Introduction
Liver fibrosis refers to the abnormal proliferation
of intrahepatic fibrous connective tissue caused by several liver
injury factors, which induce an imbalance between the generation
and degradation of the extracellular matrix in liver and may
progress to liver cirrhosis and liver failure (1–3). Early
liver fibrosis may be reversible; thus, identifying the key factors
involved in the occurrence of liver fibrosis and understanding the
molecular mechanism are crucial for its treatment (4).
Previous studies have demonstrated that long
non-coding (lnc)RNAs are involved in a wide range of biological
processes and regulate gene expression at multiple levels (5–7). Thus,
investigating the molecular mechanisms and role of lncRNAs in liver
fibrosis may provide novel therapeutic approaches for its clinical
prevention and treatment. lncRNAs are transcribed RNA molecules
with a length of >200 nucleotides that have no capacity of
protein coding (8). Previous
studies have demonstrated that lncRNAs regulate important
biological processes, including cell proliferation, survival,
apoptosis and differentiation (8–10).
Numerous lncRNAs are associated with liver fibrosis (11–13).
For example, lnc-MALAT1 can reverse liver fibrosis and the
activation of hepatic stellate cells (HSCs) (14). A previous study revealed that the
upregulation of lncRNA ENSMUST00000158992 [lncRNA-MBI-52
(lnc-MBI-52)] could promote the progression of liver fibrosis in a
mouse model (15). However, the
roles of lncRNA ENSMUST00000158992 in human liver fibrosis have not
yet been investigated.
MicroRNAs (miRNAs/miRs) are a family of small RNAs
that play crucial roles in the occurrence of and protection from
liver fibrosis (16). For example,
Lan et al (17) demonstrated
that the process of liver fibrosis is inhibited by upregulating
miR-19b-3p and downregulating C-C chemokine receptor type 2
expression. Similarly, miR-181-5p can activate autophagy by
modifying the exosomes of adipose-derived mesenchymal stem cells,
thereby preventing liver fibrosis (18). In addition, miR-34a-5p inhibits
liver fibrosis by downregulating SMAD4 (19). However, the molecular mechanism
underlying the role of miR-466g in liver fibrosis remains
unclear.
The present study was undertaken to investigate the
roles of lnc-MBI-52 in liver fibrosis using both in vivo and
in vitro assays. The expression levels of mRNAs and miRNAs
were detected using reverse transcription-quantitative PCR
(RT-qPCR). The expression levels of proteins were determined using
western blotting. The interaction between miR-466g and
lnc-MBI-52/SMAD4 was verified using dual luciferase reporter and
RNA pull-down assays. Cellular functions were detected using a Cell
Counting Kit-8 (CCK-8) assay.
Materials and methods
Cell culture and transfection
The human HSC line, LX-2, was purchased from
MilliporeSigma (Merck KGaA). Cells were maintained in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin, at 37°C in 5% CO2. TGF-β1 (5
ng/ml; PeproTech, Inc.) was used to induce LX-2 cells and cultured
24 h before transfection.
lnc-MBI-52 small interfering RNA (si-lnc-MBI-52) and
scrambled si-Control [si-negative control (NC)], were synthesized
by Shanghai GenePharma Co., Ltd. HSCs (6×105 cells) were
transfected with si-lnc-MBI-52 1# (5′-GCAGAACCATAAAGATGGTCCA-3′),
si-lnc-MBI-52 2# (5′-UGGUAAUGGUGGAGGAAGAUU-3′), si-NC
(5′-UUCUCCGAACGUGUCACGUTT-3′), lnc-MBI-52 overexpression
(lnc-MBI-52) plasmids (lnc-MBI-52), pcDNA3.1 (vector)
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′), miR-466g mimic
(5′-UAUGUGUGUGUACAUGUACAUA-3′), mimic NC
(5′-UUCUCCGAACGUGUCACGUTT-3′), miR-466g inhibitor
(5′-GTGTTGCGTGTATGTGTA-3′) or miRNA inhibitor NC
(5′-GTGTAACACGTCTATACGCCCA-3′ (Shanghai GenePharma Co., Ltd.) at a
final concentration of 50 nM using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48
h. After 48 h, cells were used in subsequent experiments.
CCl4 liver injury
model
The present study was approved by the Animal Care
and Use Committee of Tangdu Hospital of Air Force Medical
University [approval no. TDYY(2019)033; Xi'an, China]. All
experiments were performed in accordance with the approved National
Institutes of Health Guidelines for the Care and Use of Laboratory
Animals (20). C57BL/6 J male mice
(n=12; 6 weeks old,18–22 g) were obtained from the Institute of
Laboratory Animal Sciences (Chinese Academy of Medical Sciences;
Peking Union Medical College). The mice were maintained under the
following conditions:40–60% humidity at 18–23°C, 12-h light-dark
cycle (light on from 8:00 am to 8:00 pm) and with free access to
food and water. Following acclimation for 1 week, a liver fibrosis
mouse model was constructed via intraperitoneal injections of
CCl4 (7 µl/g body weight; Sigma-Aldrich; Merck KGaA)
every week for 7 weeks, which was dissolved in corn oil. After 21
days, all mice were intraperitoneally euthanized with 3% sodium
pentobarbital (160 mg/kg; Sigma-Aldrich; Merck KGaA) using a
standard acceptable euthanasia method. Liver specimens and serum
samples were obtained for analyses.
Histological analysis
Liver tissues were fixed with 10% buffered formalin
at room temperature for 24 h and embedded in paraffin. Then, the
sections (5-µm) were stained with Sirius Red at room temperature
for 2 h (Sigma-Aldrich; Merck KGaA) and visualized under a light
microscope (magnification, ×200).
RT-qPCR
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse-transcribed into cDNA using a
PrimeScript™ RT-qPCR kit (Takara Bio, Inc.) according to the
manufacturer's protocols at 37°C for 75 min. qPCR was subsequently
performed using SYBR Green mixture (Takara Bio, Inc.). The
thermocycling conditions were as follows: 95°C for 30 sec, followed
by 40 cycles of 95°C for 15 sec and 60°C for 35 sec. Relative
expression was calculated using the 2−ΔΔCq method
(21). Relative expression levels
were normalized to the internal reference genes U6 or GAPDH. The
primer sequences used were as follows: lnc-MBI-52 forward (F),
5′-GTCCAGGGACCTCTGACCTA-3′ and reverse (R),
5′-CTGGAGAATCACCCCGACTG-3′; miR-466g F,
5′-CACTAGTGGTTCCGTTTAGTAG-3′ and R, 5′-TTGTAGTCACTAGGGCACC-3′;
α-SMA F, 5′-CACCATCGGGAATGAACGCTTC-3′ and R,
5′-CTGTCAGCAATGCCTGGGTA-3′; Col-1 F, 5′-GGTCATTCTCTTCGCAGACAG-3′
and R, 5′-CCACCGGATACTTGGTCTCCA-3′; SMAD4 F,
5′-CTCATGTGATCTATGCCCGTC-3′ and R, 5′-AGGTGATACAACTCGTTCGTAGT-3′;
U6 F, 5′-CTCGCTTCGGCAGCACA-3′ and R, 5′-AACGCTTCACGAATTTGCGT-3; and
GAPDH F, 5′-AAGGTGAAGGTCGGAGTCA-3′ and R,
5′-GGAAGATGGTGATGGGATTT-3′.
Western blotting
Transfected HSCs cells were harvested after 48 h and
lysed using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA). Protein
concentration was calculated using a BCA kit (Pierce; Thermo Fisher
Scientific, Inc.). Protein (30 µg/lane) was separated via SDS-PAGE
on a 12% gel, transferred onto PVDF membranes and subsequently
blocked with 5% non-fat milk at room temperature for 80 min. The
membranes were incubated overnight at 4°C with primary antibodies
(1:1,000) against Col-1 (cat. no. ab34710), α-SMA (cat. no.
ab5694), SMAD4 (cat. no. ab40759) and GAPDH (cat. no. ab9485 (all
purchased from Abcam). Following the primary antibody incubation,
membranes were incubated with goat anti-rabbit antibody (cat. no.
ab205718; 1:2,000; Abcam) at room temperature for 2 h. Protein
bands were visualized using BeyoECL Plus (Beyotime Institute of
Biotechnology).
CCK-8 assay
After transfection, cells were seeded into a 96-well
plate (2×103 cells/well). Then, cells were collected and
washed with PBS. After culture for 48 h, cells were incubated with
10 µl CCK solution at 37°C for 2 h. The absorbance values were
determined with a microplate reader at 450 nm.
Dual-luciferase reporter assay
The targets of lnc-MBI-52 and miR-466g were
predicted using miRDB (http://mirdb.org)
and TargetScan 7.2 (http://www.targetscan.org/vert_72). The wild-type (WT)
or mutant (MUT) sequences of the SMAD4 and lnc-MBI-52 prediction
region were generated by PCR and cloned into the pGL3 luciferase
reporter vector (Promega Corporation) located at KpnI and
BamHI sites. The pGL3 vectors containing SMAD4 and
lnc-MBI-52 WT or MUT predicted binding regions were co-transfected
with the WT or MUT 3′-untranslated region of lnc-MBI-52 (or SMAD4)
and miR-466g mimics or mimic NC into HSCs using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 48 h. After 48 h, luciferase activity
was detected via a dual-luciferase assay kit (Promega Corporation).
Firefly luciferase activity was normalized to Renilla
luciferase activity.
RNA pull-down assay
The biotin-labeled miR-466g and control probes were
synthesized by Sangon Biotech Co., Ltd. The probes were
co-incubated with streptavidin-coated microspheres (Invitrogen;
Thermo Fisher Scientific, Inc.) at 25°C for 2 h. HSCs were
collected and lysed using Pierce IP lysis buffer (Thermo Fisher
Scientific, Inc.). Cellular lysates (50 µl) were incubated with
miR-466g or control probes overnight at 4°C. The immunoprecipitate
was obtained via magnetic forces and centrifugation at 1,000 × g
for 20 min at room temperature, and then washed using the Pierce™
Magnetic RNA Pull-Down kit (Thermo Fisher Scientific, Inc.). 40 µl
streptavidin magnetic beads were isolated from the supernatant
after centrifugation (2,500 × g; 5 min; 4°C) and washed with
washing buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2 M NaCl and 0.1%
Tween-20), followed by another centrifugation step (2,500 × g; 5
min; 4°C). The beads (100 µl; Sigma-Aldrich; Merck KGaA) were
eluted and the complex was chilled on ice for 3 min before
separating the beads and purification using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The results
were detected using RT-qPCR as previously described. Proteins of
the RNA-protein complexes were eluted from the magnetic beads by
boiling (8 min at 100°C), and SMAD4 protein expression was examined
as aforementioned via western blotting using the antibody against
SMAD4 (1:5,000).
Statistical analysis
All experiments were performed in triplicate. Data
were analyzed using SPSS 22.0 software (IBM Corp.) and presented as
the mean ± SEM. One-way ANOVA followed by Tukey's post hoc test
were used to compare differences between multiple groups. P<0.05
was considered to indicate a statistically significant
difference.
Results
lnc-MBI-52 is upregulated in liver
fibrosis models in vivo and in vitro
To investigate the role of lnc-MBI-52 in liver
fibrosis, its expression levels were detected both in vivo
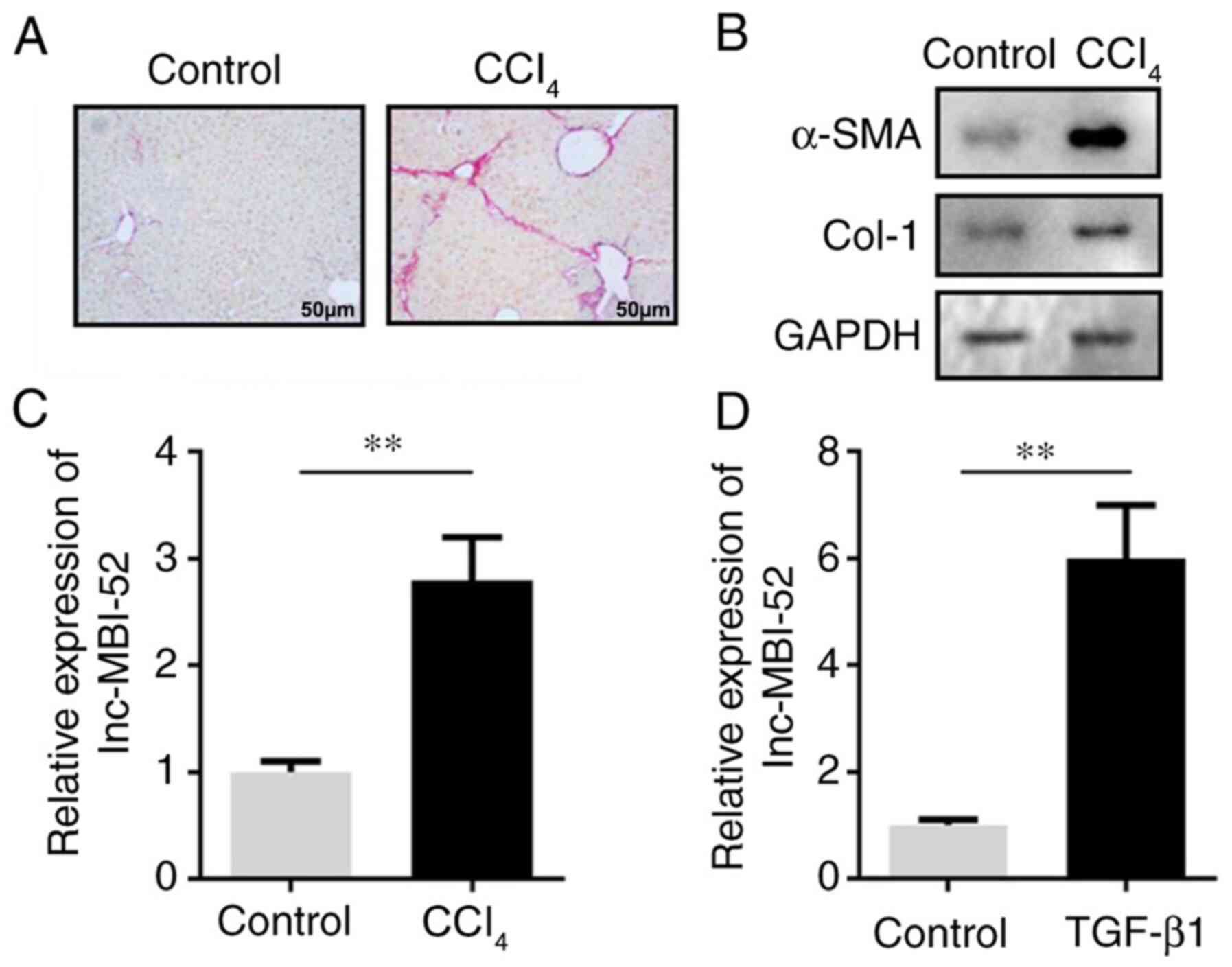
and in vitro. In the in vivo assay, CCl4
increased the Sirius Red staining area (Fig. 1A). Moreover, the protein expression
levels of α-SMA and Col-1 were increased, suggesting that the in
vivo hepatic fibrosis model was successfully established
(Fig. 1B). lnc-MBI-52 expression
was significantly upregulated in liver fibrosis mice compared with
the control group (P<0.01; Fig.
1C). This was consistent with the results of the in
vitro assay. In addition, following treatment with TGF-β1,
lnc-MBI-52 expression was significantly increased in HSCs compared
with the control group (P<0.01; Fig.
1D).
Silencing lnc-MBI-52 alleviates liver
fibrosis
To determine the role of lnc-MBI-52 in the
progression of liver fibrosis, the expression of lnc-MBI-52 was
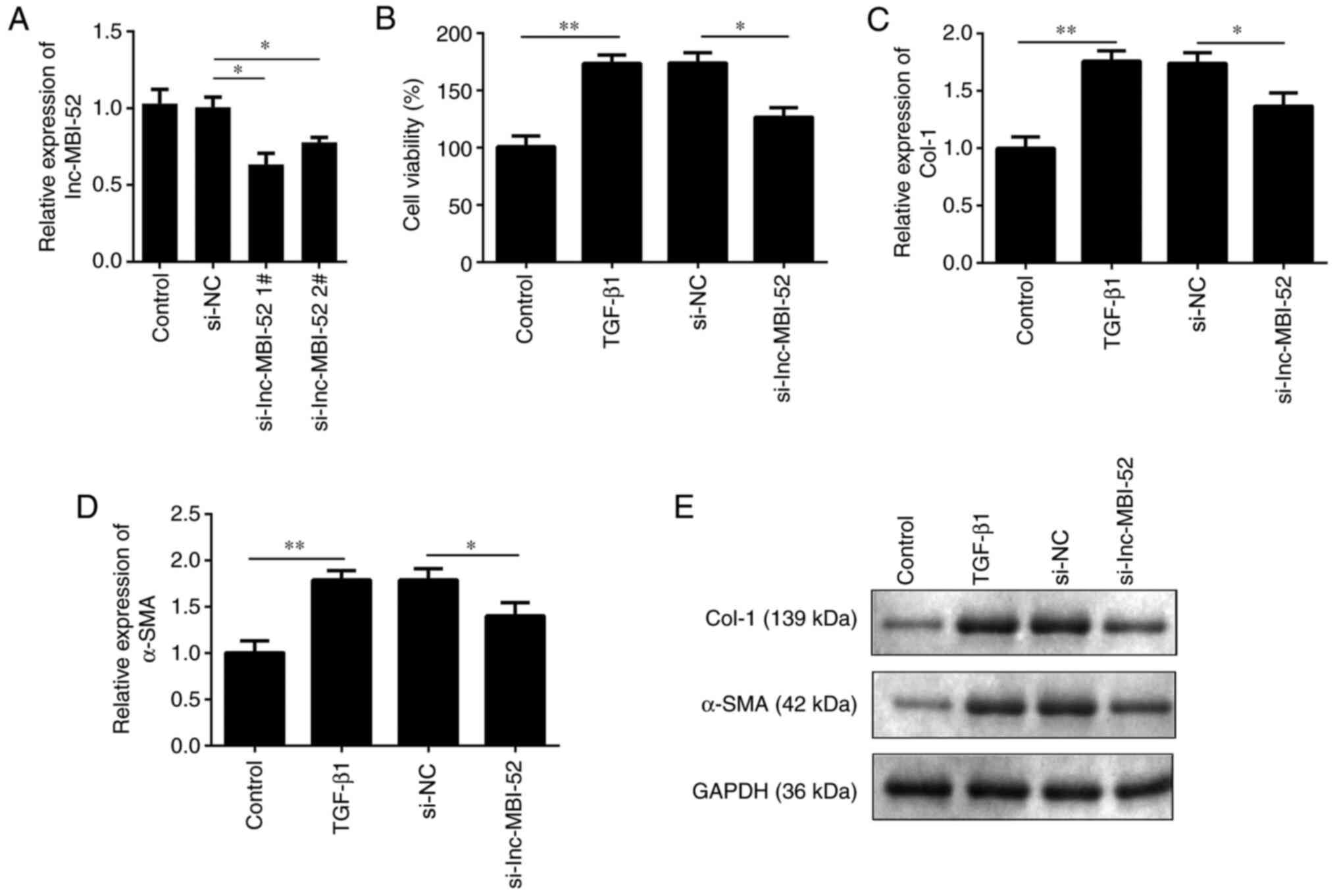
suppressed with si-lnc-MBI-52. As presented in Fig. 2A, lnc-MBI-52 expression was
significantly decreased by si-lnc-MBI-52 in cells, which was more
potent in the si-lnc-MBI-52 1# group (P<0.05). Thus,
si-lnc-MBI-52 1# was used in subsequent experiments. Cell viability
was assessed, and the results demonstrated that following treatment
with TGF-β1 viability of HSCs was increased compared with the
control group (P<0.01). Furthermore, transfection with
si-lnc-MBI-52 significantly decreased the viability of HSCs
compared with the TGF-β1 + si-NC group (P<0.05; Fig. 2B). In addition, knockdown of
lnc-MBI-52 inhibited α-SMA and Col-1 expression induced by the
si-NC group (P<0.05; Fig. 2C-E).
Taken together, these results suggested that lnc-MBI-52 knockdown
could inhibit the progression of liver fibrosis.
Interaction between lnc-MBI-52 and
miR-466g
lncRNAs modulate the expression of miRNAs by binding
to their 3′-untranslated regions (22). Thus, the present study investigated
the inhibitory effect of lnc-MBI-52 on miRNAs. miR-466g was
predicted as a potential target of lnc-MBI-52 using miRDB (Fig. 3A). Furthermore, the results of the
dual-luciferase reporter assay demonstrated that miR-466g mimics
decreased the luciferase activity of pmirGLO-lnc-MBI-52-WT without
affecting pmirGLO-lnc-MBI-52-MUT (P<0.01; Fig. 3B). To verify these results, miR-466g
expression was assessed in HSCs transfected with si-lnc-MBI-52.
lnc-MBI-52 expression was significantly upregulated by lnc-MBI-52
overexpression plasmids compared with the NC OE group (P<0.01;
Fig. 3C). The expression of
miR-466g was significantly increased by transfection with the
miR-466g mimic compared with the mimic NC group, and downregulated
following transfection with the miR-466g inhibitor compared with
the inhibitor NC group (P<0.01; Fig.
3D). The results demonstrated that lnc-MBI-52 overexpression
vector significantly decreased miR-466g expression compared with
the NC OE group (P<0.05; Fig.
3E). Conversely, miR-466g expression increased following
transfection with si-lnc-MBI-52 compared with the si-NC group
(P<0.05; Fig. 3E). The results
of the RNA pull-down assay verified the interaction between
lnc-MBI-52 and miR-466g (P<0.05; Fig. 3F). miR-466g expression was assessed
in a mouse model of CCl4-induced liver fibrosis and in
HSCs treated with TGF-β1. The results demonstrated that miR-466g
expression was significantly decreased in liver fibrosis models
in vivo and in vitro (P<0.01; Fig. 3G and H).
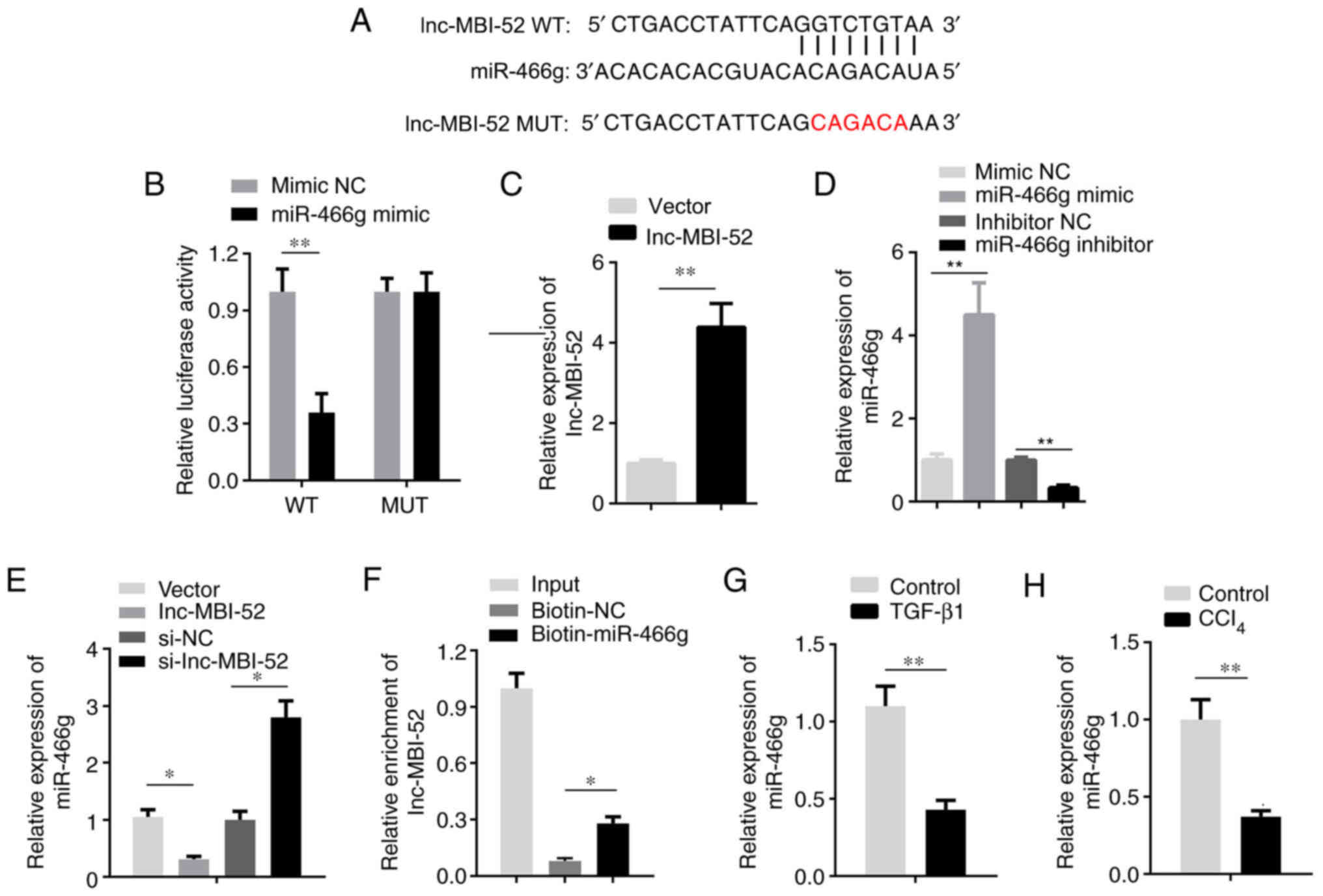 | Figure 3.lnc-MBI-52 sponges miR-466g. (A) The
binding sites between lnc-MBI-52 and miR-466g. (B) The interaction
between lnc-MBI-52 and miR-466g was verified by dual-luciferase
reporter assay. (C) The expression of lnc-MBI-52 was detected via
RT-qPCR. (D) The transfection efficiency of miR-466g mimics and
inhibitors detected via RT-qPCR. (E) lnc-MBI-52 negatively mediates
miR-466g expression. (F) The interaction between lnc-MBI-52 and
miR-466g was determined by RNA pull-down assay. (G) The expression
of miR-466g was downregulated in LX-2 cells treated with TGF-β1.
(H) The expression of miR-466g was downregulated in the
CCl4-induced hepatic fibrosis group compared with the
control group. *P<0.05, **P<0.01. lnc, long non-coding RNA;
si-, small interfering RNA; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control;
CCl4, carbon tetrachloride; WT, wild-type; MUT, mutant;
OE, overexpression. |
Knockdown of lnc-MBI-52 inhibits liver
fibrosis by interacting with miR-466g in vitro
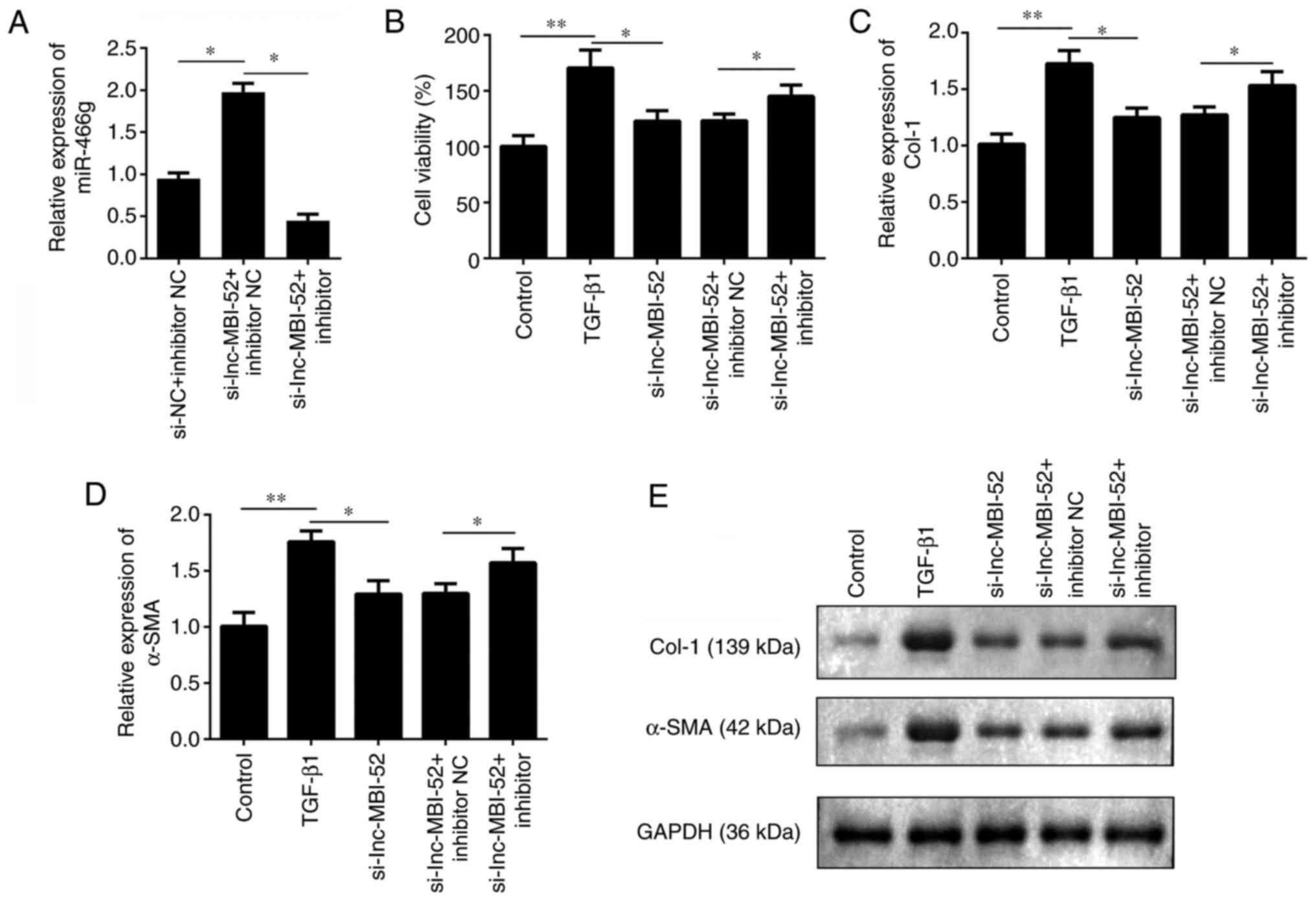
A rescue experiment was performed to investigate the
effect of miR-466g on liver fibrosis. As presented in Fig. 4A, cells were divided into three
groups: si-NC + inhibitor NC, si-lnc-MBI-52 + inhibitor NC and
si-lnc-MBI-52 + miR-466g inhibitor. si-lnc-MBI-52 significantly
increased miR-466g expression compared with the si-NC + inhibitor
NC group, which was alleviated by transfection with the miR-466g
inhibitor (P<0.05). The viability of HSCs was subsequently
assessed. After exposure to TGF-β1, cells were transfected with
si-lnc-MBI-52 and/or miR-466g inhibitor or inhibitor NC. Compared
with the TGF-β1 group, adding si-lnc-MBI-52 significantly decreased
the viability of HSCs compared with TGF-β1 group (P<0.05);
however, cell viability was partially restored following addition
of miR-466g inhibitor (P<0.05; Fig.
4B). Furthermore, knockdown of miR-466g promoted the expression
levels of α-SMA and Col-1 compared with the si-lnc-MBI-52 +
inhibitor NC group (P<0.05; Fig.
4C-E). Collectively, these results suggested that the
progression of liver fibrosis may be promoted by miR-466g
knockdown.
miR-466g activates HSCs via the SMAD4
pathway
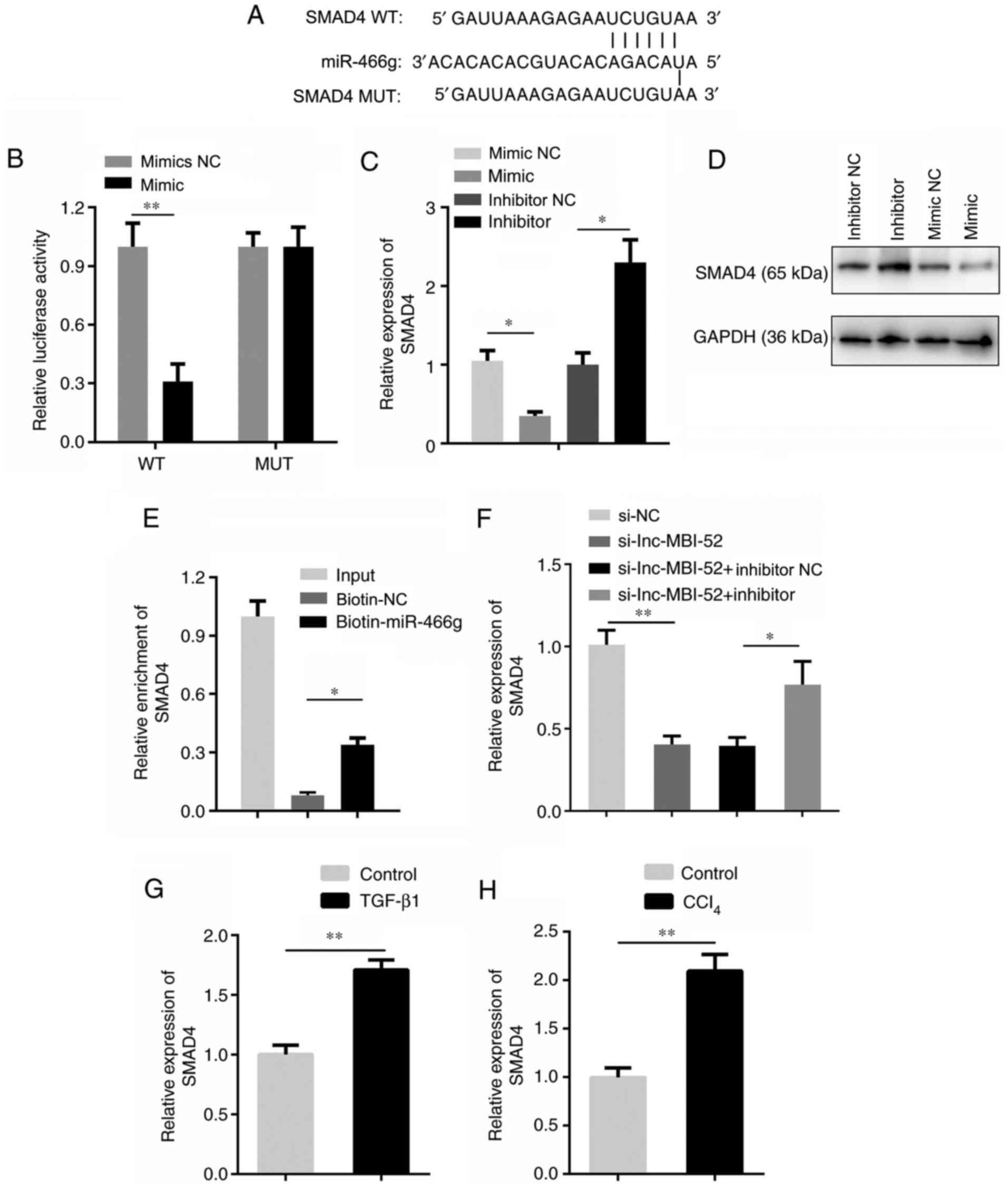
The TargetScan database was used to predict whether
SMAD4 is a target of miR-466g. The target binding region between
miR-466g and SMAD4 presented in Fig.
5A. In addition, the dual-luciferase reporter assay
demonstrated that miR-466g mimics decreased the luciferase activity
of pmirGLO-SMAD4-WT, without affecting pmirGLO-SMAD4-MUT
(P<0.01; Fig. 5B). To verify
these results, SMAD4 expression was assessed in HSCs transfected
with miR-466g mimic or miR-466g inhibitor. The results demonstrated
that transfection with the miR-466g mimic significantly decreased
SMAD4 expression compared with the mimic NC group (P<0.05;
Fig. 5C). Conversely, SMAD4
expression increased following transfection with the miR-466g
inhibitor compared with the inhibitor NC group (P<0.05; Fig. 5C). In addition, SMAD4 protein
expression increased following transfection with the miR-466g
inhibitor, and decreased following transfection with the miR-466g
mimic, compared with the corresponding NC groups (Fig. 5D). The RNA pull-down assay verified
the interaction between SMAD4 and miR-466g (P<0.05; Fig. 5E). The expression of SMAD4 was
significantly decreased by lnc-MBI-52 knockdown compared with the
si-NC group (P<0.01), which was partially restored by
transfection with the miR-466g inhibitor (P<0.05; Fig. 5F). SMAD4 expression was assessed in
mice with CCl4-induced liver fibrosis and HSCs treated
with TGF-β1. The results demonstrated that SMAD4 expression was
significantly upregulated in liver fibrosis both in vivo and
in vitro (P<0.01; Fig. 5G and
H).
Discussion
Liver fibrosis increases the incidence of cirrhosis
(23). Inhibiting the proliferation
of HSCs and promoting collagen degradation has been reported as an
effective means for treating liver fibrosis (24). Thus, understanding the molecular
mechanism underlying HSC activation may provide novel insights into
the development of more effective strategies to decrease the
incidence of liver fibrosis. lnc-MBI-52 is a newly discovered
lncRNA. The results of the present study demonstrated that
lnc-MBI-52 expression increased during the process of liver
fibrosis, both in vivo and in vitro. In addition,
following lnc-MBI-52 knockdown, the expression levels of α-SMA and
Col-1 were significantly decreased. Moreover, lnc-MBI-52 was
observed to promote liver fibrosis via the miR-466g/SAMD4 axis. To
the best of our knowledge, the present study was the first to
demonstrate that lnc-MBI-52 plays a role in promoting liver
fibrosis.
lncRNAs play crucial roles in the development of
liver fibrosis, whereby alterations in their expression levels and
mutations may promote or inhibit the occurrence of liver fibrosis
(25–27). It was previously demonstrated that
lnc-LFAR1 directly binds to SMAD2/3 and promotes TGF-β and Notch
pathway activation, which in turn activates HSCs and promotes liver
fibrosis (14). In addition,
lnc-H19 can promote the progression of cholestatic liver fibrosis,
mainly by promoting HSC differentiation and activation (28). Shen et al (29) reported that the MAPK signaling
pathway could be inhibited by silencing lncRNA HULC to reverse
liver fibrosis in non-alcoholic fatty liver disease in vivo
and decrease hepatocyte apoptosis. The results of the present study
demonstrated that lnc-MBI-52 expression was increased in liver
fibrosis models, both in vivo and in vitro. However,
lnc-MBI-52 knockdown inhibited the activation of HSCs and promoted
α-SMA and Col-1 degradation, which may affect the occurrence and
development of liver fibrosis (30). Thus, knockdown of lnc-MBI-52 may
protect against liver fibrosis. However, the underlying molecular
mechanisms remain unclear.
A number of studies have confirmed that lncRNAs act
as molecular sponges to regulate miRNA expression and biological
functions (31,32). miRNAs play key roles in the
occurrence and progression of liver fibrosis (33–35). A
previous study reported that miR-122 plays an inhibitory role in
liver fibrosis by inhibiting the activation of HSCs and the
expression of fibrosis-related genes (36). Another study demonstrated that
exosomes derived from adipose mesenchymal stem cells modified with
miR-181-5p prevent liver fibrosis via autophagy activation
(18). miRNAs are highly conserved
RNAs that exhibit a high degree of homology between different
species. Jia et al (37)
demonstrated that miR-466 inhibits the aggressive behavior of
hepatocellular carcinoma by directly targeting metadherin. However,
the results of the present study demonstrated that knockdown of
miR-466g reversed the activation of HSCs and the downregulation of
α-SMA and Col-1 expression induced by lnc-MBI-52 knockdown. Thus,
lnc-MBI-52 may participate in the development of liver fibrosis by
inhibiting miR-466g expression.
Accumulating evidence suggests that miRNAs interact
with their targets to participate in the development of liver
fibrosis (33,38). In the present study, SMAD4 was
predicted and proven to be a target of miR-466g. SMAD4 regulates a
considerable number of fundamental cellular processes, such as
protein synthesis and cell proliferation (39). For example, SMAD4 interacts with
SMAD2/3 in cells and participates in mediating the effects of the
TGF-β signaling pathway (40). It
has been demonstrated that overexpression of miR-34 in HSCs can
reverse the development of liver fibrosis by targeting SMAD4
(19). A previous study
demonstrated that, following SMAD4 gene knockout, liver fibrosis in
mice significantly decreased, suggesting that SMAD4 knockdown can
delay the progression of liver fibrosis (41). SMAD4 plays key roles in fibrotic
diseases; thus, inhibition of SMAD4 may decrease fibrosis by
decreasing the activity of the SMAD3 responsive promoter. The
results of the present study demonstrated that downregulation of
SMAD4 inhibited the activation of HSCs and the expression levels of
α-SMA and Col-1, and that miR-466g may participate in the
development of liver fibrosis by inhibiting SMAD4 expression.
However, there are limitations in this study. This
study mainly focused on the competing endogenous RNA potential of
lnc-MBI-52. However, lncRNAs may interact with RNA binding proteins
to regulate gene expression and biological processes, which
requires further study. Further studies will use clinical samples
to verify the potentials of lnc-MBI-52 in liver fibrosis.
In conclusion, the results presented herein
indicated the role of the lnc-MBI-52/miR-466g/SMAD4 signaling
cascade in liver fibrosis and further highlighted the promoting
effect of lnc-MBI-52 in this disease.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL drafted the manuscript and worked with PL to
perform the experiments, collect the data and interpret the data.
FW conceived and designed the study and revised the manuscript. PL
and FW confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Use Committee of Tangdu Hospital of Air Force Medical
University [approval no. TDYY(2019)033; Xi'an, China]. All
experiments were performed in accordance with the approved National
Institutes of Health Guidelines for the Care and Use of Laboratory
Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Aydin MM and Akcali KC: Liver fibrosis.
Turk J Gastroenterol. 29:14–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parola M and Pinzani M: Liver fibrosis:
Pathophysiology, pathogenetic targets and clinical issues. Mol
Aspects Med. 65:37–55. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun M and Kisseleva T: Reversibility of
liver fibrosis. Clin Res Hepatol Gastroenterol. 39 (Suppl
1):S60–S63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robinson EK, Covarrubias S and Carpenter
S: The how and why of lncRNA function: An innate immune
perspective. Biochim Biophys Acta Gene Regul Mech. 1863:1944192020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jathar S, Kumar V, Srivastava J and
Tripathi V: Technological developments in lncRNA biology. Adv Exp
Med Biol. 1008:283–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu J, Fu H, Wu Y and Zheng X: Function of
lncRNAs and approaches to lncRNA-protein interactions. Sci China
Life Sci. 56:876–885. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murillo-Maldonado JM and Riesgo-Escovar
JR: The various and shared roles of lncRNAs during development. Dev
Dyn. 248:1059–1069. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bian EB, Xiong ZG and Li J: New advances
of lncRNAs in liver fibrosis, with specific focus on lncRNA-miRNA
interactions. J Cell Physiol. 234:2194–2203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He Z, Yang D, Fan X, Zhang M, Li Y, Gu X
and Yang M: The roles and mechanisms of lncRNAs in liver fibrosis.
Int J Mol Sci. 21:14822020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hanson A, Wilhelmsen D and DiStefano JK:
The role of long non-coding RNAs (lncRNAs) in the development and
progression of fibrosis associated with nonalcoholic fatty liver
disease (NAFLD). Noncoding RNA. 4:182018.PubMed/NCBI
|
|
14
|
Zhang K, Han X, Zhang Z, Zheng L, Hu Z,
Yao Q, Cui H, Shu G, Si M, Li C, et al: The liver-enriched
lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch
pathways. Nat Commun. 8:1442017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang K, Han Y, Hu Z, Zhang Z, Shao S, Yao
Q, Zheng L, Wang J, Han X, Zhang Y, et al: SCARNA10, a
nuclear-retained long non-coding RNA, promotes liver fibrosis and
serves as a potential biomarker. Theranostics. 9:3622–3638. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Z, Lin CY and Cheng K: siRNA- and
miRNA-based therapeutics for liver fibrosis. Transl Res. 214:17–29.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lan T, Li C, Yang G, Sun Y, Zhuang L, Ou
Y, Li H, Wang G, Kisseleva T, Brenner D and Guo J: Sphingosine
kinase 1 promotes liver fibrosis by preventing miR-19b-3p-mediated
inhibition of CCR2. Hepatology. 68:1070–1086. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu Y, Zhang Q, Cai X, Li F, Ma Z, Xu M and
Lu L: Exosomes derived from miR-181-5p-modified adipose-derived
mesenchymal stem cells prevent liver fibrosis via autophagy
activation. J Cell Mol Med. 21:2491–2502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feili X, Wu S, Ye W, Tu J and Lou L:
MicroRNA-34a-5p inhibits liver fibrosis by regulating TGF-β1/Smad3
pathway in hepatic stellate cells. Cell Biol Int. 42:1370–1376.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Petersen BW, Harms TJ, Reynolds MG and
Harrison LH: Use of vaccinia virus smallpox vaccine in laboratory
and health care personnel at risk for occupational exposure to
orthopoxviruses-recommendations of the advisory committee on
immunization practices (ACIP), 2015. MMWR Morb Mortal Wkly Rep.
65:257–262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khatun M, Sur S, Steele R, Ray R and Ray
RB: Inhibition of long noncoding RNA linc-pint by hepatitis C virus
in infected hepatocytes enhances lipogenesis. Hepatology. 74:41–54.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Campana L and Iredale JP: Regression of
liver fibrosis. Semin Liver Dis. 37:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang CY, Yuan WG, He P, Lei JH and Wang
CX: Liver fibrosis and hepatic stellate cells: Etiology,
pathological hallmarks and therapeutic targets. World J
Gastroenterol. 22:10512–10522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao Y, Liu R, Li X, Gurley EC, Hylemon
PB, Lu Y, Zhou H and Cai W: Long noncoding RNA H19 contributes to
cholangiocyte proliferation and cholestatic liver fibrosis in
biliary atresia. Hepatology. 70:1658–1673. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peng H, Wan LY, Liang JJ, Zhang YQ, Ai WB
and Wu JF: The roles of lncRNA in hepatic fibrosis. Cell Biosci.
8:632018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen MJ, Wang XG, Sun ZX and Liu XC:
Diagnostic value of LncRNA-MEG3 as a serum biomarker in patients
with hepatitis B complicated with liver fibrosis. Eur Rev Med
Pharmacol Sci. 23:4360–4367. 2019.PubMed/NCBI
|
|
28
|
Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang
X, Gurley EC, Liang G, Chen W, Lai G, et al: Cholangiocyte-derived
exosomal long noncoding RNA H19 promotes hepatic stellate cell
activation and cholestatic liver fibrosis. Hepatology.
70:1317–1335. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shen X, Guo H, Xu J and Wang J: Inhibition
of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis
by inhibiting the MAPK signaling pathway in rats with nonalcoholic
fatty liver disease. J Cell Physiol. 234:18169–18179. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng J, Wang C, Liu T, Li J, Wu L, Yu Q,
Li S, Zhou Y, Zhang J, Chen J, et al: Procyanidin B2 inhibits the
activation of hepatic stellate cells and angiogenesis via the
Hedgehog pathway during liver fibrosis. J Cell Mol Med.
23:6479–6493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang Y: The novel regulatory role of
lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med.
22:5768–5775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen L, Zhou Y and Li H: LncRNA, miRNA and
lncRNA-miRNA interaction in viral infection. Virus Res. 257:25–32.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsay HC, Yuan Q, Balakrishnan A, Kaiser M,
Mobus S, Kozdrowska E, Farid M, Tegtmeyer PK, Borst K, Vondran FWR,
et al: Hepatocyte-specific suppression of microRNA-221-3p mitigates
liver fibrosis. J Hepatol. 70:722–734. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Caviglia JM, Yan J, Jang MK, Gwak GY, Affo
S, Yu L, Olinga P, Friedman RA, Chen X and Schwabe RF: MicroRNA-21
and Dicer are dispensable for hepatic stellate cell activation and
the development of liver fibrosis. Hepatology. 67:2414–2429. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Calvente CJ, Tameda M, Johnson CD, Del
Pilar H, Lin YC, Adronikou N, De Mollerat Du Jeu X, Llorente C,
Boyer J and Feldstein AE: Neutrophils contribute to spontaneous
resolution of liver inflammation and fibrosis via microRNA-223. J
Clin Invest. 129:4091–4109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zeng C, Wang YL, Xie C, Sang Y, Li TJ,
Zhang M, Wang R, Zhang Q, Zheng L and Zhuang SM: Identification of
a novel TGF-β-miR-122-fibronectin 1/serum response factor signaling
cascade and its implication in hepatic fibrogenesis. Oncotarget.
6:12224–12233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jia C, Tang D, Sun C, Yao L, Li F, Hu Y,
Zhang X and Wu D: MicroRNA466 inhibits the aggressive behaviors of
hepatocellular carcinoma by directly targeting metadherin. Oncol
Rep. 40:3890–3898. 2018.PubMed/NCBI
|
|
38
|
Abdel-Al A, El-Ahwany E, Zoheiry M, Hassan
M, Ouf A, Abu-Taleb H, Abdel Rahim A, El-Talkawy MD and Zada S:
miRNA-221 and miRNA-222 are promising biomarkers for progression of
liver fibrosis in HCV Egyptian patients. Virus Res. 253:135–139.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McCarthy AJ and Chetty R: Smad4/DPC4. J
Clin Pathol. 71:661–664. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao M, Mishra L and Deng CX: The role of
TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 14:111–123. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu XB, He ZP, Leng XS, Liang ZQ, Peng JR,
Zhang HY, Zhang HY, Xiao M, Zhang H, Liu CL and Zhang XD: Effects
of Smad4 on liver fibrosis and hepatocarcinogenesis in mice treated
with CCl4/ethanol. Zhonghua Gan Zang Bing Za Zhi. 18:119–123.
2010.(In Chinese). PubMed/NCBI
|