Introduction
Prostate cancer (PCa) endangers the life and health
of older men. The latest statistics on cancer in the United States
revealed that the occurrence of PCa was the highest among men,
accounting for >1/5 of newly diagnosed cancer cases (1). With changes to the economy and
environment, the incidence of PCa in China has been rising
significantly (2,3). Despite the satisfactory prognosis of
PCa at early stage, most cases inevitably develop into
castration-resistant PCa (CRPC) within 2 years, which is
accompanied by a poorer prognosis (4). At present, the molecular mechanisms
of the occurrence and development of PCa and its transformation to
CRPC remain unknown.
It has been reported that changes in growth factor
receptor-related signaling pathways, especially EGFR, may
participate in the complex progression of PCa (5). Several researchers have indicated
that EGFR is involved in the transition from hormone-dependent PCa
to hormone-independent PCa, and is closely associated with the high
recurrence rate and poor prognosis of PCa after castration (5–7).
In addition, it has been confirmed that EGF can promote the process
of epithelial-mesenchymal transition (EMT) in a variety of tumors
via the EGFR pathway (8–11). EMT is a complex and vital process of
cell biology, which serves an important role in tumor development
(10–12). However, the therapeutic effect of EGFR-targeting therapy
on patients with CRPC is not satisfactory, indicating that the
regulatory mechanism of EGFR in CRPC may be more complex and is
worthy of further study (13,14).
CKLF-like Marvel transmembrane domain containing
family member 5 (CMTM5), a member of CMTM, was first reported by
the Human Disease Gene Research Center of Beijing University in
2003, and has been shown to significantly inhibit tumor cell
proliferation, adhesion and migration (15,16). Other members of the CMTM family,
including CMTM3, CMTM7 and CMTM8, have been revealed to inhibit
tumor cell proliferation or migration by regulating EGFR signaling
pathways (9,17–19). However, it remains unknown whether
CMTM5 has a similar regulatory effect in hormone-independent PCa
cells, such as CRPC. Therefore, the present study aimed to
investigate the role and molecular mechanism of CMTM5 in the
development of PCa.
Materials and methods
Patients and tissue samples
Tissue samples were collected from 70 patients at
the Wenzhou People's Hospital between July 2018 and July 2019. All
patients were male and the mean age was 61.52±8.31 (range, 51–82)
years. The distance between the healthy and the cancer tissue was
<5 cm. The inclusion criteria were as follows: Patients with
pathologically confirmed primary PCa with different metastasis. The
exclusion criteria were as follows: Recurrent PCa, treatment with
androgen-deprivation, chemotherapy or radiotherapy prior to
sampling, other infectious disease, malignant tumor, severe liver
and kidney disease, pulmonary fibrosis, bone metabolic disease,
secondary renal hypertension, systemic immune disease or malignant
tumor complications. Adjacent tissues in the same patients were
also collected as the normal control. The Sixth Edition of the TNM
Classification of Malignant Tumors (Union for International Cancer
Control) (20) was used for TNM
staging. This study was approved by the Ethics Committee of the
Wenzhou People's Hospital [Ethics Review (2018) No. (11)], and oral and written informed
consent was obtained from all patients according to the committee's
regulations. All tissue samples and data were collected and shared
in accordance with good medical practices and local laws. The
clinical characteristics of all patients are shown in Table I.
 | Table I.Association between CMTM5 expression
and clinicopathological features of patients with prostate
cancer. |
Table I.
Association between CMTM5 expression
and clinicopathological features of patients with prostate
cancer.
|
|
| CMTM5
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Variables | Cases (n=70) | High (n=16)
(%) | Low (n=54) (%) | χ2 | P-value |
|---|
| Age, years |
|
|
| 0.102 | 0.750 |
|
<70 | 43 | 11 (25.6) | 32 (74.4) |
|
|
|
≥70 | 27 | 6 (22.2) | 21 (77.8) |
|
|
| PSA, ng/ml |
|
|
| 0.324 | 0.850 |
|
<4 | 13 | 9 (69.2) | 4 (30.8) |
|
|
|
4-10 | 8 | 6 (75.0) | 2 (25.0) |
|
|
|
>10 | 49 | 32 (65.3) | 17 (34.7) |
|
|
| AFP, ng/ml |
|
|
| 0.251 | 0.616 |
|
<20 | 32 | 6 (18.38) | 26 (81.2) |
|
|
|
≥20 | 38 | 9 (23.7) | 29 (76.3) |
|
|
| Gleason score |
|
|
| 3.424 | 0.064 |
|
<7 | 46 | 22 (47.8) | 24 (52.2) |
|
|
| ≥7 | 24 | 6 (25.0) | 18 (75.0) |
|
|
| Status of primary
tumor |
|
|
| 0.015 | 0.901 |
|
T1-T2 | 21 | 5 (23.8) | 16 (76.2) |
|
|
|
T3-T4 | 49 | 11 (22.4) | 38 (77.6) |
|
|
| Clinical stage |
|
|
| 0.004 | 0.947 |
| II | 33 | 11 (33.3) | 22 (66.7) |
|
|
|
III–IV | 47 | 16 (34.0) | 31 (66.0) |
|
|
| Lymph node
metastasis |
|
|
| 0.102 | 0.749 |
| + | 28 | 9 (32.1) | 19 (67.9) |
|
|
| - | 42 | 12 (28.6) | 30 (71.4) |
|
|
| Distant
metastasis |
|
|
| 0.188 | 0.665 |
| + | 19 | 7 (36.8) | 12 (63.2) |
|
|
| - | 51 | 16 (36.8) | 35 (31.4) |
|
|
Cell culture and transfection
Full-length CMTM5 cDNA (Sino Biological, Inc.) was
used to construct the overexpressing CMTM5 lentivirus vector, which
was packed using 1.5 µg CMTM5 pLVX–IRES-ZsGreen1 plasmid (YouBio),
according to manufacturer's protocol (pMDL:VSV-G:REV=5:3:2,
multiplicity of infection=20). A 3rd generation system and 293 cell
line (Shenzhen Haodi Huatuo Biotechnology Co., Ltd.) were used. The
PCa cell line, DU145 (Shenzhen Haodi Huatuo Biotechnology Co.,
Ltd.), was transfected with the overexpressing CMTM5 lentiviral
vector (CMTM5-overexpressing cells) and empty vector (vector
control cells) to construct stable transfection. The duration of
transduction into cells of interest was 12 h. A flow cytometer
(cat. no. 175487; Beckman Coulter, Inc.) was used to detect the
expression of CMTM5. Cells were then cultured in the mixture of
RPMI-1640 medium (HyClone; Cytiva) containing 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.), penicillin (100 U/ml) and
streptomycin (100 U/ml) at 37°C with 5% CO2. Subsequent
experiments were performed at 24 h post-transfection.
Immunohistochemical analysis of tissue
samples
Immunohistochemical analysis was performed to detect
the cellular localization and expression of CMTM5 and EGFR. Tissue
samples were fixed in 4% paraformaldehyde for 24 h at room
temperature, embedded in paraffin, sectioned to 4–5 µm thickness
and analyzed via immunohistochemistry. Samples were deparaffinized
with xylene, dehydrated with alcohol and subjected to antigen
retrieval by boiling samples in decreasing concentrations of EDTA
buffer (pH 9.0) at 120°C for 2 min, followed by cooling to room
temperature for 30 min. Then, the sections were soaked in 3%
peroxide solution for 15 min to quench endogenous peroxidase. After
washing in PBS buffer, the sections were blocked with 10% normal
goat serum (Sigma-Aldrich; Merck KGaA) for 10 min (pH 7.5) at room
temperature and incubated with rabbit polyclonal to human CMTM5
antibody (1:100; cat. no. ab187980; Abcam) and primary antibodies
against EGFR (1:50; cat. no. MA5-13070; Thermo Fisher Scientific,
Inc.) at 4°C overnight. On the following day, the sections were
washed with PBS for 5 min and incubated with biotin-labeled goat
anti-rabbit IgG (cat. no. ab205718; 1:5,000; Abcam) at 37°C for 30
min. The slides were washed and visualized with substrate
diaminobenzidine tetrahydrochloride, and then counterstained with
hematoxylin at 37°C for 1 min. The stained samples were imaged
under a light microscope (×20, ×50 and ×100 magnification; Olympus
CX21; Olympus Corporation). According to the percentage of positive
cells (0, <10%; 1, 10–24%; 2, 25–50%; and 3, >50%), the
grades of staining intensity were 0 (no staining), 1 (weak
staining), 2 (moderate staining) and 3 (strong staining). The CMTM5
score was calculated by the percentage of positive cells × staining
intensity, and the range was 0–9. For all cases, a total score of
≥4 was high expression, a score of 0–3 was low expression.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.) from tissue samples. RNA
purity was measured via ultraviolet spectrophotometry, and RNA
samples were stored at −80°C for later use. Subsequently, cDNA was
synthesized from 1 µg RNA using a PrimeScript RT Master Mix kit
(Takara Bio, Inc.) in accordance with manufacturer's protocols. The
mRNA expression levels of CMTM5, EGFR, AKT, PI3K, Bcl-2 and Bax
were quantitatively analyzed using SYBR Green qRT PCR Master mix
(Thermo Fisher Scientific, Inc.) on ABI 7300 Plus real-time PCR
system (Thermo Fisher Scientific, Inc.). Thermocycling conditions
were as follows: Initial hold at 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec. The mRNA expression
level was normalized to β-actin expression in the same sample. The
primers were as follows: CMTM5 forward, 5′-CTTCCTCACCTCCCACAAG-3′
and reverse, 5′-AGATGGAAACCAGGATGATG-3′; EGFR forward,
5′-TTGCCGCAAAGTGTGTAACG-3′ and reverse, 5′-GTCACCCCTAAATGCCACCG-3′;
AKT forward, 5′-AGCGACGTGGCTATTGTGAAG-3′ and reverse,
5′-GCCATCATTCTTGAGGAGGAAGT-3′; PI3K forward,
5′-TATTTGGACTTTGCGACAAGACT-3′ and reverse,
5′-TCGAACGTACTGGTCTGGATAG-3′; Bcl-2 forward,
5′-GGTGGGGTCATGTGTGTGG-3′ and reverse,
5′-CGGTTCAGGTACTCAGTCATCC-3′; Bax forward,
5′-CCCGAGAGGTCTTTTTCCGAG-3′ and reverse,
5′-CCAGCCCATGATGGTTCTGAT-3′; and β-actin (as an internal control)
forward, 5′-GGCACTCTTCCAGCCTTCC-3′ and reverse,
5′-GAGCCGCCGATCCACAC-3′. The 2−ΔΔCq method was used to
calculate relative expression of CLCA2 mRNA (21).
Western blotting assay
Total protein was extracted from the tissue samples
using RIPA buffer (cat. no. BYL40825; JRDUN Biotechnology)
containing protease inhibitor and phosphatase inhibitors, followed
by centrifugation at 16,000 g for 30 min at 4°C to collect the
supernatant. Then, samples were separated via 10% SDS-PAGE and
transferred onto a PVDF membrane, and 40 µg protein/lane was
separated via 8–12% SDS-PAGE. The membrane was blocked with 5%
non-fat milk in TBS at 37°C for 2 h, and then incubated with
antibodies against CMTM5 (1:500; cat. no. ab187980; Abcam), EGFR
(1:500; cat. no. MA5-13070; Thermo Fisher Scientific, Inc.), p-EGFR
(1:200; cat. no. 44–788G; Thermo Fisher Scientific, Inc.) and
β-actin (1:1,000; cat. no. ab8226; Abcam) at 4°C overnight. After
three washes in 0.05% TBS-Tween-20, the membrane was incubated with
HRP-conjugated goat anti-rabbit secondary antibody (1:5,000; cat.
no. sc2030; Santa Cruz Biotechnology, Inc.) and HRP-conjugated
Affinipure Goat Anti-Mouse IgG (1:2,500, cat. no. SA00001-1;
ProteinTech Group, Inc.), at room temperature for 1 h. According to
the manufacturer's instructions, subsequent detection was performed
using the ECL western blotting system (Amersham; Cytiva).
Western blotting was also used to detect the
relative protein expression level of CMTM5 in CMTM5-overexpressing
cells and vector control cells. The procedure was repeated as
aforementioned.
Moreover, primary antibodies against β-actin
(1:1,000; Abcam; cat. no. ab8226), AKT (1:800; cat. no. 4691; Cell
Signaling Technology, Inc.), phosphorylated (p)-AKT (1:800; cat.
no. 4051; Cell Signaling Technology, Inc.), PI3K (1:800; cat. no.
ab86714; Abcam), Bcl-2 (1:200; cat. no. ab117115; Abcam) and Bax
(1:200; cat. no. ab32503, Abcam) were used detect cell apoptosis.
The procedure was repeated as described previously.
ImageJ software (version 1.46; National Institutes
of Health) was used to measure the grey values of protein
bands.
Wound healing assay
A wound healing assay was performed to assess the
effect of CMTM5 on cell lateral migration. CMTM5-overexpressing
cells and vector control cells were placed in 6-well plates (500
l/well). A scratch was created using a 100 µl-sterile micropipette
tip. Cells were then washed twice with PBS to remove cellular
debris and cultured at 37°C in serum-free DMEM for 24 h. Cells were
imaged using an inverted light microscope (×20 magnification, DM
IL; Leica Microsystems GmbH) at 0 and 48 h, and the wound width was
analyzed using ImageJ software 1.46 (National Institutes of
Health).
Transwell migration and invasion
assays
The Transwell assay was performed to assess the
efficacy of CMTM5 on cell longitudinal migration and invasion.
CMTM5-overexpressing cells and vector control cells were placed in
common Transwell plate (Corning, Inc.) chambers to evaluate the
migratory ability. Cells were resuspended in 200 µl serum-free
DMEM, and the cell suspension was then added into the upper
chambers (1×105 cells/well) for 24 h, and DMEM
containing 10% FBS was added to the lower chambers (600 µl/well).
After culturing at 37°C in 5% CO2 for 48 h, the membrane
was washed with PBS, fixed in 4% paraformaldehyde at room
temperature for 30 min and stained with 0.1% crystal violet at room
temperature for 10 min. Cells that did not pass through the
polycarbonate membrane were scraped away gently with a cotton swab,
and the number of migrated cells was counted using an inverted
light microscope (MSHOT/MC30 Digital imaging system; Guangzhou
Micro-shot Technology Co., Ltd.) under a ×200 magnification.
The invasive ability of cells was determined in the
same approach, but the cells were added into upper chambers
precoated with Matrigel (MilliporeSigma) at room temperature for 15
min.
Cell Counting Kit-8 (CCK-8) assay
CMTM5-overexpressing cells and vector control cells
were inoculated in 96-well plates in triplicate at the density of
2×103 cells/well, and cultured at 37°C. Then, 10 µl
CCK-8 solution (Tomos Biotools Shanghai Co., Ltd.) was added to
each well at the indicated time points according to the
manufacturer's instructions following transfection for 2 h at 37°C.
Cells were incubated for 2 h in a 5% CO2 incubator at
37°C. The number of viable cells was calculated via absorbance
measurements at 570 nm using a microplate reader (Bio-Rad
Laboratories, Inc.).
Plate clone formation assay
CMTM5-overexpressing cells and vector control cells
in the logarithmic phase were selected and digested with 0.05%
trypsin into single cells. Cells were suspended in RPMI-1640 medium
containing 10% FBS, and the cell concentration was adjusted to
1×103 cells/ml. In total, 200 µl CMTM5-overexpressing
cell and vector control cell suspensions were seeded into culture
dishes (60-mm; 200 cells per dish) and complete medium was added to
10 ml. The plates were incubated in a cell incubator with 5%
CO2 at 37°C for 2–3 weeks. Then, cells were fixed in 5
ml methanol for 15 min at room temperature and dyed with Giemsa
stain for 20 min at room temperature. Then, 0.1% crystal violet
staining solution was used for staining for 30 min at room
temperature, and then the stain was washed away with double
distilled water and samples were allowed to dry naturally. A
transparent film with grids was placed under a culture dish and the
number of colonies (>50 cells) was counted using a light
microscope (magnification, ×100; Olympus Corporation).
Immunofluorescence
CMTM5-overexpressing cells and vector control cells
were subjected grown on glass slides, washed with PBS and fixed in
using 4% buffered paraformaldehyde dissolved in 0.2 M phosphate
buffer (pH 7.3) for 15 min at room temperature. The samples were
then blocked with 2% BSA (Sigma-Aldrich; Merck KGaA) for 1 h at
room temperature and incubated overnight with p-EGFR (1:200; cat.
no. 44–788G; Thermo Fisher Scientific, Inc.) or E-cadherin primary
antibodies (1:200; cat. no. 14–3249-82; Thermo Fisher Scientific,
Inc.). After washing three times with PBS, the samples were
incubated in a dark chamber with fluorochrome-conjugated secondary
antibody (1:400; cat. no. 4408S; Cell Signaling Technology, Inc.)
for 2 h at room temperature. The samples were washed with PBS and
cover slips were mounted with 90% glycerol in PBS. DAPI (Roche
Diagnostics) was used for nuclear staining for 10 min at room
temperature. The fluorescent images were observed under a
fluorescence microscope (magnification, ×100, Nikon TE-2000U; Nikon
Corporation). The total area of target marker-positive cells was
then evaluated using ImageJ software (version 1.8.0, National
Institutes of Health).
FITC-Annexin V/PI double staining
CMTM5-overexpressing cells and vector control cells
were digested with trypsin and both were either stimulated with or
without the AKT inhibitor MK-2206 (50 nmol/l; MedChem Express) for
2 h at room temperature. On this basis, the four samples were
plated in 96-well plates at a density of 1×104
cells/well in triplicate. The apoptosis rate of cells was evaluated
via Annexin V FITC/PI staining using corresponding apoptosis
detection kits (BD Biosciences). The cells were stained with 5 µl
Annexin V FITC and 5 µl PI and incubated at room temperature in the
dark for 15 min. The extent of apoptosis was analyzed using a flow
cytometer (FACScan; BD Biosciences). Data were analyzed using
FlowJo software V10.7.1 (FlowJo, LLC).
Statistical analysis
Each experiment was repeated three times. SPSS 20.0
statistical software (IBM Corp.) and GraphPad Prism software
(version 8.0.2; GraphPad Software, Inc.) were used for data
analysis. ImageJ software (v1.53a, National Institutes of Health)
was used to analyze the image results of cell experiments. All data
are presented as the mean ± SD. The results were accessed using a
χ2 test, paired t-test, unpaired t-test and one-way
ANOVA followed by Bonferroni correction comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
CMTM5 expression is downregulated in
PCa tissues
CMTM5 was predominately localized in the cytoplasm
(Fig. 1A). The negative
expression, weak expression and medium or strong expression of
CMTM5 in prostate tumor tissue samples were 47.1% (33/70), 35.7%
(25/70) and 17.1% (12/70), respectively. By contrast, CMTM5 in the
70 normal prostate tissue samples was either strongly (37/70;
52.9%) or moderately (30/70; 42.9%) expressed, with only a few with
weakly positive staining (3/70; 4.3%). This indicated that the high
expression of CMTM5 was significantly lower in PCa tissues compared
with that in normal prostate tissues (P<0.001; Fig. 1B). Western blotting and RT-qPCR
results suggested that the protein and mRNA expression levels of
CMTM5 were significantly downregulated in PCa tissues compared with
adjacent tissues (both P<0.001; Fig. 1C-E).
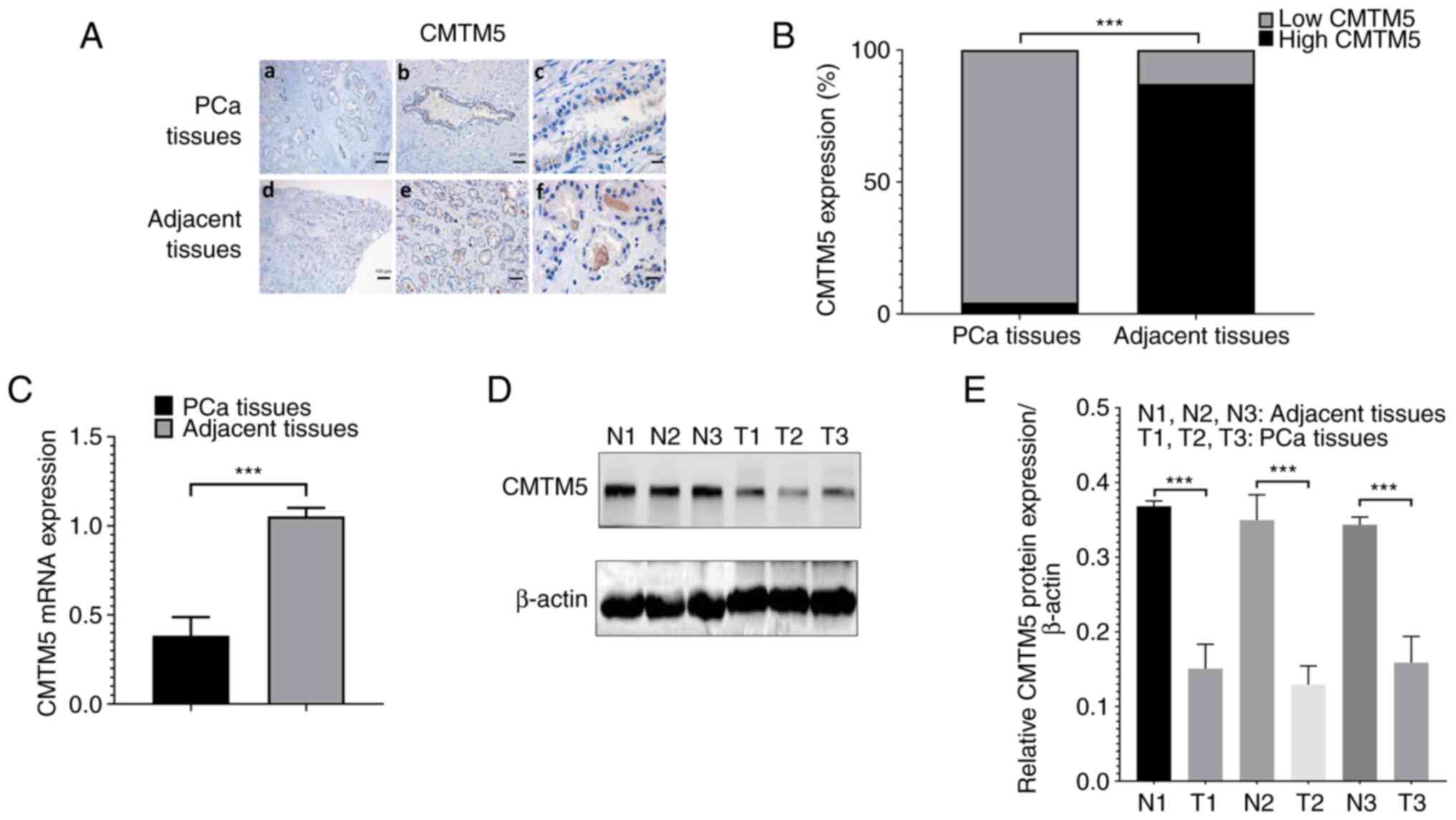 | Figure 1.CMTM5 expression is downregulated in
PCa tissues. (A) Representative images of CMTM5 staining in PCa and
adjacent prostate tissues, (a) PCa tissues in ×20, (b) PCa tissues
in ×40, (c) PCa tissues in ×100, (d) adjacent prostate tissues in
×20, (e) adjacent prostate tissues in ×40, (f) adjacent tissues in
×100 magnification. Scale bars, 100, 50 and 20 µm. (B) The ratio of
high CMTM5 expression and low CMTM5 expression between PCa and
adjacent prostate tissues. (C) CMTM5 mRNA expression in PCa and
adjacent prostate tissues was detected via reverse
transcription-quantitative PCR. (D) Western blotting results of
CMTM5 protein expression in PCa and adjacent prostate tissues. (E)
The gray degree values of CMTM5 western blotting results.
***P<0.001. PCa, prostate cancer; CMTM5, CKLF-like Marvel
transmembrane domain containing family member 5; N, normal; T,
tumor. |
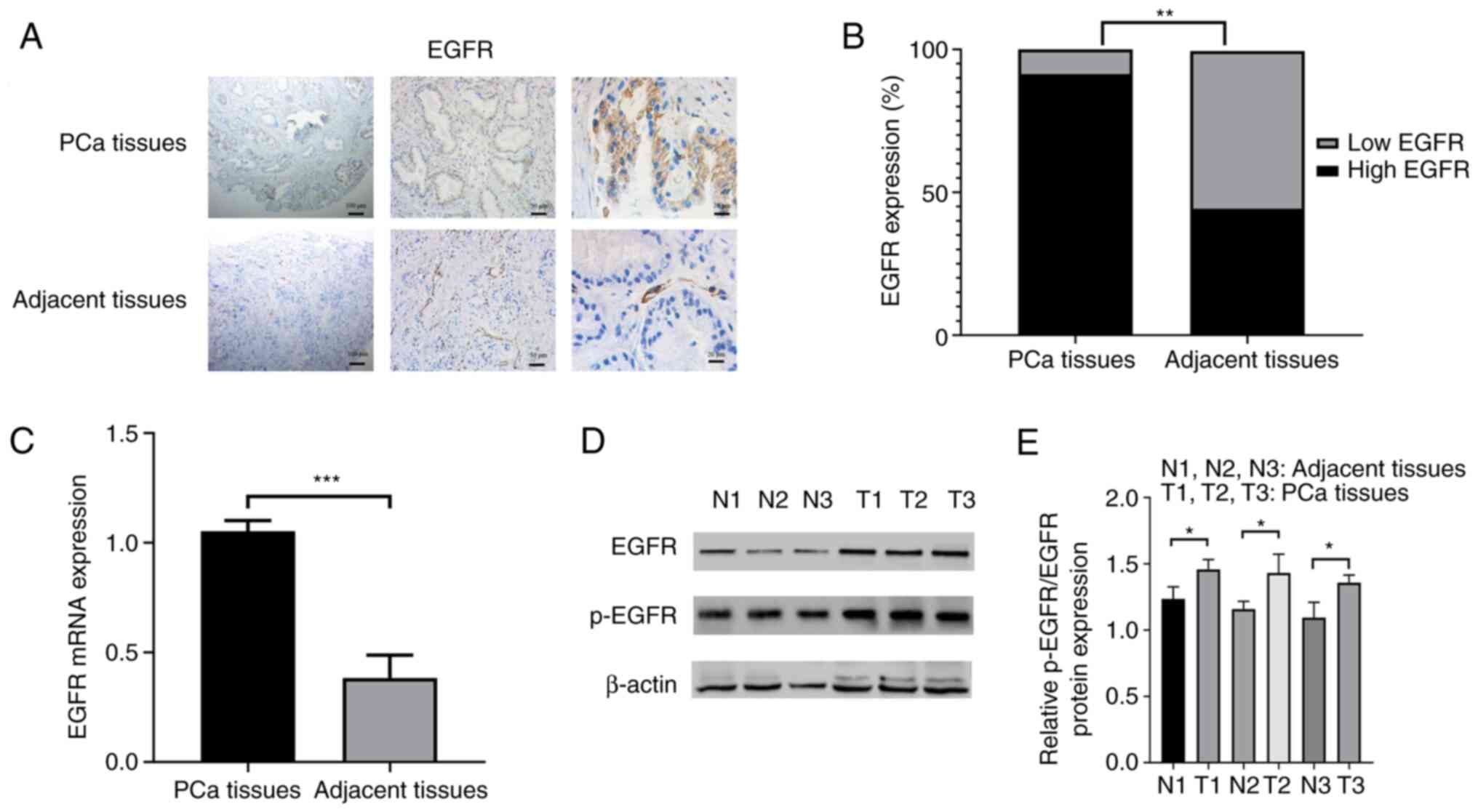
EGFR expression is upregulated in PCa
tissues
The immunohistochemistry results identified that
EGFR was mainly localized on the cell biofilm system, such as
plasma membrane, endoplasmic reticulum and vacuolar, as well as a
small amount on the nuclear and cytoplasmic matrix (Fig. 2A). In the 70 PCa tissues, 64
samples (91.4%) showed high EGFR expression and only 6 samples
(8.6%) showed low EGFR expression. However, in the 70 adjacent
prostate tissues, low EGFR expression was found in 39 samples
(55.7%) and high EGFR expression in 31 samples (44.3%) (P<0.01;
Fig. 2B). The RT-qPCR results
demonstrated that EGFR mRNA expression was upregulated in PCa
tissues (P<0.001; Fig. 2C). To
further confirm this finding, EGFR and p-EGFR expression was
detected using western blotting, and found that the protein
expression levels of EGFR and p-EGFR were upregulated in PCa
tissues (both P<0.05; Fig. 2D and
E).
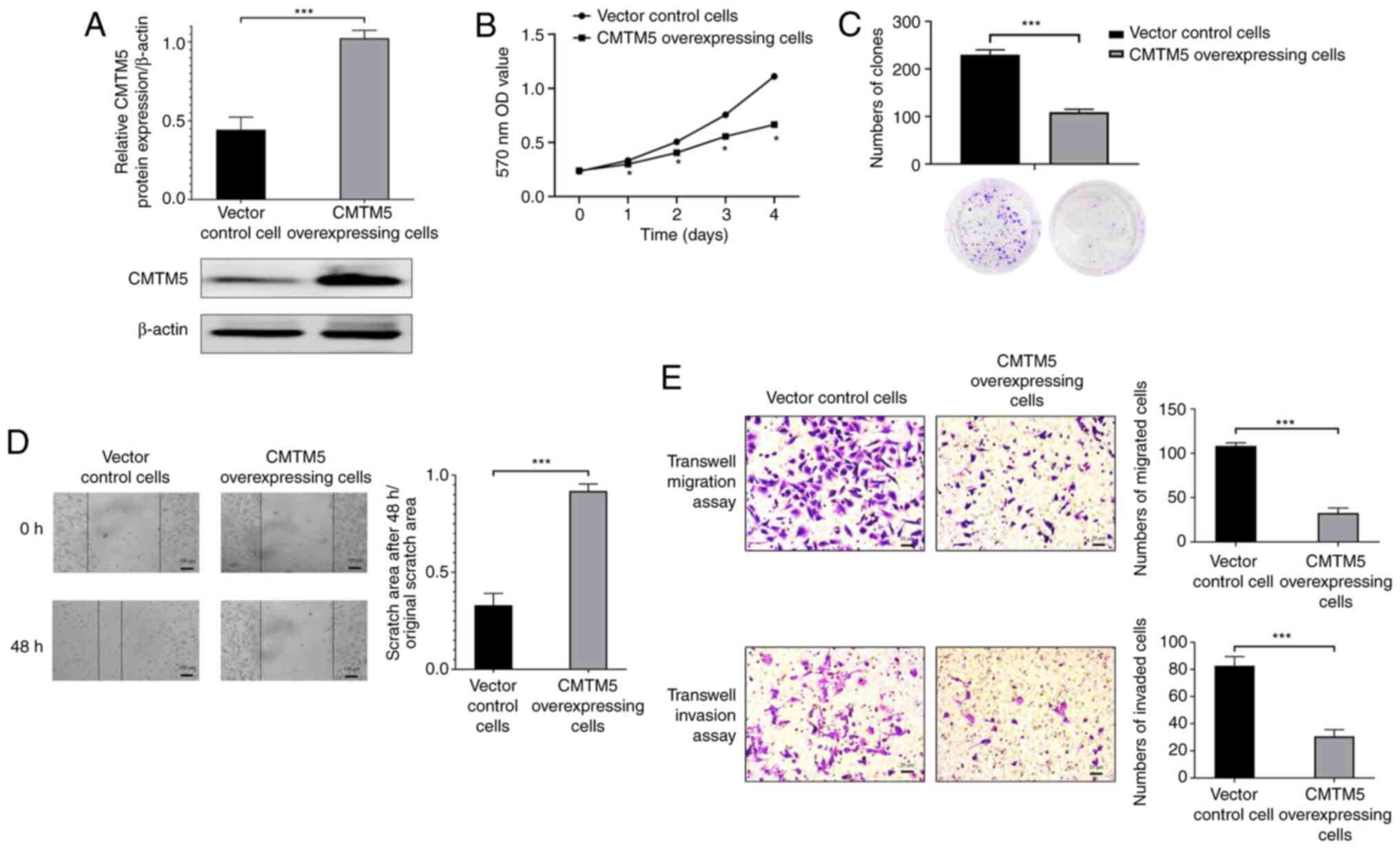
CMTM5 inhibits the proliferation,
migration and invasion of PCa cells
To determine the biological function of CMTM5 in
pathogenesis of PCa, CMTM5 overexpression was induced in DU145
cells. As shown in Fig. 3A, the
lentivirus-mediated overexpression was established, and a higher
expression of CMTM5 protein was detected in CMTM5-overexpressing
cells (P<0.001). CCK-8 and clone formation assays indicated that
the proliferation and colony number of cells were significantly
decreased in CMTM5 overexpression group (P<0.05 and P<0.001,
respectively; Fig. 3B and C).
Moreover, the wound healing assay revealed that
CMTM5-overexpressing cells exhibited a lower scratch closure rate
compared with that of vector control cells (P<0.001; Fig. 3D). Similarly, the results of the
Transwell migration assay indicated that CMTM5 overexpression
significantly reduced the number of migrated cells (P<0.001;
Fig. 3E). Furthermore, the
Transwell invasion assay was conducted to evaluate the effect of
CMTM5 overexpression on the invasive ability of DU145 cells, and it
was found that CMTM5 overexpression inhibited cell invasion
(P<0.001; Fig. 3E). Taken
together, these results suggested that CMTM5 inhibited cell
proliferation, migration and invasion in vitro.
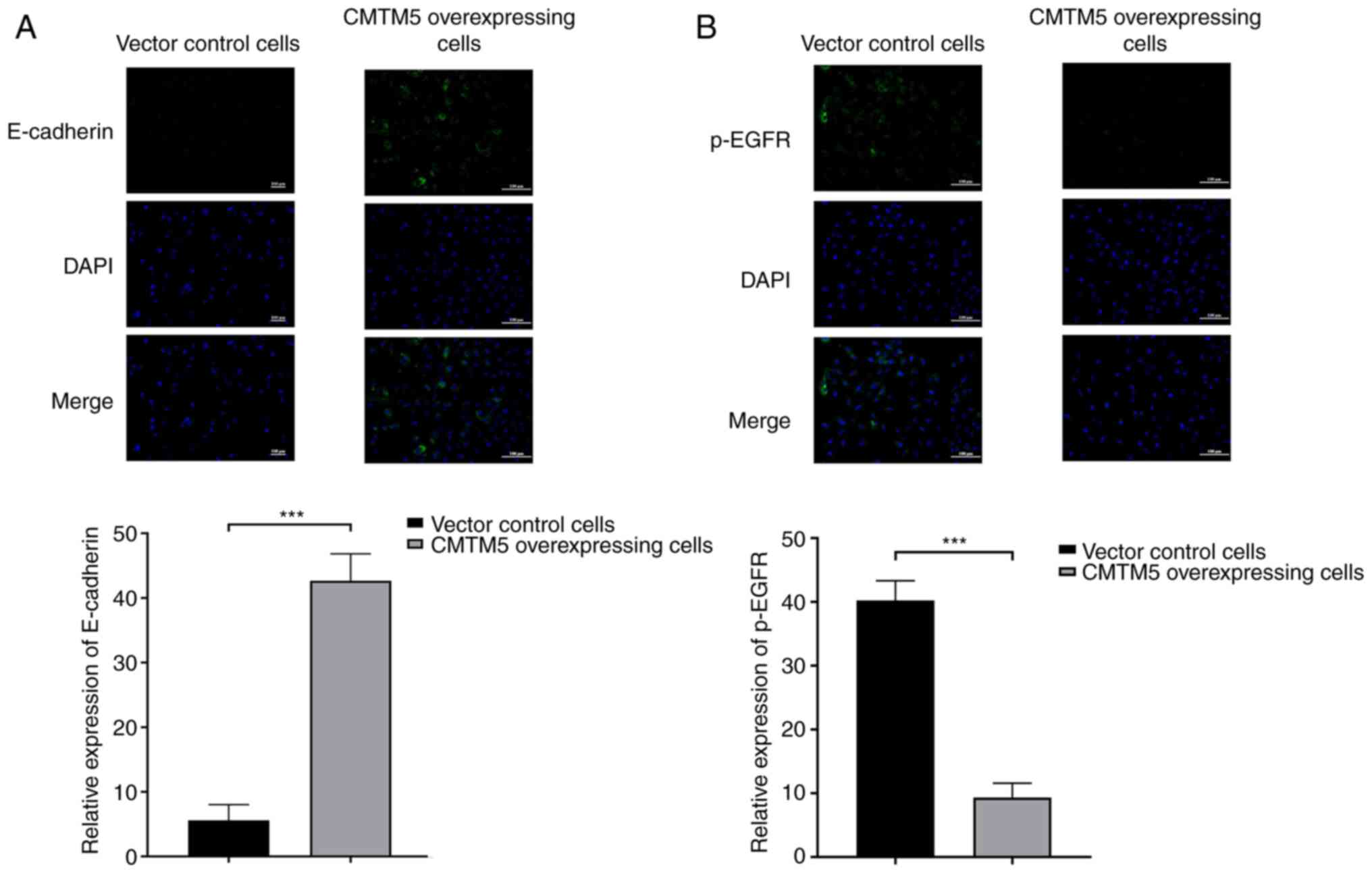
CMTM5 overexpression increases
E-cadherin and decreases p-EGFR in PCa cells
As detected via immunofluorescence, CMTM5
overexpression led to a significant upregulation of E-cadherin
expression when compared with control group (Fig. 4A). On the contrary, p-EGFR was
negatively expressed in CMTM5-overexpressing cells, but positively
expressed in vector control cells (Fig. 4B). Both the results indicated that
CMTM5 may inhibit metastasis of PCa according to the expression of
the analyzed marker proteins.
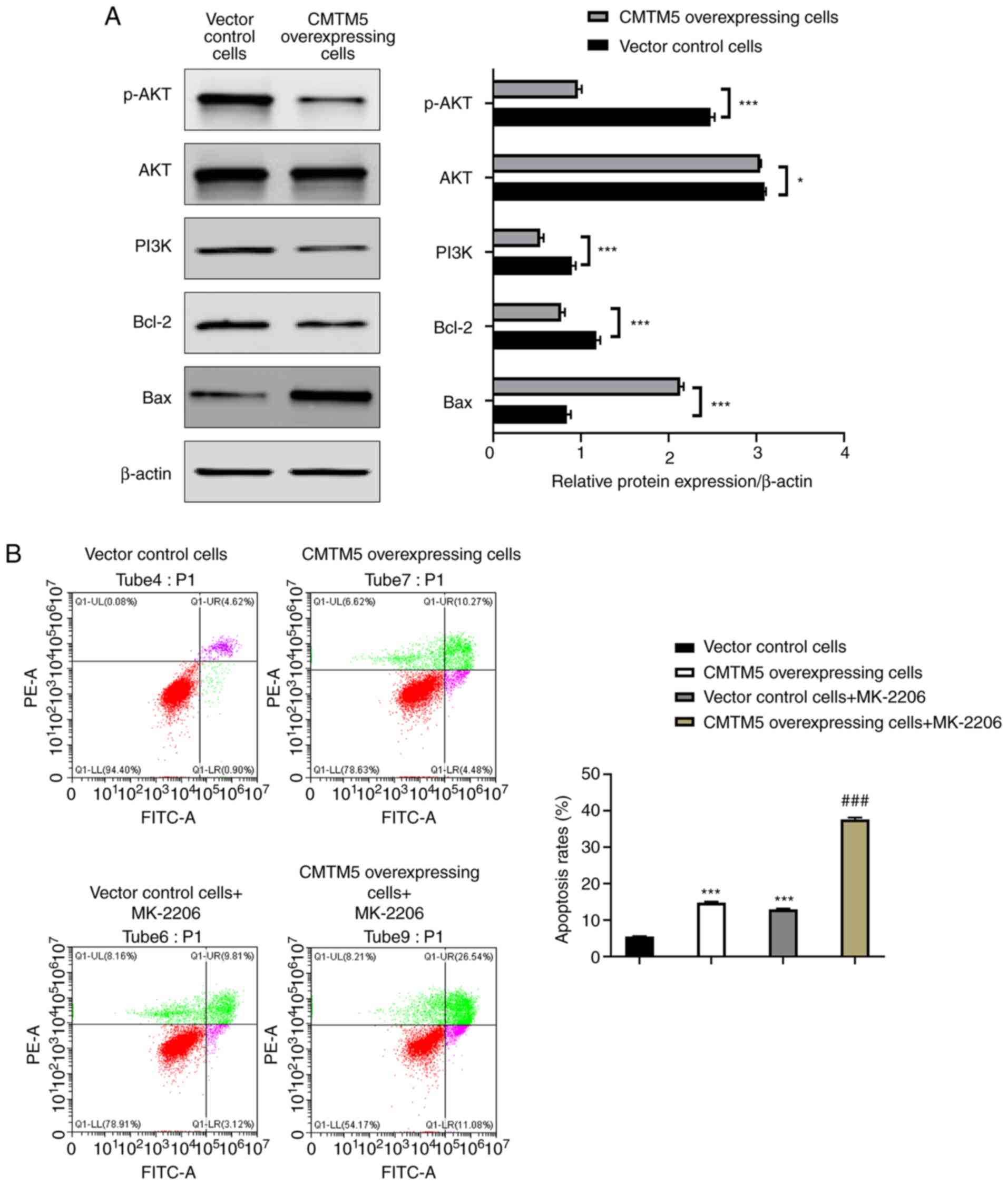
CMTM5 promotes PCa cell apoptosis by
downregulating the PI3K/AKT signaling pathway
As shown in Fig.
5A, CMTM5 overexpression significantly downregulated the
protein expression levels of p-AKT, PI3K and Bcl-2 and upregulated
Bax protein expression (P<0.001) when compared with the control
group. Vector control cells treated with the AKT inhibitor MK-2206
had higher early apoptosis, late apoptosis and necrosis rates
compared with those without MK-2206 (P<0.001; Fig. 5B). The late apoptosis and necrosis
rates of CMTM5-overexpressing cells treated with MK-2206 were
significantly higher compared with those without MK-2206
(P<0.001; Fig. 5B). These
findings indicated CMTM5 could promote apoptosis by downregulating
the PI3K/AKT signaling pathway components in PCa cells.
Discussion
CMTM5, located at 14q11.2, is a polygenic locus
associated with various cancer types. For example, in
nasopharyngeal carcinoma, the frequent loss of heterozygosity at
14q11.2 indicates the presence of functional tumor-suppressor gene
at this locus, suggesting that CMTM5 may be involved in
tumorigenesis (22). The present
study aimed to investigate the effect of CMTM5 on tumorigenesis of
PCa in vitro.
Previous studies have revealed that the expression
level of CMTM5 was reduced or undetectable in most tumor cell lines
or tissues (23,24). Similarly, the current study found
that CMTM5 expression was significantly downregulated in PCa
tissues when compared with normal prostate cancer tissues.
Moreover, EGFR expression was upregulated in PCa tissues. By
contrast, p-EGFR was significantly decreased in
CMTM5-overexpressing cells. Other members of the CMTM gene family
have been shown to serve a role in human cancer by influencing EGFR
and EGFR-related signaling pathways. Yuan et al (9) reported that CMTM3 inhibited
EGF-mediated tumorigenicity of gastric cancer cells by reducing
EGFR expression and promoting EGFR degradation. In addition, the
loss of CMTM7 in non-small cell lung cancer contributes to the
maintenance of aberrant EGFR-mediated oncogenic signaling (18), whereas enhanced expression of
CMTM7 inhibits the proliferation and migration of hepatoma cells
(25). Thus, we hypothesized that
CMTM5 may be able to affect the expression of EGFR in PCa.
In the present study, E-cadherin expression was
upregulated in CMTM5-transfected DU145 cells, while p-EGFR was
negatively expressed. It has been reported that EMT serves an
important role in the development and metastasis of PCa (26), and EGFR can mediate the occurrence
of EMT in human cancer types, including PCa (10,27). Moreover, the expression changes of
related markers, such as E-cadherin, are vital characteristics of
EMT (28). Aberrant EMT
activation may trigger the dissociation of cancer cells from the
primary cancer and migrate to distant organs (29). During this process, the expression
of cell adhesion molecules, including E-cadherin, is suppressed
(29). Combined with the
experimental findings of clinical samples, the current
immunofluorescence results demonstrated that CMTM5 downregulated
EGFR-related proteins in PCa, thereby inhibiting the occurrence of
the EMT response and further suppressing the proliferation and
invasion of PCa cells. In combination with previous studies (8–11),
the current study demonstrated that the ability to downregulate
EGFR may prevent the conversion of PCa to CRPC, which provided a
novel idea for the treatment of this conversion.
A previous study revealed that CMTM3 from the same
gene family as CMTM5 can inhibit migration, invasion and
proliferation of the PCa cell line LNCaP (30). A study conducted by Guan et
al (31) examined the
relationship between CMTM5 and the proliferation, migration and
invasion of hepatocellular carcinoma cells (HCC), and found that
the inhibition of CMTM5 expression by microRNA-10b-3p promoted the
progression of HCC cells, whereas CMTM5 overexpression reduced cell
proliferation, migration and invasion (31). Consistently, the present findings
revealed that the proliferation, migration and invasion of DU145
cells were decreased after transfection with CMTM5, demonstrating
that CMTM5 had a regulatory effect on PCa cell proliferation,
migration and invasion. At the same time, PCa cells transfected
with the CMTM5 overexpressing vector had a higher apoptotic rate
compared with cells transfected with empty vectors. Several
researches have shown that CMTM5 can induce the apoptosis of renal
and pancreatic cancer cells (23,32), which was consistent with the
current findings.
It has been reported that the PI3K/AKT signaling
pathway is involved in the occurrence and development of PCa, and
ectopic expression of CMTM5 in PCa cells can decrease Akt activity
(33,34). Phosphorylation of PI3K activates
the phosphorylation of Akt, which have central roles in regulating
cell proliferation and survival (35). In the present study, the western
blotting results suggested that the protein expression levels of
p-AKT, AKT and PI3K were downregulated in CMTM5-overexpressing
cells. In addition, both CMTM5-overexpressing cells and vector
control cells had significantly increased apoptosis, and the
apoptosis of CMTM5-overexpressing cells was further enhanced by the
addition of the AKT inhibitor MK-2206. This indicated that CMTM5
inhibited PCa progression via the inhibition of the PI3K/AKT
signaling pathway.
Compared with other studies, to the best of our
knowledge, the present study was the first to examine how CMTM5
exerts its anticancer effect on PCa by inhibiting EMT, providing a
new direction for the future clinical treatment of PCa and
prevention of the conversion of PCa to CRPC. However, the current
study did not conduct in vivo experiments, and the in
vivo results may differ from those in this study. In addition,
it remains difficult to analyze the association of both EGFR and
CMTM5 between the same individuals. These issues will be addressed
in future research, when more in-depth studies will be conducted.
Moreover, the correlation between the expression of CMTM5 and the
corresponding characteristics of tumors in various PCa cell lines
will be investigated, with the aim of elucidating the biologically
relevant functions of CMTM5.
In conclusion, the present study provided evidence
to demonstrate that CMTM5 expression was downregulated in PCa and
exerts an anticancer role by regulating the EGFR/PI3K/AKT signaling
pathways. Thus, CMTM5 may be the starting point of a novel
therapeutic strategy for the treatment of PCa.
Acknowledgements
Not applicable.
Funding
This work was supported by the Medical Health and Scientific
Program of Zhejiang province (grant no. 2019RC277).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL, YH and RF conceived the study. DC developed the
methodology. JZ performed the experiments. WB, XX, HC and WC
analyzed and interpretated data. All authors have read and approved
the final manuscript. LL and YH confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the Wenzhou People's Hospital. Informed consent was obtained from
all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller K and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhuo L, Cheng Y, Pan Y, Zong J, Sun W, Xu
L, Soriano-Gabarró M, Song Y, Lu J and Zhan S: Prostate cancer with
bone metastasis in Beijing: An observational study of prevalence,
hospital visits and treatment costs using data from an
administrative claims database. BMJ Open. 9:e0282142019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jia Y, Zhu LY, Xian YX, Sun XQ, Gao JG,
Zhang XH, Hou SC, Zhang CC and Liu ZX: Detection rate of prostate
cancer following biopsy among the northern Han Chinese population:
A single-center retrospective study of 1022 cases. World J Surg
Oncol. 15:1652017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu G, Huang S, Nastiuk KL, Li J, Gu J, Wu
M, Zhang Q, Lin H and Wu D: Variant allele of HSD3B1 increases
progression to castration-resistant prostate cancer. Prostate.
75:777–782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mimeault M and Batra SK: Recent advances
on multiple tumorigenic cascades involved in prostatic cancer
progression and targeting therapies. Carcinogenesis. 27:1–22. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peraldo-Neia C, Migliardi G, Mello-Grand
M, Montemurro F, Segir R, Pignochino Y, Cavalloni G, Torchio B,
Mosso L, Chiorino G and Aglietta M: Epidermal Growth Factor
Receptor (EGFR) mutation analysis, gene expression profiling and
EGFR protein expression in primary prostate cancer. BMC Cancer.
11:312011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mitsunari K, Miyata Y, Asai A, Matsuo T,
Shida Y, Hakariya T and Sakai H: Human antigen R is positively
associated with malignant aggressiveness via upregulation of cell
proliferation, migration, and vascular endothelial growth factors
and cyclooxygenase-2 in prostate cancer. Transl Res. 175:116–128.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan M, Schinke H, Luxenburger E, Kranz G,
Shakhtour J, Libl D, Huang Y, Gaber A, Pavšič M, Lenarčič B, et al:
EpCAM ectodomain EpEX is a ligand of EGFR that counteracts
EGF-mediated epithelial-mesenchymal transition through modulation
of phospho-ERK1/2 in head and neck cancers. PLoS Biol.
16:e20066242018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yuan W, Liu B, Wang X, Li T, Xue H, Mo X,
Yang S, Ding S and Han W: CMTM3 decreases EGFR expression and
EGF-mediated tumorigenicity by promoting Rab5 activity in gastric
cancer. Cancer Lett. 386:77–86. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsai PC, Fu YS, Chang LS and Lin SR:
Taiwan cobra cardiotoxin III suppresses EGF/EGFR-mediated
epithelial-to-mesenchymal transition and invasion of human breast
cancer MDA-MB-231 cells. Toxicon. 111:108–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clapéron A, Mergey M, Nguyen Ho-Bouldoires
TH, Vignjevic D, Wendum D, Chrétien Y, Merabtene F, Frazao A,
Paradis V, Housset C, et al: EGF/EGFR axis contributes to the
progression of cholangiocarcinoma through the induction of an
epithelial-mesenchymal transition. J Hepatol. 61:325–332. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Hu J, Wang Y, Ye W, Zhang X, Ju H,
Xu D, Liu L, Ye D, Zhang L, et al: EGFR activation induced
Snail-dependent EMT and myc-dependent PD-L1 in human salivary
adenoid cystic carcinoma cells. Cell Cycle. 17:1457–1470. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jathal MK, Steele TM, Siddiqui S, Mooso
BA, D'Abronzo LS, Drake CM, Whang YE and Ghosh PM: Dacomitinib, but
not lapatinib, suppressed progression in castration-resistant
prostate cancer models by preventing HER2 increase. Br J Cancer.
121:237–248. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bender R and Gabhann FM: Dysregulation of
the vascular endothelial growth factor and semaphorin
ligand-receptor families in prostate cancer metastasis. BMC Syst
Biol. 9:552015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han W, Ding P, Xu M, Wang L, Rui M, Shi S,
Liu Y, Zheng Y, Chen Y, Yang T and Ma D: Identification of eight
genes encoding chemokine-like factor superfamily members 1-8
(CKLFSF1-8) by in silico cloning and experimental validation.
Genomics. 81:609–617. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, Guo X, Shao L, Plate M, Mo X, Wang Y
and Han W: CMTM5-v1, a four-transmembrane protein, presents a
secreted form released via a vesicle-mediated secretory pathway. J
Biochem Mol Biol. 43:182–187. 2010.PubMed/NCBI
|
|
17
|
Liu B, Su Y, Li T, Yuan W, Mo X, Li H, He
Q, Ma D and Han W: CMTM7 knockdown increases tumorigenicity of
human non-small cell lung cancer cells and EGFR-AKT signaling by
reducing Rab5 activation. Oncotarget. 6:41092–41107. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Li J, Su Y, Fan Y, Guo X, Li L, Su
X, Rong R, Ying J, Mo X, et al: A novel 3p22.3 gene CMTM7 represses
oncogenic EGFR signaling and inhibits cancer cell growth. Oncogene.
33:3109–3118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Both J, Krijgsman O, Bras J, Schaap GR,
Baas F, Ylstra B and Hulsebos TJ: Focal chromosomal copy number
aberrations identify CMTM8 and GPR177 as new candidate driver genes
in osteosarcoma. PLoS One. 9:e1158352014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang SS and Amin MB: Utilizing the
tumor-node-metastasis staging for prostate cancer: The sixth
edition, 2002. CA Cancer J Clin. 58:54–59. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shao L, Cui Y, Li H, Liu Y, Zhao H, Wang
Y, Zhang Y, Ng KM, Han W, Ma D and Tao Q: CMTM5 exhibits tumor
suppressor activities and is frequently silenced by methylation in
carcinoma cell lines. Clin Cancer Res. 13:5756–5762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo X, Li T, Wang Y, Shao L, Zhang Y, Ma D
and Han W: CMTM5 induces apoptosis of pancreatic cancer cells and
has synergistic effects with TNF-alpha. Biochem Biophys Res Commun.
387:139–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shao L, Guo X, Plate M, Li T, Wang Y, Ma D
and Han W: CMTM5-v1 induces apoptosis in cervical carcinoma cells.
Biochem Biophys Res Commun. 379:866–871. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang ZM, Li PL, Yang P, Hou XD, Yang YL,
Xu X and Xu F: Overexpression of CMTM7 inhibits cell growth and
migration in liver cancer. Kaohsiung J Med Sci. 35:332–340.
2019.PubMed/NCBI
|
|
26
|
Montanari M, Rossetti S, Cavaliere C,
D'Aniello C, Malzone MG, Vanacore D, Di Franco R, La Mantia E,
Iovane G, Piscitelli R, et al: Epithelial-mesenchymal transition in
prostate cancer: An overview. Oncotarget. 8:35376–3589. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang L, Song G, Tan W, Qi M, Zhang L, Chan
J, Yu J, Han J and Han B: MiR-573 inhibits prostate cancer
metastasis by regulating epithelial-mesenchymal transition.
Oncotarget. 6:35978–35990. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lai Y, Kong Z, Zeng T, Xu S, Duan X, Li S,
Cai C, Zhao Z and Wu W: PARP1-siRNA suppresses human prostate
cancer cell growth and progression. Oncol Rep. 39:1901–1909.
2018.PubMed/NCBI
|
|
29
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu F, Yuan W, Wang X, Sheng Z, Yuan Y, Qin
C, He C and Xu T: CMTM3 is reduced in prostate cancer and inhibits
migration, invasion and growth of LNCaP cells. Clin Transl Oncol.
17:632–639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guan L, Ji D, Liang N, Li S and Sun B:
Up-regulation of miR-10b-3p promotes the progression of
hepatocellular carcinoma cells via targeting CMTM5. J Cell Mol Med.
22:3434–3441. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cai B, Xiao Y, Li Y and Zheng S: CMTM5
inhibits renal cancer cell growth through inducing cell-cycle
arrest and apoptosis. Oncol Lett. 14:1536–1542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen H, Zhou L, Wu X, Li R, Wen J, Sha J
and Wen X: The PI3K/AKT pathway in the pathogenesis of prostate
cancer. Front Biosci (Landmark Ed). 21:1084–1091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiao Y, Yuan Y, Zhang Y, Li J, Liu Z,
Zhang X, Sheng Z, Xu T and Wang X: CMTM5 is reduced in prostate
cancer and inhibits cancer cell growth in vitro and in vivo. Clin
Transl Oncol. 17:431–437. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu JS and CUI W: Proliferation, survival
and metabolism: The role of PI3K/AKT/mTOR signalling in
pluripotency and cell fate determination. Development.
143:3050–3060. 2016. View Article : Google Scholar : PubMed/NCBI
|