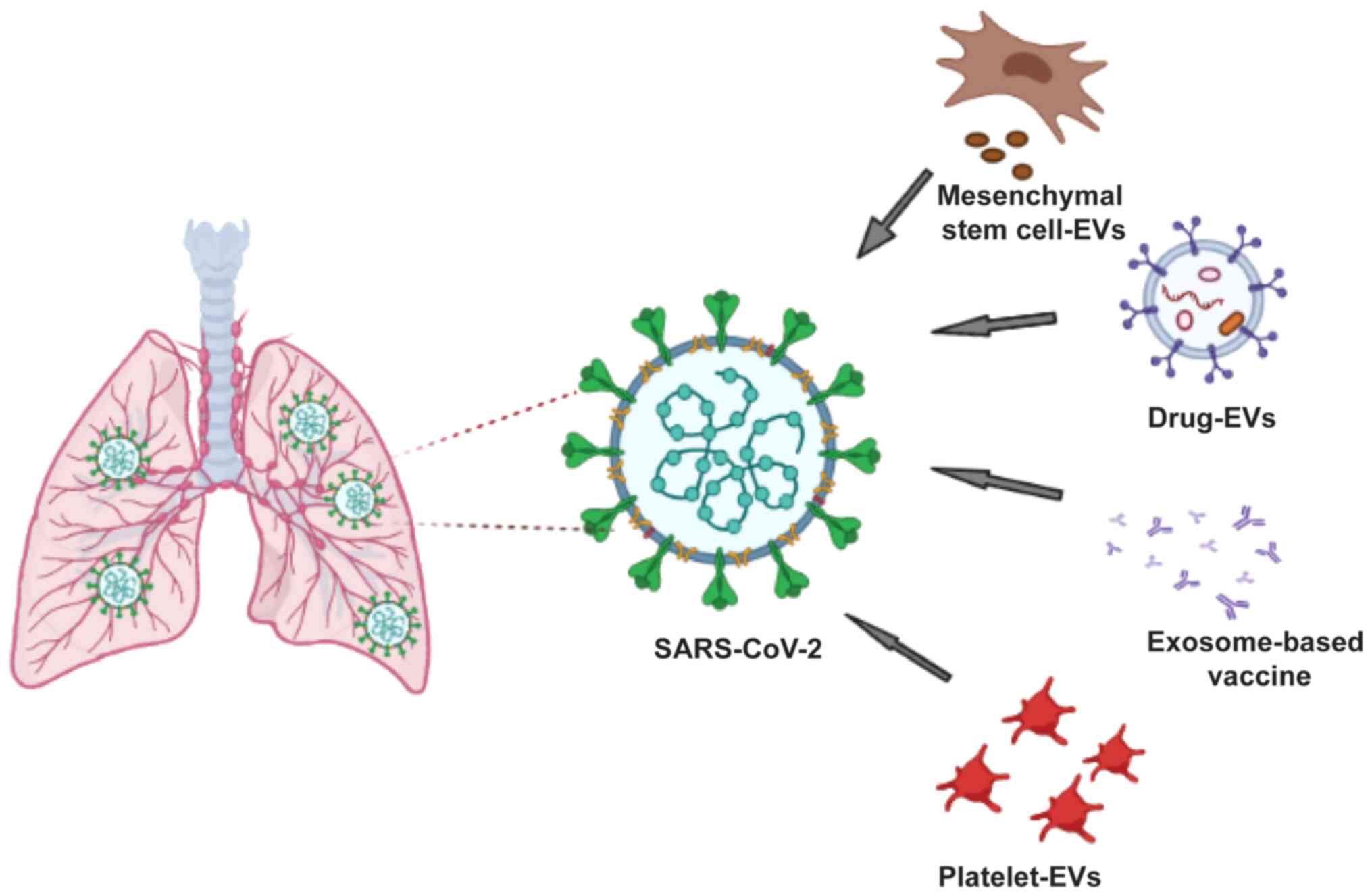
Introduction
Coronavirus disease 2019 (COVID-19), first reported
in Wuhan, China, in December 2019, caused a viral epidemic, and was
declared a pandemic by the World Health Organization (WHO) in March
2020 due to a rapid surge of cases worldwide. As a pandemic, the
COVID-19 outbreak may have a long-lasting impact on public health
if not properly controlled. The WHO
(worldometers.info/coronavirus/) confirmed 241,915,631 COVID-19
cases, including 4,921,308 deaths, worldwide up to October 19,
2021.
Severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2), a positive-sense, single-stranded RNA virus, is the
causative agent of COVID-19. This virus belongs to the coronavirus
family, a group of enveloped viruses that primarily cause
respiratory illness (1). Other
viruses in this family are Middle East respiratory syndrome (MERS)
and SARS viruses. SARS-CoV-2 shares 79.6% of its sequence with
SARS-CoV, which caused an epidemic in 2003 (2). SARS-CoV-2 is more transmissible than
SARS-CoV and MERS-CoV because it can easily spread via liquid
droplets while speaking, coughing or sneezing. Infection can lead
to acute respiratory syndrome, which primarily affects the lungs
and can result in septic shock, pneumonia and death (3).
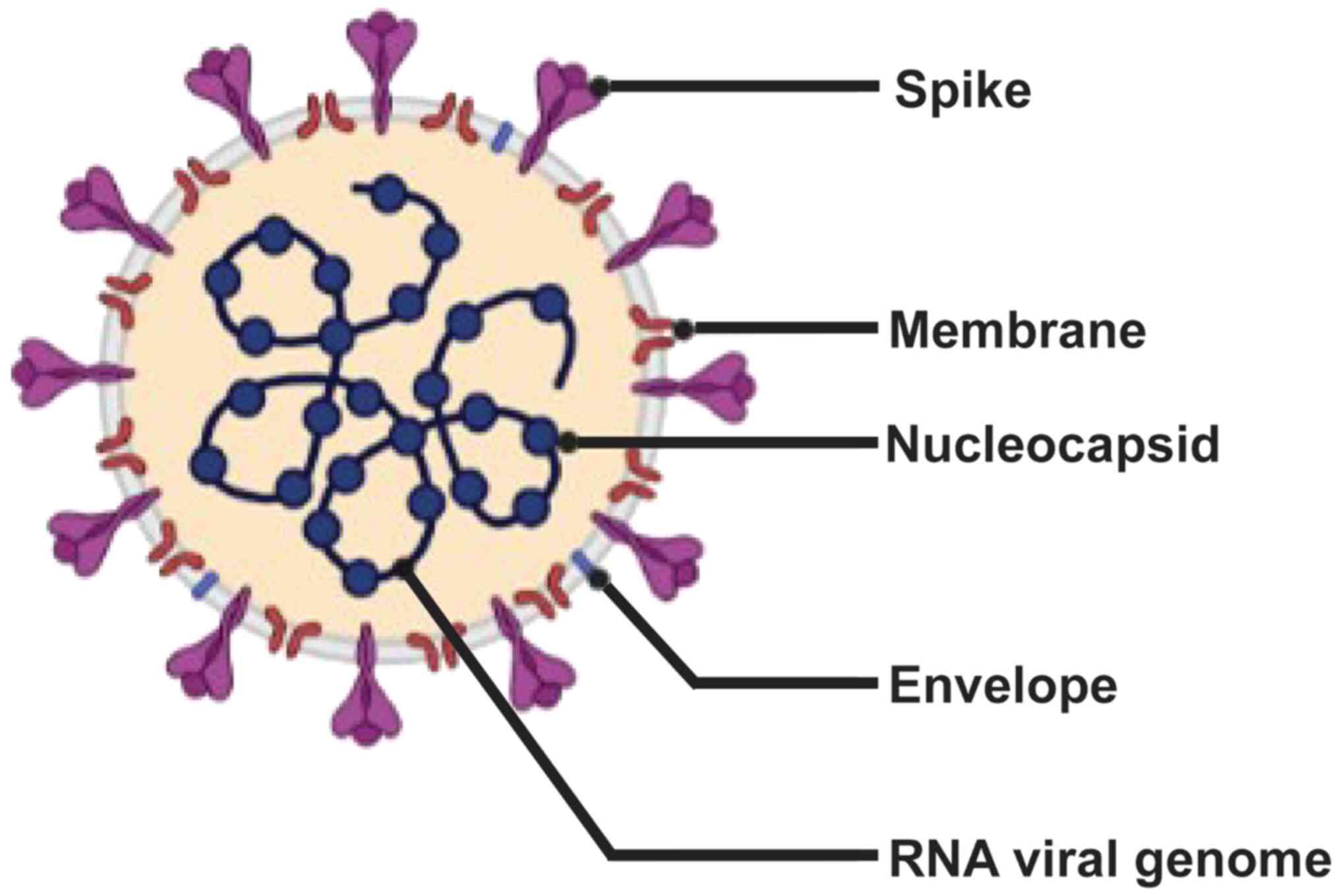
The SARS-CoV-2 genome contains 30 kb RNA, five major
open reading frames and four primary structural proteins [spike
(S), envelope (E), membrane (M) and nucleocapside (N)], all of
which elicit immune responses (Fig.
1) (4–7). The S protein attaches to the host
cell by binding to the angiotensin converting enzyme 2 receptor
(8). Transmembrane serine
protease 2, a host cell serine protease, mediates the
internalization of S protein, followed by its integration into the
host cell (8).
In the current context of the COVID-19 pandemic,
vaccination is an effective measure to restrict widespread viral
infection, and may also prevent future outbreaks. The vaccines
currently used against SARS-CoV-2 primarily target either the S
protein or its receptor-binding domain (RBD) (1). The ongoing vaccine development trial
involves classical molecular strategies that are based upon
inactivated, modified live or attenuated virus, single peptides or
viral vectors (1). Although such
vaccines have been used for long periods against several viral
diseases, they still present multiple issues, such as the risk of
reversion to virulence, inability to provide long-lasting
protection and limited protective immunity (9). Therefore, to overcome the
aforementioned drawbacks of existing vaccines, an alternative
strategy is required to design vaccines that are safer, exhibit
effective antigen presentation and can impart long-term
immunity.
Extracellular vesicles (EVs) and their
significance
EVs are lipid bilayer membrane vesicles derived from
endosomes, which can be produced by all types of cell, including
prokaryotic or eukaryotic and healthy or malignant cells (10). Per the guidelines stated by the
International Society of Extracellular Vesicles (10), EVs can be categorized into three
types: Microvesicles, apoptotic bodies and exosomes. They are
categorized based on size, biogenesis pathway and content. Exosomes
are heterogeneous membrane-bound vesicles 30–150 nm in size
(10). Exosomes were previously
believed to function in the disposal of unwanted materials from the
cell; however, later studies identified a key role in intercellular
signaling and the pathogenesis of cancer and infectious disease
(11,12). Exosomes are unique from other EVs
in that they are released from multi-vesicular bodies (late
endosomes) via exocytosis. Once released from the parent cell, they
fuse with the plasma membrane of target cells and deliver packaged
cargo into the parent cell cytosol (12). The packaged cargo constitutes
nucleic acids, lipids and proteins involved in multiple functions,
such as the surface display of protein (major histocompatibility
complex molecule), epigenetic modification and antigen transfer to
dendritic cells for cross-presentation to T cells (13). Among these lipids, exosomes
usually carry ceramides, sphingolipids, phosphoglycerides and
cholesterol, which serve essential roles in cargo sorting and
internalization (14).
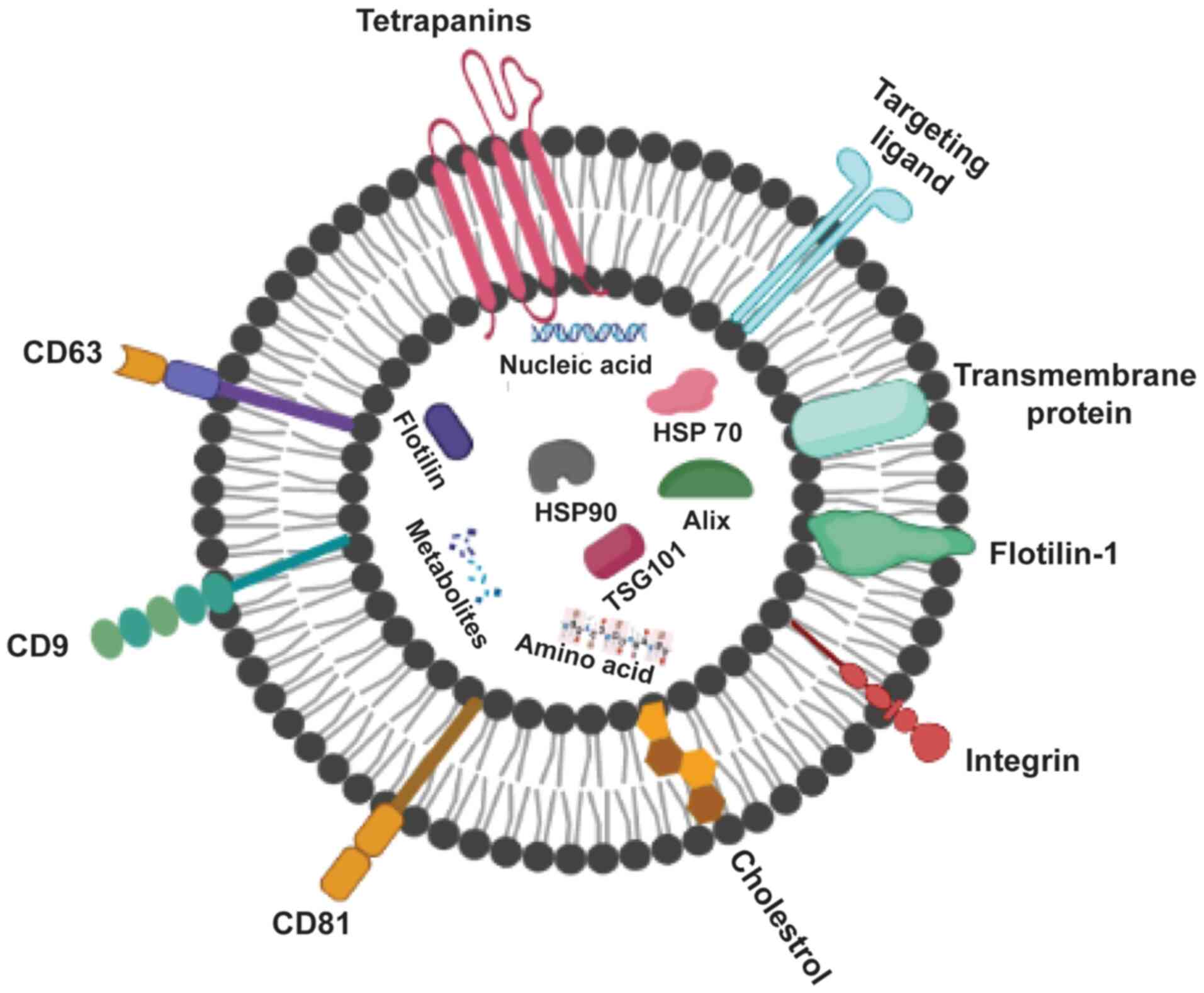
Exosomes (Fig. 2)
can be recognized by the type of proteins they contain; these
include GTPase, which are membrane transport proteins; CD63, CD81,
CD82 and CD9, which are molecular scaffolds; endosomal sorting
complex required for transport (ESCRT), which is a
biogenesis-associated protein; and heat shock protein (HSP)60,
HSP90 and HSP70 (15). Healthy
cells release exosomes under normal physiological conditions to
mediate intercellular communication against growth or stress
response (16). In addition,
exosomes are released by various types of cell, including cancer,
mesenchymal stem and immune cells (17,18). They are also present in a variety
of body fluids, such as plasma, urine, semen, saliva and breast
milk (19,20).
Exosomes derived from apoptotic cells, also known as
apoptotic exosomes (ApoExos), are newly discovered types of EVs
formed in a caspase-dependent pathway and secreted during
apoptosis. Unlike exosomes, ApoExos are produced via
sphingosine-1-phosphate receptor (S1P) signaling independently of
ESCRT (21). ApoExos share
similar characteristics with exosomes in terms of size, protein
expression and role in intercellular communication (21) and exhibit exosome-specific markers
such as HSP70, lysosomal-associated membrane protein 1 and CD63.
ApoExos consist of distinctive marker proteins (S1P receptors 1 and
3) that induce inflammation in mouse macrophages (22). ApoExos have two primary functions:
Apoptotic cell clearance and cell signaling. Similar to exosomes,
the role of ApoExos in cell communication involves immune
modulation, such as antigen presentation during autoimmunity.
Exosome-based vaccines
Exosome-based vaccines may be considered as the
future of therapeutics owing to their involvement in disease
progression and their role in inhibiting viral infection and
triggering host immune response (Fig.
3). There is similarity between viruses and exosomes in terms
of size, biochemical composition, mechanisms of biomolecule
transfer, facilitation of entry into host cells and biogenesis and
multiplication of viruses in host cells. Human immunodeficiency
virus-1 is a notable example, wherein the virus hijacks EV
biogenesis to enhance its spread into the host body by exploiting
the ESCRT pathway (23,24). Furthermore, changes in EV cargo
during viral infection, such as the transfer of viral particles
into uninfected cells and immune response modulation, have led
researchers to characterize EVs and investigate their therapeutic
potential (such as a drug delivery system) or use in antigen
presentation for safe vaccine design (25).
To design an efficacious vaccine, multiple criteria
should be considered (26). Given
that EVs efficiently carry cargo, thus acting as natural delivery
vehicles, they constitute a specific and efficient delivery system
in terms of antigen presentation (27). Moreover, characteristics of EVs,
such as high vascular permeability, stability, solubility and
bio-distribution, make them ideal candidates for vaccines (28). An appropriate approach to develop
a safe vaccine is important. Numerous in vivo studies have
evaluated the toxicity and immunogenicity of EVs (28,29). In one study,
CD81+/CD9+/CD63+ EVs derived from
human embryonic kidney Expi293F cells caused no change in mRNA
expression levels of HepG2 cells; moreover, no hepatotoxicity or
inflammation induction was noted in BALB/c mice (29). In another study, the
administration of CD63+/TSG101+ EVs from
human embryonic kidney 293T cells did not result in toxicity or
immune response in mice, affirming the safety of EVs in an in
vivo model (30).
The key features of EV-based vaccines, including
their ability to induce poor immunogenicity, mean EVs can be safely
and efficiently used in vaccine development. The ability of EVs to
preserve naïve antigen conformation and access to all organs via
bodily fluids give an added advantage compared with other delivery
agents, such as lipid-based nanoparticles (LNPs) or viral vectors
(31). Therefore, engineered EVs
fulfill the criteria for an efficacious vaccine due to their
efficient antigen-presenting system, and high biosafety.
Prospects of exosome-based COVID-19
vaccines
With the increasing global prevalence of COVID-19,
the development of an effective vaccine is imperative to contain
the pandemic. No specific antiviral treatment is currently
available for public use. However, since the SARS-CoV-2 genomic
sequence was identified, >100 vaccine studies have been
performed, ~50 of which have reached human experimentation and a
number of vaccines are currently being administered to certain
sections of the population (ourworldindata.org/covid-vaccinations).
Vaccines approved by medical regulators for use in the US, Europe
and the UK include BNT162b2 and mRNA-1273. ChAdOx1 nCoV-19, a
vaccine produced by AstraZeneca, has been approved by the UK
authorities. Additionally, BNT162b2, a Pfizer-BioNTech COVID-19
vaccine, has recently received US Food and Drug Administration
approval for individuals aged ≥16 years.
Currently available SARS-CoV-2 vaccines are based on
the classical approach of viral vectors, particularly adenoviruses.
Although adenovirus-based vaccines are well-characterized, they are
limited by pre-existing immunity of the virus vector employed in
the vaccine design, which may restrict the immune response against
COVID-19 antigens, thereby decreasing their efficacy (9).
Another point of concern is the risk of re-infection
with emerging viruses in the community due to lack of long-lasting
immunity. Thus, viral infection may become endemic (similar to the
influenza virus endemic), necessitating yearly vaccination
programs. Multiple immunizations with such viral vectors, if not
effective, could lead to more complicated form of the disease, such
as antibody-dependent enhancement (ADE), increasing the disease
burden (32). ADE, a theoretical
exaggeration of disease severity, usually occurs in an infected
individual during viral infection or following vaccination against
viral-based antigens when an antibody against a pathogen amplifies
the infection instead of protecting an individual against it. Such
adverse effects may pose more danger to an individual than the
original disease. Notably, certain recipients of the AstraZeneca
COVID-19 vaccine, which has received approval by the UK
authorities, exhibited a rare blood-clotting disorder; hence, the
safety of these vaccines is questionable (33).
EV-based vaccines constitute an innovative approach
for an efficient virus-free, human-derived vaccine design; this
eliminates the aforementioned virus vector-based vaccine drawbacks
associated with pre-exiting immunity. Due to this advantage of EVs
over virus-based vectors, a number of biotechnology companies are
focusing on vaccine development using EVs as a platform against
SARS-CoV-2. Capricor Therapeutics, for example, developed two
distinct SARS-CoV-2 vaccines using an EV-based platform. First,
they developed an EV-display vaccine comprising 293T cells
transfected with vectors expressing the four structural SARS-CoV-2
proteins (S, E, M and N proteins). Secondly, they developed an
exosome-based mRNA vaccine encoding the N and S proteins of
immunogenic SARS-CoV-2. Moreover, they recently reported that
mRNA-loaded exosome vaccines elicit long-lasting cellular and
humoral responses to both the N and S proteins and result in fewer
adverse effects than currently available COVID-19 vaccines
(34). Similarly, Polak et
al (35) recently reported
induction of neutralizing antibody (NAb) and cellular response by
EV-based vaccines enclosing viral envelope proteins in mice,
thereby eliminating the need for adjuvants. Furthermore, Codiak
BioSciences has reported the safety and tolerability profile of
EV-derived vaccines displaying the anti-tumor cytokine IL-12 from
293T cells in a Phase 1 trial (36). Table
I shows the comparative advantages and disadvantages of
currently available and exosome-based vaccines.
 | Table I.Advantages and disadvantages of
vaccine approaches. |
Table I.
Advantages and disadvantages of
vaccine approaches.
| Vaccine type | Advantages | Disadvantages |
|---|
| Adenoviral
vector |
-
• Direct production of
antigen in the cell of interest
-
• Multiple epitopes can
be included
-
• Scalable production
globally
-
• More immunogenic than
other types of viral vector
|
-
• Pre-existing
anti-adenovirus immunity and potential adverse events, such as
dangerous blood clots
-
• Lack of strong,
long-lasting immunity after single dose
-
• Vaccine-induced
thrombotic thrombocytopenia
|
| DNA |
-
• Stimulation of both
humoral and cell-mediated immunity
-
• Construction of a
vector encoding different antigens in a single vaccine
-
• Efficient large-scale,
low-cost, production and high storage stability
|
|
| RNA |
-
• Ease and rapidity of
assembling novel mRNA sequences into existing vaccine
formulations
-
• Non-toxic and
non-immunogenic
-
• Variant-specific
boosters not required
-
• No risk of integration
with host cell genome
|
-
• Rare, severe
anaphylactic reactions
-
• Long-term immunity
issue
-
• Expensive to
manufacture
|
| Recombinant
protein |
-
• Easy to produce at
large-scale (cost-efficient).
-
• Can be produced in
different expression systems
-
• Well-defined
composition
|
-
• Expression of only
fragment of the protein (not whole protein)
-
• More prone to be
impacted by antigenic drift
-
• Usually elicits weak
immune responses
-
• Need adjuvant
|
| Extracellular
vesicle-based |
-
• Excellent carriers for
viral antigens; present antigens in their native state
-
• Can self-present
antigens (surface major histocompatibility complex molecules)
-
• Can generate
protective immune responses
-
• Can pass through the
blood–brain barrier
|
|
Immunological perspective of present and
future vaccine development strategies
With the global spread of emerging COVID-19 variants
(37), it is critical to develop
a protective vaccine that is effective against multiple strains of
SARS-CoV-2. Moreover, to develop an effective strategy, the
mechanism of the natural immune response against SARS-CoV-2 needs
to be thoroughly understood and targeted.
Ongoing trials for SARS-CoV-2 vaccine construction
are based on the principle of eliciting NAbs against the S protein,
thereby interfering with viral-receptor binding (38,39). To date, research associated with
COVID-19 vaccine development has focused primarily on antibody
titers and the ability of antibodies to neutralize viral particles
(40). A number of studies have
focused on inducing NAb production against the S protein; this
overlooks cell-mediated immunity, a key aspect of adaptive immunity
(39,41).
Both viruses and vaccines induce virus-specific T
cell responses, in addition to antibody responses (42). The potential to elicit
virus-specific T cell response should be exploited for enhanced
immune protection. A recent study reported long-lasting memory T
cell immunity specific to the original SARS-CoV up to 17 years
after initial infection; these SARS-CoV-specific T cells were
almost exclusively directed against the N protein (43). Studies on SARS-CoV, SARS-CoV-2 and
MERS-CoV have confirmed the induction of both CD4+ and
CD8+ T cell responses against the S protein (44,45). The S gene of SARS-CoV-2 shares a
76% amino acid similarity with the S gene of SARS-CoV (44,46) and is more prone to mutation
(47,48). By contrast, the N gene is more
stable and conserved, with 90% amino acid homology and fewer
mutations over time (49–52). The abundance of the N protein,
along with its high immunogenicity, makes it a key target for both
antibody- and cell-mediated immunity to elicit a strong adaptive
immune response (52). A previous
study detected IgG antibodies against the N protein in sera of
patients with SARS (53); the
presence of SARS-specific T cell proliferation and cytotoxic
activity have also been detected, indicating that the N protein is
the primary antigen for T cell-mediated immunity (54,55).
Grifoni et al (44) demonstrated the presence of
SARS-CoV-2 CD8+ T cells against the S and M proteins in
COVID-19 convalescent patients. Ferretti et al (56) validated the presence of maximal
common epitopes on ORF1ab, N, M and ORF3ab protein, but very few on
the S protein; only one epitope was found in the RBD of the S
protein. These results provide better understanding of the
CD8+ T cell response in patients with COVID-19, as well
as a route for designing and developing next-generation vaccines
(56). A recently published study
by Zollner et al (57)
reported that the N protein is a potent T cell inducer. These
results indicate the significance of CD8+ T cell
response in SARS-CoV-2 infection, which may be another vaccine
target (57).
Contrary to the S and N proteins of SARS-CoV-2, the
M and E proteins do not possess strong immunogenicity to trigger
antibody-mediated responses (58). However, the sequence identity of
both the M and E proteins among SARS-CoV, MERS-CoV, and SARS-CoV-2
is greater than that of the S protein, suggesting them as a
potential target to induce T cell-mediated immune response.
Furthermore, a previous study on SARS-CoV and MERS-CoV immunity
reported that several T cell epitopes were found in the M and E
proteins (45). These findings
suggest that including all these proteins as antigens for the
development of vaccines may improve protection against SARS-CoV-2
infection by imparting both T cell- and antibody-based protection;
moreover, this may protect an individual against SARS-CoV
variants.
T cell-mediated immunity may resolve the challenges
associated with providing long-term immunity against SARS-CoV-2 by
existing vaccines, which target only the S protein to produce Nabs
(39–41). As previously reported, individuals
who recover from SARS-CoV infection show virus-specific memory
CD8+ T cells that last for 6–11 years, whereas memory B
cells and antiviral antibodies are not detected for such periods of
time in these individual (59).
Similarly, antibody response decreases within 3 months in patients
with COVID-19 (60). In addition,
certain vaccines are ineffective against the delta variant, which
is more infectious and highly transmissible that other COVID-19
variants (61). Therefore,
vaccines solely based on NAbs cannot provide long-term immunity
against COVID-19; furthermore, the relevance of cell-mediated
immunity should be acknowledged and considered for vaccine
development amid the ongoing pandemic.
Safety aspect of 293T cells for
exosome-based vaccine production
293T cells are utilized for production of EV
vaccines because of their high transfection efficiency and rapid
growth (62). 293T cells are used
in the intermediate and final production of exosome-based vaccines,
as well as multiple types of COVID-19 vaccine. For example, the
Oxford-AstraZeneca adenovirus-based COVID-19 vaccine is derived
from 293T cells (63). Moreover
exosome-based vaccines developed by Capricor Therapeutics, Inc. are
made of 293T cells transfected with vectors that express the four
structural SARS-CoV-2 proteins. However, due to difficulties in
large-scale production of purified exosomes, the company has been
unable to market this vaccine (36).
CKV21, an exosome-based virus like particle vaccine,
is manufactured by Korean biotechnology company CK-Exogene, Inc.
This vaccine uses S, M, E and N protein genes cloned into 293T
cells using a viral vector. Following mass culture of these cells,
exosomes containing S, M, E and N proteins are produced.
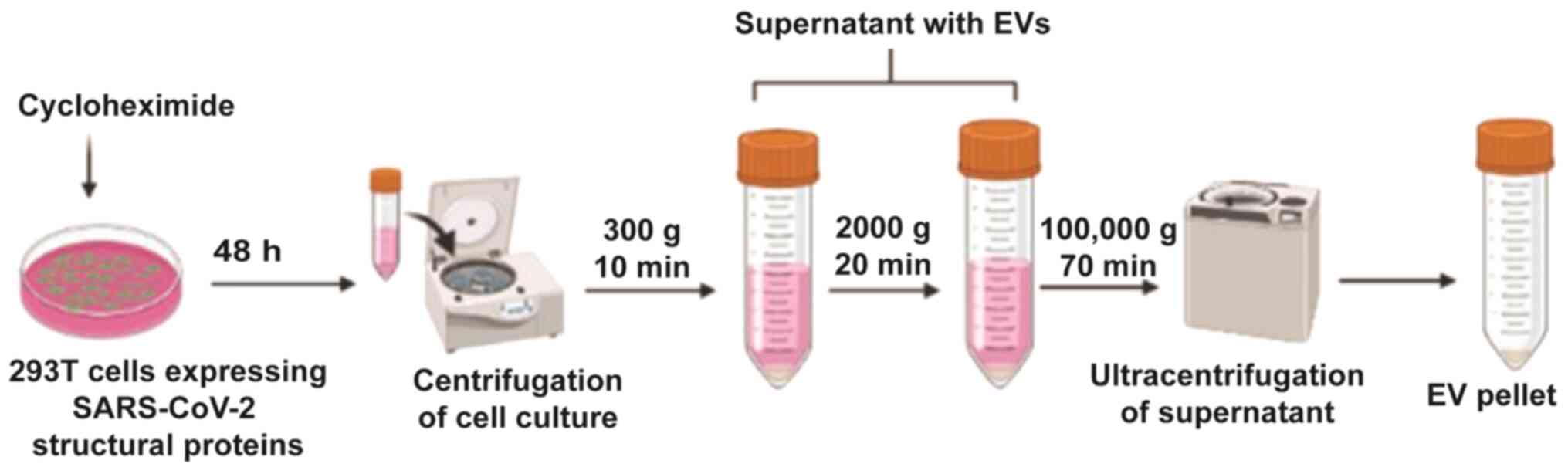
Cyclohexamide is added prior to centrifugation; this promotes
exosome secretion after 48 h (Fig.
4). In addition, due to the nature of exosomes, they are easily
absorbed into human cells without a special delivery medium
(64). As a result, this vaccine
is non-adjuvanted, virus-free and comprises all four structural
proteins (with the N protein being minimal); thus, the vaccine aims
to induce strong immunogenicity against SARS-CoV-2 mutant
variants.
Unlike other companies, such as Capricor
Therapeutics and Codiak Biosciences that have failed to produce
exosomes on a mass commercial scale, CK-Exogene, Inc. manufactures
exosomes on a large scale. Patented technology is able to produce a
quantity of exosomes ≥1,000 times higher than that produced by
existing technology. Fig. 4
depicts the purification strategy for the mass production of highly
purified and concentrated exosomes (Korean patent application no.
10-2020-0062365). Production of apoptotic exosomes was induced
using Golgi bodies; such exosomes are more concentrated and
purified than normal exosomes. The CKV21 vaccine is awaiting
approval from the Ministry of Food and Drug Safety (Korean Food and
Drug Administration), prior to commercialization.
In exosome-based vaccine, it is important to examine
and characterize the safety of EV-producing cells that are employed
for therapeutic purposes to avoid any potential side effects.
Numerous studies have reported that 293T cells are a safe source
for EV production in therapeutics because their cargo does not
exhibit disease or tumor marker (62,63). To the best of our knowledge,
however, few studies have highlighted other aspects of 293T cells
(63,65).
Regarding the safety of use of 293T cells, SV40 T
antigen was found to trigger both in vitro and in
vivo transformation of human and rat cells; notably,
SV40-transformed 293T cells reportedly cause tumors when
administered to nude mice (66).
Therefore, EVs derived from 293T cells may pose a safety concern
due to the presence of SV40 T antigens (66).
Shen et al (67) revealed a significant effect of the
passage number of 293T cells on tumorigenicity in nude mice; tumor
induction was observed when the cell passage exceeded 65 within 2
weeks. By contrast, no tumor production was found after injecting
mice with 293T cells with a low passage number (<52) under
identical circumstances. These findings were validated using PCR,
isoenzyme and histological analysis in nude mice. Therefore, 293T
cells with a low passage number (<52) may be safely used in EV
production, whereas those with a high passage number (>65) may
pose a safety concern, especially when used for therapeutic
purposes such as gene therapy or vaccine production.
Conclusions
Immunologically, a vaccine that targets the
mutation-prone S protein as well as the more stable and conserved
N, M, and E proteins is required to surmount the immune escape
characteristics exhibited by SARS-CoV-2 variants (37). The use of exosome-based vaccines
displaying SARS-CoV-2 structural proteins is a novel approach to
overcome the shortcomings of existing vaccines and contain
escalating cases of COVID-19 (62). Exosome-based vaccines comprising
all four-target antigens (S, M, E and N proteins) induce strong NAb
and T cell responses, thereby conferring prolonged immunity with no
risk of reversion of vaccination-induced virulence and pre-existing
immunity. Moreover, incorporating these immunogens with an
efficient delivery vehicle, such as exosomes, which are virus-free
and exhibit lower immunogenicity and higher absorption rate than
exiting vehicles such as LNPs or adenoviruses, would fulfill the
requirements of an ideal vaccine that eliminates the need for
booster doses (63). These
advantages of exosome-based vaccines over conventional vaccines fit
the requirement of a vaccine targeting SARS-CoV-2 and emerging
SARS-CoV-2 variants.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Data sharing not applicable to this article, as no
datasets were generated or analyzed during the current study.
Authors' contributions
KHY, NT, BJK, JOL, YNJ, YJC and JK made substantial
contributions to study conception and design, data acquisition and
data analysis and interpretation and wrote and critically revised
the manuscript for important intellectual content. Data
authentication is not applicable. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The purification strategy for the mass production of
highly purified and concentrated exosomes is subject to Korean
patent application no. 10-2020-0062365, associated with CK-Exogene,
Inc. JK and NT are employees of CK-Exogene, Inc. The other authors
(KHY, BJK, JOL, YNJ and YJC) are not associated with CK-Exogene,
Inc. and declare that they have no competing interests.
References
|
1
|
Cascella M, Rajnik M, Aleem A, Dulebohn S
and Di Napoli R: Features, Evaluation, and Treatment of Coronavirus
(COVID-19). StatPearls. 2021.
|
|
2
|
Zhou P, Yang XL, Wang XG, Hu B, Zhang L,
Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al: A pneumonia outbreak
associated with a new coronavirus of probable bat origin. Nature.
579:270–273. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mousavizadeh L and Ghasemi S: Genotype and
phenotype of COVID-19: Their roles in pathogenesis. J Microbiol
Immunol Infect. 54:159–163. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wrapp D, Wang N, Corbett KS, Goldsmith JA,
Hsieh CL, Abiona O, Graham BS and McLellan JS: Cryo-EM structure of
the 2019-nCoV spike in the prefusion conformation. Science.
367:1260–1263. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Walls AC, Park YJ, Tortorici MA, Wall A,
McGuire AT and Veesler D: Structure, function, and antigenicity of
the SARS-CoV-2 spike glycoprotein. Cell. 181:281–292.e6. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jaimes JA, André NM, Chappie JS, Millet JK
and Whittaker GR: Phylogenetic analysis and structural modeling of
SARS-CoV-2 spike protein reveals an evolutionary distinct and
proteolytically sensitive activation loop. J Mol Biol.
432:3309–3325. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kang S, Yang M, Hong Z, Zhang L, Huang Z,
Chen X, He S, Zhou Z, Zhou Z, Chen Q, et al: Crystal structure of
SARS-CoV-2 nucleocapsid protein RNA binding domain reveals
potential unique drug targeting sites. Acta Pharm Sin B.
10:1228–1238. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280.e8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ghaebi M, Osali A, Valizadeh H, Roshangar
L and Ahmadi M: Vaccine development and therapeutic design for
2019-nCoV/SARS-CoV-2: Challenges and chances. J Cell Physiol.
235:9098–9109. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Théry C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extracellular vesicles 2018 (MISEV2018): A position statement of
the International Society for Extracellular Vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kowal J, Tkach M and Théry C: Biogenesis
and secretion of exosomes. Curr Opin Cell Biol. 29:116–125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Minciacchi VR, Freeman MR and Di Vizio D:
Extracellular vesicles in cancer: Exosomes, microvesicles and the
emerging role of large oncosomes. Semin Cell Dev Biol. 40:41–51.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Choi DS, Kim DK, Kim YK and Gho YS:
Proteomics, transcriptomics and lipidomics of exosomes and
ectosomes. Proteomics. 13:1554–1571. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Record M, Carayon K, Poirot M and
Silvente-Poirot S: Exosomes as new vesicular lipid transporters
involved in cell-cell communication and various pathophysiologies.
Biochim Biophys Acta. 1841:108–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Yu M and Tian W: Physiological
and pathological impact of exosomes of adipose tissue. Cell Prolif.
49:3–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yarana C and St Clair DK:
Chemotherapy-induced tissue injury: An insight into the role of
extracellular vesicles-mediated oxidative stress responses.
Antioxidants. 6:752017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng
J and Fan S: The roles of tumor-derived exosomes in non-small cell
lung cancer and their clinical implications. J Exp Clin Cancer Res.
37:2262018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu B, Zhang X and Li X: Exosomes derived
from mesenchymal stem cells. Int J Mol Sci. 15:4142–4157. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zlotogorski-Hurvitz A, Dayan D, Chaushu G,
Korvala J, Salo T, Sormunen R and Vered M: Human saliva-derived
exosomes: Comparing methods of isolation. J Histochem Cytochem.
63:181–189. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Park SJ, Kim JM, Kim J, Hur J, Park S, Kim
K, Shin HJ and Chwae YJ: Molecular mechanisms of biogenesis of
apoptotic exosome-like vesicles and their roles as
damage-associated molecular patterns. Proc Natl Acad Sci USA.
115:E11721–E11730. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weichand B, Weis N, Weigert A, Grossmann
N, Levkau B and Brüne B: Apoptotic cells enhance
sphingosine-1-phosphate receptor 1 dependent macrophage migration.
Eur J Immunol. 43:3306–3313. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lorizate M, Sachsenheimer T, Glass B,
Habermann A, Gerl MJ, Kräusslich HG and Brügger B: Comparative
lipidomics analysis of HIV-1 particles and their producer cell
membrane in different cell lines. Cell Microbiol. 15:292–304. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lorizate M and Kräusslich HG: Role of
lipids in virus replication. Cold Spring Harb Perspect Biol.
3:a0048202011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dogrammatzis C, Waisner H and Kalamvoki M:
Cloaked viruses and viral factors in cutting edge exosome-based
therapies. Front Cell Dev Biol. 8:3762020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nainu F, Abidin RS, Bahar MA, Frediansyah
A, Emran TB, Rabaan AA, Dhama K and Harapan H: SARS-CoV-2
reinfection and implications for vaccine development. Hum Vaccin
Immunother. 16:3061–3073. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao X, Wu D, Ma X, Wang J, Hou W and
Zhang W: Exosomes as drug carriers for cancer therapy and
challenges regarding exosome uptake. Biomed Pharmacother.
128:1102372020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun D, Zhuang X, Xiang X, Liu Y, Zhang S,
Liu C, Barnes S, Grizzle W, Miller D and Zhang HG: A novel
nanoparticle drug delivery system: The anti-inflammatory activity
of curcumin is enhanced when encapsulated in exosomes. Mol Ther.
18:1606–1614. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saleh AF, Lázaro-Ibáñez E, Forsgard MA,
Shatnyeva O, Osteikoetxea X, Karlsson F, Heath N, Ingelsten M, Rose
J, Harris J, et al: Extracellular vesicles induce minimal
hepatotoxicity and immunogenicity. Nanoscale. 11:6990–7001. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu X, Badawi M, Pomeroy S, Sutaria DS,
Xie Z, Baek A, Jiang J, Elgamal OA, Mo X, Perle K, et al:
Comprehensive toxicity and immunogenicity studies reveal minimal
effects in mice following sustained dosing of extracellular
vesicles derived from HEK293T cells. J Extracell Vesicles.
6:13247302017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Walker S, Busatto S, Pham A, Tian M, Suh
A, Carson K, Quintero A, Lafrence M, Malik H, Santana MX, et al:
Extracellular vesicle-based drug delivery systems for cancer
treatment. Theranostics. 9:8001–8017. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Arvin AM, Fink K, Schmid MA, Cathcart A,
Spreafico R, Havenar-Daughton C, Lanzavecchia A, Corti D and Virgin
HW: A perspective on potential antibody-dependent enhancement of
SARS-CoV-2. Nature. 584:353–363. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Greinacher A, Thiele T, Warkentin TE,
Weisser K, Kyrle PA and Eichinger S: Thrombotic thrombocytopenia
after ChAdOx1 nCov-19 vaccination. N Engl J Med. 384:2092–2101.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsai SJ, Guo C, Atai NA and Gould SJ:
Exosome-mediated mRNA delivery for SARS-CoV-2 vaccination. bioRxiv.
Nov 6–2021.(Epub ahead of print). doi:
10.1101/2020.11.06.371419.
|
|
35
|
Polak K, Greze N, Lachat M, Merle D,
Chiumento S, Bertrand-Gaday C, Trentin B and Mamoun R:
Extracellular vesicle-based vaccine platform displaying native
viral envelope proteins elicits a robust anti-SARS-CoV-2 response
in mice. bioRxiv. Oct 28–2020.(Epub ahead of print). doi:
org/10.1101/2020.10.28.357137.
|
|
36
|
Lewis ND, Sia CL, Kirwin K, Haupt S,
Mahimkar G, Zi T, Xu K, Dooley K, Jang SC, Choi B, et al: Exosome
surface display of IL12 results in tumor-retained pharmacology with
superior potency and limited systemic exposure compared with
recombinant IL12. Mol Cancer Ther. 20:523–534. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Garcia-Beltran WF, Lam EC, St Denis K,
Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory
DJ, Poznansky MC, et al: Multiple SARS-CoV-2 variants escape
neutralization by vaccine-induced humoral immunity. Cell.
184:2372–2383. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vabret N, Britton GJ, Gruber C, Hegde S,
Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, et al:
Sinai Immunology Review Project, Immunology of COVID-19: current
state of the science. Immunity. 52:910–941. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Niu L, Wittrock KN, Clabaugh GC,
Srivastava V and Cho MW: A structural landscape of neutralizing
sntibodies sgainst SARS-CoV-2 receptor binding fomain. Front
Immunol. 12:6479342021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garcia-Beltran WF, Lam EC, Astudillo MG,
Yang D, Miller TE, Feldman J, Hauser BM, Caradonna TM, Clayton KL,
Nitido AD, et al: COVID-19-neutralizing antibodies predict disease
severity and survival. Cell. 184:476–488. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Prompetchara E, Ketloy C, Tharakhet K,
Kaewpang P, Buranapraditkun S, Techawiwattanaboon T,
Sathean-Anan-Kun S, Pitakpolrat P, Watcharaplueksadee S, Phumiamorn
S, et al: DNA vaccine candidate encoding SARS-CoV-2 spike proteins
elicited potent humoral and Th1 cell-mediated immune responses in
mice. PLoS One. 16:e02480072021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Khoury DS, Cromer D, Reynaldi A, Schlub
TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA and
Davenport MP: Neutralizing antibody levels are highly predictive of
immune protection from symptomatic SARS-CoV-2 infection. Nat Med.
27:1205–1211. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Le Bert N, Tan AT, Kunasegaran K, Tham CY,
Hafezi M, Chia A, Chng MH, Lin M, Tan N, Linster M, et al:
SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS,
and uninfected controls. Nature. 584:457–462. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Grifoni A, Weiskopf D, Ramirez SI, Mateus
J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L,
Jadi RS, et al: Targets of T cell responses to SARS-CoV-2
coronavirus in humans with COVID-19 disease and unexposed
individuals. Cell. 181:1489–1501.e15. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu WJ, Zhao M, Liu K, Xu K, Wong G, Tan W
and Gao GF: T-cell immunity of SARS-CoV: Implications for vaccine
development against MERS-CoV. Antiviral Res. 137:82–92. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Grifoni A, Sidney J, Zhang Y, Scheuermann
RH, Peters B and Sette A: A sequence homology and bioinformatic
approach can predict candidate targets for immune responses to
SARS-CoV-2. Cell Host Microbe. 27:671–680.e2. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang ZY, Werner HC, Kong WP, Leung K,
Traggiai E, Lanzavecchia A and Nabel GJ: Evasion of antibody
neutralization in emerging severe acute respiratory syndrome
coronaviruses. Proc Natl Acad Sci USA. 102:797–801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ruan YJ, Wei CL, Ee AL, Vega VB, Thoreau
H, Su ST, Chia JM, Ng P, Chiu KP, Lim L, et al: Comparative
full-length genome sequence analysis of 14 SARS coronavirus
isolates and common mutations associated with putative origins of
infection. Lancet. 361:1779–1785. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Holmes KV and Enjuanes L: Virology. The
SARS coronavirus: A postgenomic era. Science. 300:1377–1378. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rota PA, Oberste MS, Monroe SS, Nix WA,
Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen
MH, et al: Characterization of a novel coronavirus associated with
severe acute respiratory syndrome. Science. 300:1394–1399. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu Y, Liu M, Zhao W, Zhang J, Zhang X,
Wang K, Gu C, Wu K, Li Y, Zheng C, et al: Isolation of virus from a
SARS patient and genome-wide analysis of genetic mutations related
to pathogenesis and epidemiology from 47 SARS-CoV isolates. Virus
Genes. 30:93–102. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cong Y, Ulasli M, Schepers H, Mauthe M,
V'kovski P, Kriegenburg F, Thiel V, de Haan CAM and Reggiori F:
Nucleocapsid protein recruitment to replication-transcription
complexes plays a crucial role in coronaviral life cycle. J Virol.
94:e01925–e19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Leung DT, Tam FC, Ma CH, Chan PK, Cheung
JL, Niu H, Tam JS and Lim PL: Antibody response of patients with
severe acute respiratory syndrome (SARS) targets the viral
nucleocapsid. J Infect Dis. 190:379–386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gao W, Tamin A, Soloff A, D'Aiuto L,
Nwanegbo E, Robbins PD, Bellini WJ, Barratt-Boyes S and Gambotto A:
Effects of a SARS-associated coronavirus vaccine in monkeys.
Lancet. 362:1895–1896. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Okada M, Takemoto Y, Okuno Y, Hashimoto S,
Yoshida S, Fukunaga Y, Tanaka T, Kita Y, Kuwayama S and Muraki Y:
The development of vaccines against SARS corona virus in mice and
SCID-PBL/hu mice. Vaccine. 23:2269–2272. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ferretti AP, Kula T, Wang Y, Nguyen DMV,
Weinheimer A, Dunlap GS, Xu Q, Nabilsi N, Perullo CR, Cristofaro
AW, et al: Unbiased screens show CD8+ T cells of
COVID-19 patients recognize shared epitopes in SARS-CoV-2 that
largely reside outside the spike protein. Immunity.
53:1095–1107.e3. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zollner A, Watschinger C, Rössler A,
Farcet MR, Penner A, Böhm V, Kiechl SJ, Stampfel G, Hintenberger R,
Tilg H, et al: B and T cell response to SARS-CoV-2 vaccination in
health care professionals with and without previous COVID-19.
EBioMedicine. 70:1035392021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fields BN, Knipe DM and Howley PM: Fields
virology 6th edition Chapter 28. 825–858. 2013.
|
|
59
|
Ng OW, Chia A, Tan AT, Jadi RS, Leong HN,
Bertoletti A and Tan YJ: Memory T cell responses targeting the SARS
coronavirus persist up to 11 years post-infection. Vaccine.
34:2008–2014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L,
Zhu Q, Zhang X, Zheng Y, Geng C, et al: Potent neutralizing
antibodies against SARS-CoV-2 identified by high-throughput
single-cell sequencing of convalescent patients' B cells. Cell.
182:73–84.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Walsh EE, Frenck RW Jr, Falsey AR, Kitchin
N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey
R, et al: Safety and immunogenicity of two RNA-based Covid-19
vaccine candidates. N Engl J Med. 383:2439–2450. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kim J, Song Y, Park CH and Cho C: Platform
technologies and human cell lines for the production of therapeutic
exosomes. Extracell Vesicles Circ Nucleic Acids. 2:3–17. 2021.
|
|
63
|
van Doremalen N, Lambe T, Spencer A,
Belij-Rammerstorfer S, Purushotham JN, Port JR, Avanzato VA,
Bushmaker T, Flaxman A, Ulaszewska M, et al: ChAdOx1 nCoV-19
vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature.
586:578–582. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Song L, Tang S, Han X, Jiang Z, Dong L,
Liu C, Liang X, Dong J, Qiu C, Wang Y, et al: KIBRA controls
exosome secretion via inhibiting the proteasomal degradation of
Rab27a. Nat Commun. 10:16392019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li J, Chen X, Yi J, Liu Y, Li D, Wang J,
Hou D, Jiang X, Zhang J, Wang J, et al: Identification and
characterization of 293T cell-derived exosomes by profiling the
protein, mRNA and MicroRNA components. PLoS One. 11:e01630432016.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Merten OW, Hebben M and Bovolenta C:
Production of lentiviral vectors. Mol Ther Methods Clin Dev.
3:160172016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shen C, Gu M, Song C, Miao L, Hu L, Liang
D and Zheng C: The tumorigenicity diversification in human
embryonic kidney 293 cell line cultured in vitro. Biologicals.
36:263–268. 2008. View Article : Google Scholar : PubMed/NCBI
|