Introduction
A hemangioma is a common benign vascular tumor that
often occurs in childhood and more frequently affects females
(1). Hemangiomas typically follow
a characteristic pattern of a proliferative growth phase, followed
by a slow involuting phase, during which the majority of
hemangiomas stop growing and begin to shrink (2). During the proliferative growth
phase, hemangiomas present with several symptomatic clinical
manifestations, including ulcerations, bleeding, physical
functional limitations and breakdown in the surface of the skin,
and current treatment options comprise medical therapy, laser
therapy and surgical resection (2,3).
However, cases of severe hemangioma are challenging to treat in the
clinic. Hence, further understanding of the mechanisms underlying
hemangioma progression is required.
Long non-coding (lnc) RNAs are a group of non-coding
transcripts of >200 nucleotides in length, which comprise 4–9%
of mammalian transcriptomes (4).
lncRNAs have been discovered to be involved in numerous
physiological and pathobiological processes. For example, previous
studies report that the expression levels of lncRNAs are
dysregulated and thereby implicated in several processes during
malignancy, including malignant cell proliferation, invasion,
migration, epithelial-mesenchymal transition (EMT) and drug
resistance (5–7). Notably, lncRNA maternally expressed
8, small nucleolar RNA host gene (MEG8), as a member of the
lncRNA family, has been found to serve an oncogenic role in several
types of malignancy, including hepatocellular carcinoma (HCC),
non-small cell lung cancer (NSCLC) and pancreatic cancer (8–11).
For instance, lncRNA MEG8 promotes NSCLC cell proliferation,
migration and invasion via targeting the microRNA
(miRNA/miR)-107/CDK6 axis (9). In
addition, lncRNA MEG8 targets several miRNAs
(miR-34a/miR-200b/miR-203) to promote cell
morphological changes and enhance cell motility in tumors (10). In another study, lncRNA
MEG8 regulates the Notch signaling pathway to mediate the EMT
of hepatic stellate cells, thereby serving a role in the
pathological processes of liver fibrosis as well as HCC (12). Of note, the Notch signaling
pathway is a conserved ligand-receptor signaling pathway that
modulates cell-cell interactions and the activity of VEGF, which
has been shown to serve a role in the pathogenesis of hemangioma
(13). Therefore, it was
hypothesized that lncRNA MEG8 may be involved in the
pathological process of hemangiomas via targeting possible miRNAs
(miR-34a/miR-200b/miR-203) and Notch signaling
pathway; however, the underlying mechanism remains to be
elucidated.
The current study aimed to investigate the effect of
lncRNA MEG8 knockdown on cell proliferation, apoptosis and
invasion, as well as to determine its possible molecular mechanism
in hemangioma.
Materials and methods
Cell lines and culture
Human hemangioma endothelial cells (HemECs) were
isolated from 3 individuals (2 females and 1 male, mean age of 14
months with range 9–24 months) with proliferating-phase hemangioma
as previously described (14).
The HemECs were cultured in Human Endothelial Serum Free medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and maintained in a
humidified incubator with 5% CO2 at 37°C. The
experimental protocol was approved by the Ethics Committee of
Handan Seventh Hospital (approval no. 2019[K]016) and written
informed consent was obtained from the parents/guardians of each
patient prior to participation.
Cell transfection
lncRNA MEG8 small interfering RNA (siRNA),
negative control (NC) siRNA and miR-203 inhibitor were
purchased from Shanghai GenePharma Co., Ltd. The sequences for
lncRNA MEG8 siRNA, NC siRNA, miR-203 inhibitor and NC inhibitor
were as follows: lncRNA MEG8 sense, 5′GGCCAGCUGAUUUAAUAAUUU3′,
lncRNA MEG8 antisense, 5′AUUAUUAAAUCAGCUGGCCUU3′; NC siRNA sense,
5′GAAUUAAUUAAAGAUGGCCCGUUGUAC3′, NC siRNA antisense,
5′UCAUCGAAGUUAUAGGGAUACAUUACGUGAUC3′; miR-203 inhibitor,
5′CUAGUGGUCCUAAACAUUUCAC3′; NC inhibitor,
5′CAGUACUUUUGUGUAGUACAA3′. A total of 2×105 HemECs were
seeded into 6-well plates 24 h prior to transfection. Following
incubation, the HemECs were transfected with 50 pM NC siRNA, 50 pM
lncRNA MEG8 siRNA, 50 pM NC inhibitor, 50 pM miR-203
inhibitor or 50 pM lncRNA MEG8 siRNA + 50 pM miR-203
inhibitor using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) for 6 h at 37°C, according to the
manufacturer's protocol. Untransfected HemECs were used as the
control.
miR-203 transfection efficiency
assessment
NC inhibitor, NC mimic and miR-203 mimic were
purchased from Shanghai GenePharma Co. The NC mimic, miR-203
mimic, NC inhibitor and miR-203 inhibitor were transfected
into HemECs with the application of Lipofectamine 2000 reagent as
aforementioned. The HemECs without transfection were served as
normal control. The transfection efficacy was validated by
detecting miR-203 expression at 24 h post transfection, as
shown in Fig. S1.
Reverse transcription-quantitative
(RT-q) PCR
At 24 h post-transfection, 2×106 HemECs
were harvested and underwent RT-qPCR. Briefly, total RNA was
extracted from cells using a RNeasy Mini kit (Qiagen GmbH) and
reverse transcribed into cDNA using a cDNA Reverse Transcription
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according
to the kit's instructions. qPCR was subsequently performed using a
SYBR™ Green PCR Master mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) by following the kit's protocol. The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 95°C for 10 min; followed by 40 cycles of 95°C for
15 sec and 61°C for 1 min. LncRNA (lncRNA MEG8) and mRNA
[jagged canonical notch ligand 1 (JAG1), NOTCH1] were
normalized to β-actin and miRNAs (miR-34a, miR-200b,
miR-203) were normalized to U6. The experiments were
repeated 3 times. These gene expression levels were quantified
using the 2−ΔΔCq method (15). The primers used for the qPCR are
listed in Table I.
 | Table I.Primers. |
Table I.
Primers.
| Gene | Forward
(5′→3′) | Reverse
(5′→3′) |
|---|
| lncRNA
MEG8 |
GCCACCAGCCTTATGATTGC |
TCCTAACACAGAGAACCAACCAT |
| JAG1 |
TGGTTAATGGTTATCGCTGTATCTG |
ATAGTCACTGGCACGGTTGTAG |
| NOTCH1 |
CAGAGGCGTGGCAGACTATG |
GGCAGTGGCAGATGTAGGAG |
| β-actin |
TCGTGCGTGACATTAAGGAGAAG |
AGGACTCCATGCCCAGGAA |
| miR-34a |
ACACTCCAGCTGGGTGGCAGTGTCTTAGCT |
TGTCGTGGAGTCGGCAATTC |
|
miR-200b |
ACACTCCAGCTGGGCATCTTACTGGGCAGC |
TGTCGTGGAGTCGGCAATTC |
| miR-203 |
ACACTCCAGCTGGGGTGAAATGTTTAGGAC |
TGTCGTGGAGTCGGCAATTC |
| U6 |
GCTCGCTTCGGCAGCACATA |
AATATGGAACGCTTCACGAATTTGC |
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was analyzed using a CCK-8 assay
(Sigma-Aldrich; Merck KGaA) at 0, 24, 48 or 72 h post-transfection.
Briefly, CCK-8 reagent was added to the cells at each time point
and incubated for a further 2 h. The optical density value was
measured using a microplate reader (BioTek Instruments, Inc.) at
450 nm.
Flow cytometric analysis of cell
apoptosis
HemECs were harvested for apoptosis analysis at 48 h
post-transfection. Briefly, HemECs were centrifuged (800 g, 5 min,
4°C) and resuspended, then incubated with Annexin V-FITC
(eBioscience; Thermo Fisher Scientific, Inc.) and PI (eBioscience;
Thermo Fisher Scientific, Inc.) in the dark for 20 min at room
temperature. Apoptotic HemECs were analyzed using a flow cytometer
(FACSCalibur; BD Biosciences) and a FlowJo 7.6 software (BD
Biosciences). The apoptosis rate was defined as percentage of early
plus late apoptotic cells.
Transwell assay
The invasive ability of HemECs was analyzed using a
Transwell assay. At 24 h post-transfection, cells were collected
and seeded into Matrigel basement membrane matrix-coated Transwell
inserts (Corning, Inc.). The medium in upper and lower chambers was
Human Endothelial Serum Free medium (Gibco; Thermo Fisher
Scientific, Inc.) and 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.), respectively. After incubation for another 24 h, the cells
remaining in the top of the inserts were removed and the cells in
the bottom were fixed with 10% formalin (Sigma-Aldrich; Merck KGaA)
for 15 min at room temperature and stained with crystal violet
(Sigma-Aldrich; Merck KGaA) for 20 min at room temperature.
Invasive cells were observed using an inverted microscope (Nikon
Corporation) at the magnification of ×200, with 5 fields being
chosen randomly.
Western blotting
Total protein was extracted from HemECs at 48 h
post-transfection using RIPA lysis buffer (Sigma-Aldrich; Merck
KGaA). Total protein was quantified using a BCA Protein assay kit
(Thermo Fisher Scientific, Inc.) and 20 µg protein/lane was
separated using 4–20% precast polyacrylamide gels (Sigma-Aldrich;
Merck KGaA). The separated proteins were subsequently transferred
onto nitrocellulose filter membranes (Pall Life Sciences) and
blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) for 1.5 h. The
membranes were then incubated with primary antibodies at 4°C
overnight. Following the primary antibody incubation, the membranes
were incubated with a secondary antibody for 1.5 h. Protein bands
were visualized using ECL western blotting substrate (Thermo Fisher
Scientific, Inc.). The protein density was evaluated with ImageJ
(Version 1.8.0, National Institutes of Health). The antibodies used
for western blotting are listed in Table II.
 | Table II.Antibodies. |
Table II.
Antibodies.
| Antibody | Company | Dilution |
|---|
| Primary
antibodies |
|
|
| Rabbit
polyclonal to Cleaved caspase-3 | Abcam | 1:500 |
| Rabbit
monoclonal to Cleaved caspase-7 | Abcam | 1:1,000 |
| Rabbit
polyclonal to JAG1 | Invitrogen; Thermo
Fisher Scientific, Inc. | 1:1,000 |
| Rabbit
polyclonal to NOTCH1 | Invitrogen; Thermo
Fisher Scientific, Inc. | 1:2,000 |
| Rabbit
monoclonal to β-actin | Invitrogen; Thermo
Fisher Scientific, Inc. | 1:2,000 |
| Secondary
antibody |
|
|
| Goat
Anti-Rabbit IgG H&L (HRP) | Invitrogen; Thermo
Fisher Scientific, Inc. | 1:50,000 |
Dual luciferase reporter gene
assay
The wild-type (WT) or mutant type (MT) lncRNA
MEG8 sequences were cloned into a pGLuc plasmid (Beyotime
Institute of Biotechnology) to generate WT or MT plasmids. NC mimic
or miR-203 mimic were co-transfected with WT or MT plasmid into
HemECs with the application of Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Following 48
h of transfection, the cells were harvested and the relative
luciferase activity was measured using a Dual Luciferase Reporter
Gene assay kit (Beyotime Institute of Biotechnology).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 7.02 software (GraphPad Software, Inc.) and presented as the
mean ± standard deviation. Statistical differences between two
groups were determined using an unpaired Student's t-test, while
the comparisons among multiple groups were performed using a
one-way ANOVA followed by a Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Effect of lncRNA MEG8 knockdown on the
proliferation, apoptosis and invasion of HemECs
The relative expression levels of lncRNA MEG8
were downregulated following the transfection with lncRNA
MEG8 siRNA (P<0.001), which suggested the successful
transfection of the siRNA (Fig.
1A). In addition, lncRNA MEG8 siRNA also decreased cell
proliferation following 48 (P<0.05) or 72 h (P<0.05) of
transfection (Fig. 1B).
Conversely, cell apoptosis was increased by transfection with
lncRNA MEG8 siRNA (P<0.01; Fig. 1C-E). Furthermore, the number of
invasive cells was decreased following the transfection with
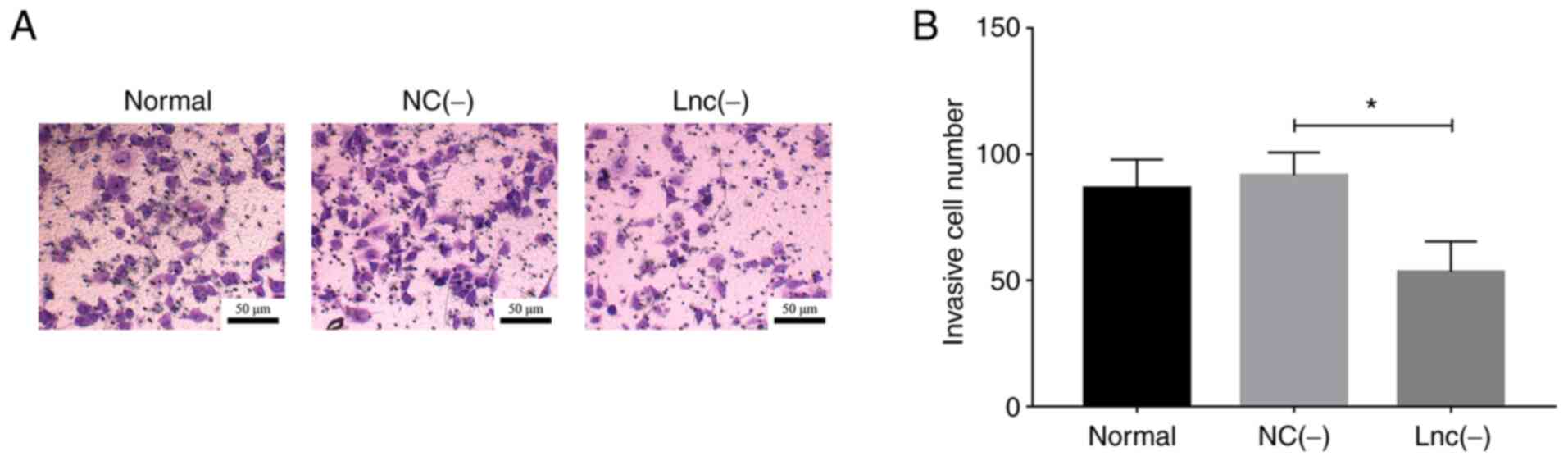
lncRNA MEG8 siRNA (P<0.05; Fig. 2A and B).
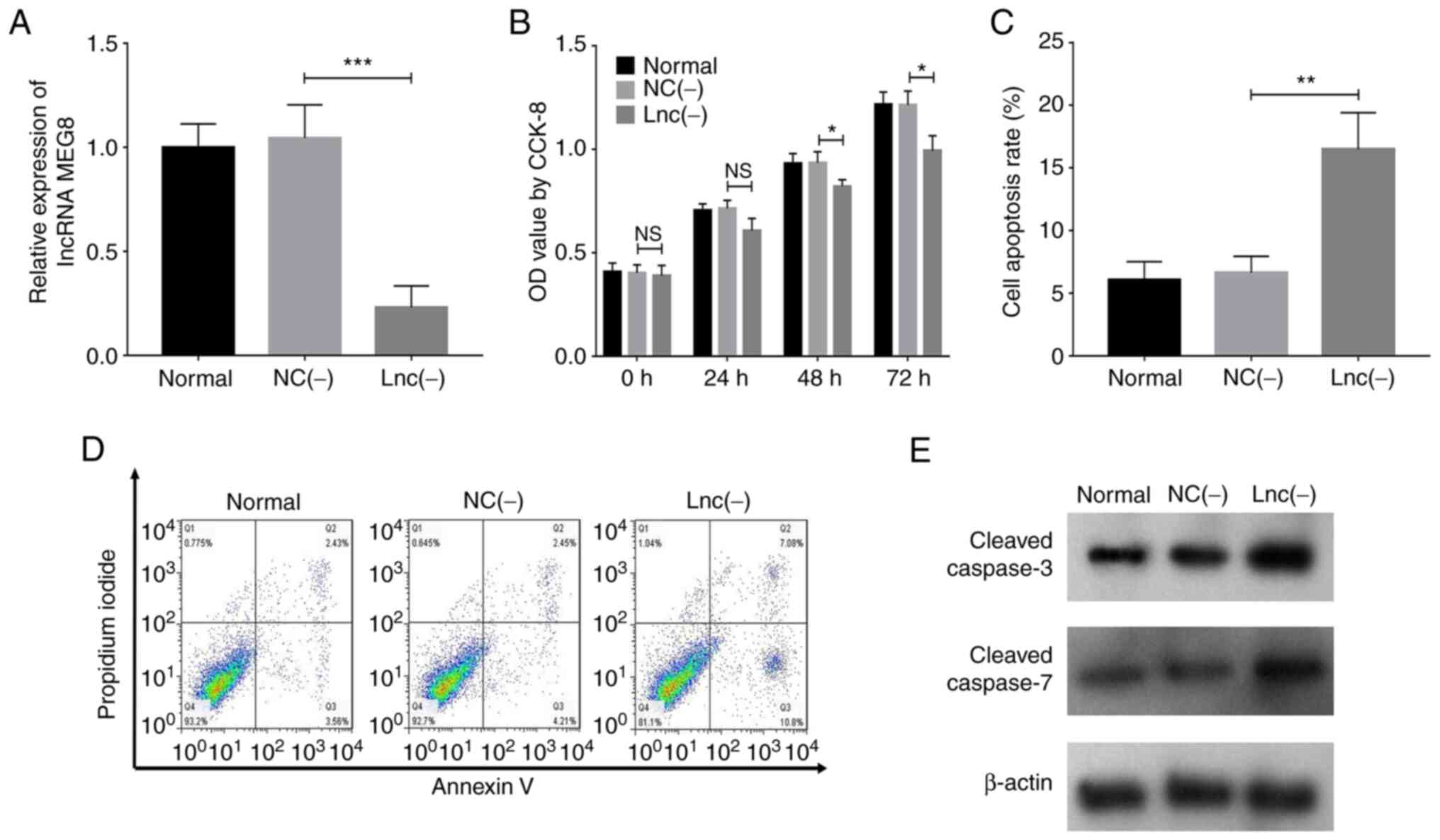 | Figure 1.lncRNA MEG8 knockdown inhibits
the proliferation but promotes the apoptosis of hemangioma
endothelial cells. (A) Analysis of lncRNA MEG8 expression
levels, (B) cell proliferation, (C and D) cell apoptosis and (E)
cleaved caspase-3 and caspase-7 protein expression levels in the
NC(−) and lnc(−) groups. Statistical differences among groups were
determined using a one-way ANOVA followed by a Tukey's post hoc
test. All experiments were conducted in triplicate. *P<0.05,
**P<0.01, ***P<0.001. lncRNA, long non-coding RNA;
MEG8, maternally expressed 8, small nucleolar RNA host gene;
NC, negative control; lnc (−), cells transfected with lncRNA
MEG8 small interfering RNA; OD, optical density. |
Effect of lncRNA MEG8 knockdown on
miR-34a, miR-200b and miR-203 expression levels in HemECs
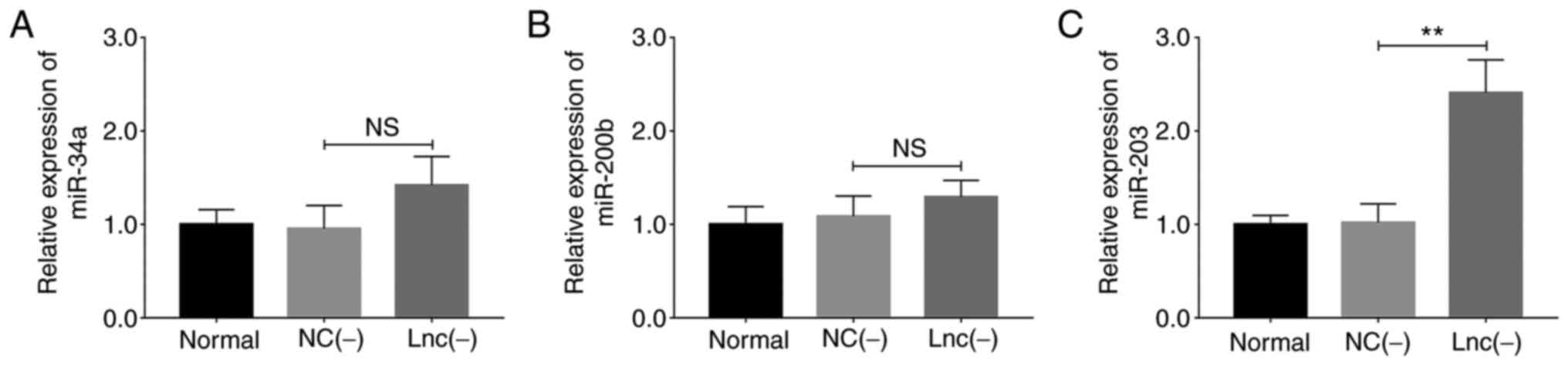
Transfection with lncRNA MEG8 siRNA exerted
no significant effect on miR-34a (P>0.05; Fig. 3A) and miR-200b (P>0.05;
Fig. 3B) expression levels;
however, miR-203 expression levels were upregulated
(P<0.01; Fig. 3C).
Association between lncRNA MEG8 and
miR-203 in HemECs
The binding site between WT lncRNA MEG8 and
miR-203 is shown in Fig.
4A. The results of the dual luciferase reporter gene assay
indicated that the knockdown of miR-203 decreased the
relative luciferase activity of the lncRNA MEG8 WT vector
(P<0.01), but did not affect the relative luciferase activity of
the lncRNA MEG8 MT vector (P>0.05; Fig. 4B). Transfection with the
miR-203 inhibitor did not significantly alter lncRNA
MEG8 expression levels (P>0.05; Fig. 4C), whereas the transfection with
lncRNA MEG8 siRNA upregulated miR-203 expression
levels (P<0.001; Fig. 4D).
These findings indicated that lncRNA MEG8 may negatively
regulate miR-203 expression.
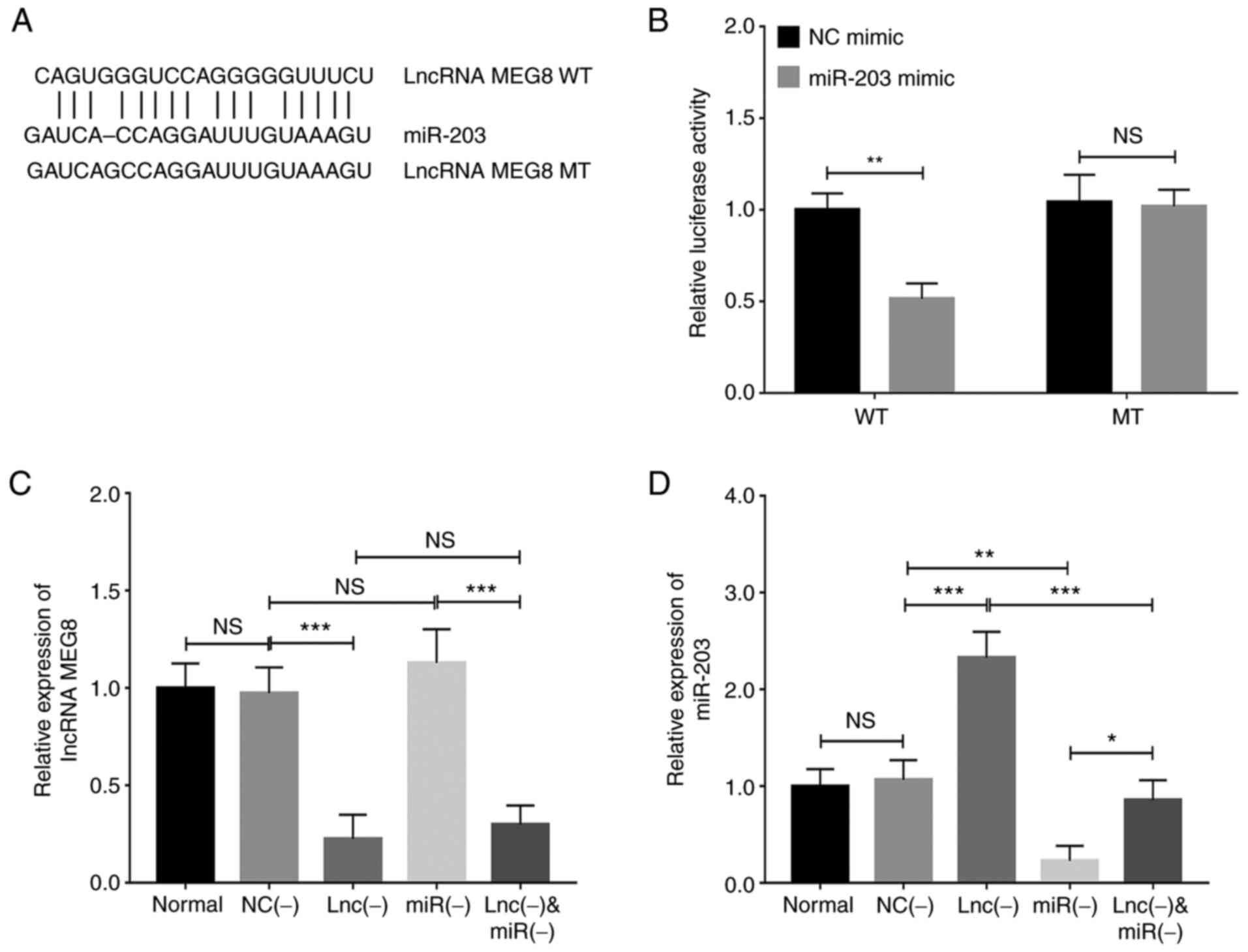 | Figure 4.lncRNA MEG8 knockdown
upregulates miR-203 expression in hemangioma endothelial
cells. (A) Binding sites between WT lncRNA MEG8 and
miR-203 are shown. (B) Relative luciferase activity in cells
co-transfected with miR-203 mimic or NC mimic and WT or MT
vectors. (C) lncRNA MEG8 and (D) miR-203 expression
levels were analyzed in the normal, NC (−), lnc (−), miR (−) and
lnc (−) + miR (−) groups. Statistical differences between two
groups were determined using an unpaired Student's t-test, while
statistical differences between multiple groups were determined
using a one-way ANOVA followed by a Tukey's post hoc test. All
experiments were conducted in triplicate. *P<0.05, **P<0.01,
***P<0.001. lncRNA, long non-coding RNA; MEG8, maternally
expressed 8, small nucleolar RNA host gene; miR, microRNA; WT,
wild-type; MT, mutant type; NC, negative control; lnc (−), cells
transfected with lncRNA MEG8 small interfering RNA; miR (−),
cells transfected with the miR-203 inhibitor; NS, not
significant. |
Knockdown of lncRNA MEG8 regulates
miR-203-induced regulation of proliferation and apoptosis in
HemECs
Transfection with the miR-203 inhibitor
increased cell proliferation at 48 (P<0.05) and 72 h
(P<0.05), whereas the co-transfection with the miR-203
inhibitor reversed the effect of lncRNA MEG8 siRNA on cell
proliferation at 48 (P<0.05) and 72 h (P<0.05; Fig. 5A). Furthermore, transfection with
the miR-203 inhibitor suppressed cell apoptosis (P<0.001)
and reversed the effect of lncRNA MEG8 siRNA on cell
apoptosis (P<0.001; Fig.
5B-D).
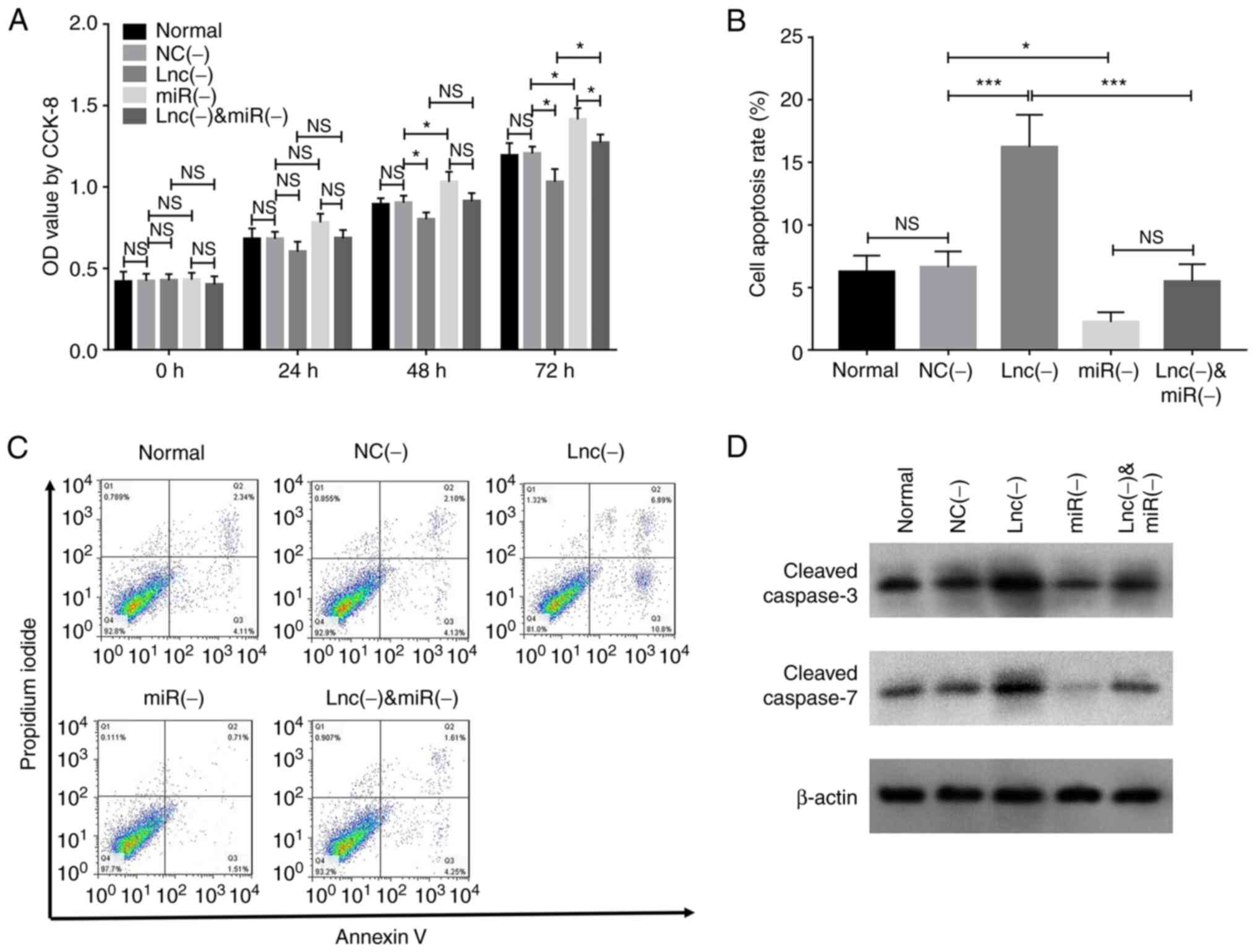 | Figure 5.miR-203 inhibitor rescues the
effects of lncRNA MEG8 knockdown on the proliferation and
apoptosis of hemangioma endothelial cells. (A) Cell proliferation,
(B and C) cell apoptosis and (D) cleaved caspase-3 and caspase-7
protein expression levels in the normal, NC (−), lnc (−), miR (−)
and lnc (−) + miR (−) groups were analyzed. Statistical differences
among groups were determined using a one-way ANOVA followed by a
Tukey's post hoc test. All experiments were conducted in
triplicate. *P<0.05, ***P<0.001. lncRNA, long non-coding RNA;
MEG8, maternally expressed 8, small nucleolar RNA host gene;
miR, microRNA; NC, negative control; lnc (−), cells transfected
with lncRNA MEG8 small interfering RNA; miR (−), cells
transfected with the miR-203 inhibitor; OD, optical density;
NS, not significant. |
Knockdown of lncRNA MEG8 regulates the
miR-203-induced regulation of invasion in HemECs
Transfection with the miR-203 inhibitor
promoted cell invasion (P<0.01) and reversed the effect of
lncRNA MEG8 siRNA on cell invasion (P<0.01; Fig. 6A and B).
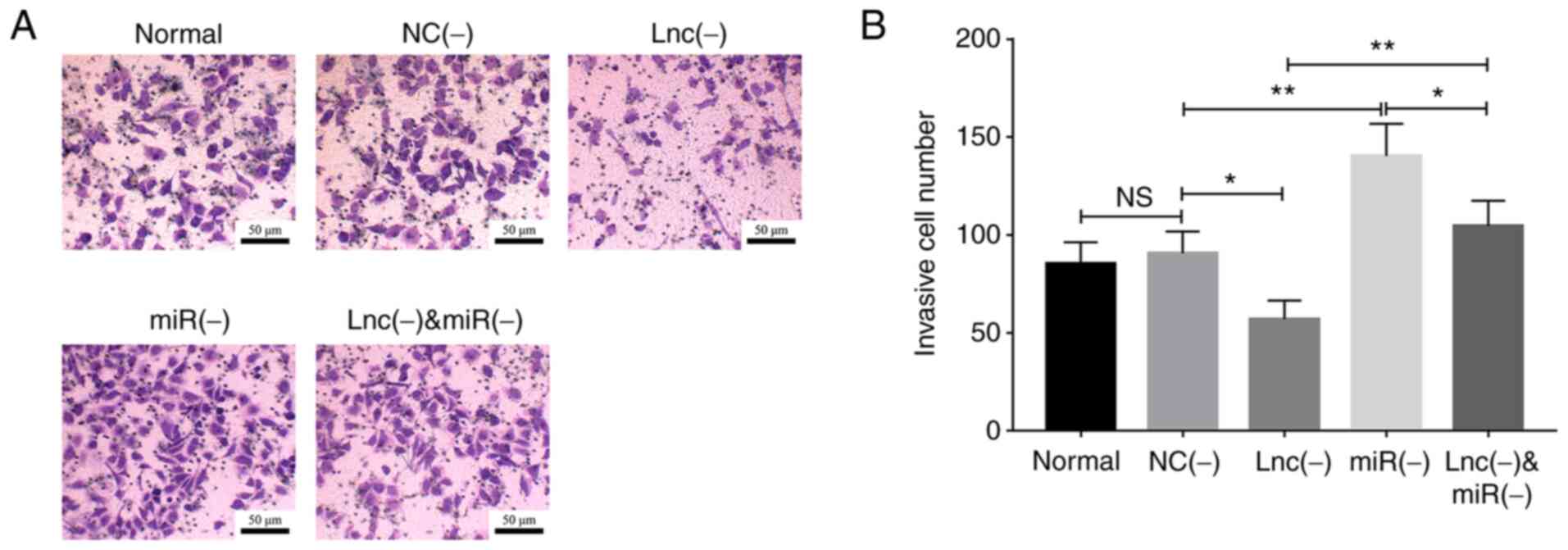 | Figure 6.miR-203 inhibitor rescues the
effects of lncRNA MEG8 knockdown on the invasion of
hemangioma endothelial cells. (A and B) Cell invasion was analyzed
in the normal, NC (−), lnc (−), miR (−) and lnc (−) + miR (−)
groups. Statistical differences among groups were determined using
a one-way ANOVA followed by a Tukey's post hoc test. All
experiments were conducted in triplicate. *P<0.05, **P<0.01.
lncRNA, long non-coding RNA; MEG8, maternally expressed 8,
small nucleolar RNA host gene; miR, microRNA; NC, negative control;
lnc (−), cells transfected with lncRNA MEG8 small
interfering RNA; miR (−), cells transfected with the miR-203
inhibitor; NS, not significant. |
Knockdown of lncRNA MEG8 regulates the
miR-203-induced mediation of the Notch signaling pathway in
HemECs
Transfection with lncRNA MEG8 siRNA
downregulated JAG1 mRNA expression levels (P<0.05), while
the miR-203 inhibitor upregulated JAG1 mRNA
expression levels (P<0.01) and reversed the effect of lncRNA
MEG8 siRNA on JAG1 mRNA expression in HemECs (P<0.01;
Fig. 7A). Moreover, transfection
with lncRNA MEG8 siRNA downregulated Notch1 mRNA
expression levels (P<0.05); however, the miR-203
inhibitor upregulated the mRNA expression levels of Notch1
(P<0.01) and reversed the effect of lncRNA MEG8 siRNA on
Notch1mRNA expression (P<0.01; Fig. 7B). These data were further
validated by western blotting (Fig.
7C).
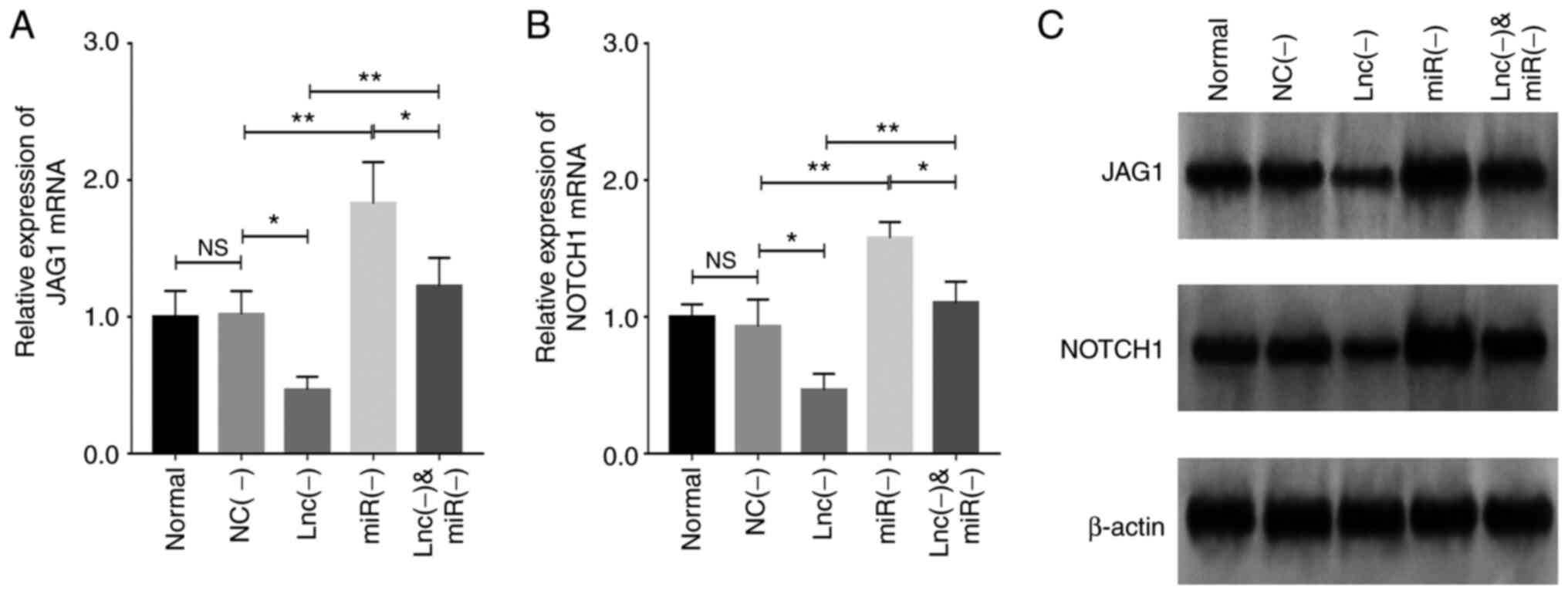 | Figure 7.miR-203 inhibitor rescues the
effects of lncRNA MEG8 knockdown on the Notch signaling
pathway in hemangioma endothelial cells. (A) JAG1 and (B)
Notch1 mRNA expression levels were analyzed in the normal,
NC (−), lnc (−), miR (−) and lnc (−) + miR (−) groups. (C) Jag1 and
Notch1 protein expression levels were analyzed in the normal, NC
(−), lnc (−), miR (−) and lnc (−) + miR (−) groups. Statistical
differences among groups were determined using a one-way ANOVA
followed by a Tukey's post hoc test. All experiments were conducted
in triplicate. *P<0.05, **P<0.01. lncRNA, long non-coding
RNA; miR, microRNA; MEG8, maternally expressed 8, small
nucleolar RNA host gene; NC, negative control; lnc (−), cells
transfected with lncRNA MEG8 small interfering RNA; miR (−), cells
transfected with miR-203 inhibitor; JAG1, jagged
canonical notch ligand 1; NS, not significant. |
Discussion
lncRNA MEG8 has been reported to serve a role in the
tumorigenesis of several tumor types (8–12).
For example, the expression levels of lncRNA MEG8 are
upregulated in HCC tissues and cells and the knockdown of lncRNA
MEG8 represses the proliferative, migratory and invasive
abilities of NSCLC cells via the miR-107/CDK6 signaling axis
(9). Another study revealed that
in HCC, lncRNA MEG8 regulates the TGF-β-mediated
epigenetic progression of EMT and further promotes EMT-related cell
morphological changes and migration in lung and pancreatic cancer
cells (10). Furthermore,
lncRNA MEG8 has a regulatory role over the Notch signaling
pathway in HCC and activation of the Notch signaling pathway serves
an important role in the proliferative and involuted phases of
hemangioma (16). Regarding the
role of lncRNA MEG8 in hemangioma, only one previous
microarray analysis has reported that the expression levels of
lncRNA MEG8 are upregulated in hemangioma tumor tissues
compared with adjacent normal specimens (17). However, to the best of the
authors' knowledge, the detailed underlying mechanism of the role
of lncRNA MEG8 in hemangioma has not been investigated and
was therefore the focus of the current study.
The findings of the present study revealed that
lncRNA MEG8 knockdown inhibited the proliferation and
invasion, but promoted the apoptosis, of HemECs. The results
observed in the present study may be due to several different
reasons. For example, according to previous studies, lncRNA
MEG8 knockdown inhibits adipogenesis via regulating peroxisome
proliferator activated receptor (PPAR)-α expression and
adipogenic differentiation-related genes (such as PPAR-γ),
further suppressing the proliferation and invasion, but enhancing
the apoptosis of HemECs (18,19). Furthermore, as lncRNA MEG8
was previously demonstrated to modulate the activation of hepatic
stellate cells and the EMT of hepatocytes via the Notch signaling
pathway (12), and based on the
evidence that Notch ligands were found to be involved in the
stimulation of VEGF signaling and angiogenesis in hemangioma
(13), lncRNA MEG8
knockdown may inhibit the development and progression of hemangioma
via regulating Notch signaling. Finally, lncRNA MEG8
knockdown may serve as a competing endogenous RNA of tumor-related
miRNAs (such as miR-34a, miR-200b and miR-203)
(9,11,20), and regulate oncogenic signaling
pathways, thereby promoting HemEC proliferation and invasion, but
inhibiting apoptosis. However, further investigations are
required.
Furthermore, lncRNA MEG8 has been found to
target several miRNAs (such as miR-34a, miR-200b and
miR-203) to promote cellular morphological changes and
enhance cell motility in lung and pancreatic cancer (10). The results of the present study
were that lncRNA MEG8 knockdown upregulated miR-203
expression; however, miR-203 silencing did not affect
lncRNA MEG8 expression, suggesting that lncRNA MEG8
may be involved in the pathological process of hemangioma via
targeting miR-203. Moreover, in the current study,
miR-203 silencing could reverse the effects of lncRNA
MEG8 knockdown on cell proliferation, apoptosis and invasion.
According to a previous study, lncRNA MEG8 knockdown was
associated with the recruitment of enhancer of zeste 2 polycomb
repressive complex 2 subunit (EZH2), which is involved in
the transcriptional repression of targeting tumor-suppressor genes
and regulated the EZH2-containing polycomb repressive
complex 2, subsequently upregulating miR-203 expression and
EMT activity, which inhibits the progression of hemangioma
(9,21). These findings may explain the
results obtained in the present study; however, further
experimental studies are required for verification.
In addition, it is known that Notch receptors bind
with their ligands to activate the Notch signaling pathway and the
disrupted expression of core components of the Notch signaling
pathway has been found to induce abnormal angiogenesis in
hemangioma (16). To determine
the effects of lncRNA MEG8 and miR-203 on the Notch
signaling pathway, experiments were conducted and the results
revealed that lncRNA MEG8 knockdown inactivated the Notch
signaling pathway, while miR-203 silencing reversed the
inactivating effect of lncRNA MEG8 knockdown on the Notch
signaling pathway in HemECs, suggesting that lncRNA MEG8
knockdown inactivated the Notch signaling pathway via targeting
miR-203 in hemangioma. As lncRNA MEG8 was found to
exert a regulatory effect on TGF-β expression, which belongs
to the growth factor family and is closely associated with the
Notch signaling pathway, and considering that miR-203
attenuates the TGF-β signaling pathway in breast and ovarian
cancer (10), it was hypothesized
that lncRNA MEG8 knockdown may regulate
TGF-β-associated components and mediate their
anti-angiogenic effects via targeting miR-203, thereby
inactivating the Notch signaling pathway in hemangioma (10,22–24). However, further experimental
studies are required to explore this hypothesis and determine
whether this was the mechanism underlying the results of the
present study.
However, the present study still has some
limitations. Previous evidence indicates that lncRNA MEG8 is
upregulated in infantile hemangioma tissues compared with adjacent
non-tumor tissues (17) and other
studies observe that lncRNA MEG8 serves as an oncogenic gene
in several tumors (9–11) Therefore, the present study did not
detect the effect of lncRNA MEG8 overexpression on pro-tumor
properties of hemangioma cells. The information might be
informative, which needed exploration in further studies.
In conclusion, lncRNA MEG8 knockdown
inhibited cell proliferation and invasion, but promoted apoptosis
in hemangioma via miR-203-induced mediation of the Notch
signaling pathway. This suggested that targeting lncRNA MEG8
might mediate the miR-203-induced Notch signaling pathway,
inhibiting hemangioma progression and providing novel treatment
targets for hemangioma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by Program of Hebei Medical
Science Research (grant no. 20200582).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
ZH and XL contributed to the study design and
manuscript writing. ZH, XL, JG, LZ, YC and HY conducted literature
research and isolated and cultured hemangioma endothelial cells. ZH
and XL contributed to the data acquisition and analysis. The
figures are the authors' own work. All authors read and approved
the final manuscript. ZH and XL confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Handan Seventh Hospital and written informed consent
was obtained from the parents/guardians of each patient prior to
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeHart A and Richter G: Hemangioma: Recent
advances. F1000Res. 8:F1000 Faculty Rev-1926. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li X, Chen B, Chi D, Zhang Y and Jiang W:
lncRNA CASC9 regulates cell migration and invasion in hemangioma
endothelial cells by targeting miR-125a-3p/Nrg1. Onco Targets Ther.
12:423–432. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Valdebran M and Wine Lee L:
Hemangioma-related syndromes. Curr Opin Pediatr. 32:498–505. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin H, Wang J, Wang T, Wu J, Wang P, Huo
X, Zhang J, Pan H and Fan Y: The LncRNA MIR503HG/miR-224-5p/TUSC3
signaling cascade suppresses gastric cancer development via
modulating ATF6 Branch of unfolded protein response. Front Oncol.
11:7085012021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cen X, Huang Y, Lu Z, Shao W, Zhuo C, Bao
C, Feng S, Wei C, Tang X, Cen L, et al: LncRNA IGFL2-AS1 promotes
the proliferation, migration, and invasion of colon cancer cells
and is associated with patient prognosis. Cancer Manag Res.
13:5957–5968. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lou J, Yan W, Li QY, Zhu AK, Tan BQ, Dong
R, Zou XZ and Liu T: LncRNA MEG8 plays an oncogenic role in
hepatocellular carcinoma progression through
miR-367-3p/14-3-3ζ/TGFβR1 axis. Neoplasma. 68:273–282. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Li L, Shang P and Song X: LncRNA
MEG8 promotes tumor progression of non-small cell lung cancer via
regulating miR-107/CDK6 axis. Anticancer Drugs. 31:1065–1073. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Terashima M, Ishimura A, Wanna-Udom S and
Suzuki T: MEG8 long noncoding RNA contributes to epigenetic
progression of the epithelial-mesenchymal transition of lung and
pancreatic cancer cells. J Biol Chem. 293:18016–18030. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo K, Qi D and Huang B: LncRNA MEG8
promotes NSCLC progression by modulating the
miR-15a-5p-miR-15b-5p/PSAT1 axis. Cancer Cell Int. 21:842021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen T, Lin H, Chen X, Li G, Zhao Y, Zheng
L, Shi Z, Zhang K, Hong W and Han T: LncRNA Meg8 suppresses
activation of hepatic stellate cells and epithelial-mesenchymal
transition of hepatocytes via the Notch pathway. Biochem Biophys
Res Commun. 521:921–927. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji Y, Chen S, Li K, Li L, Xu C and Xiang
B: Signaling pathways in the development of infantile hemangioma. J
Hematol Oncol. 7:132014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khan ZA, Melero-Martin JM, Wu X, Paruchuri
S, Boscolo E, Mulliken JB and Bischoff J: Endothelial progenitor
cells from infantile hemangioma and umbilical cord blood display
unique cellular responses to endostatin. Blood. 108:915–921. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang H, Wei T, Johnson A, Sun R, Richter
G and Strub GM: NOTCH pathway activation in infantile hemangiomas.
J Vasc Surg Venous Lymphat Disord. 9:489–496. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu X, Lv R, Zhang L, Xu G, Bi J, Gao F,
Zhang J, Xue F, Wang F, Wu Y, et al: Long noncoding RNA expression
profile of infantile hemangioma identified by microarray analysis.
Tumour Biol. Oct 5–2016.(Epub ahead of print). doi:
10.1007/s13277-016-5434-y. View Article : Google Scholar
|
|
18
|
Zhang B, Dong Y and Zhao Z: LncRNA MEG8
regulates vascular smooth muscle cell proliferation, migration and
apoptosis by targeting PPARα. Biochem Biophys Res Commun.
510:171–176. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan SM, Guo Y, Xu Y, Wang M, Chen HN and
Shen WM: The adipogenesis in infantile hemangioma and the
expression of adipogenic-related genes. Int J Clin Exp Pathol.
10:11596–11602. 2017.PubMed/NCBI
|
|
20
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eich ML, Athar M, Ferguson JE III and
Varambally S: EZH2-targeted therapies in cancer: Hype or a reality.
Cancer Res. 80:5449–5458. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carmeliet P and Jain RK: Molecular
mechanisms and clinical applications of angiogenesis. Nature.
473:298–307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He S, Zhang G, Dong H, Ma M and Sun Q:
MiR-203 facilitates tumor growth and metastasis by targeting
fibroblast growth factor 2 in breast cancer. Onco Targets Ther.
9:6203–6210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang B, Li X, Zhao G, Yan H, Dong P,
Watari H, Sims M, Li W, Pfeffer LM, Guo Y and Yue J: MiR-203
inhibits ovarian tumor metastasis by targeting BIRC5 and
attenuating the TGFβ pathway. J Exp Clin Cancer Res. 37:2352018.
View Article : Google Scholar : PubMed/NCBI
|