Introduction
Osteoarthritis (OA) is one of the main causes of
disability among the middle-aged and elderly population (1). Its main pathological characteristics
include the gradual loss of articular cartilage and formation of
osteophytes, with symptoms including joint pain, deformity and loss
of joint function, leading to a decline in the quality of life of
patients (2). According to the
latest demographic data, the prevalence of knee OA in China ranges
from 1.3-11.1% (2). By contrast,
for individuals aged ≥60 years the prevalence of symptomatic knee
OA is 19.4% (3). IL-1β is
frequently used to establish the OA model in vitro (4). At present, treatment methods for OA
include surgery, physiotherapy and drug therapy (5,6).
However, they can only relieve the symptoms of arthritis instead of
effectively curing the condition in addition to being limited by
the occurrence of adverse reactions (7). Therefore, it is necessary to develop
novel effective treatment methods to prevent the occurrence of
OA.
Long non-coding RNAs (lncRNAs) are a type of RNA
transcripts that do not encode proteins and are typically >200
nucleotides in length (8).
lncRNAs have been reported to be involved in cartilage destruction
during OA pathogenesis (9). The
mechanism underlying the role of lncRNA in OA, especially through
its reported epigenetic regulatory effects, has become a topic of
intense research over the past decade (10). A previous study demonstrated that
expression levels of the lncRNA family with sequence similarity 201
member A (FAM201A) are decreased in necrotic femoral head samples
(11). Whereas, FAM201A
upregulation has been observed to promote the migration and
invasion of cancer cells (10,12,13). However, the role of FAM10A in OA
remains poorly understood. A previous study reported that FAM201A
could inhibit the expression of microRNA (miR)-146a-5p, which is
increased in knee OA (14).
Furthermore, it has been reported that miR-146a expression is
upregulated in IL-1β-treated chondrocytes (15). Smad4 can be knocked down to weaken
the effects of the TGF-β signaling pathway to increase apoptosis,
whilst promoting the development of OA (16). Inhibition of Smad4 can also
promote the expression of proinflammatory factors and MMP13
(17). Through ENCORI database
analysis, it was previously found that miR-146a-5p can target POU
class 2 homeobox 1 (POU2F1). A previous study has shown that POU2F1
expression is downregulated in the articular cartilage tissues of
patients with OA (18). In
addition, inflammation can lead to the downregulation of POU2F
expression in the liver (18).
Therefore, it was hypothetically speculated in the present study
that the lncRNA FAM201A/miR-146a-5p/POU2F1 system can regulate
IL-1β-induced chondrocyte apoptosis, inflammatory response and
extracellular matrix degradation, thereby aggravating OA
injury.
Materials and methods
Bioinformatics analysis
ENCORI database (https://starbase.sysu.edu.cn/) was used to predict the
binding sites between FAM201A and miR-146a-5p as well as
miR-146a-5p and POU2F1. The site of interaction between POU2F1 and
FAM201A promoter was predicted by JASPAR database (https://jaspar.genereg.net/).
Cell culture and treatment
The human C-28/I2 chondrocytes were obtained from
BeNa Culture Collection (Beijing Beina Chunglian Institute of
Biotechnology). The cells were cultured in DMEM/F12 medium
(Hyclone; Cytiva) containing 10% FBS (Thermo Fisher Scientific,
Inc.) and 1% penicillin/streptomycin in a 5% CO2
incubator at 37°C. Cells in logarithmic phase were used for
subsequent experiments. Cells in model group were stimulated with
IL-1β (Sigma-Aldrich; Merck KGaA) (10 ng/ml) at 37°C for 24 h
(19). Untreated cells were
considered as control.
Cell transfection
Small interfering RNAs (siRNAs) against POU2F1
(siRNA-POU2F1), siRNA negative control (siRNA-NC), miR-146-5p
mimic, miR-146-5p inhibitor and inhibitor/mimic NC were synthesized
by Shanghai GenePharma Co., Ltd. Full length of FAM201A and POU2F1
was inserted into pcDNA3.1 plasmid (Abcam) to construct
overexpression (Ov)-FAM201A and Ov-POU2F1. An empty pcDNA3.1
plasmid was used as the negative control (Ov-NC). Cells were seeded
into 6-well plates at a density of 3×105 cells/well and
cultured for 24 h at 37°C. In accordance with the instructions,
these aforementioned indicated plasmids or RNA sequences (2–4
µg/ml) were transfected into the cells using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at
37°C for 48 h. Transfection efficiency was detected using reverse
transcription-quantitative PCR (RT-qPCR) at 48 h after
transfection. The sequences used were as follows: siRNA-POU2F1
sense, 5′-GGUGUUUGUAGACUAUAUATT-3′ and antisense,
5′-UAUAUAGUCUACAAACACCTT-3′; siRNA-NC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-146a-5p mimic sense,
5′-UGAGAACUGAAUUCCAUGGGUU-3′ and antisense,
5′-AACCCAUGGAAUUCAGUUCUCA-3′; NC mimic sense,
5′-UUUGUACUACACAAAAGUACUG-3′ and antisense,
5′-CAGUACUUUUGUGUAGUACAAA-3′; miR-146a-5p inhibitor,
5′-AACCCAUGGAAUUCAGUUCUCA-3′; and NC inhibitor,
5′-CAGUACUUUUGUGUAGUACAAA-3′.
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.). RNA
was synthesized into cDNA using a reverse transcription kit
(Promega Corporation) according to the manufacturer's protocol. The
qPCR reaction was performed using SYBR Green Supermix (Applied
Biosystems; Thermo Fisher Scientific, Inc.) in the ABI 7500 PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 94°C for 30 sec, 55°C for
30 sec and 72°C for 30 sec (22 cycles). The relative expression
levels of target genes were calculated using the 2−ΔΔCq
method (20) and normalized to
those of the housekeeping genes GAPDH or U6. The sequences used
were as follows: FAM201A forward, 5′-CTCGGGACCTCTAGCCAGT-3′ and
reverse, 5′-TGTGGGCAGATGTGGTTTCC-3′; miR-146a-5p forward,
5′-ACACTCCAGCTGGGTGAGAACTGAATTCCA-3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′; POU2F1 forward,
5′-GTAGTACCTTCTTTCCCCACCC-3′ and reverse,
5′-TTGGTTTGTGTGCCTGTGTTG-3′; GAPDH forward,
5′-AATGGGCAGCCGTTAGGAAA-3′ and reverse, 5′-GCGCCCAATACGACCAAATC-3′;
and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
TUNEL assay
Apoptosis was detected using the TUNEL Apoptosis
Assay kit (cat. no. C1088; Beyotime Institute of Biotechnology).
Briefly, cells were washed with PBS and fixed with 4%
paraformaldehyde at room temperature for 20 min. Subsequently,
cells were treated with 0.1% Triton X-100 for 10 min, and then
incubated with TUNEL reagent at 37°C in the dark for 60 min. The
nuclei were stained with 5 µg/ml DAPI at room temperature for 5
min. The morphological changes of apoptotic cells were observed
under a fluorescence microscope in three fields of view (Olympus
FV500; Olympus Corporation). Bright green fluorescence was
considered to indicate apoptotic cells.
ELISA
According to the manufacturer's protocols,
corresponding ELISA kits (Nanjing Jiancheng Bioengineering
Institute) were used to analyze the levels of IL-6 (IL-6 Assay Kit,
cat. no. H007-1-1), TNF-α (TNF-α Assay Kit, cat. no. H052-1) and
IL-1β (IL-1β Assay Kit, cat. no. H002) in the cell culture
supernatant. The samples in each group were analyzed using an
Automatic Microplate Reader (Syngene).
Luciferase reporter assay
The C-28/I2 cells were inoculated into 96-well
plates at a density of 2×104 cells per well. When the
cell confluence reached 60%, the cells were co-transfected with the
reporter construct with the Renilla luciferase expression
vector pRL-TK (Promega Corporation), encoding either the wild-type
(WT) POU2F1 or FAM201A 3′-untranslated region (3′-UTR) or the
mutant (MUT) POU2F1 or FAM201A 3′-UTR plasmid generated by a
Phusion Site-directed Mutagenesis kit (Thermo Fisher Scientific,
Inc.; cat. no. F541) and miR-146-5p mimics or miR-NC using
Lipofectamine® 2000 reagent. At 48 h after transfection,
the relative luciferase intensity of each group was detected using
a Dual-Luciferase assay kit (Promega Corporation) by comparison
with Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was performed using a ChIP assay kit
(Pierce™ Agarose ChIP kit; Thermo Fisher Scientific, Inc.; cat. no.
26156). Briefly, the chromatin fragments derived from C-28/I2 cells
that extracted by the lysis buffer included in the ChIP kit (Thermo
Fisher Scientific, Inc.) were immunoprecipitated with 10 µg
antibody against FAM201A (cat. no. ab184572; Abcam) or 5 µg mouse
IgG (cat. no. sc-2025; Santa Cruz Biotechnology, Inc.) at 4°C
overnight. All antibodies were mixed with the dilution buffer and
250 µg magnetic beads (Thermo Fisher Scientific, Inc.) followed by
incubation for 1 h at 4°C. DNA fragments and these antibodies were
incubated at 4°C overnight. The next day, products of
immunoprecipitation were treated with the ChIP elution buffer and
centrifuged at 6,000 × g for 1 min. Subsequently, precipitated DNA
samples were detected via RT-qPCR.
Western blot analysis
Total RNA was extracted from cells using RIPA buffer
(cat. no. P0013B; Beyotime Institute of Biotechnology). The protein
concentration was detected using a bicinchoninic acid protein
quantitative kit (cat. no. P0012; Beyotime Institute of
Biotechnology). A total of 20 µg protein was separated by 10%
SDS-PAGE and subsequently transferred onto PVDF membranes. These
membranes were blocked with 5% skimmed milk at room temperature for
30 min and incubated with primary antibodies (1:1,000) against
POU2F1 (cat. no. ab178869; Abcam), IL-6 (cat. no. ab233706; Abcam),
TNF-α (cat. no. ab183218; Abcam), IL-1β (cat. no. ab254360; Abcam),
MMP1 (cat. no. ab137332; Abcam), MMP13 (cat. no. ab39012; Abcam),
collagen II (cat. no. ab34712; Abcam), Aggrecan (cat. no. ab3778;
Abcam), Bcl-2 (cat. no. ab182858; Abcam), Bax (cat. no. ab182733;
Abcam), caspase 3 (cat. no. ab32150; Abcam), cleaved caspase 3
(cat. no. ab2302; Abcam), caspase 9 (cat. no. ab65608; Abcam),
cleaved caspase 9 (cat. no. ab2324; Abcam) and GAPDH (cat. no.
ab9485; Abcam) overnight at 4°C. Subsequently, the membranes were
incubated with the corresponding HRP-conjugated Goat Anti-Rabbit
IgG H&L secondary antibody (cat. no. ab205718; Abcam) at 37°C
for 2 h at 1:10,000. An ECL Plus kit (cat. no. P0018; Beyotime
Institute of Biotechnology) was used to visualize the protein
bands. The densitometry analysis was performed using ImageJ
software (version 1.8.0; National Institutes of Health).
Statistical analysis
All data were analyzed using GraphPad Prism 7
software (GraphPad Software, Inc.). All experiments in this study
were repeated three times. The measurement data are presented as
the mean ± standard deviation. Unpaired Student's t-test was used
to compare two groups of data. Comparisons among multiple groups
were performed using one-way ANOVA followed by Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
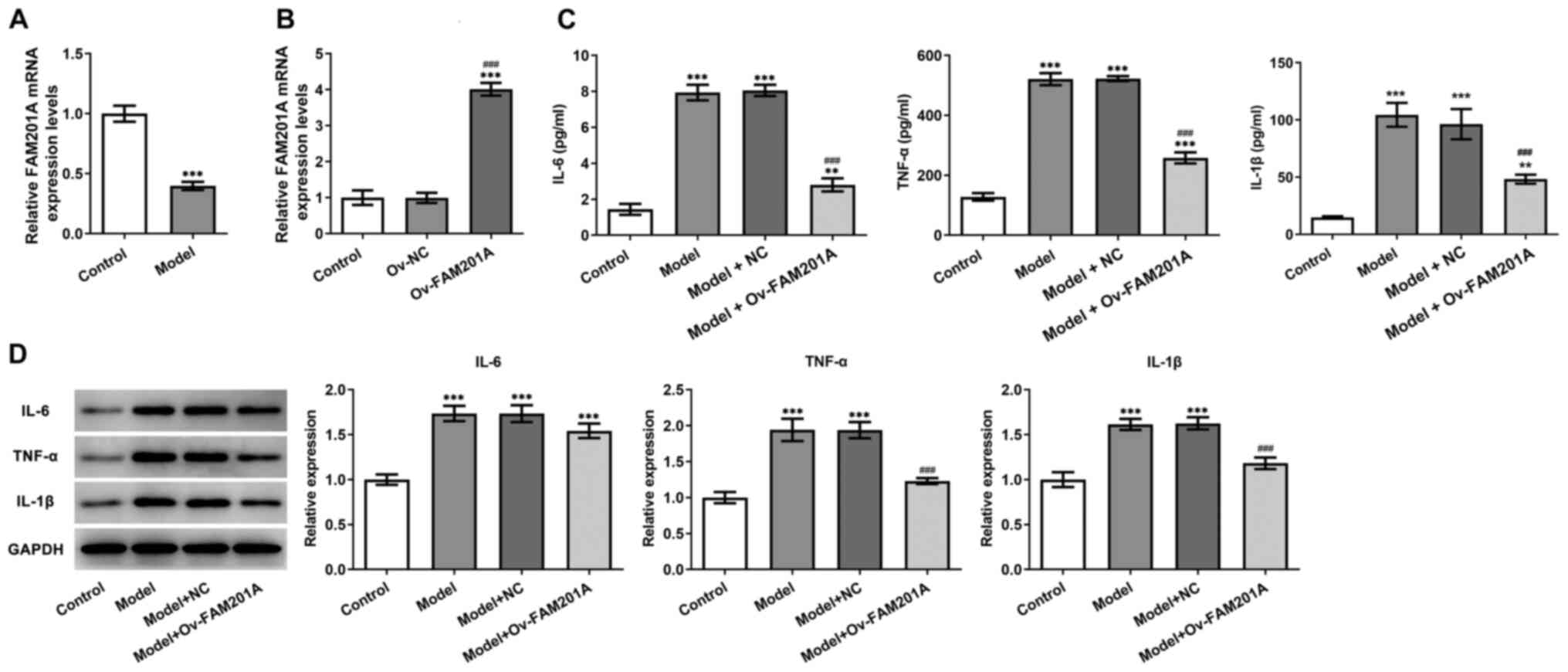
FAM201A expression is downregulated by
IL-1β treatment in C-28/I2 cells
To simulate OA in vitro, C-28/I2 cells were
stimulated with 10 ng/ml IL-1β for 24 h, then FAM201A expression
was detected by RT-qPCR assay, results revealed that FAM201A
expression in C-28/I2 cells was significantly decreased in the
model group compared with that in the normal C-28/I2 cells
(Fig. 1A).
Overexpression of FAM201A attenuates
the inflammatory response, apoptosis and matrix degradation of
IL-1β-treated C-28/I2 cells
The physiological function of FAM201A in
IL-1β-treated C-28/I2 cells was next investigated. As shown in
Fig. 1B, the successful
overexpression of FAM210A in C-28/I2 cells following transfection
with the Ov-FAM201A plasmid was confirmed via RT-qPCR.
Subsequently, ELISA results showed that the secretion levels of
IL-6, TNF-α and IL-1β were significantly decreased in the model +
Ov-FAM210A group compared with those in the model group (Fig. 1C). Results from western blotting
showed that overexpression of FAM201A reduced the inflammatory
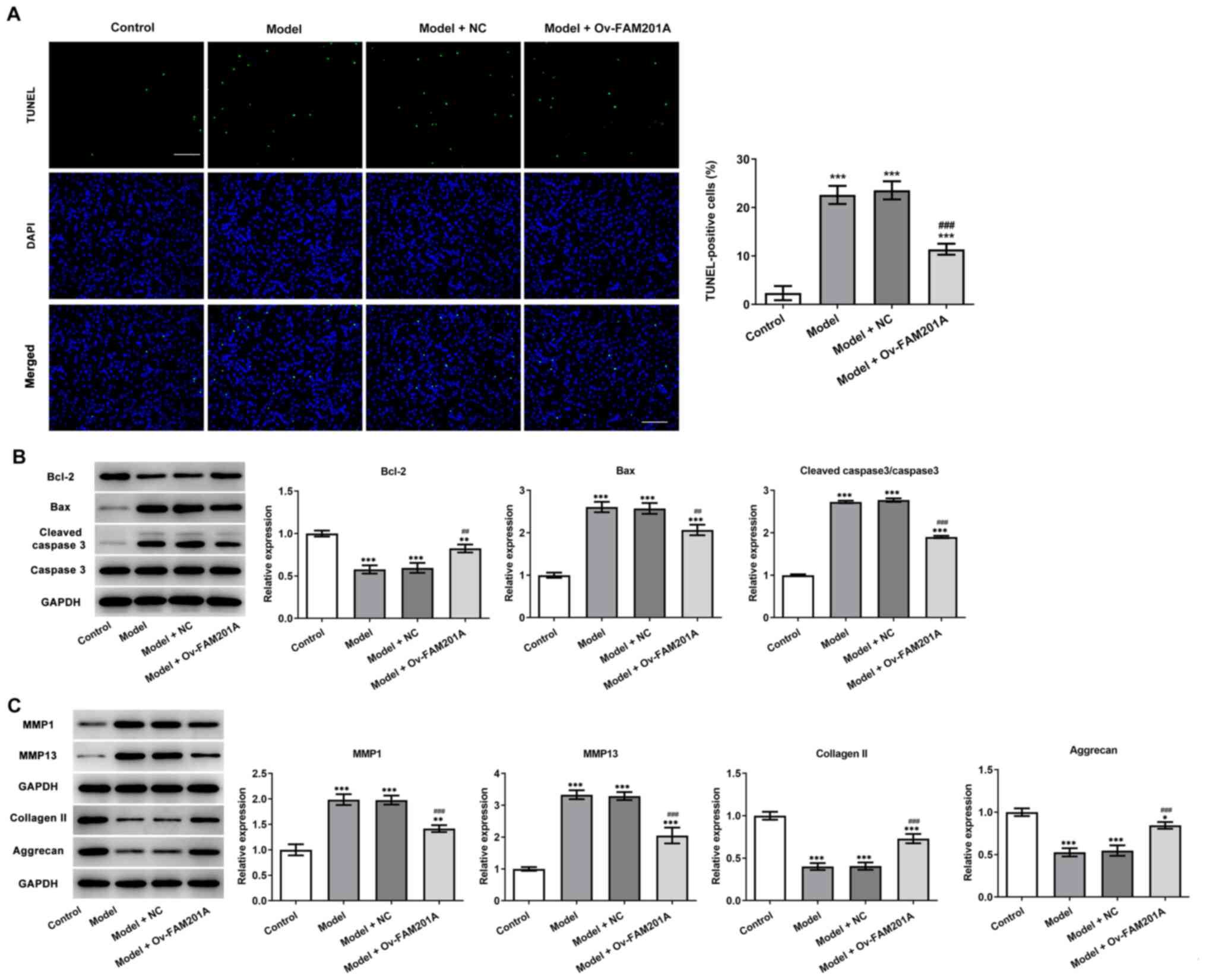
response in the model group compared with the model group (Fig. 1D). TUNEL assay (Fig. 2A) and western blotting (Fig. 2B) results showed that
IL-1β-induced C-28/I2 cell apoptosis was significantly suppressed
in the model + Ov-FAM210A group compared with the model group,
while the expression levels of apoptosis-related proteins were also
significantly reduced and anti-apoptosis protein Bcl-2 was
increased. These findings suggested that the overexpression of
FAM201A could attenuate the apoptosis of IL-1β-treated C-28/I2
cells. The expression levels of proteins associated with
extracellular matrix degradation and components of the
extracellular matrix were subsequently detected via western
blotting. MMP1 and MMP13 expression levels were significantly
decreased, while collagen II and Aggrecan expression levels were
significantly increased in the model + Ov-FAM210A group compared
with the model group (Fig.
2C).
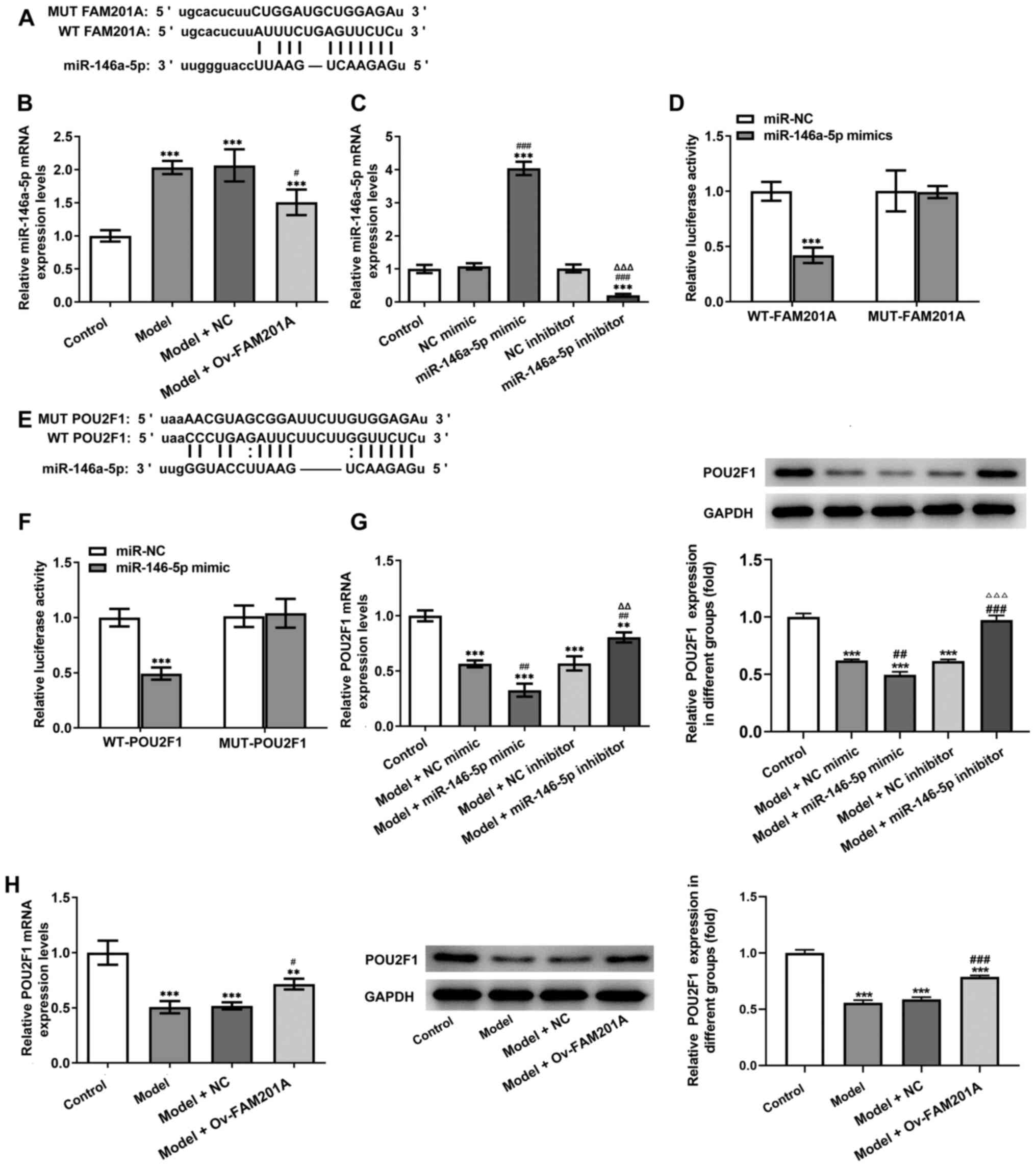
FAM201A directly targets
miR-146a-5p
To further study the mechanism underlying the
function of FAM201A in IL-1β-treated C-28/I2 cells, miR-146a-5p was
predicted to be a target of FAM201A through the ENCORI database
(Fig. 3A). The expression of
miR-146a-5p was found to be significantly decreased in the model +
Ov-FAM210A group compared with the model group (Fig. 3B).
Subsequently, miR-146a-5p mimic and miR-146a-5p
inhibitor were successfully used to modify the expression of
miR-146a-5p in C-28/I2 cells (Fig.
3C). Results of the dual-luciferase reporter assays showed that
compared with that in the miR-NC group, co-transfection with the
miR-16-5p mimic significantly decreased the luciferase activity of
the WT-FAM201A plasmid, but not that of the MUT-FAM201A plasmid
(Fig. 3D). Therefore, these
findings suggested that FAM201A could negatively target
miR-16-5p.
miR-146a-5p directly targets
POU2F1
The downstream target genes of miR-146a-5p were
subsequently predicted by using the ENCORI database. As shown in
Fig. 3E, POU2F1 was predicted to
be a target gene of miR-146a-5p. Results of the dual-luciferase
reporter assays showed that compared with that in the miR-NC group,
miR-146a-5p mimic significantly decreased the luciferase activity
of the WT-POU2F1 plasmid, but not that of the MUT-POU2F1 plasmid
(Fig. 3F). In addition, the
expression of POU2F1 was significantly inhibited in the model +
miR-146a-5p mimic group compared with the model + NC mimic group,
whereas the expression of POU2F1 was significantly increased in the
model + miR-146a-5p inhibitor group compared with the model + NC
inhibitor group (Fig. 3G). The
expression levels of POU2F1 after FAM201A overexpression was then
detected by RT-qPCR. As shown in Fig.
3H, expression of POU2F1 was significantly increased in the
model + Ov-FAM210A group compared with the model + NC group.
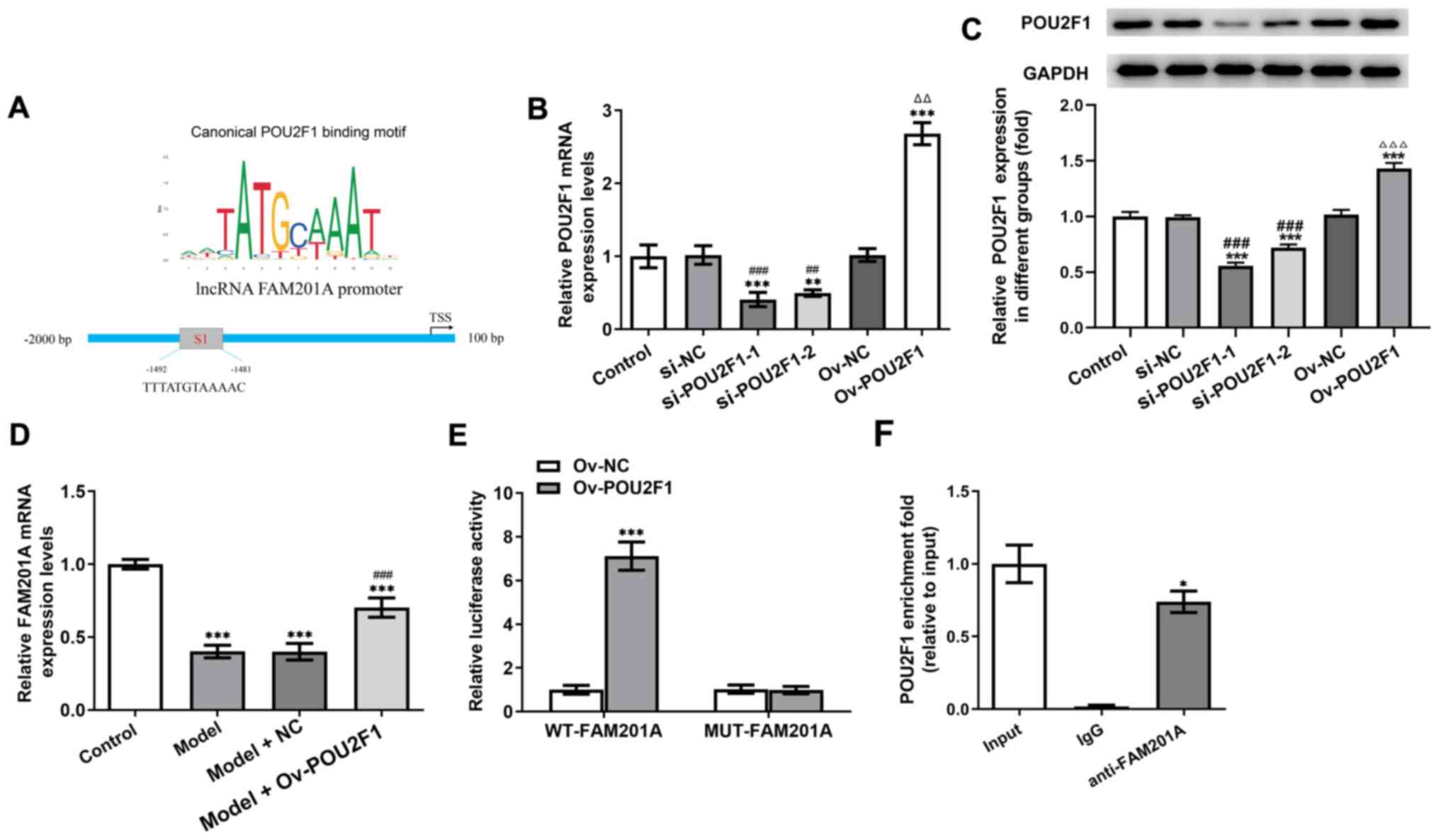
POU2F1 and FAM201A promoters combine
to form a positive feedback loop
It was predicted using the JASPAR database that
there may be a site of interaction (S1) between POU2F1 and FAM201A
(Fig. 4A). Therefore, the
relationships among FAM201A, miR-146a-5p and POU2F1 were studied
further. The expression of POU2F1 in cells was manipulated by
transfection with either siRNA-POU2F1 or pcDNA-POU2F1. As shown in
Fig. 4B and C, compared with that
in the control group, the expression of POU2F1 in the group
transfected with shRNA-POU2F1-1 was the lowest, whereas that in the
pcDNA-POU2F1 group was the highest. In addition, the expression of
FAM201A was significantly promoted in the model + Ov-POU2F1 group
compared with the model group (Fig.
4D). Results of the dual-luciferase reporter assays showed that
compared with that in the Ov-NC group, overexpression of POU2F1
significantly enhanced the luciferase activity of the WT-FAM201A
plasmid, but not that of the MUT-FAM201A plasmid (Fig. 4E). Results of the ChIP assay
revealed the enrichment of POU2F1 on the promoter of FAM201A
(Fig. 4F).
Overexpression of FAM201A upregulates
POU2F1 expression and attenuates the inflammatory response,
apoptosis and matrix degradation in IL-1β-treated C-28/I2
cells
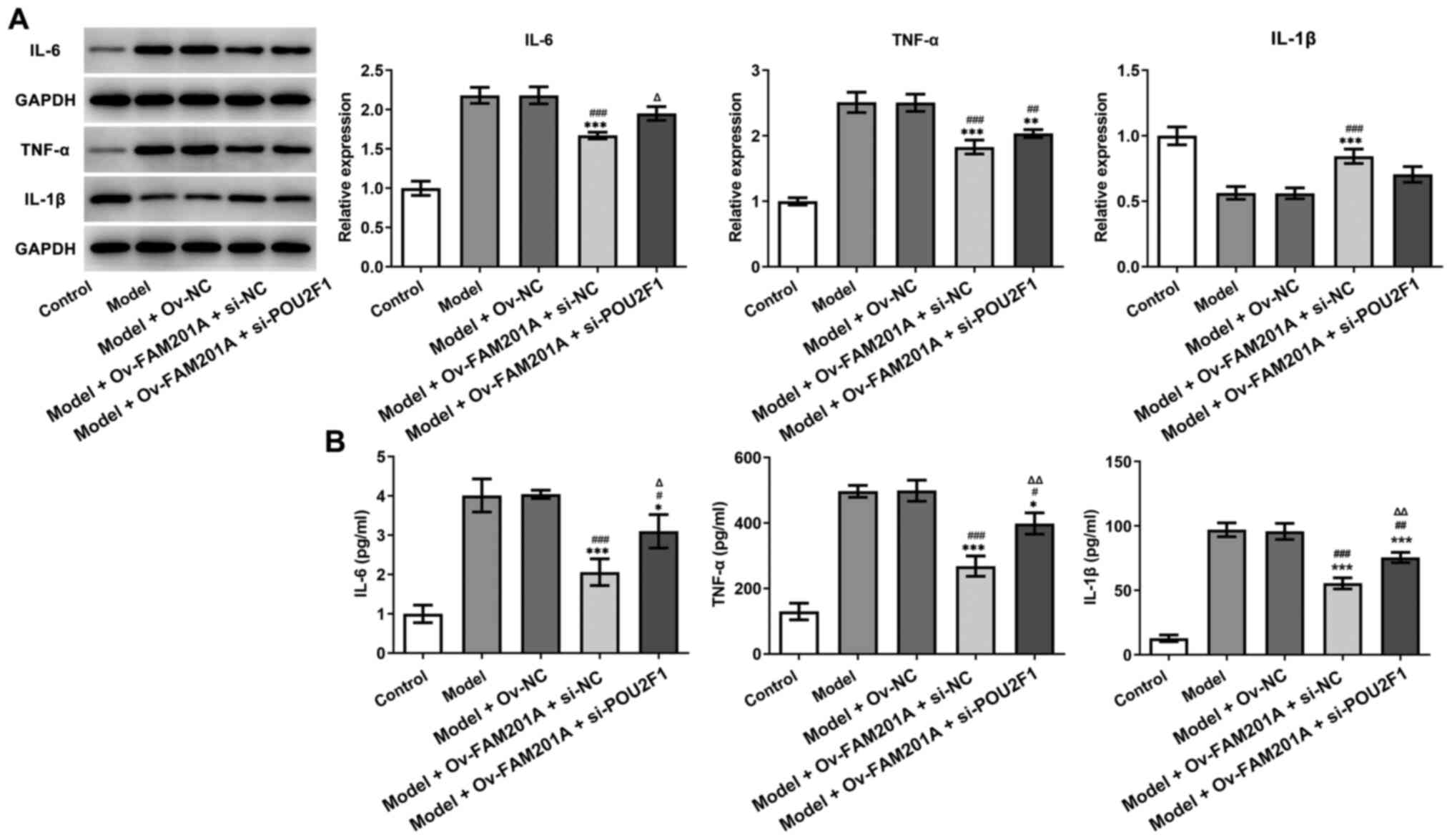
As shown in Fig. 5A
and B, western blotting and ELISA demonstrated that the
expression levels of inflammatory-related proteins, including IL-6,
TNF-α and IL-1β, were upregulated in the model + Ov-FAM201A +
si-POU2F1 group compared with the model + Ov-FAM201A + si-NC. As
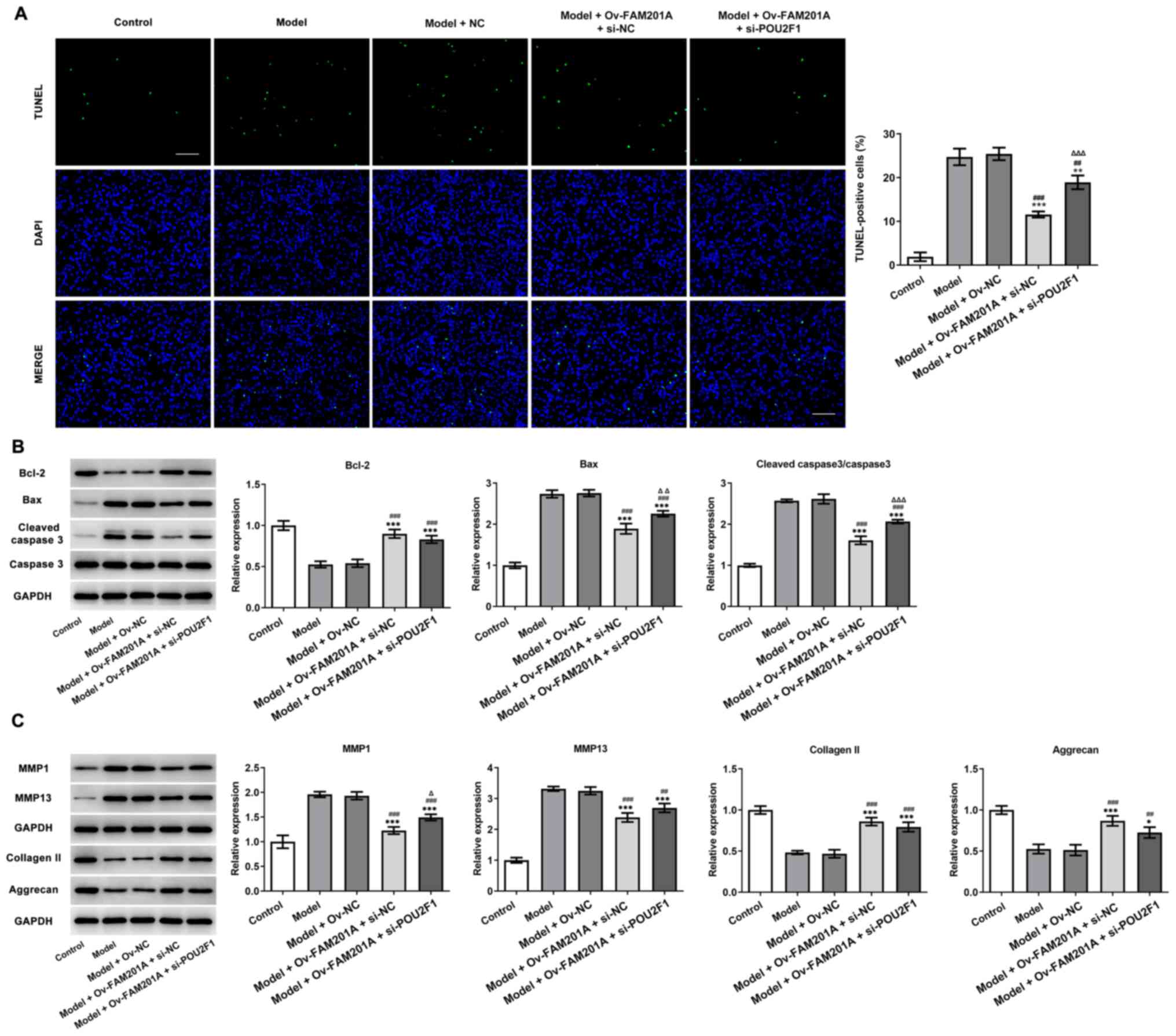
shown in Fig. 6A, TUNEL revealed
that POU2F1 knockdown partially reversed the inhibitory effect of
FAM201A overexpression on the apoptosis of IL-1β-treated C-28/I2
cells. Consistently, western blotting also demonstrated that the
changes in the expression levels of Bcl-2, Bax and cleaved caspase
3/caspase 3 observed in the model + Ov-FAM201A + si-NC were
markedly blocked in the model + Ov-FAM201A + si-POU2F1 group
(Fig. 6B). Furthermore, the
expression of extracellular matrix-related proteins, including
MMP1, MMP13, collagen II and Aggrecan, was measured by western
blotting. The results showed that POU2F1 knockdown partially
reversed the effects of FAM201A overexpression on these proteins
expression in IL-1β-treated C-28/I2 cells (Fig. 6C).
Discussion
OA is one of the most common joint diseases and is
becoming a public health concern worldwide (1). At present, the treatment strategy
for OA is mainly focused on relieving the symptoms (5). However, the underlying mechanisms of
OA development remain to be fully elucidated, forcing the majority
of patients with advanced OA to eventually require joint
replacement surgery. Therefore, studies on OA have focused on
investigating the epigenetic regulation of its pathogenesis and the
discovery of potential therapeutic targets, with non-coding RNAs,
including lncRNAs and miRNAs, of interest (6,21).
Previous studies have reported that lncRNAs serve a key role in the
development of bone and cartilage tissues (9,22).
This suggests that lncRNAs may regulate the balance between
anabolic and catabolic processes in the articular cartilage, which
could be utilized for the diagnosis and treatment of OA.
Furthermore, recent studies have shown that lncRNAs can be used as
personalized therapeutic biomarkers to prevent, halt or even
reverse the progression of OA (23–26). These lncRNAs may be used to
investigate the underlying molecular mechanism involved in
cartilage damage and may be beneficial for the development of novel
therapies for OA. Huang et al (11) recently found that FAM201A
expression was associated with the occurrence of femoral head
necrosis. However, to the best of our knowledge, no relevant
studies on the potential effects of lncRNA FAM201A on OA currently
exists. In the present study, it was found that the expression of
the lncRNA FAM201A was decreased in IL-1β-treated chondrocytes. In
addition, after exploring the function of FAM201A in these
chondrocytes, overexpression of FAM201A alleviated the inflammatory
damage caused by IL-1β, suggesting the protective role of FAM201A
against OA.
Recently, the competing endogenous RNA network
pathway has been widely reported to be the classical mechanism of
action mediated by lncRNAs (27),
which exerts its influence on biological processes by regulating
the expression of miRNAs. A study previously conducted by Hu et
al (26) showed that HOX
transcript antisense RNA promoted the development of OA by
regulating the miR-17-5p/fucosyltransferase 2/β-catenin axis. In
addition, the lncRNA plasmacytoma variant translocation 1 mediates
inflammatory injury by targeting miR-27b-3p/TNF receptor-associated
factor 3 in chondrocytes (28).
The current study subsequently investigated the possible target
miRNA of FAM201A, and the results demonstrated the interaction
between FAM201A and miR-146a-5p. Of note, miR-146a-5p has been
reported to be involved in IL-1β-induced chondrocyte injury and OA
through multiple mechanisms (29–31). Further experiments in the
present study demonstrated that miR-146a-5p inhibited the
expression of FAM201A and overexpression of miR-146a-5p reversed
the protective effects of FAM201A on IL-1β-treated chondrocytes.
These data revealed that FAM201A may exhibit its protective role
against OA via targeting miR-146a-5p.
The current study next demonstrated that POU2F1 was
a downstream target gene of miR-146a-5p, and POU2F1 was
subsequently found to bind to the promoter of FAM201A. In a
previous study, the positive feedback loop LINC00511/miR-150-5p/SP1
was reported to modulate chondrocyte apoptosis and proliferation in
OA (32). The existence of the
FAM201A/miR-146a-5p/POU2F1 positive feedback loop that regulates OA
progression was therefore hypothesized. In accordance with this
hypothesis, results showed that silencing of POU2F1 markedly
blocked the effects of FAM201A overexpression on IL-1β-induced
chondrocytes inflammation and apoptosis. Thus, these results
implied that lncRNA FAM201A could directly regulate the expression
of POU2F1 by binding to miR-146a-5p to exert protective effects
against inflammation in injured chondrocytes. However, the
potential downstream mechanisms that are regulated by the
FAM201A/miR-146a-5p/POU2F1 axis during OA has not been covered in
this study. miR-146a-5p has been reported to promote IL-1β-induced
chondrocyte apoptosis via NF-κB signaling (15). NF-κB signaling can be activated in
response to IL-1β stimulation, and has been widely accepted to
contribute to OA upon activation (33,34). Therefore, future research may
explore the involvement of NF-κB signaling in
FAM201A/miR-146a-5p/POU2F1 axis-mediated chondrocyte injury.
In conclusion, the results from the present study
demonstrated that the positive feedback loop of
FAM201A/miR-146a-5p/POU2F1 could regulate IL-1β-induced
inflammation and apoptosis in chondrocytes. This study may provide
novel targets for the therapeutic treatment of OA.
Acknowledgements
Not applicable.
Funding
This study was supported by The Natural Science Foundation of
Ningbo (grant no. 2019A610247), The Natural Science Foundation of
Zhejiang (grant no. LQ21H060002), The Project of the Science and
Technology Plan for Zhejiang Province (grant no. LGF21F020022), The
Natural Science Foundation of Ningbo (grant no. 202003N4324), The
Natural Science Foundation of Ningbo (grant no. 2019A610252) and
The Ningbo Scientific Project of Public Welfare Plan (grant no.
2019C50056).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Jin L and KG conceived and designed the study. KG,
WW, Jie L and DX performed the experiments. YL, MB and LL analyzed
and interpreted the data. Jin L and KG drafted the manuscript and
revised it for critically important intellectual content. Jin L and
KG confirm the authenticity of all the raw data. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mandl LA: Osteoarthritis year in review
2018: Clinical. Osteoarthritis Cartilage. 27:359–364. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alonso B, Bravo B, Mediavilla L, Gortazar
AR, Forriol F, Vaquero J and Guisasola MC: Osteoarthritis-related
biomarkers profile in chronic anterior cruciate ligament injured
knee. Knee. 27:51–60. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi D, Dai J, Xu Z, Chen D and Jiang Q:
Update on basic and clinical aspects of osteoarthritis. Ann Transl
Med. 3:1422015.PubMed/NCBI
|
|
4
|
Nasi S, Ea HK, So A and Busso N:
Revisiting the Role of Interleukin-1 Pathway in Osteoarthritis:
Interleukin-1α and −1β, and NLRP3 Inflammasome Are Not Involved in
the Pathological Features of the Murine Menisectomy Model of
Osteoarthritis. Front Pharmacol. 8:2822017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng W, Gan D, Hu Y, Zheng Z, Zeng Q, Li
L, Wang X, Zhang Y, Xu Z, Qin L, et al: The effect and mechanism of
QufengZhitong capsule for the treatment of osteoarthritis in a rat
model. J Orthop Translat. 28:65–73. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He M, Zhong X, Li Z, Shen K and Zeng W:
Progress in the treatment of knee osteoarthritis with high tibial
osteotomy: A systematic review. Syst Rev. 10:562021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Glyn-Jones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ou F, Su K, Sun J, Liao W, Yao Y, Zheng Y
and Zhang Z: The LncRNA ZBED3-AS1 induces chondrogenesis of human
synovial fluid mesenchymal stem cells. Biochem Biophys Res Commun.
487:457–463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang SD, Lu J, Deng ZH, Li YS and Lei GH:
Long noncoding RNAs in osteoarthritis. Joint Bone Spine.
84:553–556. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He W, Qiao ZX and Ma B: Long noncoding RNA
FAM201A mediates the metastasis of lung squamous cell cancer via
regulating ABCE1 expression. Eur Rev Med Pharmacol Sci.
23:10343–10353. 2019.PubMed/NCBI
|
|
11
|
Huang G, Zhao G, Xia J, Wei Y, Chen F,
Chen J and Shi J: FGF2 and FAM201A affect the development of
osteonecrosis of the femoral head after femoral neck fracture.
Gene. 652:39–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia H, Wu D, Zhang Z and Li S:
TCF3-activated FAM201A enhances cell proliferation and invasion via
miR-186-5p/TNKS1BP1 axis in triple-negative breast cancer. Bioorg
Chem. 104:1043012020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang J, Yu Q, Zhou Y, Chu Y, Jiang F and
Wang Q: FAM201A knockdown inhibits proliferation and invasion of
lung adenocarcinoma cells by regulating miR-7515/GLO1 axis. J Cell
Physiol. 236:5620–5632. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rousseau JC, Millet M, Croset M,
Sornay-Rendu E, Borel O and Chapurlat R: Association of circulating
microRNAs with prevalent and incident knee osteoarthritis in women:
The OFELY study. Arthritis Res Ther. 22:22020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shao J, Ding Z, Peng J, Zhou R, Li L, Qian
Q and Chen Y: miR-146a-5p promotes IL-1β-induced chondrocyte
apoptosis through the TRAF6-mediated NF-kB pathway. Inflamm Res.
69:619–630. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Huang J, Dai L, Yu D, Chen Q, Zhang
X and Dai K: miR-146a, an IL-1β responsive miRNA, induces vascular
endothelial growth factor and chondrocyte apoptosis by targeting
Smad4. Arthritis Res Ther. 14:R752012. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Wang C, Zhao J, Xu J, Geng Y, Dai
L, Huang Y, Fu SC, Dai K and Zhang X: miR-146a facilitates
osteoarthritis by regulating cartilage homeostasis via targeting
Camk2d and Ppp3r2. Cell Death Dis. 8:e27342017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tian H: Detection of differentially
expressed genes involved in osteoarthritis pathology. J Orthop Surg
Res. 13:492018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Q, Xu X, Kang Z, Zhang Z and Li Y:
Paeonol prevents IL-1β-induced inflammatory response and
degradation of type II collagen in human primary chondrocytes.
Artif Cells Nanomed Biotechnol. 47:2139–2145. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schaukowitch K and Kim TK: Emerging
epigenetic mechanisms of long non-coding RNAs. Neuroscience.
264:25–38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun H, Peng G, Ning X, Wang J, Yang H and
Deng J: Emerging roles of long noncoding RNA in chondrogenesis,
osteogenesis, and osteoarthritis. Am J Transl Res. 11:16–30.
2019.PubMed/NCBI
|
|
23
|
Cen X, Huang XQ, Sun WT, Liu Q and Liu J:
Long noncoding RNAs: A new regulatory code in osteoarthritis. Am J
Transl Res. 9:4747–4755. 2017.PubMed/NCBI
|
|
24
|
Wang Z, Hao J and Chen D: Long Noncoding
RNA Nuclear Enriched Abundant Transcript 1 (NEAT1) Regulates
Proliferation, Apoptosis, and Inflammation of Chondrocytes via the
miR-181a/Glycerol-3-Phosphate Dehydrogenase 1-Like (GPD1L) Axis.
Med Sci Monit. 25:8084–8094. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu J and Xu Y: The lncRNA MEG3
downregulation leads to osteoarthritis progression via miR-16/SMAD7
axis. Cell Biosci. 7:692017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu J, Wang Z, Shan Y, Pan Y, Ma J and Jia
L: Long non-coding RNA HOTAIR promotes osteoarthritis progression
via miR-17-5p/FUT2/β-catenin axis. Cell Death Dis. 9:7112018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ju C, Liu R, Zhang YW, Zhang Y, Zhou R,
Sun J, Lv XB and Zhang Z: Mesenchymal stem cell-associated lncRNA
in osteogenic differentiation. Biomed Pharmacother. 115:1089122019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu X, Yu Y, Yin F, Yang C, Li B, Lin J and
Yu H: Knockdown of PVT1 inhibits IL-1β-induced injury in
chondrocytes by regulating miR-27b-3p/TRAF3 axis. Int
Immunopharmacol. 79:1060522020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meng Q, Liang C, Hua J, Zhang B, Liu J,
Zhang Y, Wei M, Yu X, Xu J and Shi S: A miR-146a-5p/TRAF6/NF-κB p65
axis regulates pancreatic cancer chemoresistance: Functional
validation and clinical significance. Theranostics. 10:3967–3979.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao G and Gu W: Effects of miR-146a-5p on
chondrocyte interleukin-1β-induced inflammation and apoptosis
involving thioredoxin interacting protein regulation. J Int Med
Res. 48:3000605209695502020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Zheng W, Li D and Zheng J:
miR-146a-5p Promotes Chondrocyte Apoptosis and Inhibits Autophagy
of Osteoarthritis by Targeting NUMB. Cartilage. Jul 27–2012.(Epub
ahead of print). doi: 10.1177/19476035211023550.
|
|
32
|
Zhang Y, Dong Q and Sun X: Positive
Feedback Loop LINC00511/miR-150-5p/SP1 Modulates Chondrocyte
Apoptosis and Proliferation in Osteoarthritis. DNA Cell Biol.
39:1506–1512. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jimi E, Fei H and Nakatomi C: NF-κB
Signaling Regulates Physiological and Pathological Chondrogenesis.
Int J Mol Sci. 20:62752019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rigoglou S and Papavassiliou AG: The NF-κB
signalling pathway in osteoarthritis. Int J Biochem Cell Biol.
45:2580–2584. 2013. View Article : Google Scholar : PubMed/NCBI
|