Introduction
Flavin-containing monooxygenases (FMOs) form a
family of microsomal antioxidant defense enzymes responsible for
nicotinamide adenine dinucleotide phosphate-dependent oxygenation
of soft nucleophiles (1,2). Five functional isoforms of FMO have
been identified in humans (FMO1-5) (2). FMO3, primarily located in the
liver, is the second most common FMO that metabolizes various
nitrogen- and sulfur-containing drugs and exhibits a broad range of
substrates (3–5). The FMO3 gene is clustered on
chromosome 1 (q24.3) and contains nine exons ranging from 80 to 705
bp (2). Several genetic
polymorphisms have been identified in this region (2). Moreover, previous studies have
reported genetic polymorphisms of FMO3 that affect the
enzyme activity and plasma concentrations of certain medications,
and diseases such as trimethylaminuria (6–8).
Of these polymorphisms, the c.855C>T (N285N, rs909530),
c.441C>T (S147S, rs1800822), c.923A>G (E308G, rs2266780) and
c.472G>A (E158K, rs2266782) mutations are commonly detected
single nucleotide polymorphisms (SNPs) in East Asian populations
(9–12). Considering their clinical
importance and prevalence, there is a need to investigate the
differences in the allelic frequencies of these polymorphisms
between various ethnic groups and develop a reliable method for
such analysis, which could be applied for optimal subject group
targeting in clinical practice (8).
In the present study, a rapid and reliable
pyrosequencing method was developed to detect SNPs of the
FMO3 gene, including two synonymous (c.855C>T and
c.441C>T) and two non-synonymous (c.923A>G and c.472G>A)
variants, all of which are clinically important and common in the
Korean population (13,14). Additionally, this study aimed to
compare the allelic frequencies of these SNPs in a Korean
population with those reported in other ethnic groups.
Materials and methods
Subjects and methods
This study was conducted in Korea University Anam
Hospital (Seoul, Korea) between April 2017 and February 2020.
Genomic DNA was extracted from the blood samples of 122 unrelated
healthy Korean subjects (age: 20–45, all male participants) who
provided written informed consent to participate in this study. The
protocol for this assay was approved by the institutional review
board of Anam Hospital, Korea University Medical Center (IRB
approval no. 2017AN0117, Seoul, South Korea).
Polymerase chain reaction (PCR)
conditions and FMO3 genotyping using pyrosequencing
Genomic DNA was extracted from peripheral blood
leukocytes as previously described (15). GeneAll® Exgene Blood SV
kit (GeneAll) was used according to the manufacturer's
instructions. DNA quantification was processed by using
Biospec-Nano (Shimadzu, Kyoto, Japan). A pyrosequencing method was
developed to detect the functional SNPs of the FMO3 gene:
c.855C>T, c.441C>T, c.923A>G and c.472G>A. PCR primers
used for FMO3 genotyping and pyrosequencing are listed in
Table I. PCR was performed to
amplify the specific sequences and detect each SNP of FMO3
using the newly developed primer sets after tagging the 5′ end of
each forward (or reverse) primer with biotin using the PSQ Assay
Design software (version 2.0; Qiagen GmbH).
 | Table I.Oligonucleotide primers used for PCR
and pyrosequencing to detect FMO3 SNPs. |
Table I.
Oligonucleotide primers used for PCR
and pyrosequencing to detect FMO3 SNPs.
| SNP | Primer | Sequences | Size, bp | PCR Tm, °C |
|---|
| FMO3
c.855C>T (rs909530) | Forward | B
5′-TTGGGTCATTTTTTCCTTCCTTAT-3′ | 261 | 60 |
|
| Reverse |
5′-ACCCTGTTGCAAAGATTACACAGT-3′ |
|
|
|
| Sequencing |
5′-TTGCTGGGAGCTCAT-3′ |
|
|
| FMO3
c.441C>T (rs1800822) | Forward | B
5′-CCACTGAAAGGGATGGTAAAAA-3′ | 125 | 60 |
|
| Reverse |
5′-AGCAGCTTAAATTTTGGCCTTAC-3′ |
|
|
|
| Sequencing |
5′-TGGGATACACATGATGTC-3′ |
|
|
| FMO3
c.923A>G (rs2266780) | Forward |
5′-AGCATTCTGTGTGGCATTGT-3′ | 144 | 60 |
|
| Reverse | B
5′-AAGGAAGGGGTAGGCAAAACTAT-3′ |
|
|
|
| Sequencing |
5′-CGTGAAGGAATTCACAG-3′ |
|
|
| FMO3
c.472G>A (rs2266782) | Forward | B
5′-ATGGTAAAAAAGAATCGGCTGTC-3′ | 132 | 60 |
|
| Reverse |
5′-TTTTGTCAGTTATGTGGCTAGCAG-3′ |
|
|
|
| Sequencing |
5′-GCCTTACCTGGAAAGGACT-3′ |
|
|
The PCR mixture (30 µl) comprised genomic DNA (30
ng), 10X PCR buffer (Intron Biotechnology, Inc.), dNTPs (0.25 mM),
10 pmol primers (1 µl each) and 5 units Taq polymerase (Intron
Biotechnology, Inc.). PCR was performed with an initial
denaturation step at 95°C for 3 min, followed by 45 cycles of
denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, and
extension at 72°C for 30 sec. The final termination step was
performed at 72°C for 5 min. For pyrosequencing reactions, 25 µl
PCR template in a single well was immobilized by incubation (with
continuous shaking at 1,400 rpm for 10 min at room temperature)
with a mixture of 5 µl streptavidin beads (Streptavidin Sepharose™
High Performance; Cytiva) and 40 µl annealing buffer containing 0.4
µM sequencing primer incorporated into each well. For strand
separation, the liquid component was removed using a vacuum prep
workstation (Qiagen GmbH). The beads captured on the probes were
treated in 70% ethanol, and the solution was passed through a
filter for 5 sec. The beads were then treated with a denaturing
solution (0.2 M NaOH), and the solution was passed through a filter
for 5 sec. Thereafter, a wash buffer (10 mM Tris-acetate, pH 7.6)
was used to rinse the beads for 5 sec. The liquid component was
completely removed from the probes, and the beads were placed into
a PSQ 96 Plate Low (Pyrosequencing AB) containing the sequencing
primer. The prepared PSQ 96 Plate Low was heated at 85°C for 2 min,
and the reactions were allowed to cool to room temperature. The
resulting mixture was analyzed using the PSQ 96MA pyrosequencer
(Pyrosequencing AB). The accuracy of pyrosequencing was validated
by direct DNA sequencing of randomly selected samples using the
same genomic DNA. The analyzed allelic frequencies were then
compared with those of other ethnic groups and those reported in
the HapMap database (https://www.ncbi.nlm.nih.gov/snp).
Statistical analysis
Genetic equilibrium and linkage disequilibrium (LD)
were tested according to the Hardy-Weinberg equation (HWE)
(16) using SNPalyzer software
(version 9.0; DYNACOM Co., Ltd.). A chi-square test was performed
to assess the deviation of the pyrosequencing results from the HWE.
The detected genotype frequencies were then compared to the
expected frequencies. P<0.05 (two-tailed) was considered to
indicate a statistically significant difference. D′ and
r2 are standard measurements for the LD (17). D′ values were calculated as
D/Dmax, where D is the coefficient of LD ranging from
−0.25 to 0.25. In general, the standardized value of D′ is
preferred because D is often affected by allelic frequencies
(18).
Results
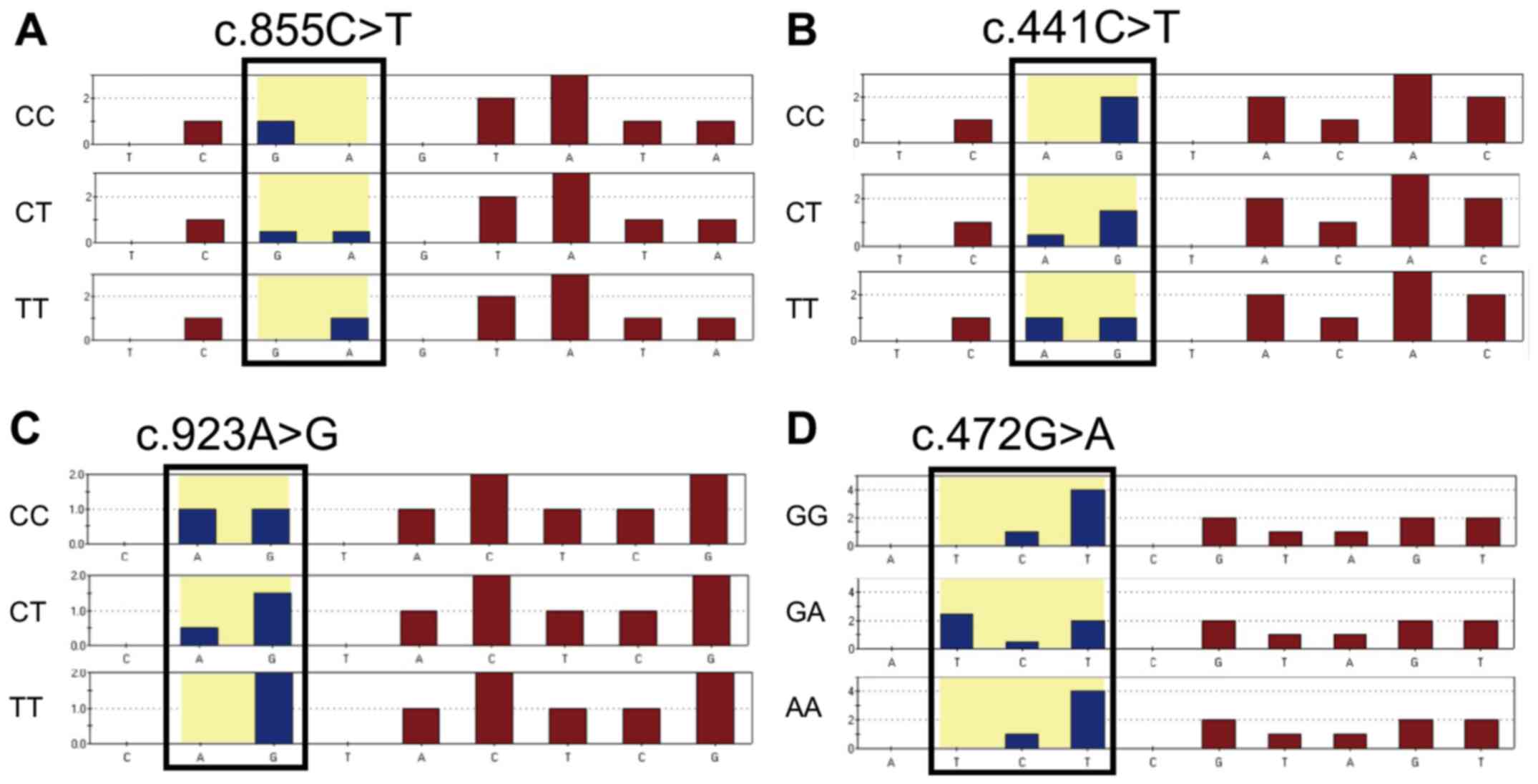
Each FMO3 SNP, including c.855C>T, c.441C>T,
c.923A>G and c.472G>A, was successfully detected, as shown in
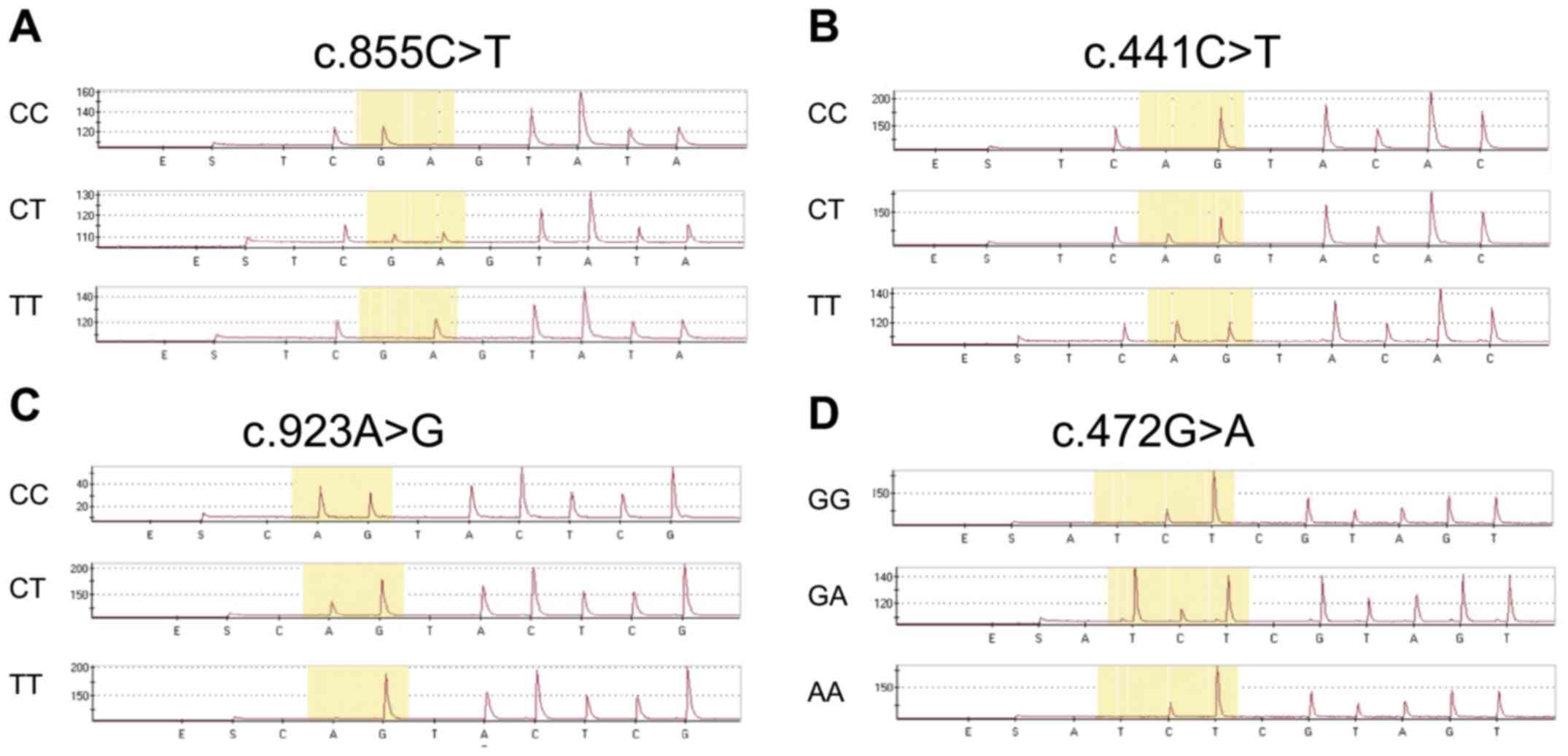
the predicted pyrosequencing histogram (Fig. 1). Representative peaks for each
SNP are shown in Fig. 2. The
sequenced data obtained using the pyrosequencing method were
randomly selected and validated by direct DNA sequencing. The
results were 100% concordant with the pyrosequencing data,
indicating 100% specificity and sensitivity (data not shown).
The allelic frequencies of FMO3 SNPs in the
Korean population obtained using our pyrosequencing method were as
follows: i) 44.7% for c.855C>T; ii) 23.4% for c.441C>T; iii)
23.0% for c.923A>G; and iv) 27.1% for c.472G>A (Table II). The allelic frequencies
obtained in these genetic analyses did not deviate from the
Hardy-Weinberg equilibrium (χ2=0.1843, 0.1201, 0.0318
and 0.4729 for c.855C>T, c.441C>T, c.923A>G and
c.472G>A, respectively; P=0.6677, 0.7290, 0.8584 and 0.4917 for
c.855C>T, c.441C>T, c.923A>G and c.472G>A,
respectively); however, the LD analysis revealed that c.923A>G
and c.472G>A exhibited strong LD (D′=0.8289,
r2=0.5332; Table
SI).
 | Table II.Genotyping and allelic frequencies of
FMO3 SNPs identified in this study. |
Table II.
Genotyping and allelic frequencies of
FMO3 SNPs identified in this study.
| A, c.855C>T |
|---|
|
|---|
| Genotype | Counts | Genotyping
frequency | Allele | Allelic
frequency | χ2 | P-value |
|---|
| G/G | 36 | 0.2951 | G | 0.5533 | 0.1843 | 0.6677 |
| G/A | 63 | 0.5164 | A | 0.4467 |
|
|
| A/A | 23 | 0.1885 |
|
|
|
|
| B,
c.441C>T |
|
|
Genotype | Counts | Genotyping
frequency | Allele | Allelic
frequency |
χ2 | P-value |
|
| G/G | 70 | 0.5738 | G | 0.7664 | 0.1201 | 0.7290 |
| G/A | 47 | 0.3852 | A | 0.2336 |
|
|
| A/A | 5 | 0.0410 |
|
|
|
|
|
| C,
c.923A>G |
|
|
Genotype | Counts | Genotyping
frequency | Allele | Allelic
frequency |
χ2 | P-value |
|
| A/A | 72 | 0.5901 | A | 0.7705 | 0.0318 | 0.8584 |
| A/G | 44 | 0.3607 | G | 0.2295 |
|
|
| G/G | 6 | 0.0492 |
|
|
|
|
|
| D,
c.472G>A |
|
|
Genotype | Counts | Genotyping
frequency | Allele | Allelic
frequency |
χ2 | P-value |
|
| C/C | 63 | 0.5164 | C | 0.7295 | 0.4729 | 0.4917 |
| C/T | 52 | 0.4262 | T | 0.2705 |
|
|
| T/T | 7 | 0.0574 |
|
|
|
|
The ethnic differences of the SNPs were described in
Table III. Although the data
were limited, particularly for the European and African
populations; however, the trend of the allelic frequencies for
FMO3 SNPs obtained in the present study was similar to that
previously reported in a Japanese population (12). In particular, the allelic
frequencies of c.923A>G and c.472G>A appeared to be similar
to those in the Chinese population (3). The SNP c.923A>G frequency
exhibited some similarity to the minor allele frequency (MAF) of
the HapMap data of Europeans (Utah residents with Northern and
Western European ancestry from the CEPH collection reported by the
National Center for Biotechnology Information SNP database;
HapMap-CEU; http://www.ncbi.nlm.nih.gov/snp), whereas the
frequencies of other SNPs exhibited remarkable differences from the
MAF of this population.
 | Table III.Comparisons between FMO3
allele frequencies obtained in this study and those in other ethnic
groups. |
Table III.
Comparisons between FMO3
allele frequencies obtained in this study and those in other ethnic
groups.
| A, c.855C>T |
|---|
|
|---|
| Population | Number, n | MAF, % | Refs. |
|---|
| Korean | 122 | 44.7 | Present study |
| Japanese | 3,552 | 38.8 | (12) |
| Chinese | 285 | 26.1 | (3) |
| European | 226 | 27.9 | HapMap-CEU
database |
| Sub-Saharan
African | 226 | 54.0 | HapMap-YRI
database |
|
| B,
c.441C>T |
|
|
Population | Number,
n | MAF, % | Refs. |
|
| Korean | 122 | 23.4 | Present study |
| Japanese | 3,552 | 19.9 | (12) |
| Chinese | 285 | 5.8 | (3) |
| European | 226 | 6.6 | HapMap-CEU
database |
| Sub-Saharan
African | 226 | 3.1 | HapMap-YRI
database |
|
| C,
c.923A>G |
|
|
Population | Number,
n | MAF, % | Refs. |
|
| Korean | 122 | 23.0 | Present study |
| Japanese | 3,552 | 19.8 | (12) |
| Chinese | 285 | 19.8 | (3) |
| European | 170 | 35.9 | (31) |
| Sub-Saharan
African | 226 | 1.3 | HapMap-YRI
database |
|
| D,
c.472G>A |
|
|
Population | Number,
n | MAF, % | Refs. |
|
| Korean | 122 | 27.1 | Present study |
| Japanese | 3,552 | 21.0 | (12) |
| Chinese | 285 | 16.5 | (3) |
| European | 224 | 42.0 | HapMap-CEU
database |
|
African-American | 133 | 41.9 | (32) |
Discussion
The results of the present study indicated that this
newly developed rapid pyrosequencing method for analyzing the
c.855C>T, c.441C>T, c.923A>G and c.472G>A SNPs of the
FMO3 gene was a reliable and accurate technique. The allelic
frequencies obtained in 122 Korean subjects using this method
revealed that these frequencies were most similar to those reported
in the Japanese population (12).
To the best of our knowledge, this was the first study to analyze
FMO3 SNPs using a pyrosequencing method.
Various methods have been proposed to analyze the
targeted SNPs. For example, FMO3-related SNPs have been
detected by using PCR-restriction fragment length polymorphism
analysis (19), real-time PCR
(20) and direct sequencing
methods (21). Sequencing
technology was first conceptualized and developed in the 1970s by
Sanger et al (22). The
principle of this method is based on the use of dideoxynucleotide
triphosphates for DNA sequence termination. The pyrosequencing
method that was designed to analyze FMO3 SNPs in the current
study was based on the solution-based pyrosequencing method
suggested by Ronaghi et al (23) in 1998. This is a simple method
that is suitable for automation as it uses apyrase, DNA polymerase
and luciferase, which eventually detect light emission through
pyrophosphate production during DNA synthesis (23). The major advantages of this method
are its simplicity, reliability, high sensitivity and specificity
compared with conventional sequencing systems (24). Therefore, it was speculated that
the method described in the present study could be suitable for
precise, rapid and cost-effective assessment of SNP frequencies in
a relatively large sample set.
SNPs are the most frequently occurring sequence
variations in the human genome and often vary among different
ethnic groups (1,2). The allelic frequencies of selected
FMO3 SNPs observed in this study were comparable to those
reported in the Japanese population (9), whereas the frequency of each
genotype in the Chinese population was generally lower than that in
the Korean or Japanese populations (3,12).
FMO3 c.855C>T was the most commonly detected SNP in the
current study; this result was consistent with that previously
reported in a smaller Korean population previously (n=41,
MAF=0.329) (13). The frequencies
of the c.855C>T and c.472G>A SNPs were higher in the African
population (HapMap-YRI database; http://www.ncbi.nlm.nih.gov/snp,32) than in the Asian
populations; however, the frequencies of c.441C>T and
c.923A>G in the African population were markedly lower (<5%)
(HapMap-YRI database; http://www.ncbi.nlm.nih.gov/snp). Therefore,
FMO3 appears to exhibit a large interethnic difference
(3,9,13).
FMO3 genetic polymorphisms have been the
focus of considerable interest in research; these findings can be
applied to various studies on the pharmacokinetics of various
medications, including anti-diabetics (e.g., teneligliptin)
(5,6), antibiotics (e.g., voriconazole)
(20,25) and non-steroidal anti-inflammatory
drugs (e.g., sulindac) (4,13,14),
as well as human diseases, such as cardiovascular disorders
(2,7). FMO3 increases plasma trimethylamine
N-oxide (TMAO) levels by catalyzing the conversion of
trimethylamine (TMA) derived from the gut microbiome (26,27). Therefore, SNPs responsible for
FMO3 loss-of-function seem to result in increased plasma TMA levels
(9). At a clinical level, TMAO is
associated with atherosclerosis (28), and a recent study demonstrated
that higher plasma TMAO levels were associated with poor
cardiovascular outcomes, while the FMO3 SNP (c.472G>A)
has been shown to reduce TMAO levels in the Asian population
(7).
FMO3 also affects the levels of several
clinically important medications, and its polymorphisms are
associated with drug toxicity (25,29,30). The c.923A>G SNP has been shown
to increase voriconazole concentrations by reducing FMO3 enzyme
activity (25), while c.855C>T
SNP can increase the concentration of teneligliptin (6). FMO3 c.441C>T and
c.855C>T have been associated with fast tacrolimus elimination
in Chinese patients (30).
Studies by Park et al (13) and Sung et al (14) demonstrated that the SNPs
c.855C>T and c.472G>A affected the pharmacokinetics of
sulindac in women who underwent preterm labor. Febrile neutropenia,
myelosuppression and agranulocytosis related to these SNPs have
also been reported previously (25,29,30).
Considering the relatively high frequency of
FMO3 genetic polymorphisms in the population, the functional
defects in FMO3 enzymes associated with these SNPs may have notable
clinical implications, such as the variations in drug exposure
followed by toxicity or delayed elimination of toxic substances.
Therefore, the development of a faster and more precise method to
identify FMO3 SNPs could be clinically beneficial when
purposed for optimal treatment (e.g., suggesting lower dosage in
the patients with FMO3 genetic polymorphism to reduce the
drug toxicity and adverse events). However, evidence should be
accumulated through clinical studies.
The ethnic and interindividual differences in SNPs
and their suspected clinical manifestations, personalized dosing,
pharmacokinetics and pharmacodynamics studies of drugs based on
FMO3 SNPs may present a novel research direction. Thus, the
pyrosequencing method developed in this study could be applied
directly to analyze individual FMO3 SNPs for research in
this domain.
In conclusion, the pyrosequencing method developed
in the present study was successfully applied to detect the SNPs
c.855C>T, c.441C>T, c.923A>G and c.472G>A of the
FMO3 gene. In Korean subjects, c.855C>T was the most
frequent among the four FMO3 SNPs.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to information that
could compromise the privacy of research participants, but are
available from the corresponding author upon reasonable
request.
Authors' contributions
JWP was responsible for data acquisition, analysis
and interpretation, and drafting of the article. JYP conceptualized
and co-designed the study, critically screened the revised article
for important intellectual content, and provided final approval of
the submitted manuscript. KAK designed the study, and performed
data analysis and interpretation. IHP, JMK and JHN were responsible
for data acquisition and analysis. JWP and JYP confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The protocol for this assay was approved by the
Institutional Review Board of Anam Hospital, Korea University
Medical Center (Seoul, South Korea). Subjects provided written
informed consent to participate in this study.
Patients consent for publication
Not applicable.
Competing interests
The author declare that they have no competing
interests.
References
|
1
|
Phillips IR and Shephard EA: Drug
metabolism by flavin-containing monooxygenases of human and mouse.
Expert Opin Drug Metab Toxicol. 13:167–181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Phillips IR and Shephard EA:
Flavin-containing monooxygenase 3 (FMO3): Genetic variants and
their consequences for drug metabolism and disease. Xenobiotica.
50:19–33. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu M, Bhatt DK, Yeung CK, Claw KG,
Chaudhry AS, Gaedigk A, Pearce RE, Broeckel U, Gaedigk R, Nickerson
DA, et al: Genetic and nongenetic factors associated with protein
abundance of flavin-containing monooxygenase 3 in human liver. J
Pharmacol Exp Ther. 363:265–274. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang YJ, Hu K, Huang WH, Wang CZ, Liu Z,
Chen Y, Ouyang DS, Tan ZR, Zhou HH and Yuan CS: Effects of FMO3
polymorphisms on pharmacokinetics of sulindac in Chinese healthy
male volunteers. BioMed Res Int. 2017:41896782017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ceriello A, De Nigris V, Iijima H, Matsui
T and Gouda M: The unique pharmacological and pharmacokinetic
profile of teneligliptin: Implications for clinical practice.
Drugs. 79:733–750. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park JW, Kim KA, Kim JM, Park IH and Park
JY: Influence of FMO3 and CYP3A4 polymorphisms on the
pharmacokinetics of teneligliptin in humans. Front Pharmacol.
12:7363172021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei H, Zhao M, Huang M, Li C, Gao J, Yu T,
Zhang Q, Shen X, Ji L, Ni L, et al: FMO3-TMAO axis modulates the
clinical outcome in chronic heart-failure patients with reduced
ejection fraction: evidence from an Asian population. Front Med.
Jun 22–2021.(Epub ahead of print). doi: 10.1007/s11684-021-0857-2.
View Article : Google Scholar
|
|
8
|
Scimone C, Alibrandi S, Donato L, Giofrè
SV, Rao G, Sidoti A and D'Angelo R: Antiretroviral treatment
leading to secondary trimethylaminuria: Genetic associations and
successful management with riboflavin. J Clin Pharm Ther.
46:304–309. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shimizu M, Allerston CK, Shephard EA,
Yamazaki H and Phillips IR: Relationships between flavin-containing
mono-oxygenase 3 (FMO3) genotype and trimethylaminuria phenotype in
a Japanese population. Br J Clin Pharmacol. 77:839–851. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimizu M, Kobayashi Y, Hayashi S, Aoki Y
and Yamazaki H: Variants in the flavin-containing monooxygenase 3
(FMO3) gene responsible for trimethylaminuria in a Japanese
population. Mol Genet Metab. 107:330–334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shimizu M, Yoda H, Igarashi N, Makino M,
Tokuyama E and Yamazaki H: Novel variants and haplotypes of human
flavin-containing monooxygenase 3 gene associated with Japanese
subjects suffering from trimethylaminuria. Xenobiotica.
49:1244–1250. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shimizu M, Yoda H, Nakakuki K, Saso A,
Saito I, Hishinuma E, Saito S, Hiratsuka M and Yamazaki H: Genetic
variants of flavin-containing monooxygenase 3 (FMO3) derived from
Japanese subjects with the trimethylaminuria phenotype and
whole-genome sequence data from a large Japanese database. Drug
Metab Pharmacokinet. 34:334–339. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park S, Lee NR, Lee KE, Park JY, Kim YJ
and Gwak HS: Effects of single-nucleotide polymorphisms of FMO3 and
FMO6 genes on pharmacokinetic characteristics of sulindac sulfide
in premature labor. Drug Metab Dispos. 42:40–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sung JW, Yun HY, Park S, Kim YJ, Yee J,
Lee KE, Song B, Chung JE and Gwak HS: Population pharmacokinetics
of sulindac and genetic polymorphisms of FMO3 and AOX1 in women
with preterm labor. Pharm Res. 37:442020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kwak HD, Kim SH, Seo YS and Song KJ:
Detecting hepatitis B virus in surgical smoke emitted during
laparoscopic surgery. Occup Environ Med. 73:857–863.
2016.PubMed/NCBI
|
|
16
|
Abramovs N, Brass A and Tassabehji M:
Hardy-weinberg equilibrium in the large scale genomic sequencing
era. Front Genet. 11:2102020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du FX, Clutter AC and Lohuis MM:
Characterizing linkage disequilibrium in pig populations. Int J
Biol Sci. 3:166–178. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sunderaraman P, Cosentino S, Schupf N,
Manly J, Gu Y and Barral S: MEF2C common genetic variation is
associated with different aspects of cognition in non-hispanic
white and caribbean hispanic non-demented older adults. Front
Genet. 12:6423272021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shimizu M, Mizugaki A, Koibuchi N, Sango
H, Uenuma Y and Yamazaki H: A series of simple detection systems
for genetic variants of flavin-containing monooxygenase 3 (FMO3)
with impaired function in Japanese subjects. Drug Metab
Pharmacokinet. 41:1004202021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chuwongwattana S, Jantararoungtong T,
Prommas S, Medhasi S, Puangpetch A and Sukasem C: Impact of
CYP2C19, CYP3A4, ABCB1, and FMO3 genotypes on plasma voriconazole
in Thai patients with invasive fungal infections. Pharmacol Res
Perspect. 8:e006652020. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
D'Angelo R, Scimone C, Esposito T,
Bruschetta D, Rinaldi C, Ruggeri A and Sidoti A: Fish odor syndrome
(trimethylaminuria) supporting the possible FMO3 down expression in
childhood: A case report. J Med Case Reports. 8:3282014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sanger F, Nicklen S and Coulson AR: DNA
sequencing with chain-terminating inhibitors. Proc Natl Acad Sci
USA. 74:5463–5467. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ronaghi M, Uhlén M and Nyrén P: A
sequencing method based on real-time pyrophosphate. Science.
281:363–365. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Siqueira JF Jr..Fouad AF and Rôças IN:
Pyrosequencing as a tool for better understanding of human
microbiomes. J Oral Microbiol. Jan 23–2012.(Epub ahead of print).
doi: 10.3402/jom.v4i0.10743. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Zhao J, Wen T, Liao X and Luo B:
Predictive value of FMO3 variants on plasma disposition and adverse
reactions of oral voriconazole in febrile neutropenia.
Pharmacology. 106:202–210. 2021.PubMed/NCBI
|
|
26
|
Fennema D, Phillips IR and Shephard EA:
Trimethylamine and trimethylamine N-Oxide, a flavin-containing
monooxygenase 3 (FMO3)-mediated host-microbiome metabolic Axis
implicated in health and disease. Drug Metab Dispos. 44:1839–1850.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Weng Z, Liu Q, Shao W, Guo W, Chen
C, Jiao L, Wang Q, Lu Q, Sun H, et al: FMO3 and its metabolite TMAO
contribute to the formation of gallstones. Biochim Biophys Acta Mol
Basis Dis. 1865:2576–2585. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Su C, Jiang Z, Yang Y, Zhang Y, Yang
M, Zhang X, Du Y, Zhang J, Wang L, et al: Berberine attenuates
choline-induced atherosclerosis by inhibiting trimethylamine and
trimethylamine-N-oxide production via manipulating the gut
microbiome. NPJ Biofilms Microbiomes. 7:362021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren W, Zhou C, Liu Y, Su K, Jia L, Chen L,
Li M, Ma J, Zhou W, Zhang S, et al: Genetic associations of
docetaxel-based chemotherapy-induced myelosuppression in Chinese
Han population. J Clin Pharm Ther. 45:354–364. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He YY, Hasan AME, Zhang Q, Li SQ, Yang JS,
Yan CX, Chen P, Liu Y, Nadeem A and Zhang B: Novel association
between flavin-containing monooxygenase 3 gene polymorphism and
antithyroid drug-induced agranulocytosis in the han population. Ann
Nutr Metab. 74:200–206. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ren L, Teng M, Zhang T, Zhang X, Sun B,
Qin S, Zhong L, Peng Z and Fan J: Donors FMO3 polymorphisms affect
tacrolimus elimination in Chinese liver transplant patients.
Pharmacogenomics. 18:265–275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chenoweth MJ, Zhu AZX, Sanderson Cox L,
Ahluwalia JS, Benowitz NL and Tyndale RF: Variation in P450
oxidoreductase (POR) A503V and flavin-containing monooxygenase
(FMO)-3 E158K is associated with minor alterations in nicotine
metabolism, but does not alter cigarette consumption. Pharmacogenet
Genomics. 24:172–176. 2014. View Article : Google Scholar : PubMed/NCBI
|