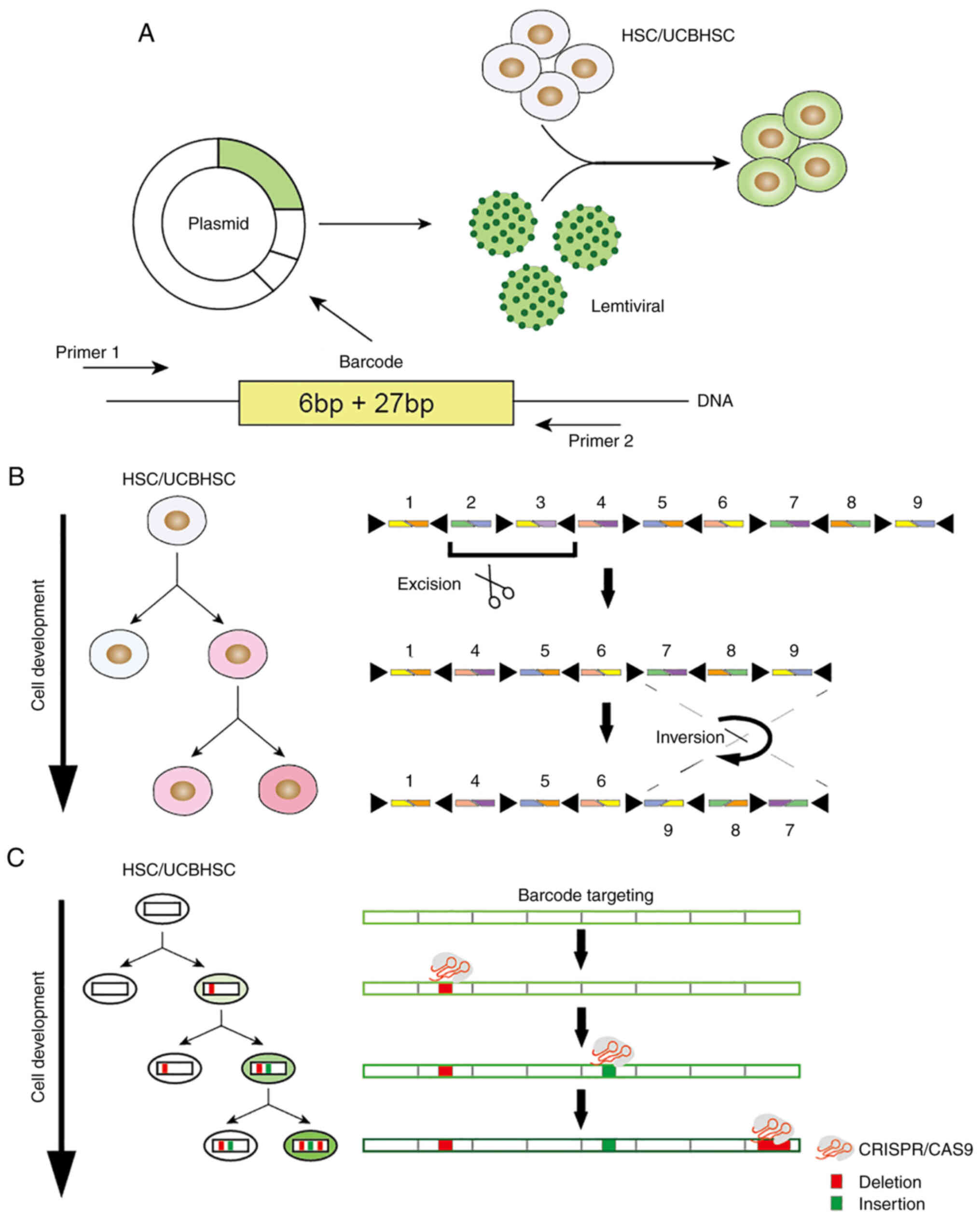
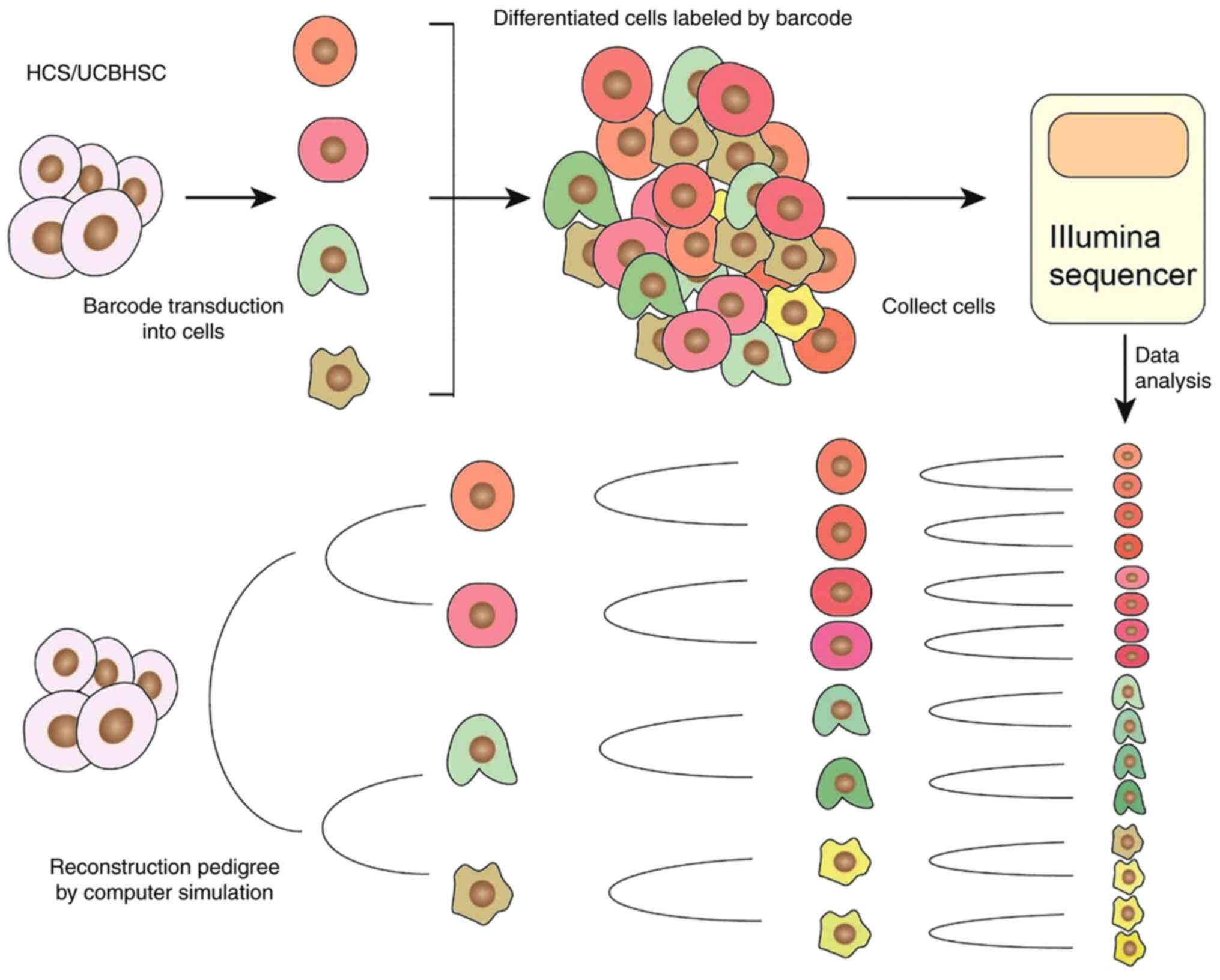
|
1
|
Broxmeyer HE, Douglas GW, Hangoc G, Cooper
S, Bard J, English D, Arny M, Thomas L and Boyse EA: Human
umbilical cord blood as a potential source of transplantable
hematopoietic stem/progenitor cells. Proc Natl Acad Sci USA.
86:3828–3832. 1989. View Article : Google Scholar
|
|
2
|
Buzańska L, Jurga M and Domańska-Janik K:
Neuronal differentiation of human umbilical cord blood neural
stem-like cell line. Neurodegener Dis. 3:19–26. 2006. View Article : Google Scholar
|
|
3
|
Zhang J, Huang X, Guo B, Cooper S,
Capitano ML, Johnson TC, Siegel DR and Broxmeyer HE: Effects of
eupalinilide E and UM171, alone and in combination on cytokine
stimulated ex-vivo expansion of human cord blood hematopoietic stem
cells. Blood Cells Mol Dis. 84:1024572020. View Article : Google Scholar
|
|
4
|
Sunitha MM, Srikanth L, Kumar PS,
Chandrasekhar C and Sarma P: Down-regulation of PAX2 promotes in
vitro differentiation of podocytes from human CD34+
cells. Cell Tissue Res. 370:477–488. 2017. View Article : Google Scholar
|
|
5
|
Alatyyat SM, Alasmari HM, Aleid OA,
Abdel-Maksoud MS and Elsherbiny N: Umbilical cord stem cells:
Background, processing and applications. Tissue Cell.
65:1013512020. View Article : Google Scholar
|
|
6
|
Francese R and Fiorina P: Immunological
and regenerative properties of cord blood stem cells. Clin Immunol.
136:309–322. 2010. View Article : Google Scholar
|
|
7
|
Fatrai S, Schepers H, Tadema H, Vellenga
E, Daenen SM and Schuringa JJ: Mucin1 expression is enriched in the
human stem cell fraction of cord blood and is upregulated in
majority of the AML cases. Exp Hematol. 36:1254–1265. 2008.
View Article : Google Scholar
|
|
8
|
Castillo-Melendez M, Yawno T, Jenkin G and
Miller SL: Stem cell therapy to protect and repair the developing
brain: A review of mechanisms of action of cord blood and amnion
epithelial derived cells. Front Neurosci. 7:1942013. View Article : Google Scholar
|
|
9
|
Cairo MS and Wagner JE: Placental and/or
umbilical cord blood: An alternative source of hematopoietic stem
cells for transplantation. Blood. 90:4665–4678. 1997. View Article : Google Scholar
|
|
10
|
Mayani H and Lansdorp PM: Biology of human
umbilical cord blood-derived hematopoietic stem/progenitor cells.
Stem Cells. 16:153–165. 1998. View Article : Google Scholar
|
|
11
|
Liu G, Ye X, Zhu Y, Li Y, Sun J, Cui L and
Cao Y: Osteogenic differentiation of GFP-labeled human umbilical
cord blood derived mesenchymal stem cells after cryopreservation.
Cryobiology. 63:125–128. 2011. View Article : Google Scholar
|
|
12
|
Zheng JH, Zhang JK, Kong DS, Song YB, Zhao
SD, Qi WB, Li YN, Zhang ML and Huang XH: Quantification of the
CM-Dil-labeled human umbilical cord mesenchymal stem cells migrated
to the dual injured uterus in SD rat. Stem Cell Res Ther.
11:2802020. View Article : Google Scholar
|
|
13
|
Kebschull JM and Zador AM: Cellular
barcoding: Lineage tracing, screening and beyond. Nat Methods.
15:871–879. 2018. View Article : Google Scholar
|
|
14
|
Wagner DE and Klein AM: Lineage tracing
meets single-cell omics: Opportunities and challenges. Nat Rev
Genet. 21:410–427. 2020. View Article : Google Scholar
|
|
15
|
Mitchell KE, Weiss ML, Mitchell BM, Martin
P, Davis D, Morales L, Helwig B, Beerenstrauch M, Abou-Easa K,
Hildreth T, et al: Matrix cells from Wharton's jelly form neurons
and glia. Stem Cells. 21:50–60. 2003. View Article : Google Scholar
|
|
16
|
Fu YS, Shih YT, Cheng YC and Min MY:
Transformation of human umbilical mesenchymal cells into neurons in
vitro. J Biomed Sci. 11:652–660. 2004. View Article : Google Scholar
|
|
17
|
Rodrigues LP, Iglesias D, Nicola FC,
Steffens D, Valentim L, Witczak A, Zanatta G, Achaval M, Pranke P
and Netto CA: Transplantation of mononuclear cells from human
umbilical cord blood promotes functional recovery after traumatic
spinal cord injury in Wistar rats. Braz J Med Biol Res. 45:49–57.
2012. View Article : Google Scholar
|
|
18
|
Wang HS, Hung SC, Peng ST, Huang CC, Wei
HM, Guo YJ, Fu YS, Lai MC and Chen CC: Mesenchymal stem cells in
the Wharton's jelly of the human umbilical cord. Stem Cells.
22:1330–1337. 2004. View Article : Google Scholar
|
|
19
|
Kakinuma S, Tanaka Y, Chinzei R, Watanabe
M, Shimizu-Saito K, Hara Y, Teramoto K, Arii S, Sato C, Takase K,
et al: Human umbilical cord blood as a source of transplantable
hepatic progenitor cells. Stem Cells. 21:217–227. 2003. View Article : Google Scholar
|
|
20
|
Tang XP, Zhang M, Yang X, Chen LM and Zeng
Y: Differentiation of human umbilical cord blood stem cells into
hepatocytes in vivo and in vitro. World J Gastroenterol.
12:4014–4019. 2006. View Article : Google Scholar
|
|
21
|
Mayani H, Wagner JE and Broxmeyer HE: Cord
blood research, banking, and transplantation: Achievements,
challenges, and perspectives. Bone Marrow Transplant. 55:48–61.
2020. View Article : Google Scholar
|
|
22
|
Shetty P, Cooper K and Viswanathan C:
Comparison of proliferative and multilineage differentiation
potentials of cord matrix, cord blood, and bone marrow mesenchymal
stem cells. Asian J Transfus Sci. 4:14–24. 2010. View Article : Google Scholar
|
|
23
|
Han JY, Goh RY, Seo SY, Hwang TH, Kwon HC,
Kim SH, Kim JS, Kim HJ and Lee YH: Cotransplantation of cord blood
hematopoietic stem cells and culture-expanded and
GM-CSF-/SCF-transfected mesenchymal stem cells in SCID mice. J
Korean Med Sci. 22:242–247. 2007. View Article : Google Scholar
|
|
24
|
Hutton JF, D'Andrea RJ and Lewis ID:
Potential for clinical ex vivo expansion of cord blood haemopoietic
stem cells using non-haemopoietic factor supplements. Curr Stem
Cell Res Ther. 2:229–237. 2007. View Article : Google Scholar
|
|
25
|
Demerdash Z, El-Baz HG, Maher K, Hassan S,
Salah F, Hassan M, Elzallat M, El-Shafei M and Taha T: Effect of
repeated passaging and cell density on proliferation and
differentiation potential of cord blood unrestricted somatic stem
cells. New Horiz Transl Med. 2:672015.
|
|
26
|
Esmaeili M, Niazi V, Pourfathollah AA,
Hosseini MKM, Nakhlestani M, Golzadeh K, Taheri M, Ghafouri-Fard S
and Atarodi K: The impact of parathyroid hormone treated
mesenchymal stem cells on ex-vivo expansion of cord blood
hematopoietic stem cells. Gene Rep. 17:1004902019. View Article : Google Scholar
|
|
27
|
Mokhtari S, Baptista PM, Vyas DA, Freeman
CJ, Moran E, Brovold M, Llamazares GA, Lamar Z, Porada CD, Soker S
and Almeida-Porada G: Evaluating interaction of cord blood
hematopoietic stem/progenitor cells with functionally integrated
three-dimensional microenvironments. Stem Cells Transl Med.
7:271–282. 2018. View Article : Google Scholar
|
|
28
|
Chaurasia P, Gajzer DC, Schaniel C,
D'Souza S and Hoffman R: Epigenetic reprogramming induces the
expansion of cord blood stem cells. J Clin Invest. 124:2378–2395.
2014. View Article : Google Scholar
|
|
29
|
Li Q, Zhao D, Chen Q, Luo M, Huang J, Yang
C, Wang F, Li W and Liu T: Wharton's jelly mesenchymal stem
cell-based or umbilical vein endothelial cell-based serum-free
coculture with cytokines supports the ex vivo expansion/maintenance
of cord blood hematopoietic stem/progenitor cells. Stem Cell Res
Ther. 10:3762019. View Article : Google Scholar
|
|
30
|
Zhang B, Wu X, Zhang X, Sun Y, Yan Y, Shi
H, Zhu Y, Wu L, Pan Z, Zhu W, et al: Human umbilical cord
mesenchymal stem cell exosomes enhance angiogenesis through the
Wnt4/β-catenin pathway. Stem Cells Transl Med. 4:513–522. 2015.
View Article : Google Scholar
|
|
31
|
Rim YA, Nam Y and Ju JH: Application of
cord blood and cord blood-derived induced pluripotent stem cells
for cartilage regeneration. Cell Transplant. 28:529–537. 2019.
View Article : Google Scholar
|
|
32
|
Zheng YL, Sun YP, Zhang H, Liu WJ, Jiang
R, Li WY, Zheng YH and Zhang ZG: Mesenchymal stem cells obtained
from synovial fluid mesenchymal stem cell-derived induced
pluripotent stem cells on a matrigel coating exhibited enhanced
proliferation and differentiation potential. PLoS One.
10:e01442262015. View Article : Google Scholar
|
|
33
|
Zhou RQ, Wu JH, Gong YP, Guo Y and Xing
HY: Transcription factor SCL/TAL1 mediates the phosphorylation of
MEK/ERK pathway in umbilical cord blood CD34+ stem cells
during hematopoietic differentiation. Blood Cells Mol Dis.
53:39–46. 2014. View Article : Google Scholar
|
|
34
|
Ajami M, Soleimani M, Abroun S and Atashi
A: Comparison of cord blood CD34 + stem cell expansion in coculture
with mesenchymal stem cells overexpressing SDF-1 and
soluble/membrane isoforms of SCF. J Cell Biochem. 120:15297–15309.
2019. View Article : Google Scholar
|
|
35
|
Naka K, Muraguchi T, Hoshii T and Hirao A:
Regulation of reactive oxygen species and genomic stability in
hematopoietic stem cells. Antioxid Redox Signal. 10:1883–1894.
2008. View Article : Google Scholar
|
|
36
|
Bonifazi F, Dan E, Labopin M, Sessa M,
Guadagnuolo V, Ferioli M, Rizzi S, De Carolis S, Sinigaglia B,
Motta MR, et al: Intrabone transplant provides full stemness of
cord blood stem cells with fast hematopoietic recovery and low GVHD
rate: Results from a prospective study. Bone Marrow Transplant.
54:717–725. 2019. View Article : Google Scholar
|
|
37
|
Lee YH: Clinical utilization of cord blood
over human health: Experience of stem cell transplantation and cell
therapy using cord blood in Korea. Korean J Pediatr. 57:110–116.
2014. View Article : Google Scholar
|
|
38
|
Li X, Ma X, Chen Y, Peng D, Wang H, Chen
S, Xiao Y, Li L, Zhou H, Cheng F, et al: Coinhibition of activated
p38 MAPKα and mTORC1 potentiates stemness maintenance of HSCs from
SR1-expanded human cord blood CD34+ cells via inhibition
of senescence. Stem Cells Transl Med. 9:1604–1616. 2020. View Article : Google Scholar
|
|
39
|
Fares I, Chagraoui J, Gareau Y, Gingras S,
Ruel R, Mayotte N, Csaszar E, Knapp DJ, Miller P, Ngom M, et al:
Cord blood expansion. Pyrimidoindole derivatives are agonists of
human hematopoietic stem cell self-renewal. Science. 345:1509–1512.
2014. View Article : Google Scholar
|
|
40
|
Seghatoleslam M, Jalali M, Alamdari DH,
Nikravesh MR, Hosseini SM and Fazel AR: Effect of incubation time
on the in vitro labeling of umbilical cord blood hematopoietic stem
cells with bromodeoxyuridine (BrdU). Clin Biochem. 44
(Suppl):S1532011. View Article : Google Scholar
|
|
41
|
Walsh C and Cepko CL: Widespread
dispersion of neuronal clones across functional regions of the
cerebral cortex. Science. 255:434–440. 1992. View Article : Google Scholar
|
|
42
|
Gerrits A, Dykstra B, Kalmykowa OJ, Klauke
K, Verovskaya E, Broekhuis MJ, de Haan G and Bystrykh LV: Cellular
barcoding tool for clonal analysis in the hematopoietic system.
Blood. 115:2610–2618. 2010. View Article : Google Scholar
|
|
43
|
Zorita E, Cuscó P and Filion GJ: Starcode:
Sequence clustering based on all-pairs search. Bioinformatics.
31:1913–1919. 2015. View Article : Google Scholar
|
|
44
|
Schepers K, Swart E, van Heijst JW,
Gerlach C, Castrucci M, Sie D, Heimerikx M, Velds A, Kerkhoven RM,
Arens R and Schumacher TN: Dissecting T cell lineage relationships
by cellular barcoding. J Exp Med. 205:2309–2318. 2008. View Article : Google Scholar
|
|
45
|
Kristiansen TA, Jaensson Gyllenbäck E,
Zriwil A, Björklund T, Daniel JA, Sitnicka E, Soneji S, Bryder D
and Yuan J: Cellular barcoding links B-1a B cell potential to a
fetal hematopoietic stem cell state at the single-cell level.
Immunity. 45:346–357. 2016. View Article : Google Scholar
|
|
46
|
Lu R, Neff NF, Quake SR and Weissman IL:
Tracking single hematopoietic stem cells in vivo using
high-throughput sequencing in conjunction with viral genetic
barcoding. Nat Biotechnol. 29:928–933. 2011. View Article : Google Scholar
|
|
47
|
Naik SH, Perié L, Swart E, Gerlach C, van
Rooij N, de Boer RJ and Schumacher TN: Diverse and heritable
lineage imprinting of early haematopoietic progenitors. Nature.
496:229–232. 2013. View Article : Google Scholar
|
|
48
|
Verovskaya E, Broekhuis MJ, Zwart E,
Ritsema M, van Os R, de Haan G and Bystrykh LV: Heterogeneity of
young and aged murine hematopoietic stem cells revealed by
quantitative clonal analysis using cellular barcoding. Blood.
122:523–532. 2013. View Article : Google Scholar
|
|
49
|
Keller G, Paige C, Gilboa E and Wagner EF:
Expression of a foreign gene in myeloid and lymphoid cells derived
from multipotent haematopoietic precursors. Nature. 318:149–154.
1985. View Article : Google Scholar
|
|
50
|
Lemischka IR, Raulet DH and Mulligan RC:
Developmental potential and dynamic behavior of hematopoietic stem
cells. Cell. 45:917–927. 1986. View Article : Google Scholar
|
|
51
|
Ludwig LS, Lareau CA, Ulirsch JC,
Christian E, Muus C, Li LH, Pelka K, Ge W, Oren Y, Brack A, et al:
Lineage tracing in humans enabled by mitochondrial mutations and
single-cell genomics. Cell. 176:1325–1339.e22. 2019. View Article : Google Scholar
|
|
52
|
Wagner DE, Weinreb C, Collins ZM, Briggs
JA, Megason SG and Klein AM: Single-cell mapping of gene expression
landscapes and lineage in the zebrafish embryo. Science.
360:981–987. 2018. View Article : Google Scholar
|
|
53
|
Guo C, Kong W, Kamimoto K, Rivera-Gonzalez
GC, Yang X, Kirita Y and Morris SA: CellTag Indexing: Genetic
barcode-based sample multiplexing for single-cell genomics. Genome
Biol. 20:902019. View Article : Google Scholar
|
|
54
|
Bramlett C, Jiang D, Nogalska A, Eerdeng
J, Contreras J and Lu R: Clonal tracking using embedded viral
barcoding and high-throughput sequencing. Nat Protoc. 15:1436–1458.
2020. View Article : Google Scholar
|
|
55
|
Pei W, Feyerabend TB, Rössler J, Wang X,
Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C,
et al: Polylox barcoding reveals haematopoietic stem cell fates
realized in vivo. Nature. 548:456–460. 2017. View Article : Google Scholar
|
|
56
|
Pei W, Wang X, Rössler J, Feyerabend TB,
Hofer T and Rodewald HR: Using Cre-recombinase-driven Polylox
barcoding for in vivo fate mapping in mice. Nat Protoc.
14:1820–1840. 2019. View Article : Google Scholar
|
|
57
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA and Zhang F:
Multiplex genome engineering using CRISPR/Cas systems. Science.
339:819–823. 2013. View Article : Google Scholar
|
|
58
|
McKenna A, Findlay GM, Gagnon JA, Horwitz
MS, Schier AF and Shendure J: Whole-organism lineage tracing by
combinatorial and cumulative genome editing. Science.
353:aaf79072016. View Article : Google Scholar
|
|
59
|
Frieda KL, Linton JM, Hormoz S, Choi J,
Chow KK, Singer ZS, Budde MW, Elowitz MB and Cai L: Synthetic
recording and in situ readout of lineage information in single
cells. Nature. 541:107–111. 2017. View Article : Google Scholar
|
|
60
|
Perli SD, Cui CH and Lu TK: Continuous
genetic recording with self-targeting CRISPR-Cas in human cells.
Science. 353:aag05112016. View Article : Google Scholar
|
|
61
|
Kalhor R, Mali P and Church GM: Rapidly
evolving homing CRISPR barcodes. Nat Methods. 14:195–200. 2017.
View Article : Google Scholar
|
|
62
|
Kalhor R, Kalhor K, Mejia L, Leeper K,
Graveline A, Mali P and Church GM: Developmental barcoding of whole
mouse via homing CRISPR. Science. 361:eaat98042018. View Article : Google Scholar
|
|
63
|
Loveless TB, Grotts JH, Schechter MW,
Forouzmand E, Carlson CK, Agahi BS, Liang G, Ficht M, Liu B, Xie X
and Liu CC: DNA writing at a single genomic site enables lineage
tracing and analog recording in mammalian cells. bioRxiv.
6391202019.
|
|
64
|
Bowling S, Sritharan D, Osorio FG, Nguyen
M, Cheung P, Rodriguez-Fraticelli A, Patel S, Yuan WC, Fujiwara Y,
Li BE, et al: An engineered CRISPR-Cas9 mouse line for simultaneous
readout of lineage histories and gene expression profiles in single
cells. Cell. 181:1410–1422.e27. 2020. View Article : Google Scholar
|
|
65
|
Nguyen LV, Cox CL, Eirew P, Knapp DJ,
Pellacani D, Kannan N, Carles A, Moksa M, Balani S, Shah S, et al:
DNA barcoding reveals diverse growth kinetics of human breast
tumour subclones in serially passaged xenografts. Nat Commun.
5:58712014. View Article : Google Scholar
|
|
66
|
Naik SH, Schumacher TN and Perie L:
Cellular barcoding: A technical appraisal. Exp Hematol. 42:598–608.
2014. View Article : Google Scholar
|
|
67
|
Nguyen LV, Pellacani D, Lefort S, Kannan
N, Osako T, Makarem M, Cox CL, Kennedy W, Beer P, Carles A, et al:
Barcoding reveals complex clonal dynamics of de novo transformed
human mammary cells. Nature. 528:267–271. 2015. View Article : Google Scholar
|
|
68
|
McKenzie JL, Gan OI, Doedens M, Wang JC
and Dick JE: Individual stem cells with highly variable
proliferation and self-renewal properties comprise the human
hematopoietic stem cell compartment. Nat Immunol. 7:1225–1233.
2006. View
Article : Google Scholar
|
|
69
|
Gonzalez-Murillo A, Lozano ML, Montini E,
Bueren JA and Guenechea G: Unaltered repopulation properties of
mouse hematopoietic stem cells transduced with lentiviral vectors.
Blood. 112:3138–3147. 2008. View Article : Google Scholar
|
|
70
|
Golden JA, Fields-Berry SC and Cepko CL:
Construction and characterization of a highly complex retroviral
library for lineage analysis. Proc Natl Acad Sci USA. 92:5704–5708.
1995. View Article : Google Scholar
|
|
71
|
Adamson B, Norman TM, Jost M, Cho MY,
Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et
al: A multiplexed single-cell CRISPR screening platform enables
systematic dissection of the unfolded protein response. Cell.
167:1867–1882.e21. 2016. View Article : Google Scholar
|
|
72
|
Cheung AM, Nguyen LV, Carles A, Beer P,
Miller PH, Knapp DJ, Dhillon K, Hirst M and Eaves CJ: Analysis of
the clonal growth and differentiation dynamics of primitive
barcoded human cord blood cells in NSG mice. Blood. 122:3129–3137.
2013. View Article : Google Scholar
|
|
73
|
Belderbos ME, Jacobs S, Koster TK, Ausema
A, Weersing E, Zwart E, de Haan G and Bystrykh LV: Donor-to-donor
heterogeneity in the clonal dynamics of transplanted human cord
blood stem cells in murine xenografts. Biol Blood Marrow
Transplant. 26:16–25. 2020. View Article : Google Scholar
|
|
74
|
Sun J, Ramos A, Chapman B, Johnnidis JB,
Le L, Ho YJ, Klein A, Hofmann O and Camargo FD: Clonal dynamics of
native haematopoiesis. Nature. 514:322–327. 2014. View Article : Google Scholar
|
|
75
|
Cai WQ, Zeng LS, Wang LF, Wang YY, Cheng
JT, Zhang Y, Han ZW, Zhou Y, Huang SL, Wang XW, et al: The latest
battles between EGFR monoclonal antibodies and resistant tumor
cells. Front Oncol. 10:12492020. View Article : Google Scholar
|
|
76
|
Han ZW, Lyv ZW, Cui B, Wang YY, Cheng JT,
Zhang Y, Cai WQ, Zhou Y, Ma ZW, Wang XW, et al: Correction to: The
old CEACAMs find their new role in tumor immunotherapy. Invest New
Drugs. 38:1899–1900. 2020. View Article : Google Scholar
|
|
77
|
Wang YY, Lyu YN, Xin HY, Cheng JT, Liu XQ,
Wang XW, Peng XC, Xiang Y, Xin VW, Lu CB, et al: Identification of
putative UL54 (ICP27) transcription regulatory sequences binding to
Oct-1, v-Myb, Pax-6 and hairy in herpes simplex viruses. J Cancer.
10:430–440. 2019. View Article : Google Scholar
|
|
78
|
Jensen P and Dymecki SM: Essentials of
recombinase-based genetic fate mapping in mice. Methods Mol Biol.
1092:437–454. 2014. View Article : Google Scholar
|
|
79
|
Herring CA, Chen B, McKinley ET and Lau
KS: Single-cell computational strategies for lineage reconstruction
in tissue systems. Cell Mol Gastroenterol Hepatol. 5:539–548. 2018.
View Article : Google Scholar
|
|
80
|
Liu XQ, Xin HY, Lyu YN, Ma ZW, Peng XC,
Xiang Y, Wang YY, Wu ZJ, Cheng JT, Ji JF, et al: Oncolytic herpes
simplex virus tumor targeting and neutralization escape by
engineering viral envelope glycoproteins. Drug Deliv. 25:1950–1962.
2018. View Article : Google Scholar
|
|
81
|
Woodworth MB, Girskis KM and Walsh CA:
Building a lineage from single cells: Genetic techniques for cell
lineage tracking. Nat Rev Genet. 18:230–244. 2017. View Article : Google Scholar
|
|
82
|
Xu J, Nuno K, Litzenburger UM, Qi Y,
Corces MR, Majeti R and Chang HY: Single-cell lineage tracing by
endogenous mutations enriched in transposase accessible
mitochondrial DNA. Elife. 8:e451052019. View Article : Google Scholar
|