6-well plates (3×105
cells/well). When the cells reached 70–80% confluence, they were
considered to be in the logarithmic growth phase and were selected
for subsequent experiments.
Cell grouping and transfection
Cells in the logarithmic growth phase were seeded
into 6-well plates (2×105 cells/well). When the cells
had attached to the well and reached 30–60% confluence they were
transfected using Lipofectamine 2000® (cat. no.
11668-027; Invitrogen; Thermo Fisher Scientific, Inc.). Each
transfection construct (50 nM; Shanghai GenePharma Co., Ltd.) was
diluted using 250 µl serum-free RPMI-1640 (Thermo Fisher
Scientific, Inc.) medium, mixed and incubated at room temperature
for 5 min; 5 µl 2000 was prepared in a similar manner.
Subsequently, the transfection constructs and 2000 were mixed and
incubated at room temperature for 20 min, prior to being added to
the cells in the 6-well plates for 6 h at 37°C with saturated
humidity and 5% CO2. The medium containing the
transfection solution was then replaced with RPMI-1640 media
containing 10% FBS at 37°C for 48 h. Cells were divided into the
following groups: i) Control group (U251MG or TJ905 cells receiving
no treatment); ii) short hairpin RNA (sh)-negative control (NC)
group (U251MG or TJ905 cells transfected with the pGPU6-sh-NC
plasmid); iii) sh-LINC01138 group (U251MG or TJ905 cells
transfected with the pGPU6-sh-LINC01138 plasmid, three shRNAs); iv)
sh-LINC01138 + inhibitor-NC group (U251MG cells transfected with
sh-LINC01138 and inhibitor-NC); and v) sh-LINC01138 + miR-375
inhibitor group (U251MG cells transfected with sh-LINC01138 and
miR-375 inhibitor plasmids). All vectors were purchased from
Shanghai GenePharma Co., Ltd., which inserted the shRNAs into the
plasmids For each transfection, 1 µg of each construct was added
into each well. The sequences are shown in Table SI.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
Cells from each group were seeded into 96-well
plates (4×103 cells/well). The Cell-Light EdU Apollo488
In Vitro kit (cat. no. 100T; (Guangzhou RiboBio Co., Ltd.)
was used to assess cell proliferation until the cells reached 80%
confluence. Briefly, cells were incubated at 37°C for 2 h with 100
µl RPMI-1640 medium (50 µM the EdU solution was diluted in medium
at a ratio of 1:1,000). Cells were subsequently washed with
phosphate-buffered saline (PBS) twice (5 min/wash), fixed using 50
µl 4% paraformaldehyde at room temperature for 30 min and incubated
with 50 µl glycine (2 mg/ml) at room temperature for 5 min. Cells
were then washed with PBS for 5 min, before being incubated with
100 µl 0.5% Triton X-100 for 10 min at room temperature. Cells were
once again washed with PBS for 5 min. Subsequently, the cells were
incubated with 100 µl 1X Apollo® staining reaction
solution in the dark for 30 min at room temperature (25°C),
permeated, and decolored with methyl alcohol. After the nuclei were
stained with 4′,6-diamidino-2-phenylindole at room temperature for
5 min, the cells were observed under a confocal microscope (Leica
Microsystems GmbH).
Cell Counting Kit-8 (CCK-8) assay
U251MG or TJ905 cells were seeded into 96-well
plates (1×103 cells/well) and incubated in 100 µl medium
containing 10% FBS. The cell proliferation was quantified after 24,
48 and 72 h using a CCK-8 Kit (Dojindo Molecular Technologies,
Inc.) according to the manufacturer's protocol. Briefly, 10 µl
CCK-8 reagent was added to each well for 1 h. The optical density
was determined at 490 nm using a microplate reader.
Assessment of glucose uptake and
lactate production
Glucose uptake was assessed as previously described
(37). Briefly, U251MG or TJ905
cells were incubated at 35°C in Hank's balanced salt solution
(HBSS; 137 mM NaCl, 5.36 mM KCl, 1.26 mM CaCl2, 0.41 mM
MgSO4, 0.49 mM MgCl2, 0.63 mM
Na2HPO47H2O, 0.44 mM
KH2PO4, 4.17 mM NaHCO3 and 5.55 mM
glucose; pH 7.2). The assay was initiated by the addition of 0.1 mM
μCi/well D-[3-3H] glucose. After 15 min, the culture medium was
removed and the cells were washed twice with cold HBSS to terminate
the reaction. Cells were then lysed in a 0.5 M NaOH solution and
analyzed using a scintillation counter (LS6500 Multipurpose
Scintillation Counter; Beckman Coulter, Inc.). Each experiment was
repeated three times.
Cell supernatant lactate levels were measured using
a Lactate Colorimetric Assay kit (BioVision, Inc.). Briefly, cells
(5×105 per dish) were seeded into 60-mm culture dishes
and incubated overnight in DMEM containing 10% FBS at 37°C.
Subsequently, the medium was replaced with serum-free DMEM for 1–2
h, the supernatant was collected, and the lactate levels were
quantified using colorimetry according to the manufacturer's
protocol.
Bioinformatics analysis
The TCGA data visualization website GEPIA
(http://gepia.cancer-pku.cn/index.html) (38) was used to analyze the expression of
LINC01138 and Sp1 in glioma (n=163). The online prediction
software, lncLocator, developed by the pattern recognition and
bioinformatics research group of Shanghai Jiaotong University
(http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/)
(39) was used to predict the
subcellular localization of LINC01138, whereas the RNA22 tool
(http://cm.jefferson.edu/rna22/Precomputed/) (40) was used to predict the binding sites
of LINC01138 and miR-375. The TargetScan website (http://www.targetscan.org/vert_71/) (41) was used to predict the target gene of
miR-375. The lncLocator and RNA22 software were used according to
the default settings. The following FASTA files were obtained from
GenBank (https://www.ncbi.nlm.nih.gov/): LINC01138 (accession
no. NR_027468.3) and miR-375 (accession no. NR_029867.1).
Dual-luciferase reporter assay
Wild-type (WT) and mutant (MUT) miR-375 and SP1
3′-untranslated regions were synthesized and cloned into
PmiR-RB-REPORT™ plasmids (Guangzhou RiboBio Co., Ltd.). NC plasmids
(empty vectors) were used as a control. Following sequencing
confirmation of the WT and MUT plasmids by Sangong Bioengineering
(Shanghai) Co., Ltd., the 293T cell line (1×105
cells/well in 24-well plates) was co-transfected with the WT or MUT
plasmids (50 nM) and the mimic-NC or miR-375 mimic (50 nM) using
Lipofectamine 2000®. Following incubation for 48 h at
37°C, cells were collected and lysed with ice-cold
radioimmunoprecipitation assay (RIPA) lysis buffer (Beijing
Solarbio Science & Technology Co., Ltd.) containing 1 mmol/l
PMSF), followed by centrifugation for 3–5 min at 1,200 × g 4°C. The
cell supernatant was harvested to quantify relative luciferase
units (RLUs) using the Firefly Luciferase Reporter Gene Assay Kit
(cat. no. RG005; Beyotime Institute of Biotechnology) according to
the manufacturer's protocol. The relative fluorescence values were
obtained by dividing the RLU value of Firefly luciferase activity
by that of Renilla luciferase activity. All experiments were
repeated three times.
Subcellular fractionation
localization
Following a previous method (42), nuclear and cytoplasmic components
were separated using the PARIS™ Kit (Thermo Fisher Scientific,
Inc.). U251MG cells were harvested, washed with PBS, detached with
trypsin, and centrifuged at 500 × g at 4°C for 5 min. The
supernatant was discarded and the precipitates were washed using
PBS and 500 µl cell fractionation buffer added with gentle
agitation. Cells were incubated on ice for 5–10 min and centrifuged
at 500 × g at 4°C for 5 min. The supernatant (cytoplasm) was
transferred to a sterile nonenzymatic tube (2 ml) and centrifuged
at 500 × g at 4°C for 5 min. The precipitates (nuclei) were added
to 500 µl cell fractionation buffer and mixed via gentle agitation
before the addition of 500 µl 2X lysis/binding solution and another
gentle agitation at room temperature before being placed on ice for
5 min. Cells were mixed with 500 µl precooled cell disruption
buffer followed by 500 µl absolute ethyl alcohol. An adsorption
column was placed in the tube, 700 µl reaction solution was added,
and the tube was centrifuged at 4°C at 12,000 × g for 30 sec; the
solution in the tube was removed and these steps were repeated.
Subsequently, 40 µl elution solution was added and the samples were
centrifuged at 4°C at 12,000 × g for 3 sec. A second elution step
was performed using 10 µl elution solution. LINC01138 expression
was detected by reverse transcription-quantitative PCR (RT-qPCR).
GAPDH and U6 were used as cytoplasmic and nuclear internal
reference genes, respectively.
RNA immunoprecipitation (RIP)
assay
Following the manufacturer's instructions, Magna
RIP™ RNA-Binding Protein Immunoprecipitation Kit (cat. no. 17-700;
Merck KGaA) was used to determine the binding relationship between
LINC01138, miR-375 and Ago2. Glioma cells (U251MG,
1.0×107 cells) were washed with precooled PBS to remove
media. Cells were then lysed with ice-cold radioimmunoprecipitation
assay (RIPA) lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.; RIPA containing 1 mmol/l PMSF) and
centrifuged at 230,000 × g at 4°C for 10 min to remove the
supernatant. Co-precipitation was performed by washing and
resuspending 50 µl magnetic beads from each co-precipitation
reaction system in 100 µl RIP wash buffer, before adding 5 µg
antibody and incubating for 30 min at room temperature. The
bead-antibody complexes were washed and resuspended in 900 µl RIP
wash buffer, and 100 µl cell extraction solution was added and
incubated overnight at 4°C. Samples were placed on a magnetic seat
to collect the bead-protein complexes. RNA was extracted from the
samples following proteinase K detachment and used for subsequent
RT-qPCR. The antibodies used in the RIP assays were rabbit
anti-Ago2 (1:50; cat. no. ab186733; Abcam) and the NC rabbit
anti-immunoglobulin G (IgG) (1:100; cat. no. ab109489; Abcam). Each
experiment was repeated three times.
RNA pull-down assay
Cells were transfected with 50 nM biotin-labeled WT-
miR-375 and MUT-miR-375 (Wuhan Genecreate Bioengineering Co.,
Ltd.). Following 48 h of incubation at 37°C, the cells were
harvested, washed with PBS and incubated in specific lysis buffer
(800 µl; Ambion; Thermo Fisher Scientific, Inc.) for at 4°C 10 min.
After centrifugation at 20,000 × g at 4°C for 10 min, the
supernatant was collected. The lysate was incubated with M-280
MagneSphere/Streptavidin beads (50 µl; 6×108 magnetic
beads/ml; cat. no. 60210; Invitrogen; Thermo Fisher Scientific,
Inc.) precoated with RNase-free bovine serum albumin (0.1% BSA) and
the enzyme tRNA (TRNABAK-RO; MilliporeSigma), and incubated at 4°C
overnight. Subsequently, the magnetic beads were collected on the
magnetic frame and the supernatant was removed the beads were
washed twice with precooled lysis buffer (800 µl), three times with
low-salt buffer (800 µl; 0.1 M NaOH and 0.05 M NaCl) and once with
high-salt buffer (800 µl; 0.1 M NaCl). Following washing, the
magnetic beads were collected on the magnetic frame and the
supernatant was removed and the beads were resuspended in the
low-salt buffer. Following extraction of the bound RNA using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
LINC01138 enrichment was assessed via RT-qPCR. All experiments were
repeated three times.
RT-qPCR
Total RNA was extracted from cells and tissues using
TRIzol reagent. The RNA concentration and purity were determined,
and the extracted total RNA was reverse transcribed into cDNA using
the RevertAid First Strand cDNA Synthesis Kit (cat. no. K1621;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The qPCR primers (Shanghai Genechem Co., Ltd.) specific
for LINC01138, miR-375 and SP1 are shown in Table I. Premix Ex Taq Probe qPCR; Takara
Biotechnology Co., Ltd.) was performed to assess the expression
levels of each gene. Each qPCR reaction contained: 5.3 µl 2X Taq
Master Mix, 1 µl forward primer (5 µM), 1 µl reverse primer (5 µM),
1 µl cDNA and 11.7 µl RNase-free H2O. The following thermocycling
conditions were used for qPCR: Expression levels were quantified
using a qPCR instrument (Applied Biosystems 7500; Applied
Biosystems Thermo Fisher Scientific, Inc.). mRNA expression levels
were quantified using the 2−ΔΔCq method (43) and normalized to the internal
reference genes; U6 for miR-375, and GAPDH for LINC01138 and SP1.
All the experiments were repeated three times.
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5–3′) |
|---|
| LINC01138 | F:
ACATCGTGAGCACATTTGAGA |
|
| R:
TCTTGCTGTTCAGGGTGGTA |
| miR-375 | F:
TCGCACAAACGTCGTATCCA |
|
| R:
GTATCCAGTGCGTGTCGTGG |
| SP1 | F:
TTGAAAAAGGAGTTGGTGGC |
|
| R:
TGCTGGTTCTGTAAGTTGGG |
| U6 | F:
CGCTTCACGAATTTGCGTGTCAT |
|
| R:
GCTTCGGCAGCACATATACTAAAAT |
| GAPDH | F:
TCCCATCACCATCTTCCA |
|
| R:
CATCACGCCACAGTTTTCC |
Western blotting
Protein expression levels were determined via
western blotting. To extract total protein, tissues were harvested
and placed in centrifuge tubes, 100 µl RIPA lysis solution was
added (Beijing Solarbio Science & Technology Co., Ltd.; RIPA
containing 1 mmol/l PMSF), and the samples were centrifuged at 4°C
at 11,000 × g for 20 min until fully lysed. After lysis the tissues
were incubated on ice for 30 min at 4°C and centrifuged at 12,000 ×
g for 4 min at 4°C. The supernatant was then extracted and stored
at −80°C. Total protein was quantified using a BCA Protein Assay
Kit (Boster Biological Technology), and was adjusted to 3 µg/µl.
The extracted proteins were boiled with loading buffer at 95°C for
10 min, and 30 µg protein/lane was separated by SDS-PAGE on a 10%
gel (Beyotime Institute of Biotechnology). The separated proteins
were subsequently transferred onto a polyvinylidene fluoride
membrane (MilliporeSigma) and blocked with 5% BSA (Invitrogen;
Thermo Fisher Scientific, Inc.) for 1 h at room temperature. The
membranes were incubated with primary antibodies against the
following: HK (1:1,000; cat. no. ab209847; Abcam), PK (1:1,000;
cat. no. ab32566; Abcam), PFK (1:2,000; cat. no. ab204131; Abcam),
GLUT (1:100,000; cat. no. ab115730; Abcam), SP1 (1:10,000; cat. no.
ab231778; Abcam) and GAPDH (1:10,000; cat. no. ab181602; Abcam) at
4°C overnight. Membranes were washed with Tris-buffered
saline-Tween-20 (0.05%) three times (5 min/wash). Following the
primary antibody incubation, membranes were incubated with the
corresponding goat anti-rabbit HRP-labelled secondary antibody
(1:2,000; cat. no. ab6721; Abcam) for 1 h at room temperature.
Membranes were subsequently washed three times (5 min/wash).
Protein bands were visualized by enhanced chemiluminescence Pierce™
ECL Western Blotting Substrate (cat. no. 32209; Thermo Fisher
Scientific, Inc.). A Bio-Rad Gel Dol EZ imager (Bio-Rad
Laboratories, Inc.) was used for band development. The gray values
of the target bands were semi-quantified using ImageJ (version
1.48; National Institutes of Health) with GAPDH as the loading
control.
Xenograft tumors in nude mice
A total of 16 female BALB/c nude mice (age, 3–4
weeks; weight, 14–18 g; Nanjing Junke Biological Engineering Co.,
Ltd.) were raised in a specific pathogen-free environment at
18–22°C, with a humidity of 50–60% under a 12-h light/dark cycle
and fed sterile food. All mice had free access to food and water.
The mice were randomly divided into the sh-NC group (mice injected
with U251MG cells stably transfected with sh-NC) and the
sh-LINC01138 group (mice injected with U251MG cells stably
transfected with sh-LINC01138). After skin disinfection, each mouse
was injected with 0.2 ml cell suspension (1.0×106 cells)
via the axilla (44). The general
behavior of the mice and the condition of the local injection site
were observed. Tumor volume (V) was assessed using Vernier calipers
every other week, and was calculated as V = length ×
width2 × 0.5 (42). Mice
were euthanized through excessive administration of sodium
pentobarbital (100 mg/kg) after ~6 weeks. Following euthanasia,
tumors were extracted and quantified. The humane endpoints of the
study were when the mice experienced weight loss of >15% of
their total weight, or mice suffered from tumor load. In the
present study, when the maximum tumor length and width were as
follows: Tumor length, 1.55 cm; and width, 1.12 cm, the mice were
sacrificed.
Immunohistochemical staining
Tumor tissues from each group of mice were washed
and dehydrated in 70, 80 and 90% ethanol solution. Then, the
tissues were placed in the same amount of mixed pure alcohol and
xylene for 15 min and in xylene I and xylene II for 15 min each
until clear followed by mixed solution of xylene and paraffin in
equal amount for 15 min and in paraffin I and II for 50–60 min
each. Next, the tissues were paraffin-embedded and sliced at 5 µm,
warmed, dewaxed and dehydrated. All operating temperatures are room
temperature and reagents are from Sigma. Sections were washed with
running water for 2 min, incubated in 3%
H2O2-methyl alcohol for 20 min, washed with
distilled water for 2 min and washed with 0.1 M PBS for 3 min.
Sections were recovered in in a water bath (95–100°C) in the
antigen retrieval solution (1 mM EDTA; pH 8.0), cooled in running
water, and incubated with normal goat serum sealant (Shanghai
Haoran Biotechnology Co., Ltd.) at room temperature for 20 min.
When the sections had dried, they were incubated with primary
antibodies targeting SP1 (1:1,000: cat. no. ab231778; Abcam) at 4°C
overnight. Sections were washed three times in 0.1 M PBS (5
min/wash) and were incubated with goat anti-rabbit IgG (1:2,000:
cat. no. ab6721; Abcam) secondary antibody at 37°C for 20 min.
Sections were washed three times in PBS (5 min/wash) and were
incubated with a HRP-labeled streptavidin working solution (Imunbio
Biotechnology Co., Ltd.) at 37°C for 20 min. Subsequently, sections
were subjected to three washes in PBS (5 min/wash), stained by DAB
at room temperature for 2 min (Guangzhou Whiga Technology Co.,
Ltd.), washed with running water, counterstained with hematoxylin
(Shanghai Bogoo Biological Technology Co., Ltd.) for 1 min at room
temperature, washed with running water and reversed to blue
staining with 1% ammonium hydroxide, followed by another wash in
running water. Sections were dehydrated with an ascending ethanol
series, treated with xylene, sealed with neutral resin and observed
under a light microscope. To determine the number of positive cells
in each field, five high-power fields were randomly chosen from
each section (45).
Statistical analysis
SPSS version 21.0 (IBM Corp.) was used for data
analysis. All experiments were repeated 3 times. Data are presented
as the mean ± standard deviation. The data were normally
distributed. Student's t-test was used for comparisons between two
groups, whereas one-way or two-way analysis of variance (ANOVA) was
used for comparisons among more than two groups, followed by
Tukey's post hoc multiple comparisons test. P<0.05 was
considered to indicate a statistically significant difference.
Results
LINC01138 is highly expressed in human
glioma tissues and cells, with high expression being associated
with clinicopathological features
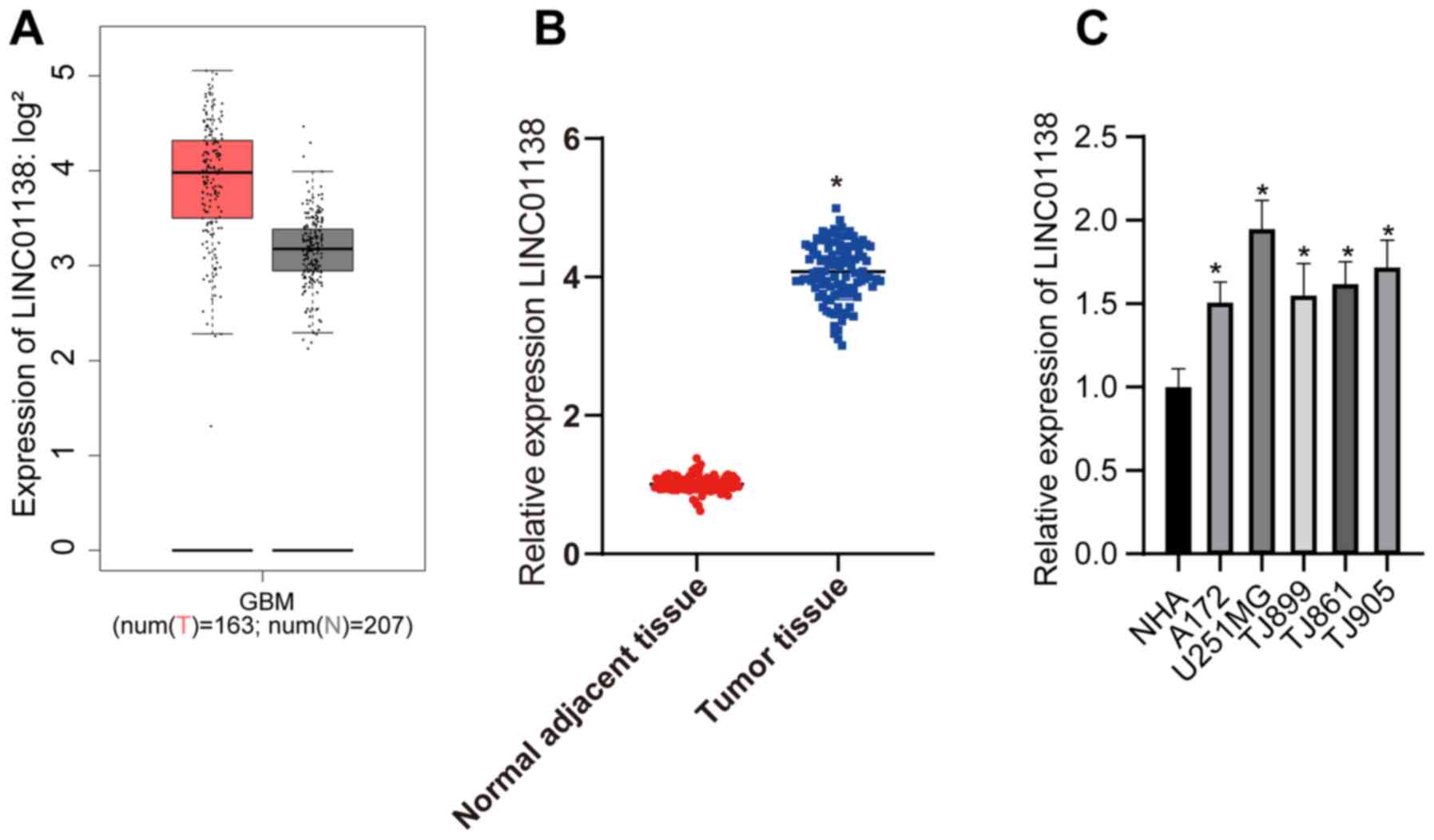
Analysis of LINC01138 expression in human glioma
tissue using The Cancer Genome Atlas revealed that LINC01138 was
markedly more highly expressed compared with that in healthy tissue
(Fig. 1A). In the present study,
LINC01138 expression was detected in the glioma tissues and
adjacent normal tissues of 108 patients with glioma. The RT-qPCR
results demonstrated that LINC01138 expression levels were
significantly higher in glioma tumor tissues compared with those in
adjacent normal tissues (Fig. 1B;
P<0.05). Furthermore, LINC01138 expression levels were
significantly elevated in human glioma cell lines compared with
those in NHAs (Fig. 1C; P<0.05).
Analysis of the relationship between LINC01138 expression and the
clinicopathological features of patients with glioma indicated that
LINC01138 expression was not associated with the age and sex of
patients (P>0.05), but was associated with WHO tumor grade,
lymph node metastasis and tumor diameter (all P<0.05; Table II).
 | Table II.LINC01138 expression and
clinicopathological features of patients with glioma. |
Table II.
LINC01138 expression and
clinicopathological features of patients with glioma.
| Clinicopathological
parameters | LINC01138 relative
expression levels | P-value |
|---|
| Age, years |
| >0.05 |
|
<50 | 4.10±0.41 |
|
|
≥50 | 4.07±0.41 |
|
| Sex |
| >0.05 |
|
Male | 4.02±0.39 |
|
|
Female | 4.14±0.42 |
|
| WHO grading |
| <0.05 |
|
I+II | 3.60±0.28 |
|
|
III+IV | 4.24±0.30 |
|
| Lymph node
metastasis |
| <0.05 |
|
Yes | 4.23±0.31 |
|
| No | 3.76±0.40 |
|
| Tumor diameter |
| <0.05 |
| ≤5
cm | 3.72±0.37 |
|
| >5
cm | 4.24±0.31 |
|
Silencing LINC01138 inhibits glioma
cell proliferation
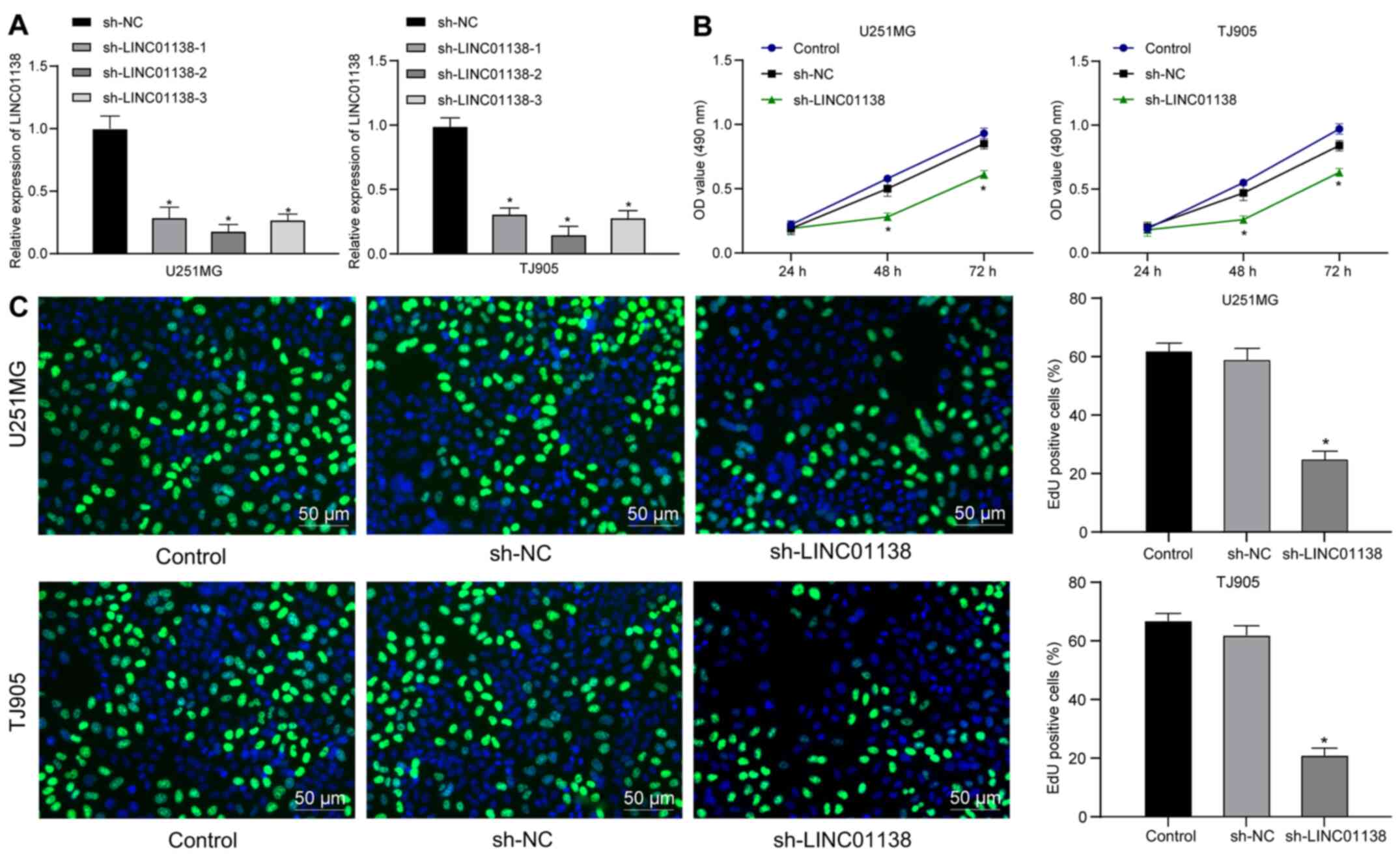
To determine the role of LINC01138 in glioma cell
proliferation, three sh-LINC01138 plasmids were constructed and
transfected into U251MG and TJ905 cells, which expressed high
levels of LINC01138. The sh-plasmids significantly reduced
LINC01138 expression. sh-LINC01138-2 exhibited the highest
silencing efficiency and was therefore selected for further
experiments (Fig. 2A). The
proliferation of U251MG and TJ905 cells with sh-LINC01138 was
determined using CCK-8 and EdU assays. The results demonstrated
that proliferation was significantly reduced in
sh-LINC01138-transfected U251MG and TJ905 cells compared with that
in sh-NC-transfected cells (Fig. 2B and
C; all P<0.05), suggesting that silencing LINC01138
inhibited human glioma cell proliferation.
Silencing LINC01138 inhibits aerobic
glycolysis in glioma cells
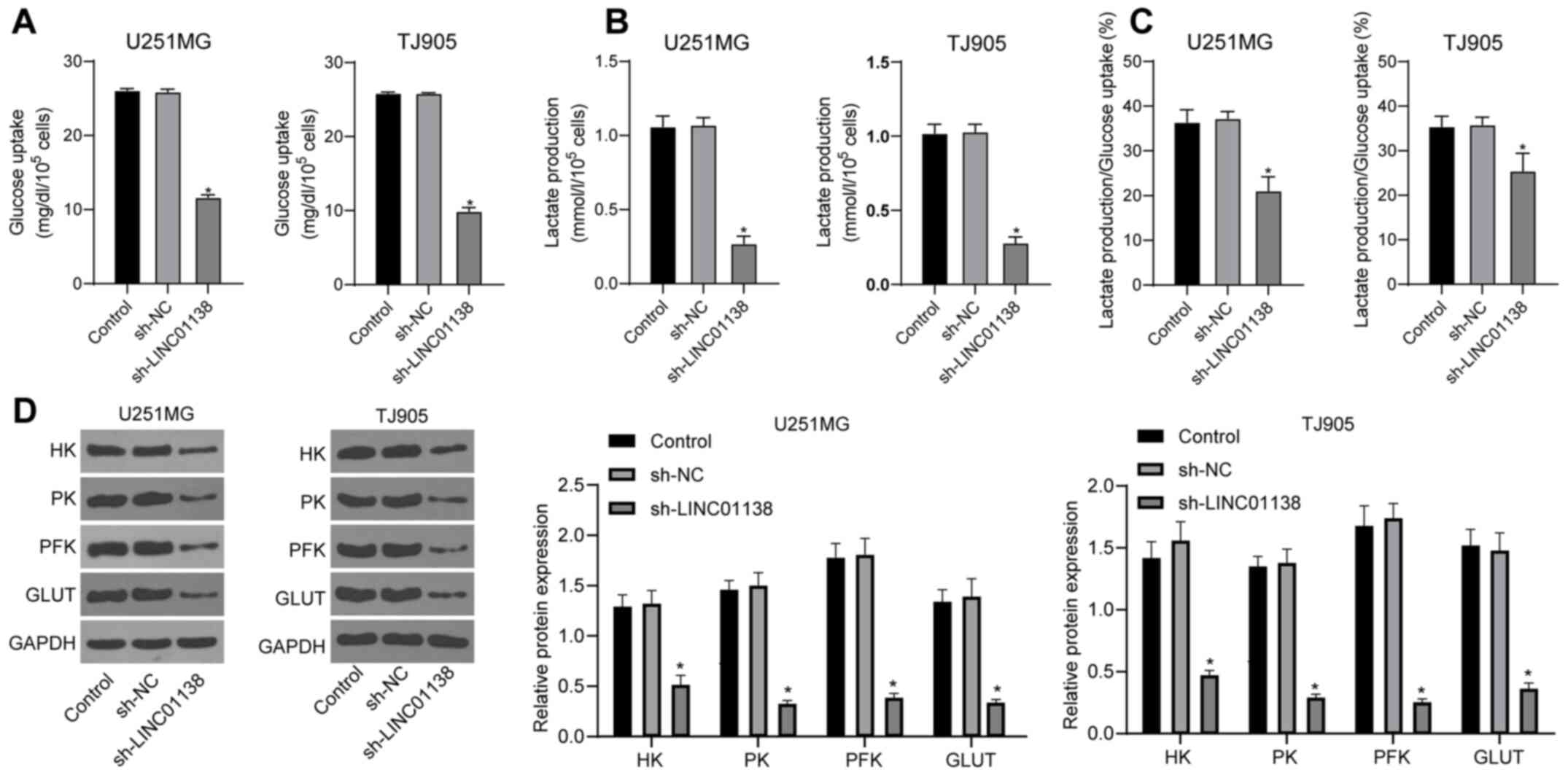
To identify the effect of LINC01138 on aerobic
glycolysis in glioma cells, cell glucose uptake and lactic acid
secretion were analyzed. The results demonstrated that both glucose
uptake and lactic acid secretion by U251MG and TJ905 cells were
significantly reduced by sh-LINC01138 compared with sh-NC (Fig. 3A and B; P<0.05). The ratio
between glucose uptake and lactic acid secretion was 1:2 during
glycolysis (46,47). Lactic acid secretion was used to
calculate the proportion of the total glucose taken up by cells
that was consumed by glycolysis. The results indicated that the
amount of glucose consumed by glycolysis, relative to the total
glucose taken up by the cells, was significantly decreased in the
sh-LINC01138 group compared with that in the sh-NC group (Fig. 3C; P<0.05). Moreover, the
expression levels of glycolysis-associated enzymes, including HK,
PK, PFK and GLUT, were detected via western blotting. The results
demonstrated that HK, PK, PFK and GLUT protein expression levels
were all significantly reduced in the sh-LINC01138 group compared
with those in the sh-NC group (Fig.
3D; P<0.05). These data indicated that silencing LINC01138
could aberrantly induce aerobic glycolysis in human glioma
cells.
LINC01138 modulates miR-375 by acting
as a ceRNA
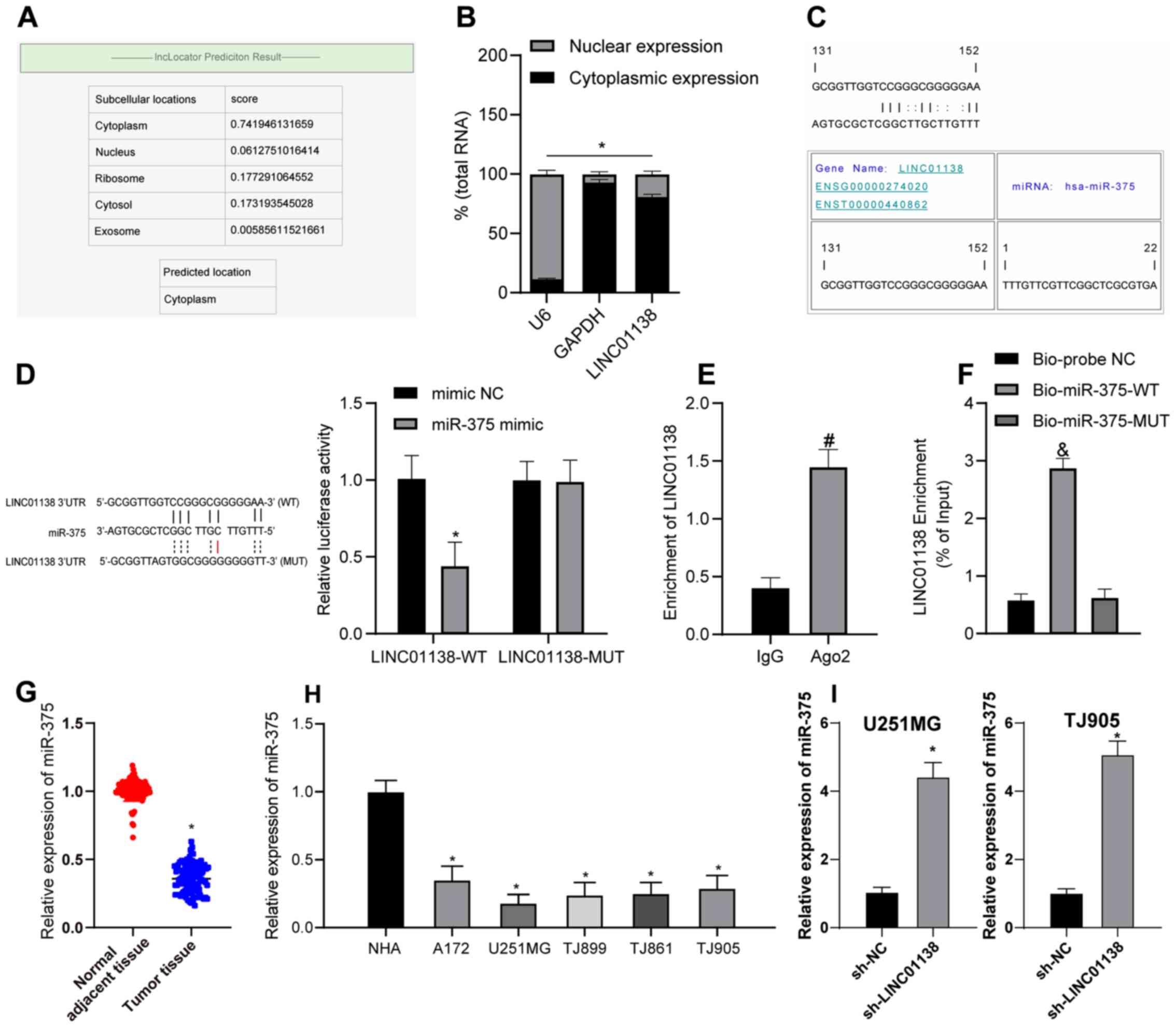
To further investigate the function of LINC01138,
its subcellular localization was predicted using lncLocator
(39). LINC01138 was determined to
be mainly localized to the cytoplasm (Fig. 4A). Analysis of the RNA in nuclear
and cytoplasmic fractions demonstrated LINC01138 was principally
distributed in the cytoplasm of U251MG (Fig. 4B) and TJ905 cells (data not shown).
These results suggested that LINC01138 served a role in glioma via
the ceRNA network by competitively binding to miRNA. RNA22, a miRNA
target discovery tool, predicted that there was a binding site
between LINC01138 and miR-375 (Fig.
4C). Furthermore, the dual-luciferase reporter assay confirmed
that, compared with in the mimic NC group, the miR-375 mimic group
exhibited significantly decreased luciferase activity following
transfection with LINC01138-WT. Luciferase activity following
transfection with LINC01138-MUT and the miR-375 mimic did not
change compared with the mimic NC (Fig.
4D). The RIP assay (Fig. 4E)
demonstrated the Ago2-targeted antibody precipitated LINC01138 and
miR-375 at significantly higher levels compared with IgG,
indicating the binding of LINC01138 and miR-375. Furthermore,
compared with the Bio-probe NC group, the RNA pull-down assay
(Fig. 4F) demonstrated that the
Bio-miR-375-WT group had significantly increased LINC01138
expression levels, whereas the Bio-miR-375-MUT group showed no
marked difference in expression. These results suggested that
miR-375 could possibly directly bind to LINC01138. The RT-qPCR
results demonstrated that miR-375 expression levels were
significantly lower in glioma tissues compared with those in
adjacent normal tissues (Fig. 4G;
P<0.05). miR-375 was also expressed at significantly lower
levels in human glioma cell lines compared with in NHAs (Fig. 4H; P<0.05). Furthermore, miR-375
expression in U251MG and TJ905 cells in which LINC01138 was knocked
down was measured by RT-qPCR. Compared with in the sh-NC group, the
sh-LINC01138 group exhibited significantly elevated miR-375
expression levels (Fig. 4I;
P<0.05). Overall, LINC01138 may competitively bind to miR-375 to
inhibit miR-375 expression in glioma.
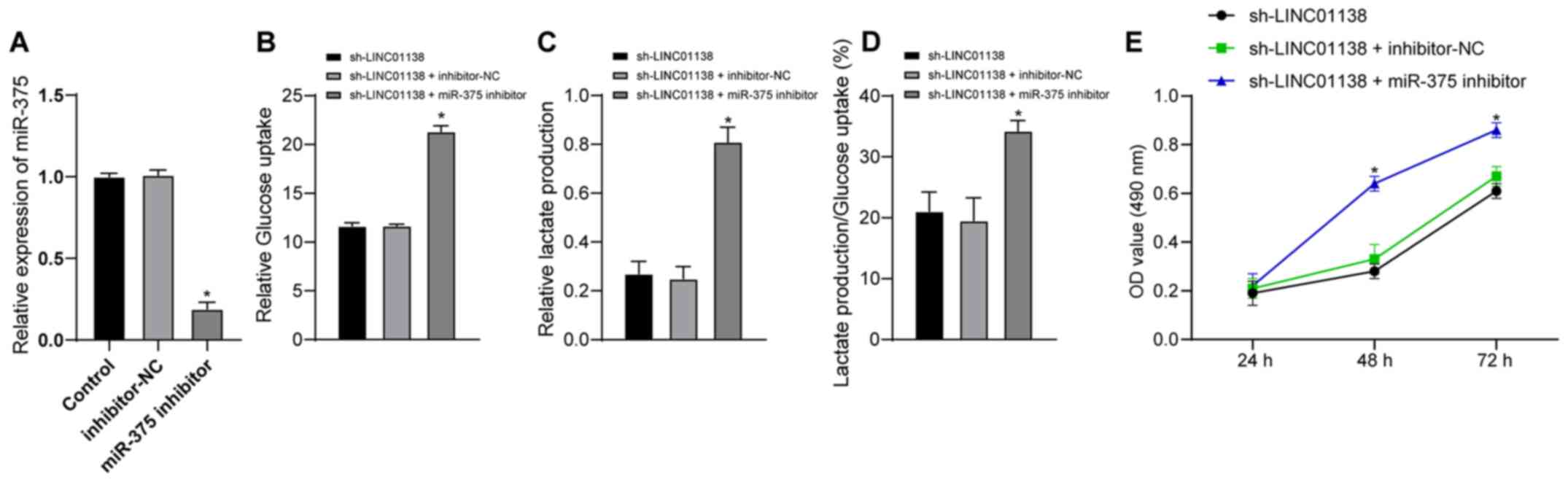
miR-375 downregulation reverses the
role of LINC01138 silencing in promoting aerobic glycolysis and
cell proliferation in human glioma
To further investigate whether LINC01138 could
regulate the function of glioma cells via miR-375, a functional
rescue assay was conducted. U251MG cells were divided into the
sh-LINC01138 group, the sh-LINC01138 + inhibitor-NC group and the
sh-LINC01138 + miR-375-inhibitor group. The transfection efficiency
of miR-375 inhibitor was verified by RT-qPCR. Inhibitor NC had no
effect on the expression of miR-375 in cells, but miR-375 inhibitor
significantly decreased the expression of miR-375 in cells compared
with inhibitor NC (Fig. 5A;
P<0.05). Compared with in the sh-LINC01138 group, the
sh-LINC01138 + miR-375-inhibitor group exhibited significantly
increased glucose uptake and lactic acid secretion, and a
significantly increased proportion of glucose consumed in
glycolysis relative to the total glucose uptake (Fig. 5B-D; P<0.05). The CCK-8 assay
demonstrated that the miR-375 inhibitor significantly reversed the
inhibitory effect of sh-LINC01138 on glioma cell proliferation
compared with the sh-LINC01138 group (Fig. 5E; P<0.05). These findings
indicated that miR-375 inhibition reversed the effect of
sh-LINC01138 on inhibiting abnormal aerobic glycolysis and cell
proliferation in human glioma.
LINC01138 can promote SP1 expression
by competing with SP1 for binding to miR-375
SP1 expression in human glioma tissues from TCGA was
analyzed; the results demonstrated that SP1 was significantly more
highly expressed in human glioma compared with that in healthy
tissue (Fig. 6A; P<0.05).
TargetScan predicted that there was a binding site between miR-375
and SP1 (Fig. 6B). The
dual-luciferase reporter assay (Fig.
6C) demonstrated that compared with the mimic NC group, the
luciferase activity in the binding region of SP1-WT and miR-375 in
the miR-375 mimic group was inhibited, but there was no significant
difference in the luciferase activity of SP1-MUT, indicating that
miR-375 could specifically bind to SP1 (P<0.05). It was
therefore hypothesized that LINC01138 might act as a ceRNA,
sponging miR-375 to upregulate SP1 expression and eventually
exacerbate the malignancy of glioma. To confirm this hypothesis,
RT-qPCR was conducted to assess SP1 mRNA expression levels in both
human glioma tissues and adjacent normal brain tissues. Compared
with those in adjacent normal tissues and normal cells, glioma
tissues and cells displayed significantly elevated SP1 mRNA
expression levels (Fig. 6D and E;
P<0.05). RT-qPCR and western blotting demonstrated that SP1 mRNA
and protein expression levels in U251MG cells in the sh-LINC01138
group were significantly lower than those in the sh-NC group.
Furthermore, SP1 mRNA and protein expression levels were recovered
in the sh-LINC01138 + miR-375-inhibitor group compared with those
in the sh-LINC01138 + inhibitor-NC group (Fig. 6F and G; P<0.05). In summary,
LINC01138 may function as a ceRNA to sponge and suppress miR-375
expression thus increasing SP1 expression.
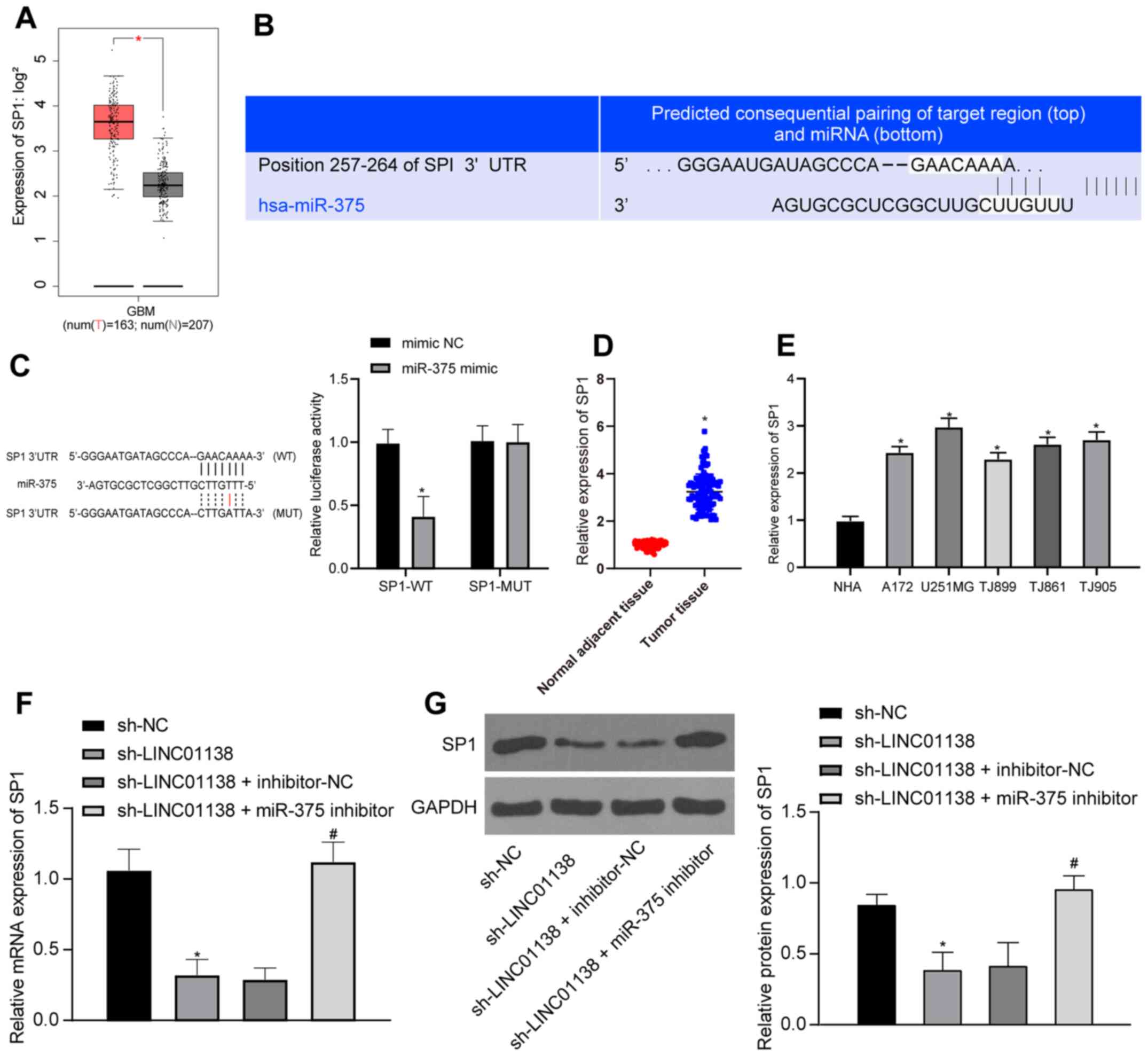 | Figure 6.LINC01138 can promote SP1 expression
by competing with SP1 for binding to miR-375. (A) SP1 expression
data in human glioma tissues from The Cancer Genome Atlas database,
red represents tumor and grey normal tissue and SP1is highly
expressed in tumor tissue. (B) miR-375 and SP1 binding site was
predicted using TargetScan software. (C) miR-375 and SP1 binding
relationship was confirmed by the dual-luciferase reporter assay.
*P<0.05 vs. mimic-NC. (D) SP1 mRNA expression levels in human
glioma tissues and adjacent normal brain tissues was determined by
RT-qPCR, n=108. *P<0.05 vs. adjacent normal tissues. (E) SP1
mRNA expression levels in NHAs and human brain glioma cell lines
were verified by RT-qPCR. *P<0.05 vs. NHAs. (F) SP1 mRNA
expression levels in differently transfected cells were measured by
RT-qPCR. (G) SP1 protein expression levels in cells were determined
by western blotting. *P<0.05 vs. sh-NC. #P<0.05
vs. sh-LINC01138 + inhibitor-NC group. Three independent repeated
cell experiments were conducted. Data are presented as the mean ±
SD. Data were analyzed using (C) two-way ANOVA and Tukey's multiple
comparisons test; (D) independent samples t-test; and (E-G) one-way
ANOVA and Tukey's multiple comparisons test. miR, microRNA; LINC,
long intergenic non-protein coding RNA; SP1, specificity protein 1;
NC, negative control; RT-qPCR, reverse transcription-quantitative
PCR; NHA, normal human astrocytes; sh, short hairpin RNA; WT,
wild-type; MUT, mutant; num(T), number of tumor samples; num(N),
number of normal adjacent tissue samples; GBM, glioblastoma. |
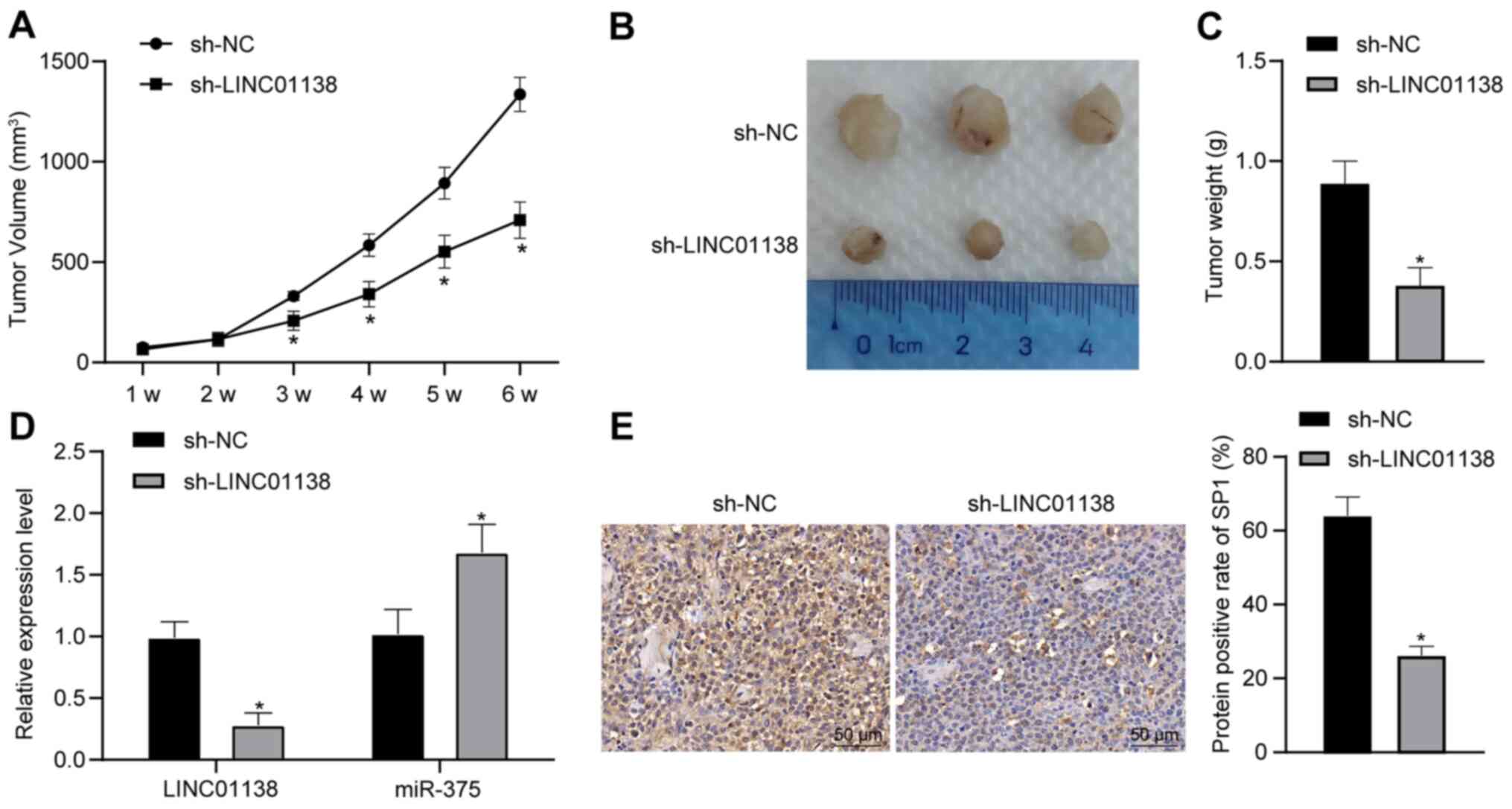
Silencing LINC01138 inhibits tumor
growth in vivo
To verify the function of LINC01138 in glioma tumor
formation, a xenograft tumor model was established in nude mice.
Compared with in the sh-NC group, the sh-LINC01138 group exhibited
significantly reduced tumor growth and weight (Fig. 7A-C; P<0.05). RT-qPCR demonstrated
that the expression levels of both LINC01138 and SP1 were
significantly downregulated in the sh-LINC01138 group compared with
those in the sh-NC group. Moreover, the expression levels of
miR-375 were significantly upregulated in the sh-LINC01138 group
compared with those in the sh-NC group (Fig. 7D; P<0.05). SP1 protein expression
levels in glioma tissues were evaluated by immunohistochemical
staining. The results demonstrated that SP1 protein expression
levels were significantly lower in the sh-LINC01138 group compared
with those in the sh-NC group (Fig.
7E; P<0.05). These findings indicated that silencing
LINC01138 inhibited glioma growth in vivo by regulating the
miR-375/SP1 axis.
Discussion
Glioma is one of the most prevalent and malignant
endocranial neoplasms, characterized by rapid infiltration and cell
growth, cellular heterogeneity, chemical resistance and a high
incidence of relapse (48).
Numerous lncRNAs are involved in the development and progression of
glioma, mediating cancer cell growth, apoptosis, invasiveness and
colony formation (49). LINC01138
has been reported to function as a biological marker of prostate
cancer, promoting tumor proliferation and reducing cancer cell
apoptosis (50), indicating the
deleterious effect of LINC01138 in tumor progression. The present
study was therefore designed to identify potential new therapeutic
targets for glioma based on LINC01138. The results demonstrated
that silencing LINC01138 attenuated aerobic glycolysis and the
proliferation of glioma cells via the LINC01138/miR-375/SP1 ceRNA
network. To the best of our knowledge, this is the first study to
report the specific mechanism by which LINC01138 functions in
glioma.
The present study demonstrated that LINC01138 was
significantly more strongly expressed in glioma cells compared with
in NHAs, and it was associated with the following
clinicopathological features of the tumor: WHO tumor grade, lymph
node metastasis and tumor diameter. High WHO tumor grade indicates
progressive and aggressive glioma with dangerous and unpredictable
consequences (51). Tumor diameter
and lymph node metastasis are both valuable prognostic indicators
for patients with glioma (52).
Recent studies have reported that LINC01138 overexpression resulted
in a poor overall survival rate and progression-free survival rate
in patients with hepatocellular carcinoma (HCC) (53,54).
LINC01138 was also revealed to be highly expressed in patients with
gastric cancer displaying increased tumor cell biological
activities, including invasiveness, viability, apoptosis evasion
and expansion (21). To the best of
our knowledge, there are currently no reports on LINC01138
expression and function in normal astrocytes and other normal
cells. Based on this aforementioned evidence, it was hypothesized
that LINC01138 was detrimental to glioma reduction.
Aerobic glycolysis is a notable characteristic of
malignant tumor growth and poor clinical outcomes (12). High-grade glioma heavily depends on
aerobic glycolysis to promote cancer growth and metabolic
activities (55). Subsequently, in
the present study LINC01138 expression was silenced in U251MG and
TJ905 cells using sh-LINC01138. Silencing LINC01138 significantly
inhibited aerobic glycolysis and the proliferation of human glioma
cells, indicated by the significantly reduced levels of glucose
uptake and lactic acid secretion, as well as significantly
decreased protein expression levels of HK, PK, PFK and GLUT. The
biological process of aerobic glycolysis in glioma has been shown
to reduce the efficacy of medical treatments (48). Du et al (56) reported that glucose uptake and
lactic acid secretion, as a result of aerobic glycolysis, greatly
exacerbated glioma progression. Furthermore, another study
demonstrated that when HK and PK levels were exhausted, glucose
levels decreased in glioma cells and effective treatment could be
facilitated (37). Moreover, PFK
activation in glioma has been shown to accelerate cell growth, and
aerobic glycolysis (57). GLUT
deficiency has also been reported to limit glioma development and
potentiate chemotherapy efficiency (58). The present study reported that
knockdown of LINC01138 decreased the levels of these aforementioned
proteins, and therefore silencing LINC01138 may be conducive to
ameliorating glioma progression.
Previous studies suggested that the interaction
between LINC01138 and protein arginine methyltransferase 5 was
involved in malignant carcinomas, such as ccRCC and HCC (19,54),
indicating the possibility of LINC01138 acting as a ceRNA. The
present study demonstrated that LINC01138 acted as a ceRNA to
sponge miR-375, and that miR-375 expression was significantly
decreased in glioma cells compared with in NHAs. It was recently
documented that miR-375 was expressed at low levels in glioma, and
inhibited tumor growth and cancer cell proliferation (59). miR-375 has been reported to function
in regulating glioma cell proliferation, with the addition of the
miR-375 inhibitor enhancing cell proliferation in both U251MG and
U87-MG cells (60). miR-375 is a
multi-functional regulator in a diverse range of cellular pathways,
which can regulate numerous functional genes (61). Ectopic expression of miR-375 is
usually associated with pathological changes and the functions of
miR-375 in immunity, such as its relevance with macrophages, T
helper cells and autoimmune diseases. For instance, miR-375 can
reduce apoptosis of macrophages in mice with liver failure; miR-375
is involved in Th1 and Th2 inflammatory and immune responses;
IL-10-deficient mice are the animal models of Th1-mediated
inflammatory bowel disease and miR-375 is notably elevated in the
mice (61). For example, miR-375
expression has been reported to be downregulated in rheumatoid
arthritis blood samples and fibroblast-like synoviocytes (62). To date, to the best of our
knowledge, there are no studies investigating the interaction
between miR-375 and astrocytes.
In the present study, functional rescue experiments
were performed using a miR-375 inhibitor to verify the role of
miR-375 in glioma. miR-375 inhibition reversed the effect of
sh-LINC01138 on aerobic glycolysis and cell proliferation in human
glioma. A recent study also demonstrated that miR-375-3p inhibited
glucose uptake and lactic acid secretion to suppress laryngeal
squamous cell carcinoma (63). The
present study revealed that LINC01138 promoted SP1 expression by
competing with SP1 for binding to miR-375. SP1 expression is often
related to poor prognosis and increased resistance of glioma to
treatments (31,34). The relationship between miR-375 and
SP1 has been detailed in numerous studies. miR-375 targeted SP1 to
attenuate inflammatory reactions and oxidative stress, relieving
the neuronal injury caused by Parkinson's disease (64). In addition, SP1 depletion was shown
to decrease glucose uptake, and inhibit cell proliferation and
invasion of U251MG cells by downregulating GLUT3 expression
(65). Moreover, Zhang et al
(66) revealed that lncRNA
RP11-626G11.3 increased SP1 expression by sponging miR-375 to
exacerbate malignant glioma. In summary, the ceRNA effect of the
LINC01138/miR-375/SP1 axis was verified in glioma.
In conclusion, the present study suggested that
silencing LINC01138 ameliorated glioma progression by sponging
miR-375 and downregulating SP1 expression. These results have
therapeutic implications for glioma treatment. Future work will
further explore the underlying mechanisms of the
LINC01138/miR-375/SP1 axis in glioma and the corresponding
potential therapeutic targets. In the in vivo experiments,
the effect of LINC01138 on tumor growth was observed; however,
glycolysis-related indicators were not investigated in these tumor
tissues. Therefore, the mechanism of aerobic glycolysis in glioma
in in vivo models will be investigated further in future
work. Although the findings of the present study have implications
for glioma treatment, the experimental results and their effective
application in clinical practice require further validation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CX and CS conceptualized the present study. HY and
XJ carried out the research, data review and writing. CS drafted
the study and revised it critically for important intellectual
content. CX and CS confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved and supervised by the
ethics committee of The First Affiliated Hospital of Soochow
University (Suzhou, China; approval no. SDU-MED-2017-085), and was
in accordance with the Declaration of Helsinki. All patients signed
written informed consent forms. Animal experiments were approved by
the Animal Ethics Committee of The First Affiliated Hospital of
Soochow University (approval no. S20200616018).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Matteoni S, Abbruzzese C, Villani V,
Malorni W, Pace A, Matarrese P and Paggi MG: The influence of
patient sex on clinical approaches to malignant glioma. Cancer
Lett. 468:41–47. 2020. View Article : Google Scholar
|
|
2
|
Kunadis E, Lakiotaki E, Korkolopoulou P
and Piperi C: Targeting post-translational histone modifying
enzymes in glioblastoma. Pharmacol Ther. 220:1077212021. View Article : Google Scholar
|
|
3
|
Angelopoulou E, Paudel YN and Piperi C:
Critical role of HOX transcript antisense intergenic RNA (HOTAIR)
in gliomas. J Mol Med (Berl). 98:1525–1546. 2020. View Article : Google Scholar
|
|
4
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar
|
|
5
|
Molinaro AM, Taylor JW, Wiencke JK and
Wrensch MR: Genetic and molecular epidemiology of adult diffuse
glioma. Nat Rev Neurol. 15:405–417. 2019. View Article : Google Scholar
|
|
6
|
Zhou X, Zhang S, Niu X, Li T, Zuo M, Yang
W, Li M, Li J, Yang Y, Wang X, et al: Risk factors for early
mortality among patients with glioma: A population-based study.
World Neurosurg. 136:e496–e503. 2020. View Article : Google Scholar
|
|
7
|
Radin DP and Tsirka SE: Interactions
between tumor cells, neurons, and microglia in the glioma
microenvironment. Int J Mol Sci. 21:84762020. View Article : Google Scholar
|
|
8
|
Hakar MH and Wood MD: Updates in pediatric
glioma pathology. Surg Pathol Clin. 13:801–816. 2020. View Article : Google Scholar
|
|
9
|
McCrorie P, Vasey CE, Smith SJ, Marlow M,
Alexander C and Rahman R: Biomedical engineering approaches to
enhance therapeutic delivery for malignant glioma. J Control
Release. 328:917–931. 2020. View Article : Google Scholar
|
|
10
|
Wank M, Schilling D, Schmid TE, Schmid TE,
Meyer B, Gempt J, Barz M, Schlegel J, Liesche F, Kessel KA, et al:
Human glioma migration and infiltration properties as a target for
personalized radiation medicine. Cancers (Basel). 10:4562018.
View Article : Google Scholar
|
|
11
|
Tech K, Deshmukh M and Gershon TR:
Adaptations of energy metabolism during cerebellar neurogenesis are
co-opted in medulloblastoma. Cancer Lett. 356:268–272. 2015.
View Article : Google Scholar
|
|
12
|
Kim MS, Huang Y, Lee J, Zhong X, Jiang WW,
Ratovitski EA and Sidransky D: Cellular transformation by cigarette
smoke extract involves alteration of glycolysis and mitochondrial
function in esophageal epithelial cells. Int J Cancer. 127:269–281.
2010.
|
|
13
|
Zoraghi R, See RH, Gong H, Lian T, Swayze
R, Finlay BB, Brunham RC, McMaster WR and Reiner NE: Functional
analysis, overexpression, and kinetic characterization of pyruvate
kinase from methicillin-resistant Staphylococcus aureus.
Biochemistry. 49:7733–7747. 2010. View Article : Google Scholar
|
|
14
|
Snášel J, Machová I, Šolínová V, Kašička
V, Krečmerová M and Pichová I: Phosphofructokinases A and B from
mycobacterium tuberculosis display different catalytic properties
and allosteric regulation. Int J Mol Sci. 22:222021. View Article : Google Scholar
|
|
15
|
Sánchez-Alvarez R, Tabernero A and Medina
JM: Endothelin-1 stimulates the translocation and upregulation of
both glucose transporter and hexokinase in astrocytes: Relationship
with gap junctional communication. J Neurochem. 89:703–714. 2004.
View Article : Google Scholar
|
|
16
|
Stieber D, Abdul Rahim SA and Niclou SP:
Novel ways to target brain tumour metabolism. Expert Opin Ther
Targets. 15:1227–1239. 2011. View Article : Google Scholar
|
|
17
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar
|
|
18
|
Xue BZ, Xiang W, Zhang Q, Wang YH, Wang
HF, Yi DY, Xiong NX, Jiang XB, Zhao HY and Fu P: Roles of long
non-coding RNAs in the hallmarks of glioma. Oncol Lett.
20:832020.
|
|
19
|
Zhang X, Wu J, Wu C, Chen W, Lin R, Zhou Y
and Huang X: The LINC01138 interacts with PRMT5 to promote
SREBP1-mediated lipid desaturation and cell growth in clear cell
renal cell carcinoma. Biochem Biophys Res Commun. 507:337–342.
2018. View Article : Google Scholar
|
|
20
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar
|
|
21
|
Dou GX, Zhang JN, Wang P, Wang JL and Sun
GB: Long intergenic non-protein-coding RNA 01138 accelerates tumor
growth and invasion in gastric cancer by regulating miR-1273e. Med
Sci Monit. 25:2141–2150. 2019. View Article : Google Scholar
|
|
22
|
Zhou Q, Liu J, Quan J, Liu W, Tan H and Li
W: MicroRNAs as potential biomarkers for the diagnosis of glioma: A
systematic review and meta-analysis. Cancer Sci. 109:2651–2659.
2018. View Article : Google Scholar
|
|
23
|
Hu C, Lv L, Peng J, Liu D, Wang X, Zhou Y
and Huo J: MicroRNA-375 suppresses esophageal cancer cell growth
and invasion by repressing metadherin expression. Oncol Lett.
13:4769–4775. 2017. View Article : Google Scholar
|
|
24
|
Zhao L, Lou G, Li A and Liu Y: lncRNA
MALAT1 modulates cancer stem cell properties of liver cancer cells
by regulating YAP1 expression via miR 375 sponging. Mol Med Rep.
22:1449–1457. 2020. View Article : Google Scholar
|
|
25
|
Xu F, Ye ML, Zhang YP, Li WJ, Li MT, Wang
HZ, Qiu X, Xu Y, Yin JW, Hu Q, et al: MicroRNA-375-3p enhances
chemosensitivity to 5-fluorouracil by targeting thymidylate
synthase in colorectal cancer. Cancer Sci. 111:1528–1541. 2020.
View Article : Google Scholar
|
|
26
|
Tang H, Huang X, Wang J, Yang L, Kong Y,
Gao G, Zhang L, Chen ZS and Xie X: circKIF4A acts as a prognostic
factor and mediator to regulate the progression of triple-negative
breast cancer. Mol Cancer. 18:232019. View Article : Google Scholar
|
|
27
|
Liu J, Zhan Y, Wang J, Wang J, Guo J and
Kong D: lncRNA-SNHG17 promotes colon adenocarcinoma progression and
serves as a sponge for miR-375 to regulate CBX3 expression. Am J
Transl Res. 12:5283–5295. 2020.
|
|
28
|
Wang X, Han L, Zhou L, Wang L and Zhang
LM: Prediction of candidate RNA signatures for recurrent ovarian
cancer prognosis by the construction of an integrated competing
endogenous RNA network. Oncol Rep. 40:2659–2673. 2018.
|
|
29
|
Lu M, Xu X, Xi B, Dai Q, Li C, Su L, Zhou
X, Tang M, Yao Y and Yang J: Molecular network-based identification
of competing endogenous RNAs in thyroid carcinoma. Genes (Basel).
9:442018. View Article : Google Scholar
|
|
30
|
Liang Y, Zhang C, Ma MH and Dai DQ:
Identification and prediction of novel non-coding and coding
RNA-associated competing endogenous RNA networks in colorectal
cancer. World J Gastroenterol. 24:5259–5270. 2018. View Article : Google Scholar
|
|
31
|
Yang WB, Hsu CC, Hsu TI, Liou JP, Chang
KY, Chen PY, Liu JJ, Yang ST, Wang JY, Yeh SH, et al: Increased
activation of HDAC1/2/6 and Sp1 underlies therapeutic resistance
and tumor growth in glioblastoma. Neuro Oncol. 22:1439–1451. 2020.
View Article : Google Scholar
|
|
32
|
Zhang X, Zhao X, Wang Y and Xing L: Long
non-coding RNA LINC00491 contributes to the malignancy of
non-small-cell lung cancer via competitively binding to
microRNA-324-5p and thereby increasing specificity protein 1
expression. Cancer Manag Res. 12:6779–6793. 2020. View Article : Google Scholar
|
|
33
|
Cong Z, Diao Y, Xu Y, Li X, Jiang Z, Shao
C, Ji S, Shen Y, De W and Qiang Y: Long non-coding RNA linc00665
promotes lung adenocarcinoma progression and functions as ceRNA to
regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis.
10:842019. View Article : Google Scholar
|
|
34
|
Guan H, Cai J, Zhang N, Wu J, Yuan J, Li J
and Li M: Sp1 is upregulated in human glioma, promotes
MMP-2-mediated cell invasion and predicts poor clinical outcome.
Int J Cancer. 130:593–601. 2012. View Article : Google Scholar
|
|
35
|
Percie du Sert N, Hurst V, Ahluwalia A,
Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl
U, et al: The ARRIVE guidelines 2.0: Updated guidelines for
reporting animal research. PLoS Biol. 18:e30004102020. View Article : Google Scholar
|
|
36
|
Villa C, Miquel C, Mosses D, Bernier M and
Di Stefano AL: The 2016 World Health Organization classification of
tumours of the central nervous system. Presse Med. 47:e187–e200.
2018. View Article : Google Scholar
|
|
37
|
Xu B, Zhang Q, Luo X, Ning X, Luo J, Guo
J, Liu Q, Ling G and Zhou N: Selenium nanoparticles reduce glucose
metabolism and promote apoptosis of glioma cells through reactive
oxygen species-dependent manner. Neuroreport. 31:226–234. 2020.
View Article : Google Scholar
|
|
38
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar
|
|
39
|
Cao Z, Pan X, Yang Y, Huang Y and Shen HB:
The lncLocator: A subcellular localization predictor for long
non-coding RNAs based on a stacked ensemble classifier.
Bioinformatics. 34:2185–2194. 2018. View Article : Google Scholar
|
|
40
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006. View Article : Google Scholar
|
|
41
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
42
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017. View Article : Google Scholar
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
44
|
Diehl KH, Hull R, Morton D, Pfister R,
Rabemampianina Y, Smith D, Vidal JM and van de Vorstenbosch C;
European Federation of Pharmaceutical Industries Association and
European Centre for the Validation of Alternative Methods, : A good
practice guide to the administration of substances and removal of
blood, including routes and volumes. J Appl Toxicol. 21:15–23.
2001. View Article : Google Scholar
|
|
45
|
Cheng R and Chen Y, Zhou H, Wang B, Du Q
and Chen Y: B7-H3 expression and its correlation with
clinicopathologic features, angiogenesis, and prognosis in
intrahepatic cholangiocarcinoma. APMIS. 126:396–402. 2018.
View Article : Google Scholar
|
|
46
|
Cerdán S, Rodrigues TB, Sierra A, Benito
M, Fonseca LL, Fonseca CP and García-Martín ML: The redox
switch/redox coupling hypothesis. Neurochem Int. 48:523–530. 2006.
View Article : Google Scholar
|
|
47
|
Jiang B: Aerobic glycolysis and high level
of lactate in cancer metabolism and microenvironment. Genes Dis.
4:25–27. 2017. View Article : Google Scholar
|
|
48
|
Han W, Shi J, Cao J, Dong B and Guan W:
Emerging roles and therapeutic interventions of aerobic glycolysis
in glioma. OncoTargets Ther. 13:6937–6955. 2020. View Article : Google Scholar
|
|
49
|
Han W, Shi J, Cao J, Dong B and Guan W:
Current advances of long non-coding RNAs mediated by wnt signaling
in glioma. Pathol Res Pract. 216:1530082020. View Article : Google Scholar
|
|
50
|
Wan X, Huang W, Yang S, Zhang Y, Pu H, Fu
F, Huang Y, Wu H, Li T and Li Y: Identification of
androgen-responsive lncRNAs as diagnostic and prognostic markers
for prostate cancer. Oncotarget. 7:60503–60518. 2016. View Article : Google Scholar
|
|
51
|
van Kessel E, Huenges Wajer IM, Ruis C,
Seute T, Fonville S, De Vos FY, Verhoeff JJ, Robe PA, van Zandvoort
MJ and Snijders TJ: Cognitive impairments are independently
associated with shorter survival in diffuse glioma patients. J
Neurol. 268:1434–1442. 2021. View Article : Google Scholar
|
|
52
|
Li K, Zhang Q, Niu D and Xing H: Mining
miRNAs expressions in glioma based on GEO database and their
effects on biological functions. BioMed Res Int.
2020:56378642020.
|
|
53
|
Jiang H, Shi X, Ye G, Xu Y, Xu J, Lu J and
Lu W: Up-regulated long non-coding RNA DUXAP8 promotes cell growth
through repressing Krüppel-like factor 2 expression in human
hepatocellular carcinoma. OncoTargets Ther. 12:7429–7436. 2019.
View Article : Google Scholar
|
|
54
|
Li Z, Zhang J, Liu X, Li S, Wang Q, Di
Chen, Hu Z, Yu T, Ding J, Li J, et al: The LINC01138 drives
malignancies via activating arginine methyltransferase 5 in
hepatocellular carcinoma. Nat Commun. 9:15722018. View Article : Google Scholar
|
|
55
|
Sperry J, Condro MC, Guo L, Braas D,
Vanderveer-Harris N, Kim KK, Pope WB, Divakaruni AS, Lai A,
Christofk H, et al: Glioblastoma utilizes fatty acids and ketone
bodies for growth allowing progression during Ketogenic Diet
Therapy. iScience. 23:1014532020. View Article : Google Scholar
|
|
56
|
Du P, Liao Y, Zhao H, Zhang J, Muyiti,
Keremu and Mu K: ANXA2P2/miR-9/LDHA axis regulates Warburg effect
and affects glioblastoma proliferation and apoptosis. Cell Signal.
74:1097182020. View Article : Google Scholar
|
|
57
|
Lee JH, Liu R, Li J, Zhang C, Wang Y, Cai
Q, Qian X, Xia Y, Zheng Y, Piao Y, et al: Stabilization of
phosphofructokinase 1 platelet isoform by AKT promotes
tumorigenesis. Nat Commun. 8:9492017. View Article : Google Scholar
|
|
58
|
Azzalin A, Brambilla F, Arbustini E,
Basello K, Speciani A, Mauri P, Bezzi P and Magrassi L: A new
pathway promotes adaptation of human glioblastoma cells to glucose
starvation. Cells. 9:12492020. View Article : Google Scholar
|
|
59
|
Li GF, Cheng YY, Li BJ, Zhang C, Zhang XX,
Su J, Wang C, Chang L, Zhang DZ, Tan CL, et al: miR-375 inhibits
the proliferation and invasion of glioblastoma by regulating Wnt5a.
Neoplasma. 66:350–356. 2019. View Article : Google Scholar
|
|
60
|
Ding P, Liang B, Shou J and Wang X: lncRNA
KCNQ1OT1 promotes proliferation and invasion of glioma cells by
targeting the miR 375/YAP pathway. Int J Mol Med. 46:1983–1992.
2020. View Article : Google Scholar
|
|
61
|
Liu Y, Wang Q, Wen J, Wu Y and Man C:
MiR-375: A novel multifunctional regulator. Life Sci.
275:1193232021. View Article : Google Scholar
|
|
62
|
Zhi L, Liang J, Huang W, Ma J, Qing Z and
Wang X: Circ_AFF2 facilitates proliferation and inflammatory
response of fibroblast-like synoviocytes in rheumatoid arthritis
via the miR-375/TAB2 axis. Exp Mol Pathol. 119:1046172021.
View Article : Google Scholar
|
|
63
|
Chang K, Wei Z and Cao H: miR-375-3p
inhibits the progression of laryngeal squamous cell carcinoma by
targeting hepatocyte nuclear factor-1β. Oncol Lett. 20:802020.
View Article : Google Scholar
|
|
64
|
Cai LJ, Tu L, Li T, Yang XL, Ren YP, Gu R,
Zhang Q, Yao H, Qu X, Wang Q, et al: Up-regulation of microRNA-375
ameliorates the damage of dopaminergic neurons, reduces oxidative
stress and inflammation in Parkinsons disease by inhibiting SP1.
Aging (Albany NY). 12:672–689. 2020. View Article : Google Scholar
|
|
65
|
Zheng C, Yang K, Zhang M, Zou M, Bai E, Ma
Q and Xu R: Specific protein 1 depletion attenuates glucose uptake
and proliferation of human glioma cells by regulating GLUT3
expression. Oncol Lett. 12:125–131. 2016. View Article : Google Scholar
|
|
66
|
Zhang Y, Mou C, Shang M, Jiang M and Xu C:
Long noncoding RNA RP11-626G11.3 promotes the progression of glioma
through miR-375-SP1 axis. Mol Carcinog. 59:492–502. 2020.
View Article : Google Scholar
|