Introduction
Proteins are synthesized in the cytosol by free ribosomes or by endoplasmic reticulum (ER)-bound ribosomes using mRNA as a template, ribosomal RNA (rRNA) as an assembler and transfer RNA as a carrier. However, all types of single-stranded RNA are created based on a DNA template in the nucleus and exported into the cytoplasm after chemical modification, including alternative splicing, 3′ cap addition and 5′ polyadenylation. Therefore, small mRNAs become structurally simple due to modification processes and carry the genetic information from nuclear DNA. Translation proceeds in three phases: Initiation, elongation and termination (1). In the co-translational translocation of eukaryotes, the entire ribosome/mRNA complex binds to the rough ER in order to synthesize new signal-peptide-containing proteins, which are anchored to the ER membrane for externalization to the plasma membrane and other organelles, or stored in the ER for future vesicle transport and secretion outside the cell (Fig. 1) (1).
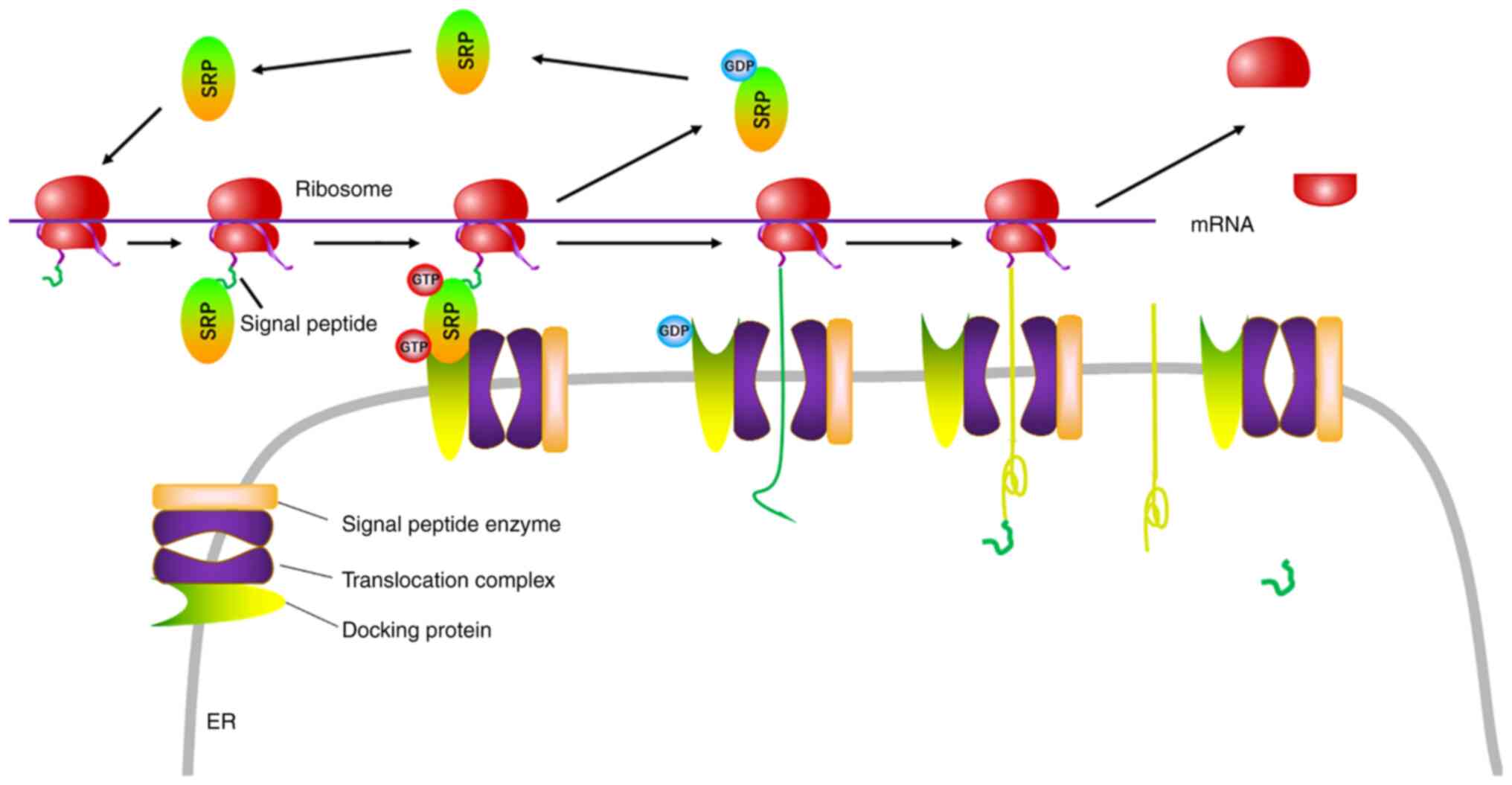 |
Figure 1.
Biosynthesis of transmembrane and secretory proteins. The ribosome is composed of 18S and 28S rRNA, in the middle of which mRNA binds to rRNA via the KOZAK sequence. In the ribosome, mRNA guides the biosynthesis of the signal peptide, which interacts with SRPs to anchor to the rough ER via docking protein. When the translation of transmembrane and secretory proteins is finished, the signal peptide is digested and left in the ER. SRP, signal recognition particle; ER, endoplasmic reticulum; rRNA, ribosomal RNA.
|
For proteins to be transported into the nucleus, their nuclear localization sequence (NLS) or other specific space structure needs to be recognized by the importin-karyopherin nuclear transport receptors. These proteins dock at the nuclear pore complex by interacting with the phenylalanine-glycine domain, and are then moved into the nucleus by importin, while exportin has the opposite function targeting nuclear export sequence (NES). Importin and exportin are hypothesized to be involved in protein shuttling between the nucleus and the cytoplasm (2). During this process, the transport receptors bind to nucleoporins and the small GTPase, Ran, which influences the interaction of transport receptors with cargo, adaptors and nucleoporins independently of GTP hydrolysis by Ran (2). Reportedly, a linker that is >120 amino acids long is needed for the interaction between NLS-carrying membrane proteins and transport factors, such as karyopherin-α (2).
After translation, new proteins then undergo post-translational modifications to form mature products in the ER and Golgi apparatus, including enzyme cleavage, chemical repertoire, phosphorylation, glycosylation, lipidation, acylation, alkylation, butyrylation, biotinylation, sumoylation and ubiquitination. Such chemical modifications alter the spatial structure that is necessary for the interaction with transport factors or their partner proteins (1–6). Transmembrane and receptor proteins undergo endocytosis via an adaptor protein and clathrin or are hydrolyzed and released into the cytosol. The membrane adaptor proteins are separated from them upon signal activation (1–6). The cytosolic proteins, especially chemically modified ones, enter the nucleus by simple diffusion, facilitated diffusion via nuclear pore action proteins and signal-mediated nuclear input via importin (Fig. 2). Meanwhile, the nuclear proteins can be exported out of the nucleus via signal-mediated nuclear output, facilitated diffusion or simple diffusion, and relocalize to membranes for signal transduction (5,6). The present review summarizes which cytosolic proteins shuttle between the plasma membrane and nucleus and how this occurs. The main nucleomembrane shuttling proteins include a variety of receptors of growth factors and cytokines, membrane adaptor proteins, adhesive proteins and nuclear proteins. The nuclear export and import proteins differ in features including their original localization, amino acid sequence (NLS or NES) and biological functions (Table I).
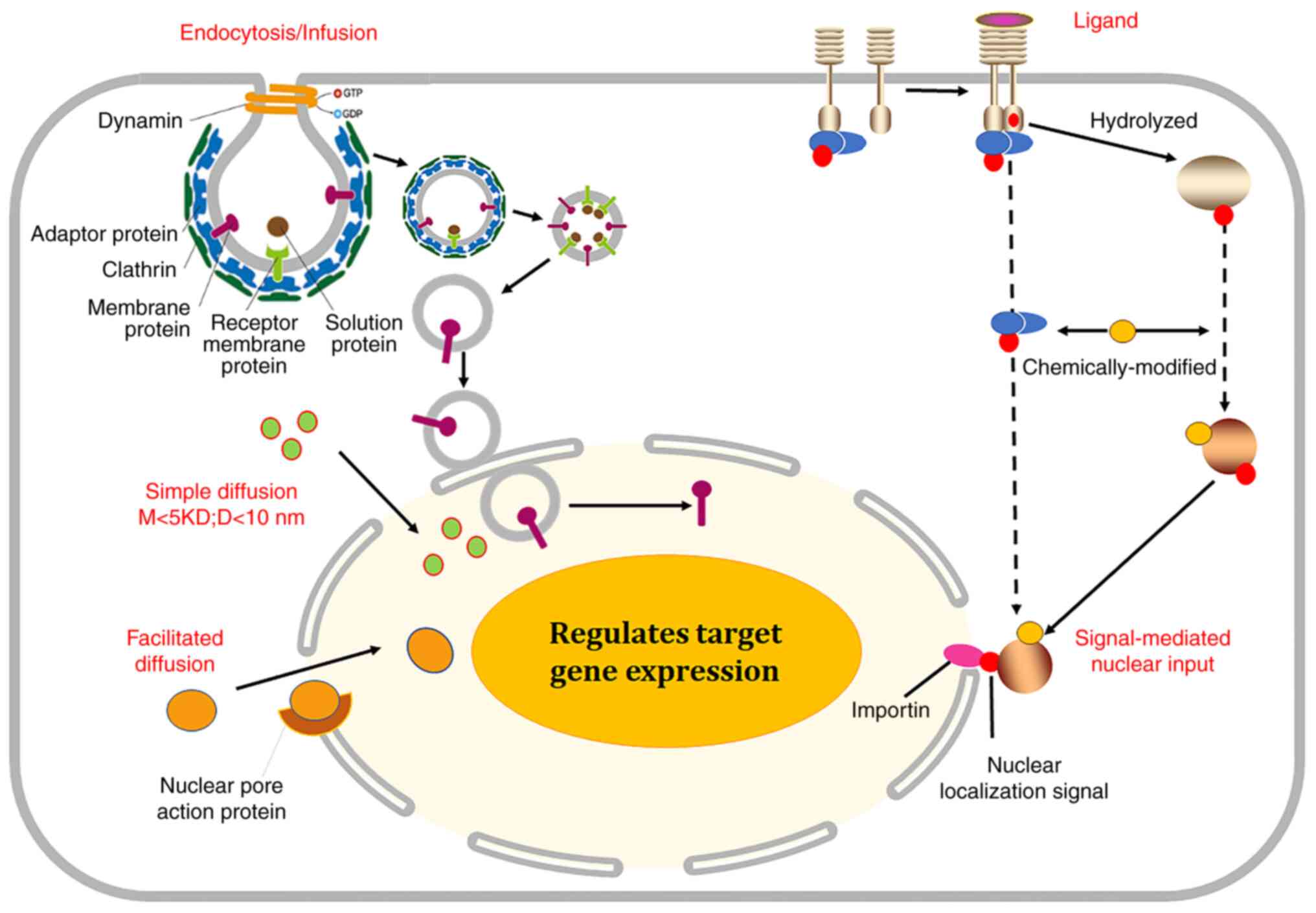 |
Figure 2.
Nuclear redistribution of the membrane proteins. Transmembrane and receptor proteins undergo endocytosis via adaptor protein and clathrin, and are degraded into cytosol. Some adaptor proteins generally bind to transmembrane proteins, but are separated from them upon signal activation. Additionally, some receptors interact with ligand and are hydrolyzed into cytosol by other proteases. The cytosolic proteins, especially chemically modified proteins, enter the nucleus by simple diffusion, facilitated diffusion via nuclear pore action protein or signal-mediated nuclear input via importin. M, molecular weight; D, diameter.
|
 |
Table I.
Classification of nucleomembrane shuttling proteins.
|
Table I.
Classification of nucleomembrane shuttling proteins.
| A, From membrane to nucleus |
|
| Type of protein |
Protein name |
| Transmembrane proteins |
Epidermal growth factor receptor, fibroblast growth factor receptor, hepatocyte growth factor receptor, insulin receptor, CD95/Fas, TNF-related apoptosis-inducing ligand-receptor, C-X-C motif chemokine receptor 4, angiotensin II receptor, CD38, sodium-calcium exchangers |
| Cell adhesion proteins |
T-cadherin, zonula occluden-2, connexin 43, β-catenin, APC, Zyxin, dystroglycan, neural cell adhesion molecules |
| Cell mobility proteins |
α-adducin, focal adhesion kinase, IQ-domain GTPase-activating protein 1 |
| Chemoresistant proteins |
P-glycoprotein, ATP-binding cassette sub-family B member 2 |
| Intracellular signal proteins |
Mechanistic target of rapamycin kinase, Ca2+/calmodulin-dependent protein kinase II, regulator of G protein signaling, SH2 domain-containing inositol polyphosphate 5-phosphatase type 2, phospholipase C, protein kinase C, SH2B adaptor protein 1, insulin receptor substrate, phospholipid scramblase 1, casein kinase 2 interacting protein-1, TNF receptor-associated factor 4, tyrosine-protein kinase 6, protease-activated receptor 2, Eps 15 homology domain protein, WD repeat domain 20 |
|
| B, From nucleus to membrane |
|
| Type of protein |
Protein name |
| Nuclear protein |
Nucleolin, histone deacetylase 3 |
Transmembrane proteins
Epidermal growth factor receptor (EGFR)
EGFR is a transmembrane protein that binds to extracellular ligands [such as EGF and transforming growth factor (TGF)]. Upon activation, the EGFR monomers polymerize into an active homodimer that stimulates downstream protein kinases, resulting in its autophosphorylation at Y992, 1045, 1068, 1148 and 1173 in its carboxyl terminal. These downstream transduction signal cascades principally include mitogen activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K)/Akt and JNK pathways for the regulation of cell migration, adhesion and proliferation. The mutation and amplification of EGFR causes its upregulation in lung cancer, glioblastoma, and head and neck squamous carcinoma. Therefore, antibodies targeting it (such as cetuximab, panitumumab, zalutumumab, nimotuzumab and matuzumab) and tyrosine kinase inhibitors (such as gefitinib, erlotinib, brigatinib, lapatinib, osimertinib, gefitinib, erlotinib and brigatinib) are used for target therapy in clinical practice. EGFR principally initiates two distinct pathways according to its subcellular localization. On one hand, EGFR activates canonical kinases at the plasma membrane, and then induces an intracellular phosphorylation cascade via MAPK and PI3K. On the other hand, it has been previously revealed that EGFR acts as a transcriptional co-activator for c-myc and cyclin D1 genes in the nucleus (7). Full-length EGFR can associate with the sec61β translocon for its nuclear translocation via the ER (7). Packham et al (8) indicated that small ubiquitin-like protein-1-modified EGFR at K37 was almost exclusively localized in the nucleus. Using dynamin and clathrin labeling, De Angelis Campos et al (9) demonstrated that EGF stimulation results in a peak of nuclear EGFR expression after 10 min. In addition, Reif et al (10) reported that phosphorylated Erb-B2 receptor tyrosine kinase 3 (ErbB3) enters the nucleus via clathrin-independent endocytosis after heregulin stimulation, and importin β1 drives the nuclear import of ErbB3, which interacts with transcription complexes to regulate gene expression. Gururaj et al (11) revealed that the accumulation of nuclear ΔEGFR promotes tumorigenicity by regulating transcription via the formation of chromatin-bound and transcription complexes (c-myc and ΔEGFR).
Fibroblast growth factor receptor (FGFR)
FGFR is a transmembrane glycoprotein that is activated by interaction with its ligand, FGF. FGFR consists of three extracellular immunoglobulin-like domains responsible for FGF-binding, a single transmembrane helix domain and an intracellular domain (12). So far, five distinct membrane FGFRs have been discovered and shown to display tyrosine kinase activity (12); they are responsible for angiogenesis, limb outgrowth, and the growth and differentiation of various cell types. FGFRs undergo internalization by endocytosis and access the nucleus via β-importin, or are alternatively hydrolyzed into a soluble form for nuclear import (12). Nuclear FGFRs interact with chromatin remodelers to change the epigenetic status for gene expression (13). Zhou et al (14) reported that FGFR3 is highly expressed in the nucleus of pancreatic cancer cells. Nuclear FGFR3 expression has been indicated to be positively correlated with a high N stage and poor prognosis of pancreatic cancer. Moreover, Narla et al (15) revealed that the expression of α7 nicotinic acetylcholine receptors results in the nuclear accumulation of FGFR1, and subsequently inhibits the proliferation of neural stem and progenitor cells in the subventricular zone.
c-Met (mesenchymal epithelial transition)
c-Met is a receptor tyrosine kinase and is also known as hepatocyte growth factor receptor; it is composed of a post-translationally cleaved and disulfide-linked extracellular α-subunit and a transmembrane β-subunit. Phosphorylated c-Met docking sites at both Y1349 and Y1356 serve as docking sites for downstream adaptor molecules, such as scaffolding protein growth factor receptor bound protein 2 (GRB2)-associated binding protein 1 (Gab1), Src and Src homology 2 domain containing protein (SHC); while phosphorylation at only Y1356 induces the interaction with GRB2, phospholipase C γ, p85 and SHC tyrosine phosphatase 2 (16).
The phosphorylation of c-Met at S985 inhibits the receptor kinase activity, while phosphorylation at Y1003 contributes to its polyubiquitination, endocytosis and degradation via ubiquitin ligase casitas B lineage lymphoma proto-oncogene. Subsequently, HGF activates Ras-MAPK, PI3K/Akt, signal transducer and activator of transcription 3, Wnt/β-catenin and Notch pathways during carcinogenesis, angiogenesis and metastasis. Cancer cells and, notably, cancer stem cells overexpressing c-Met have a greater ability to proliferate and invade in either an autocrine or a paracrine manner (16). However, HGF can activate c-Met transcription via the interaction of telomere elongation protein and hypoxia-inducible factor 1 with erythroblast transformation-specific transcription factor, activator protein-1 and hypoxia response element components of its promoter (16). Gomes et al (17) reported that exposure to HGF enhanced the rapid translocation of c-Met into the nucleus, where calcium-induced plasma membrane intrinsic protein 2 hydrolysis and inositol trisphosphate (IP3) formation took place. The nuclear translocation of c-Met was revealed to principally result from Gab1, importin β1 and calcium signals.
Insulin receptor (IR)
IR is a transmembrane tyrosine kinase receptor that can be activated by insulin, insulin-like growth factor (IGF)-I and IGF-II. Biochemically, subunits α and β of IR are encoded via either the A or the B isoform due to alternative splicing, and are composed of a leucine-rich repeat domain, a cysteine-rich region, and three fibronectin type III domains; they form homo- or heterodimers via disulfide linkages. The interaction of IR with its ligand mediates the autophosphorylation of its tyrosine residues within the intracellular tyrosine kinase domain and recruits adaptor proteins, including IR substrate (IRS) proteins, Src homology 2 B adaptor proteins 1 and 2 and protein phosphatases such as protein tyrosine phosphatase 1B.
The interaction of IR with an agonistic ligand triggers tyrosine autophosphorylation of the subunits, which generates a binding site for IRS-1. Subsequently, phosphorylated IRS-1 initiates the PI3K/Akt and GRB2/son of sevenless/Ras/MAPK pathways for glucose transporter type 4 translocation to the plasma membrane, glucose influx, glycogen biosynthesis, glycolysis and fatty acid biosynthesis (18). The insulin-IR complex is recycled back to the plasma membrane or degraded in lysosomes, although a small amount can enter the nucleus via rapid internalization (18). Kesten et al (19) demonstrated that the insulin-induced translocation of IR to the nucleus requires an NLS, which is suppressed by treatment with an inhibitor of nuclear import. Additionally, anti-IR antibodies block IR tyrosine phosphorylation and decrease the import of protein kinase C δ in the nucleus. However, mutated IR insulin-induced glucose uptake and protein kinase B phosphorylation. The addition of insulin into isolated nuclei increases IR phosphorylation but does not impact nuclear IR protein levels (19). Kabuta et al (20) indicated that GFP-IRS-3 is distributed to both the plasma membrane and the nucleus due to the presence of unique amino acid residues 192-223 in the phosphotyrosine binding domain responsible for nuclear localization. A protein involving the fusion of the Gal4 DNA binding domain and IRS-3 C-terminal region has been revealed to upregulate the activity of Gal4-binding-site-containing reporter gene, indicating that IRS-3 can be used as a transcriptional activator in the nuclear counterpart (20). Moreover, Kim et al (21) reported that the phosphorylation of Tub by insulin at Y464 is responsible for its translocation into the nucleus.
CD95
CD95 (also named Fas) has been discovered to have both apoptosis-inducing membrane-bound and soluble forms, which are produced via alternative splicing. The Fas receptor belongs to the type 1 transmembrane proteins and forms the death-inducing signaling complex upon its ligand binding (22). Fas receptor complex is internalized through the endosome, which allows the death domain of Fas to interact with the adaptor Fas-associated via death domain (FADD) for the induction of apoptosis (22). Garofalo et al (23) reported that membrane Fas/FasL complex localizes in lipid rafts to form raft-like microdomains, which are detectable in dynamic and subcompartmentalized mitochondria. Moreover, Sheikh and Huang (24) revealed that FADD is primarily distributed in the nucleus by shuttling between the nucleus and plasma membrane and is involved in genomic surveillance.
TNF-related apoptosis-inducing ligand receptor (TRAIL-R)
The interaction of TRAIL with the transmembrane TRAIL-R1/2 has the potential to kill cancer cells as an antitumor therapy via induction of apoptosis. However, a number of different types of cancer cells are resistant to TRAIL, and the loss of TRAIL-R1/R2 can promote cancer progression (25). It was previously revealed that nuclear TRAIL-R2 may speed up tumor development as a modulator of let-7 maturation (25). Mert et al (26) demonstrated that TRAIL-Rs were rapidly trafficked from the membrane to the cytosol via clathrin-dependent and TRAIL-dependent endocytosis, and subsequently to the nucleus via exportin 1/chromosome region maintenance 1 (CRM-1). In the nucleus, TRAIL-Rs have been revealed to be constitutively localized to chromatin for the regulation of gene expression (26).
C-X-C motif chemokine receptor 4 (CXCR4)
CXCR4 (also known as fusin or CD184) is an α-chemokine receptor specific for C-X-C motif chemokine ligand 12 (CXCL12), which is needed for potent chemotaxis, wound repair and regeneration, and the bone marrow homing and quiescence of hematopoietic stem cells (27). Ubiquitin binds to CXCR4 for its degradation in the proteasome. CXCR4 is upregulated in breast, ovarian and prostate cancer, and CXCR4-upregulating cancer cells are prone to distant metastasis to the lung, liver and bone marrow, which has a high level of CXCL12 (27). Don-Salu-Hewage et al (28) discovered that CXCR4 protein is localized in the nucleus of advanced types of prostate cancer due to the interaction with transportin β1 via a putative NLS (‘RPRK’). In the nucleus, CXCR4 functionally mediates the G-protein signaling pathway (28).
Angiotensin II receptor (AGTR)
AGTRs are members of the G-protein-coupled receptor family and can bind to angiotensin II, which mediates vasoconstriction via the renin-angiotensin system (RAS) (29). AGTRs also mediate the entry of COVID-19 into human target cells and blood pressure regulation. Bundalo et al (30) demonstrated that a FRD decreases levels of nuclear AGTRs, while higher angiotensin II receptor type 1 and lower angiotensin II receptor type 2 levels are be detected in the cell membrane.
CD38
CD38 is a type II transmembrane glycoprotein that is expressed on numerous immune cells (such as leukocytes) and that hydrolyzes cyclic ADP-ribose to regulate intracellular calcium. CD38 upregulation is considered to be an unfavorable marker for chronic lymphocytic leukemia due to its positive correlation with advanced disease progression, and as a target in the treatment of multiple myeloma using daratumumab (31). In a recent study, nuclear CD38 was also revealed to be present in cells without membrane CD38 (31). Orciani et al (32) revealed nuclear CD38 expression in the cells of distinct lineages and during different stages of hematopoietic differentiation, independent of the plasma membrane pool. This nuclear distribution was revealed to be responsible for calcium homeostasis and the level of NAD in the nucleus.
Sodium-calcium exchangers (NCXs)
NCXs are antiporter membrane proteins in the plasma membrane, mitochondria and ER; they pump calcium from cells and allow sodium into cells via an electrochemical gradient, finally exchanging them for each other in order to maintain calcium homeostasis (33). NCXs bind to monosialotetrahexosylganglioside (GM1) with high affinity, which is required for optimal Ca2+/Na+ exchange activity (33). Ledeen and Wu (34) demonstrated that the NCX-GM1 complex localizes at the inner membrane of the nuclear envelope for calcium transfer into the nucleus. Nuclear NCX-GM1 expression is increased during the differentiation and axonal outgrowth of neuronal cells. Cells from NCX-GM1-knockout mice exhibit a weaker ability to maintain the calcium concentration in the nucleus than wild-type mice, as a similar deficit was observed in kainite-induced seizures (34).
Cell adhesion
T-cadherin
T-cadherin is present in the apical cell membrane and mediates the functions of cell junctions and lipid rafts; it also functions as a low-density lipoprotein receptor to activate ERK 1/2 and promote the nuclear distribution of NF-κB. In addition, it enhances cell migration and the stress fiber formation of endothelial cells by activating small G-proteins, along with promoting actin reorganization. The expression of T-cadherin suppresses subcutaneous tumor growth in nude mice, and cell proliferation via p21 CDK-interacting protein 1 (CIP1)/WAF1 production and G2 arrest. The loss of T-cadherin from allelic loss or promoter hypermethylation is closely associated with the invasiveness and metastasis of basal cell carcinoma, cutaneous squamous carcinoma, and ovarian, pancreatic and colorectal cancer. T-cadherin facilitates vascular proliferation and tumor-associated angiogenesis, but attenuates the aggressiveness of gastric, colorectal and breast cancer (35). It binds to the plasma membrane via interactive partners (such as glycosylphosphatidylinositol anchor) due to the lack of a transmembrane domain (35). Andreeva et al (36) made the unexpected discovery that some T-cadherins are distributed to both the nucleus and centrosomes. In addition, the consistent co-localization of T-cadherin and the centrosomal marker γ-tubulin has been observed throughout the cell cycle in endothelial cells. T-cadherin transiently accumulates in the midbody at the telophase and is then degraded. T-cadherin increases nuclear mitosis with a higher number of multinuclear cells. It has been suggested that the deregulation of T-cadherin may disturb cytokinesis and centrosomal replication in endothelial cells (36).
Zonula occluden (ZO)-2
ZOs are tight junction (TJ) proteins that belong to the membrane-associated guanylate kinase family and are involved in the organization of epithelial and endothelial intercellular junctions. ZO proteins contain several PDZ, SH-3 and guanylate kinase-like domains, as well as NLS and NES; they can function as scaffolds that are attracted to transmembrane TJs, the cytoskeleton and signal transduction proteins. Additionally, ZOs shuttle from the TJ to the nucleus, where they may regulate gene expression (37). The cytonuclear relocalization of ZO-1 from the adherens junctions and TJs underlies epithelial-mesenchymal transition-associated invasion, as it regulates vimentin or matrix metalloproteinase (MMP)-14 expression (37). Traweger et al (38) previously reported that nuclear ZO-2 directly interacts with a heterogeneous nuclear ribonucleoprotein, scaffold attachment factor B, which contributes to chromatin organization and gene expression. The accumulation of nuclear ZO-2 upregulates M2 pyruvate kinase expression in epithelial and endothelial cells, increases their proliferation and decreases the stability of intercellular junctions.
Connexin 43 (Cx34)
Cx43 is a gap junction transmembrane protein responsible for intercellular communication in addition to cell proliferation, death and differentiation. The carboxyl terminal of Cx43 includes binding sites for transcription factors, cytoskeleton elements and other proteins, while its amino terminal includes gating channels and its transmembrane domain includes the gap junction channel, and the extracellular loops are used for appropriate channel docking. TGF-β1 may induce Cx34 expression via Smad and ERK1/2, and may mediate the differentiation of trophoblast cells into the placenta (39). Cx43 modulates the microtubule network, cell migration and polarity in the cytosol, and also suppresses apoptosis in mitochondria (39). Dang et al (40) expressed the carboxyl-terminal of Cx43 in the nucleus of HeLa cells and cardiomyocytes, and revealed that cell proliferation was inhibited. In addition, Chen et al (41) demonstrated that Cx43 interacted with AKAP95 from late G1 to metaphase, and from anaphase to late G1. Cx43 translocated to the nucleus via AKAP95 in late G1 phase.
Wnt signaling pathway
Wnt signaling is composed of the canonical and non-canonical planar cell polarity and Wnt/calcium pathways, which are activated by the interaction of Wnt ligand with the Frizzled receptor via Lrp-5/6 and receptor tyrosine kinase-like orphan receptor 2. In the plasma membrane, the formation of a complex consisting of β-catenin, p120-catenin and E-cadherin mediates cell adhesion and mobility. The destruction complex for β-catenin ubiquitination and degradation contains serine/threonine protein phosphatase 2A, glycogen synthase kinase 3 (GSK3), casein kinase Iα, adenomatous polyposis coli (APC) and Axin. Phosphorylated Dsh inhibits GSK3 activity and causes nuclear β-catenin accumulation, while β-catenin phosphorylated at S552 by Akt can cause cells to disassociate from each other and accumulate in the cytosol (42,43). 14-3-3ζ binds to β-catenin for nuclear translocation, where β-catenin can target transcription factor 7-like/anthrax toxin lethal factor promoter for transcriptional coactivation and then regulate the expression of c-myc, Cyclin D1 and interleukin-7 (42,43). Johnson et al (44) indicated that some β-catenins could rapidly shuttle from the nucleus to the plasma membrane in NIH 3T3 cells with a high turnover, but poor turnover was observed at adherens junctions of Madin-Darby canine kidney epithelial cells.
APC contains two classical NLSs and two NESs for its translocation between the nucleus and cytoplasm. In the nucleus, APC can block β-catenin-mediated transcription via the dissociation of β-catenin from the transcription complex and the interaction of APC with transcriptional corepressors, such as C-terminal-binding protein (45). p120-catenin may control Rho GTPase activity and E-cadherin stability in both the plasma membrane and the cytoplasm. Moreover, it can enter the nucleus and attach to Kaiso in order to regulate its transcription (46).
Zyxin
Zyxin is a zinc-binding phosphoprotein with one N-terminal proline-rich and SH3-binding domain and three LIM domains in its carboxyl terminal (47). As a messenger, it is involved in adhesion-stimulated alteration in gene expression, and regulates the organization of actin bundles in lamellipodia (47). Nix et al (48) demonstrated that its proline-rich region binds to α-actinin and Ena/vasodilator-stimulated phosphoprotein (VASP) in lamellipodial extensions and focal adhesions, and that its LIM region targets focal adhesions. Upon the mislocalization of full-length zyxin, at least one member of the Ena/VASP family was also revealed to be displaced, and the actin cytoskeleton organization was perturbed. Zyxin was also revealed to shuttle between the nucleus and focal adhesion sites via NLS and NES. Once nuclear export is inhibited by leptomycin B treatment, zyxin accumulation occurs in the nucleus.
Dystroglycan
The dystroglycan complex mediates the link between the intracellular cytoskeleton and the extracellular matrix (ECM). Extracellular α-dystroglycan (DG) interacts with merosin α-2 laminin in the basement membrane, while transmembrane β-DG interacts with cytoskeletal dystrophin for cytoskeletal remodeling and cell adhesion. In the cytosol, DG has been revealed to interact with caveolin-3, c-Src, tyrosine-protein kinase FYN, Grb2, NCK adaptor protein 1 and SH2 domain protein C1 (SHC1), indicating its involvement in signal transduction (49). β-DG has also revealed to be localized in the nuclear envelope via the interaction of importin α/β with its NLS (49). Gracida-Jiménez et al (50) demonstrated that β-DG is released from the ER membrane by the translocon complex Sec61 and undergoes nuclear import via its phosphorylation at Y890 by the importin translocation system.
Neural cell adhesion molecules (NCAMs)
After post-translational modification, NCAM undergoes polysialylation by polysialic acid (PSA), making it negatively charged (51). Westphal et al (52) indicated that PSA-binding NCAM is generated at the plasma membrane by MMPs and transferred to the nucleus via endosomal machinery. An NCAM fragment is imported into the nucleus after treatment with a function-triggering NCAM antibody and peptide comprising the effector domain of myristoylated alanine-rich C kinase substrate, which interacts with PSA within the plasma membrane.
Cell mobility
α-adducin
Adducins are a unique family of actin filament barbed-end capping proteins that recruit spectrin to actin filaments and promote the formation of an extended spectrin-actin network, thereby contributing to cell membrane stability, motility and junctions (53). Chen et al (54) observed nuclear α-adducin in sparsely cultured epithelial cells and its membranous counterpart in confluent cell junctions. The disruption of cell adhesion causes the nuclear distribution of adducin, while its re-establishment causes its membranous and cytoplasmic distribution due to the NLS at its carboxyl terminal and the NES at its neck region. Moreover, the phosphorylation of α-adducin at S716 inhibits its nuclear translocation. The depletion of α-adducin leads to disordered cell adhesion and weakened proliferation with mitotic spindles, chromosomal segregation and disorganized centrosomes.
Focal adhesion kinase (FAK)
FAK is a protein kinase that plays a notable role in adhesion and migration by interacting with v-Src and linking the ECM to the cytoskeleton. In focal adhesion, actin, filamin, paxillin, talin, tensin, Ras suppressor protein 1 and vinculin are present. Growth factor, integrin engagement and mitogenic neuropeptides induce the phosphorylation of FAK and the formation of focal adhesions (55). In endothelial cells, FAK is cleaved into ~90-kDa (termed ‘killer FAT’) and 130-kDa fragments by Caspase 3 at N772 (55). In RBL-2H3 cells, Jones and Stewart (56) demonstrated that the activation of FcεRI receptor mediates the nuclear translocation of N-terminal FAK fragments, which was enhanced by exposure to leptomycin B.
IQ-domain GTPase-activating protein 1 (QGAP1)
IQGAP1 is a Ras GTPase-activating-like scaffold protein that is involved in actin cytoskeleton organization, transcription, cellular adhesion and the cell cycle. IQGAP1 contains the following domains: i) A calponin homology domain for binding to actin and calponin; ii) a WW or poly-proline protein-protein domain for interaction with other proline-rich regions; iii) an IQ domain for binding to calmodulin; iv) a RasGAP-related domain responsible for interaction with Rho GTPases cell division cycle 42 (CDC42) and Rac family small GTPase 1 (RAC1); and v) a carboxyl-terminal domain for binding E-cadherin and β-catenin. IQGAP1 has been demonstrated to interact with S100 calcium-binding protein B, RAC1, protein kinase cAMP-activated catalytic subunit α, CAP-Gly domain-containing linker protein 1, CDC20 homolog 1 (CDH1), CDC42 and calmodulin 1. IQGAP1 integrates Ras-Raf-MEK to MAPK, which is stimulated by EGF, IGF-1, platelet-derived growth factor, B polypeptide and nerve growth factor (NGF) during apoptosis, cell differentiation and proliferation. IQGAP1 is a plasma membrane protein that is needed for the actin cytoskeleton, migration, polarity and adhesion (57). Johnson et al (58) indicated that nuclear IQGAP1 is observed in early S-phase, that the import into the nucleus is performed via the CRM1 export receptor and that GSK-3β inhibition promotes the progression of the cell cycle after DNA replication arrest. In addition, this study also revealed that the nuclear IQGAP1 in G1/S-arrested cells is associated with DNA replication complex factors, replication protein A2 and proliferating cell nuclear antigen (58).
Chemoresistance
P-glycoprotein (P-gp)
P-gp (also termed MDR1 or ABCB1) belongs to the membrane ABC transporters, which are responsible for the pumping of chemotherapeutics and other medications out of cells and causing chemoresistance. P-gp is expressed in the intestinal epithelium where it pumps xenobiotics back into the intestinal lumen, in liver cells where it pumps xenobiotics into bile ducts, in renal proximal tubule cells where it pumps xenobiotics into urinary filtrate and in capillary endothelial cells where it pumps xenobiotics into capillaries. P-gp contains 12 transmembrane domains, a large cytoplasmic domain and two ATP-binding sites (59). The GTPase Rab5 suppresses the endocytotic trafficking of P-gp, while the converse is true for the GTPase Rab4 (59). Tome et al (60) revealed that brain microvessels have high P-gp activity and low drug delivery to the central nervous system in rats suffering from acute peripheral inflammatory pain. P-gp was also detected in the nucleus and luminal membrane of endothelial cells of control rats in this study. Moreover, peripheral inflammatory pain strengthened luminal Pg-p immunostaining, but attenuated its nuclear expression.
ATP-binding cassette sub-family B member 2 (ABCG2)
ABCG2 is a superfamily member of the ABC transporters that acts as a xenobiotic transporter in chemoresistance. ABCG2 protects absorption at the intestinal apical membrane, blood-testis and blood-brain barriers, as well as the membranes of progenitor and stem cells; in addition, it also excretes xenobiotics at the apical membranes of both the liver and kidney, vitamins into milk in the mammary gland and urate in the gastrointestinal tract (61). Liang et al (62) indicated that nuclear ABCG2 protein interacted with the E-box of the CDH1 promoter and subsequently upregulated E-cadherin expression to reduce migration.
Intracellular signal pathways
Mechanistic target of rapamycin kinase (mTOR)
mTOR is a member of the PI3K-related serine/threonine kinase family and regulates various cellular events, including proliferation, migration, invasion and autophagy. mTOR also mediates the activation of IRs and IGF-1 receptors after growth factor binding, and maintains the actin cytoskeleton. mTOR can be inhibited by rapamycin via interaction with its receptor FK506-binding protein 12, and is activated by PI3K and PTEN loss; eventually this initiates carcinogenesis and subsequent progression via the oxygen and nutrient supply through the upregulation of HIF1α expression and promotion of angiogenesis (63,64). Zhang et al (64) observed mTOR expression in the cytoplasm of cancer cell lines, fibroblasts and myoblasts and the nucleus and cytoplasm of HEK293 cells.
Ca2+/calmodulin-dependent protein kinase II (CaMKII)
CaMKII is required for calcium homeostasis and reuptake, neurotransmitter secretion and glycogen metabolism; it has a catalytic domain for ATP and a substrate anchor, an autoinhibitory domain for the interaction of pseudosubstrate and a self-association domain (65). CaMKII is activated via phosphorylation at T286, and then phosphorylates adhesion modulation protein A (AMPA) receptors at S831 to sensitize the GluA1 channel conductance of AMPA receptors (65). Ma et al (66) reported that calcineurin triggers the nuclear translocation of γCaMKII, which induces Ca2+/CaM to activate CaMKK and its substrate CaMKIV, a cAMP-response element-binding protein kinase.
Regulator of G-protein signaling (RGS)
During G-protein signaling, transmembrane G-protein-coupled receptor is stimulated by an extracellular ligand to realize the reciprocal transformation of GTP and GDP. RGS (also known as GAP) can facilitate GTP hydrolysis in order to switch off G-protein-coupled receptors (67). Hepler (68) revealed that palmitate can assist the movement of the complex of R7BP and the RGS protein, R7-Gβ5, to the plasma membrane. However, the absence of palmitate leads to the nuclear distribution of the R7BP-R7-Gβ5 complex.
SH2 domain-containing inositol polyphosphate 5-phosphatase type 2 (SHIP2)
SHIP2 has a proline-rich PXXP at both N- and C-terminals that is recognized by the SH3 domain of Abl, Grb2, intersectin and Ras homolog family member A, and a single NPXY site for tyrosine phosphorylation at its C-terminal (69). SHIP2 is capable of binding to phosphorylated tyrosine of Shc, the immunoreceptor tyrosine-based inhibitory motif of the FcγRIIB receptor and Crk-associated substrate (69). Filamin binds to the C-terminal of SHIP2 in order to localize at membrane ruffles. Elong Edimo et al (70) revealed that serum-stimulated SHIP2 moved from a perinuclear and cytoplasmic localization to the plasma membrane and focal contacts in polarized cells; here, SHIP2 mediated the change of PIP3 into PIP2, or cell adhesion and migration. However, SHIP2 phosphorylated at S132 was detected in the nucleus and nuclear speckles, where it likely interacts with lamin A/C and controls the level of nuclear PIP2.
Phospholipase C (PLC)
PLC is a membrane enzyme that hydrolyzes phospholipids, classified into isotypes β, γ, δ, ε, ζ and η, and is activated by protein tyrosine kinases, G-proteins, phospholipids and calcium. Structurally, PLC includes a translocation-induced circling mutation barrel containing an active site, catalytic residues, calcium-binding site, PH domain, four EF hand domains and a C2 domain (71). PLC selectively hydrolyzes PIP2 into IP3 and diacylglycerol DAG by cleaving a phosphodiester bond, while histidine residues and calcium are essential for its acid/base catalysis (71). Pan et al (72) revealed that OSBP-related protein 4L (ORP4L) mediated the membrane translocation of PLCβ3 from the nucleus. First, ORP4L activates Ras-related nuclear protein, which interacts with exportin 1 for the extranuclear transport of PLCβ3. Next, ORP4L binds to vesicle-associated membrane protein-associated protein A in the nucleus and follows the movement to the plasma membrane via the cytoplasm.
Protein kinase C (PKC)
PKC is activated by DAG, calcium or phorbol ester, and is responsible for the phosphorylation of hydroxyl-containing serine and threonine. Upon its activation, PKC is translocated to the plasma membrane by the receptor for activated c kinase proteins. Low PKC is sufficient to reverse cell chirality through PI3K/Akt and alter junctional protein organization, resulting in inflammation and carcinogenesis (73). Divecha et al (74) reported that nuclear PKC facilitates the intranuclear production of DAG (a key physiological activator of enzyme as a secondary messenger) in inflammation, liver regeneration and osteosarcoma.
SH2B adaptor protein 1 (SH2B1)
The adaptor protein SH2B1 is a member of the SH2-domain family, which binds to Grb2, IR, Janus kinase 2 and TrkA; it can be recruited to the ligand-activated tyrosine kinase receptor and cytokine receptor-associated Janus kinase for downstream cascades (75). Maures et al (76) revealed that the nucleocytoplasmic shuttling of NLS-containing SH2B1β promotes NGF-dependent neurite outgrowth and the expression of NGF-responsive genes, such as urokinase plasminogen activator receptor, MMP-3 and MMP-10.
Phospholipid scramblase 1 (PLSCR1)
PLSCR1 is a calcium-binding and endofacial plasma membrane protein that is involved in the transbilayer movement of phosphatidylserine for calcium influx. PLSCR1 expression is markedly induced by interferon, and LSCR1−/− bone marrow cells exhibit defective myeloid proliferation and differentiation after stimulation by cytokines (77). PLSCR1 is principally palmitoylated, localizes in membrane lipid rafts and binds to cleavage and polyadenylation-specific factor 6, EGFR, neuraminidase 4, SHC1, secretory leukocyte protease inhibitor and TRK-fused gene (77). Wiedmer et al (78) identified that a Cys-rich sequence is necessary for PLSCR1 palmitoylation and that mutation in five cysteines or exposure to a palmitoylation inhibitor (2-bromo-palmitate) results in its translocation from the plasma membrane to the nucleus.
Casein kinase 2-interacting protein-1 (CKIP-1)
PH domain-containing CKIP-1 can interact with various partners (such as pro-inflammatory mTNF) and, therefore, contributes to cell differentiation, mobility and death, and bone formation (79). Xi et al (79) observed that the isolated PH domain of CKIP-1 was localized in the nucleus, which was determined by the net charge of its basic residues and a serine-rich motif, while this localization was counteracted by its carboxyl terminal. Therefore, the N-terminal PH domain and carboxyl-terminal auto-inhibitory region synergistically contribute to the nuclear/plasma membrane shuttling of CKIP-1.
TNF receptor-associated factor 4 (TRAF4)
TRAF4 (also known as RING finger protein 83) belongs to the TNF receptor-associated factor family. TRAF4 links IL-1R and TNFR to signaling proteins that activate NF-κB and MAPK. The complex of TRAF and neurotrophin receptor, p75 (NTR/NTSR1), inhibits NTR-induced cell death and NF-κB activation (80). TRAF4 interacts with NOD-like receptors to negatively inhibit NF-κB activation (80). Kédinger et al (81) revealed that TRAF4 assembles TJs in confluent epithelial cells, but is distributed in the cytoplasm and/or nucleus when TJs are broken. TRAF4 was detectable at the TJs of normal mammary epithelia and well-differentiated in situ adenocarcinoma, but observed in the cytoplasm and/or nucleus of poorly differentiated invasive adenocarcinoma.
Tyrosine-protein kinase 6 (PTK6)
PTK6 (also termed breast tumor kinase, Brk) is a cytoplasmic non-receptor protein kinase that sensitizes mammary epithelial cells to EGF for aggressive transformation (82). Ie Kim and Lee (83) revealed that Src myristoylation signal (Myr) or SV40 T-antigen NLS targets PTK6 to the plasma membrane or the nucleus of 293 cells, respectively. Due to the differences in cellular proteins phosphorylated by Myr-PTK6 and NLS-PTK6, membrane PTK6 promotes aggressive phenotypes, such as proliferation, survival, migration and invasion, which are impaired by nuclear PTK6.
Protease-activated receptor 2 (PAR2)
PAR2 belongs to the protease-activated seven-transmembrane receptors that couple to guanosine-nucleotide-binding proteins, and modulates inflammation, metabolism and obesity, while also functioning as a sensor of proteolytic enzymes during infection. Its activation is mediated by the proteolytic hydrolysis of its extracellular N-terminal via house dust mite protein Der P9 (84). Both cleavage fragment PAR and full-length PAR act as a tethered ligand and bind to the receptor (84). PAR2 is abundant in retinal ganglion cells and is associated with neovascularization during ischemic retinopathy (84). Joyal et al (85) revealed that stimulation results in the microtubule-dependent shuttling of PAR2 from the plasma membrane to the nucleus of retinal ganglion cells via nexin 11 sorting. In this study, PAR2 recruited the Sp1 transcription factor to trigger VEGF-A expression, followed by angiogenesis, although its membrane activation classically induces Ang1 expression.
Eps 15 homology domain protein (EHD)
EHD proteins possess a dynamin-like structural fold, and contribute to endocytic trafficking, endocytosis, internalization and the recycling of multiple receptors internalized through both clathrin-dependent and -independent pathways, including transferrin receptor, major histocompatibility complex class I and β1 integrins (86). Pekar et al (86) demonstrated that plasma-membrane-associated and NES-containing EHD2 accumulates in the nucleus upon treatment with leptomycin B. Moreover, sumoylation was demonstrated to be required for the nuclear accumulation of EHD2 and, in the nucleus, EHD2 suppressed the transcription of p21 CIP1/WAF1.
WD repeat domain 20 (WDR20)
The human deubiquitinase, ubiquitin-specific peptidase 12 (USP12), acts as a tumor suppressor and is catalytically activated by USP1-associated factor 1 (UAF1) and WDR20. WDR20 preserves and regulates the activity of the USP12-UAF1 deubiquitinating enzyme complex (87). WDR20 overexpression has been reported to suppress the phosphorylation of Akt and ERK in renal clear carcinoma cells (87). Olazabal-Herrero et al (88) revealed that WDR20 facilitates shuttling of the USP12/UAF1/WDR20 complex between the plasma membrane, cytoplasm and nucleus via the CRM1 pathway, which requires the N-terminal motif of USP12 and the novel NES of WDR20.
Nuclear proteins
Nucleolin
Nucleolin is a eukaryotic nucleolar phosphoprotein in dense fibrillar regions of the nucleolus, which promotes the synthesis and maturation of ribosomes (7); it also acts as a transcriptional coactivator with chicken ovalbumin upstream promoter transcription factor II and as a membrane receptor for the respiratory syncytial virus (89). Hovanessian et al (90) revealed that the membrane localization of nucleolin depends on the constant induction of nucleolin mRNA or environmental stress (cold or heat), but that inhibitors of RNA transcription or translation can suppress the localization of nucleolin. In addition, Chen et al (91) demonstrated that plasma membrane nucleolin functions as a receptor for DNA nanoparticles composed of pegylated polylysine and DNA.
Histone deacetylase 3 (HDAC3)
Histones are histidine-rich alkaline proteins that facilitate the packaging and ordering of DNA into nucleosomes; their electrostatic attraction and structure can be altered by the acetylation and deacetylation of lysine. HDAC3 is a class I member of the histone deacetylase superfamily and regulates both the nuclear epigenome and gene expression as an enhancer (92). Longworth and Laimins (93) demonstrated that HDAC3 is also localized to the plasma membrane as a c-Src substrate for phosphorylation in human foreskin keratinocytes and J2 mouse fibroblasts.
Conclusions
The present review should help readers to understand which cytosolic proteins shuttle between the plasma membrane and nucleus and how this occurs. It should also help researchers to establish the signal pathways and understand the immunohistochemical results regarding the subcellular localization of proteins, as well as to clarify more molecular mechanisms of therapeutic protein targets. Overall, the present study summarized current knowledge about the translocation of proteins between plasma membrane and the nucleus as follows: i) Cross-talk occurs among the aforementioned proteins, namely, the interaction of transmembrane proteins with signal proteins; ii) these proteins are responsible for various biological events, including proliferation, apoptosis, chemoresistance, adhesion and migration; iii) transmembrane proteins undergo endocytosis, inclusion into organelles or proteolysis into soluble forms for import into the nucleus, while membrane-binding adaptor proteins easily enter the nucleus; iv) translocation between the membrane and nucleus mainly occurs through export or import via nuclear membrane pores as a result of interaction with importin or exportin; v) nuclear proteins generally interact with other transcription factors, and then bind to the gene promoter for transcription, while membrane proteins are responsible for signal initiation by binding to other membrane and/or adaptor proteins; vi) protein translocation occurs in a cell-specific manner and is closely associated with cellular biological events; and vii) the shuttling of only some cellular proteins between the plasma membrane and nucleus were described, but the shuttling mechanisms and the alterations in their biological functions have been not yet been clarified.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Award for Liaoning Distinguished Professor, Natural Science Foundation of Hebei Province (grant no. 202130821010436) and The National Natural Scientific Foundation of China (grant no. 81672700).
Availability of data and materials
Not applicable.
Authors' contributions
HCZ summarized the literature and wrote the review. HMJ revised the manuscript. HCZ and HMJ were responsible for the draft. All authors have read and approved the final manuscript. Data sharing is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
ER
|
endoplasmic reticulum
|
|
NLS
|
nuclear localization sequence
|
|
NES
|
nuclear export sequence
|
|
EGFR
|
epidermal growth factor receptor
|
|
MAPK
|
mitogen-activated protein kinase
|
|
PI3K
|
phosphatidylinositol 3-kinase
|
|
ErbB3
|
Erb-B2 receptor tyrosine kinase 3
|
|
FGFR
|
fibroblast growth factor receptor
|
|
Gab1
|
GRB2-associated binding protein 1
|
|
GRB2
|
growth factor receptor bound protein 2
|
|
IR
|
insulin receptor
|
|
IRS
|
IR substrate proteins
|
|
TRAIL-R
|
TNF-related apoptosis-inducing ligand-receptor
|
|
CXCR4
|
C-X-C motif chemokine receptor 4
|
|
CXCL12
|
C-X-C motif chemokine ligand 12
|
|
AGTR
|
angiotensin II receptor
|
|
FRD
|
fructose-rich diet
|
|
RAS
|
renin-angiotensin system
|
|
NCX
|
sodium-calcium exchangers
|
|
ZO
|
zonula occludens
|
|
TJ
|
tight junction
|
|
MMP
|
matrix metalloproteinase
|
|
Cx34
|
connexin 43
|
|
TGF
|
transforming growth factor
|
|
GSK3
|
glycogen synthase kinase 3
|
|
VASP
|
vasodilator-stimulated phosphoprotein
|
|
DG
|
dystroglycan
|
|
ECM
|
extracellular matrix
|
|
NCAM
|
neural cell adhesion molecules
|
|
PSA
|
polysialic acid
|
|
FAK
|
focal adhesion kinase
|
|
QGAP1
|
IQ-domain GTPase-activating protein 1
|
|
CDC42
|
cell division cycle 42
|
|
RAC1
|
Rac family small GTPase 1
|
|
NGF
|
nerve growth factor
|
|
P-gp
|
p-glycoprotein
|
|
ABCG2
|
ATP-binding cassette sub-family B member 2
|
|
mTOR
|
mechanistic target of rapamycin kinase
|
|
CaMKII
|
Ca2+/calmodulin-dependent protein kinase II
|
|
AMPA
|
adhesion modulation protein A
|
|
SHIP2
|
SH2 domain-containing inositol polyphosphate 5-phosphatase type 2
|
|
PLC
|
phospholipase C
|
|
ORP4L
|
OSBP-related protein 4L
|
|
PKC
|
protein kinase C
|
|
SH2B1
|
SH2B adaptor protein 1
|
|
PLSCR1
|
phospholipid scramblase 1
|
|
TRAF4
|
TNF receptor-associated factor 4
|
|
PTK6
|
tyrosine-protein kinase 6
|
|
PAR2
|
protease-activated receptor 2
|
|
EHD
|
Eps 15 homology domain protein
|
|
USP12
|
ubiquitin specific peptidase 12
|
|
WDR20
|
WD repeat domain 20
|
|
HDAC3
|
histone deacetylase 3
|
|
IGF
|
insulin-like growth factor
|
|
DAG
|
diacylglycerol
|
|
APC
|
adenomatous polyposis coli
|
References
|
1
|
Alber AB and Suter DM: Dynamics of protein synthesis and degradation through the cell cycle. Cell Cycle. 18:784–794. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Soniat M and Chook YM: Nuclear localization signals for four distinct karyopherin-beta nuclear import systems. Biochem J. 468:353–362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B and Veenhoff LM: Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 333:90–93. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Borlido J, Zecchini V and Mills IG: Nuclear trafficking and functions of endocytic proteins implicated in oncogenesis. Traffic. 10:1209–1220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moroianu J: Nuclear import and export pathways. J Cell Biochem Suppl 32-33. S76–S83. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Etienne-Manneville S and Lammerding J: Connecting the plasma membrane to the nucleus by intermediate filaments. Mol Biol Cell. 28:695–696. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan D and Lin X: Epithelial growth factor receptor-activated nuclear factor κB signaling and its role in epithelial growth factor receptor-associated tumors. Cancer J. 19:461–467. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Packham S, Lin Y, Zhao Z, Warsito D, Rutishauser D and Larsson O: The nucleus-localized epidermal growth factor receptor is SUMOylated. Biochemistry. 54:5157–5166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Angelis Campos AC, Rodrigues MA, de Andrade C, de Goes AM, Nathanson MH and Gomes DA: Epidermal growth factor receptors destined for the nucleus are internalized via a clathrin-dependent pathway. Biochem Biophys Res Commun. 412:341–346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Reif R, Adawy A, Vartak N, Schröder J, Günther G, Ghallab A, Schmidt M, Schormann W and Hengstler JG: Activated ErbB3 translocates to the nucleus via clathrin-independent endocytosis, which is associated with proliferating cells. J Biol Chem. 291:3837–3847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gururaj AE, Gibson L, Panchabhai S, Bai M, Manyam G, Lu Y, Latha K, Rojas ML, Hwang Y, Liang S and Bogler O: Access to the nucleus and functional association with c-Myc is required for the full oncogenic potential of ΔEGFR/EGFRvIII. J Biol Chem. 288:3428–3438. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bryant DM and Stow JL: Nuclear translocation of cell-surface receptors: Lessons from fibroblast growth factor. Traffic. 6:947–954. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tuzon CT, Rigueur D and Merrill AE: Nuclear fibroblast growth factor receptor signaling in skeletal development and disease. Curr Osteoporos Rep. 17:138–146. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou L, Yao LT, Liang ZY, Zhou WX, You L, Shao QQ, Huang S, Guo JC and Zhao YP: Nuclear translocation of fibroblast growth factor receptor 3 and its significance in pancreatic cancer. Int J Clin Exp Pathol. 8:14640–14648. 2015.PubMed/NCBI
|
|
15
|
Narla ST, Klejbor I, Birkaya B, Lee YW, Morys J, Stachowiak EK, Prokop D, Bencherif M and Stachowiak MK: Activation of developmental nuclear fibroblast growth factor receptor 1 signaling and neurogenesis in adult brain by α7 nicotinic receptor agonist. Stem Cells Transl Med. 2:776–788. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan C, Wu Y, Li X, Li X, Li G, et al: Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 17:452018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, Bennett AM and Nathanson MH: c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 283:4344–4351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Huang L, Qi X and Chen C: Insulin receptor trafficking: Consequences for insulin sensitivity and diabetes. Int J Mol Sci. 20:50072019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kesten D, Horovitz-Fried M, Brutman-Barazani T and Sampson SR: Insulin-induced translocation of IR to the nucleus in insulin responsive cells requires a nuclear translocation sequence. Biochim Biophys Acta Mol Cell Res. 1865:551–559. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kabuta T, Hakuno F, Asano T and Takahashi S: Insulin receptor substrate-3 functions as transcriptional activator in the nucleus. J Biol Chem. 277:6846–6851. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim JW, Kim HS, Kim SD and Park JY: Insulin phosphorylates tyrosine residue 464 of tub and translocates tubby into the nucleus in HIRcB cells. Endocrinol Metab (Seoul). 29:163–168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guégan JP, Ginestier C, Charafe-Jauffret E, Ducret T, Quignard JF, Vacher P and Legembre P: CD95/Fas and metastatic disease: What does not kill you makes you stronger. Semin Cancer Biol. 60:121–131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garofalo T, Tinari A, Matarrese P, Giammarioli AM, Manganelli V, Ciarlo L, Misasi R, Sorice M and Malorni W: Do mitochondria act as ‘cargo boats’ in the journey of GD3 to the nucleus during apoptosis? FEBS Lett. 581:3899–3903. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sheikh MS and Huang Y: The FADD is going nuclear. Cell Cycle. 2:346–347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amarante-Mendes GP and Griffith TS: Therapeutic applications of TRAIL receptor agonists in cancer and beyond. Pharmacol Ther. 155:117–131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mert U, Adawy A, Scharff E, Teichmann P, Willms A, Haselmann V, Colmorgen C, Lemke J, von Karstedt S, Fritsch J and Trauzold A: TRAIL induces nuclear translocation and chromatin localization of TRAIL death receptors. Cancer (Basel). 11:11672019. View Article : Google Scholar
|
|
27
|
Adlere I, Caspar B, Arimont M, Dekkers S, Visser K, Stuijt J, de Graaf C, Stocks M, Kellam B, Briddon S, et al: Modulators of CXCR4 and CXCR7/ACKR3 function. Mol Pharmacol. 96:737–752. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Don-Salu-Hewage AS, Chan SY, McAndrews KM, Chetram MA, Dawson MR, Bethea DA and Hinton CV: Cysteine (C)-x-C receptor 4 undergoes transportin 1-dependent nuclear localization and remains functional at the nucleus of metastatic prostate cancer cells. PLoS One. 8:e571942013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kamal A, Fain C, Park A, Wang P, Gonzalez-Velez E, Leffler DA and Hutfless SM: Angiotensin II receptor blockers and gastrointestinal adverse events of resembling sprue-like enteropathy: A systematic review. Gastroenterol Rep (Oxf). 7:162–167. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bundalo M, Djordjevic A, Bursac B, Zivkovic M, Koricanac G and Stanković A: Fructose-rich diet differently affects angiotensin II receptor content in the nucleus and a plasma membrane fraction of visceral adipose tissue. Appl Physiol Nutr Metab. 42:1254–1263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hogan KA, Chini CCS and Chini EN: The multi-faceted ecto-enzyme CD38: Roles in immunomodulation, cancer, aging, and metabolic diseases. Front Immunol. 10:11872019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Orciani M, Trubiani O, Guarnieri S, Ferrero E and Di Primio R: CD38 is constitutively expressed in the nucleus of human hematopoietic cells. J Cell Biochem. 105:905–912. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sharma V and O'Halloran DM: Recent structural and functional insights into the family of sodium calcium exchangers. Genesis. 52:93–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ledeen RW and Wu G: Sodium-calcium exchangers in the nucleus: An unexpected locus and an unusual regulatory mechanism. Ann N Y Acad Sci. 1099:494–506. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Philippova M, Joshi MB, Kyriakakis E, Pfaff D, Erne P and Resink TJ: A guide and guard: The many faces of T-cadherin. Cell Signal. 21:1035–1044. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Andreeva AV, Kutuzov MA, Tkachuk VA and Voyno-Yasenetskaya TA: T-cadherin is located in the nucleus and centrosomes in endothelial cells. Am J Physiol Cell Physiol. 297:C1168–C1177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bauer H, Zweimueller-Mayer J, Steinbacher P, Lametschwandtner A and Bauer HC: The dual role of zonula occludens ZO) proteins. J Biomed Biotechnol. 2010:4025932010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Traweger A, Lehner C, Farkas A, Krizbai IA, Tempfer H, Klement E, Guenther B, Bauer HC and Bauer H: Nuclear Zonula occludens-2 alters gene expression and junctional stability in epithelial and endothelial cells. Differentiation. 76:99–106. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bonacquisti EE and Nguyen J: Connexin 43 (Cx43) in cancer: Implications for therapeutic approaches via gap junctions. Cancer Lett. 442:439–444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dang X, Doble BW and Kardami E: The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol Cell Biochem. 242:35–38. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen X, Kong X, Zhuang W, Teng B, Yu X, Hua S, Wang S, Liang F, Ma D, Zhang S, et al: Dynamic changes in protein interaction between AKAP95 and Cx43 during cell cycle progression of A549 cells. Sci Rep. 6:212242016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dzobo K, Thomford NE and Senthebane DA: Targeting the versatile Wnt/β-Catenin pathway in cancer biology and therapeutics: From concept to actionable strategy. OMICS. 23:517–538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim W, Kim M and Jho EH: Wnt/β-catenin signalling: From plasma membrane to nucleus. Biochem J. 450:9–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Johnson M, Sharma M, Jamieson C, Henderson JM, Mok MT, Bendall L and Henderson BR: Regulation of beta-catenin trafficking to the membrane in living cells. Cell Signal. 21:339–348. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Neufeld KL: Nuclear APC. Adv Exp Biol. 656:13–29. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Daniel JM: Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim Biophys Acta. 1773:59–68. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang YX, Wang DY, Guo YC and Guo J: Zyxin: A mechanotransductor to regulate gene expression. Eur Rev Med Pharmacol Sci. 23:413–425. 2019.PubMed/NCBI
|
|
48
|
Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E and Beckerle MC: Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 276:34759–34767. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hohenester E: Laminin G-like domains: Dystroglycan-specific lectins. Curr Opin Struct Biol. 56:56–63. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gracida-Jiménez V, Mondragón-González R, Vélez-Aguilera G, Vásquez-Limeta A, Laredo-Cisneros MS, Gómez-López JD, Vaca L, Gourlay SC, Jacobs LA, Winder SJ and Cisneros B: Publisher Correction: Retrograde trafficking of β-dystroglycan from the plasma membrane to the nucleus. Sci Rep. 8:177852018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Guo X, Elkashef SM, Loadman PM, Patterson LH and Falconer RA: Recent advances in the analysis of polysialic acid from complex biological systems. Carbohydr Polym. 224:1151452019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Westphal N, Loers G, Lutz D, Theis T, Kleene R and Schachner M: Generation and intracellular trafficking of a polysialic acid-carrying fragment of the neural cell adhesion molecule NCAM to the cell nucleus. Sci Rep. 7:86222017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bianchi G and Cusi D: Association and linkage analysis of alpha-adducin polymorphism: Is the glass half full or half empty? Am J Hypertens. 13((6 Pt 1)): 739–743. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen CL, Lin YP, Lai YC and Chen HC: α-Adducin translocates to the nucleus upon loss of cell-cell adhesions. Traffic. 12:1327–1340. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou J, Yi Q and Tang L: The roles of nuclear focal adhesion kinase (FAK) on cancer: A focused review. J Exp Clin Cancer Res. 38:2502019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jones G and Stewart G: Nuclear import of N-terminal FAK by activation of the FcepsilonRI receptor in RBL-2H3 cells. Biochem Biophys Res Commun. 314:39–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tanos BE, Yeaman C and Rodriguez-Boulan E: An emerging role for IQGAP1 in tight junction control. Small GTPases. 9:375–383. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Johnson M, Sharma M, Brocardo MG and Henderson BR: IQGAP1 translocates to the nucleus in early S-phase and contributes to cell cycle progression after DNA replication arrest. Int J Biochem Cell Biol. 43:65–73. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang X and Liu K: P-gp inhibition-based strategies for modulating pharmacokinetics of anticancer drugs: An update. Curr Drug Metab. 17:806–826. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tome ME, Herndon JM, Schaefer CP, Jacobs LM, Zhang Y, Jarvis CK and Davis TP: P-glycoprotein traffics from the nucleus to the plasma membrane in rat brain endothelium during inflammatory pain. J Cereb Blood Flow Metab. 36:1913–1928. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Khot MI, Downey CL, Armstrong G, Svavarsdottir HS, Jarral F, Andrew H and Jayne DG: The role of ABCG2 in modulating responses to anti-cancer photodynamic therapy. Photodiagnosis Photodyn Ther. 29:1015792020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liang SC, Yang CY, Tseng JY, Wang HL, Tung CY, Liu HW, Chen CY, Yeh YC, Chou TY, Yang MH, et al: ABCG2 localizes to the nucleus and modulates CDH1 expression in lung cancer cells. Neoplasia. 17:265–278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Crino PB: Mechanistic target of rapamycin (mTOR) signaling in status epilepticus. Epilepsy Behav. 101((Pt B)): 1065502019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang X, Shu L, Hosoi H, Murti KG and Houghton PJ: Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J Biol Chem. 277:28127–28134. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Rusciano MR, Sommariva E, Douin-Echinard V, Ciccarelli M, Poggio P and Maione AS: CaMKII activity in the inflammatory response of cardiac diseases. Int J Mol Sci. 20:43742019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ma H, Groth RD, Cohen SM, Emery JF, Li B, Hoedt E, Zhang G, Neubert TA and Tsien RW: γCaMKII shuttles Ca2+/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell. 159:281–294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
O'Brien JB, Wilkinson JC and Roman DL: Regulator of G protein signaling (RGS) proteins as drug targets: Progress and future potentials. J Biol Chem. 294:18571–18585. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hepler JR: R7BP: A surprising new link between G proteins, RGS proteins, and nuclear signaling in the brain. Sco STKE. 2005:pe382005.PubMed/NCBI
|
|
69
|
Thomas MP, Erneux C and Potter BV: SHIP2: Structure, function and inhibition. Chembiochem. 18:233–247. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Elong Edimo W, Vanderwinden JM and Erneux C: SHIP2 signalling at the plasma membrane, in the nucleus and at focal contacts. Adv Biol Regul. 53:28–37. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bill CA and Vines CM: Phospholipase C. Adv Exp Med Biol. 1131:215–242. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pan G, Cao X, Liu B, Li C, Li D, Zheng J, Lai C, Olkkonen VM, Zhong W and Yan D: OSBP-related protein 4L promotes phospholipase Cβ3 translocation from the nucleus to the plasma membrane in Jurkat T-cells. J Biol Chem. 293:17430–17441. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mérida I, Arranz-Nicolás J, Rodríguez-Rodríguez C and Ávila-Flores A: Diacylglycerol kinase control of protein kinase C. Biochem J. 476:1205–1219. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Divecha N, Treagus J, Vann L and D'santos C: Phospholipases in the nucleus. Semin Cell Dev Biol. 8:323–331. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cheng Y, Duan C and Zhang C: New perspective on SH2B1: An accelerator of cancer progression. Biomed Pharmacother. 121:1096512020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Maures TJ, Chen L and Carter-Su C: Nucleocytoplasmic shuttling of the adapter protein SH2B1beta (SH2-Bbeta) is required for nerve growth factor (NGF)-dependent neurite outgrowth and enhancement of expression of a subset of NGF-responsive genes. Mol Endocrinol. 23:1077–1091. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Venken T, Schillinger AS, Fuglebakk E and Reuter N: Interactions stabilizing the C-terminal helix of human phospholipid scramblase 1 in lipid bilayers: A computational study. Biochim Biophys Acta Biomembr. 1859:1200–1210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wiedmer T, Zhao J, Nanjundan M and Sims PJ: Palmitoylation of phospholipid scramblase 1 controls its distribution between nucleus and plasma membrane. Biochemistry. 42:1227–1233. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Xi S, Tie Y, Lu K, Zhang M, Yin X, Chen J, Xing G, Tian C, Zheng X, He F and Zhang L: N-terminal PH domain and C-terminal auto-inhibitory region of CKIP-1 coordinate to determine its nucleus-plasma membrane shuttling. FEBS Lett. 584:1223–1230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhou F, Li F, Xie F, Zhang Z, Huang H and Zhang L: TRAF4 mediates activation of TGF-β signaling and is a biomarker for oncogenesis in breast cancer. Sci China Life Sci. 57:1172–1176. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kédinger V, Alpy F, Baguet A, Polette M, Stoll I, Chenard MP, Tomasetto C and Rio MC: Tumor necrosis factor receptor-associated factor 4 is a dynamic tight junction-related shuttle protein involved in epithelium homeostasis. PLoS One. 3:e35182008. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zheng Y and Tyner AL: Context-specific protein tyrosine kinase 6 (PTK6) signalling in prostate cancer. Eur J Clin Invest. 43:397–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ie Kim H and Lee ST: Oncogenic functions of PTK6 are enhanced by its targeting to plasma membrane but abolished by its targeting to nucleus. J Biochem. 146:133–139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yau MK, Lim J, Liu L and Fairlie DP: Protease activated receptor 2 (PAR2) modulators: A patent review (2010–2015). Expert Opin Ther Pat. 26:471–483. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Joyal JS, Nim S, Zhu T, Sitaras N, Rivera JC, Shao Z, Sapieha P, Hamel D, Sanchez M, Zaniolo K, et al: Subcellular localization of coagulation factor II receptor-like 1 in neurons governs angiogenesis. Nat Med. 20:1165–1173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pekar O, Benjamin S, Weidberg H, Smaldone S, Ramirez F and Horowitz M: EHD2 shuttles to the nucleus and represses transcription. Biochem J. 444:383–394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Takahashi M, Tsukamoto Y, Kai T, Tokunaga A, Nakada C, Hijiya N, Uchida T, Daa T, Nomura T, Sato F, et al: Downregulation of WDR20 due to loss of 14q is involved in the malignant transformation of clear cell renal cell carcinoma. Cancer Sci. 107:417–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Olazabal-Herrero A, Sendino M, Arganda-Carreras I and Rodríguez JA: WDR20 regulates shuttling of the USP12 deubiquitinase complex between the plasma membrane, cytoplasm and nucleus. Eur J Cell Biol. 98:12–26. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ugrinova I, Petrova M, Chalabi-Dchar M and Bouvet P: Multifaceted nucleolin protein and its molecular partners in oncogenesis. Adv Protein Chem Struct Biol. 111:133–164. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hovanessian AG, Soundaramourty C, El Khoury D, Nondier I, Svab J and Krust B: Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS One. 5:e157872010. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chen X, Kube DM, Cooper MJ and Davis PB: Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Mol Ther. 16:333–342. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
McQuown SC and Wood MA: HDAC3 and the molecular brake pad hypothesis. Neurobiol Learn Mem. 96:27–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Longworth MS and Laimins LA: Histone deacetylase 3 localizes to the plasma membrane and is a substrate of Src. Oncogene. 25:4495–4500. 2006. View Article : Google Scholar : PubMed/NCBI
|
















