Introduction
Cells respond to the environmental stimuli through a
class of molecules on the cell surface called receptors. Receptor
tyrosine kinases (RTKs) are a superfamily of receptors that were
among the first receptors discovered and systematically studied;
more importantly, the signalling pathway was one of the first
pathways to unite various fields in developmental biology (1). Ligands of RTKs are protein molecules
classified as growth factors. This family of ligands is active
during embryonic development and in homeostasis in the adult body,
activating a series of cell signalling pathways, serving pivotal
roles in cell growth and differentiation. Research on RTK-related
diseases, such as cancer, revealed that a number of acquired
mutations in RTK receptors lead to the malfunctioning of the
receptors (2).
There are 90 unique tyrosine kinase genes that have
been identified in the human genome, of which 58 encode RTK
proteins and the remaining 32 encode non-receptor types (1–5). The
RTK family can be divided into ~20 different types of receptors
based on their ligands. Some of the well-known RTKs include
tropomyosin receptor kinase (Trk) receptor, epidermal growth factor
receptor (EGFR), platelet-derived growth factor receptor,
fibroblast growth factor receptor, vascular endothelium growth
factor receptor and insulin receptor kinase (IRK) Although RTK
ligands are diverse, their activation mechanisms are highly
conservative. After the extracellular portion of the receptor binds
to the ligand, it activates the intracellular part of the receptor,
which uses ATP as a phosphate donor to catalyse the transfer of
phosphoryl groups onto specific tyrosine residues in the
receptors.
It is now known that most RTK receptors are composed
of a polypeptide chain and can be divided into three regions: i) A
region to which the ligand binds from outside of the cell membrane;
ii) a transmembrane region that crosses the cell membrane once; and
iii) and the signal transduction region is the cytosolic side that
contains the tyrosine kinase domain. In the absence of ligand, RTK
exists in a monomeric state (Fig.
1). After binding to the ligand, RTK undergoes an
oligomerization reaction (usually dimerization), which juxtaposes
the tyrosine kinase domain in the cell membrane (4) to render the receptor active. For most
RTKs, the generally accepted view is that this juxtaposition helps
transactivation of tyrosine residues in the kinase activation loop
or near the membrane region and induces conformational changes to
stabilize the active state of the kinase (5). The resultant negatively charged
phosphotyrosine residues subsequently recruit signalling proteins,
such as through the Src homology 2 or phosphotyrosine binding
domains of the signalling proteins, to activate the downstream
signalling cascade (6,7).
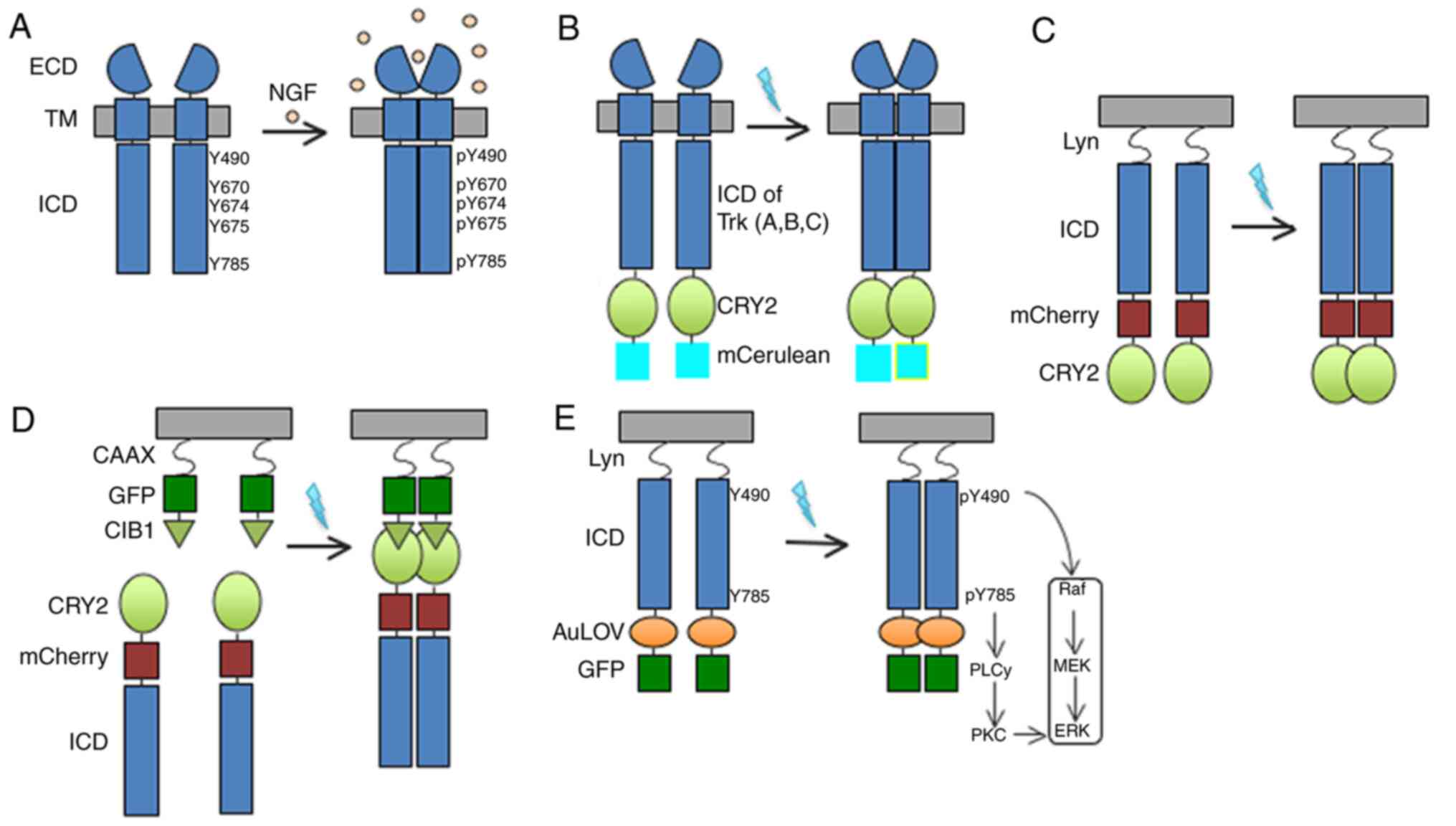 | Figure 1.Schematic illustrations of wild-type
TrkA and photoactivatable TrkA receptors. (A) TrkA has a
single-helix TM domain that anchors the receptor at the plasma
membrane. In the absence of ligand, TrkA exists in a monomeric
state. NGF binds to the ECD to promote dimerization and
phosphorylation of key tyrosines (Y490, Y670, Y674, Y675 and Y785)
within the tyrosine kinase ICD and activates downstream pathways.
(B) Full-length Trk (A, B or C) is linked to CRY2 and the monomeric
cyan fluorescent protein mCerulean. This design appends on the
photosensitive protein system, CRY2, which undergoes dimerization
by blue light illumination. Similar to GFP, mCerulean was fused to
the receptor to facilitate the characterization of receptor
expression. (C) The ECD and TM were replaced by the lipidation Lyn
moiety, but retained the normal plasma membrane activity. mCherry
was fused to the receptor to facilitate the characterization of
receptor expression. (D) The membrane targeting sequence Lyn is
replaced with CIB1-GFP-CAAX. CIB1 is the interactive partner of
CRY2. Light-sensitive TrkA is localized to the plasma membrane and
dimerizes in a light-dependent manner. (E) Similar to the design of
(C) in which the ECD and TM were replaced by the lipidation Lyn
moiety to remove ligand binding activity, this design exploited the
AuLOV domain for light induced dimerization. Blue light stimulation
promotes intracellular dimerization of the receptor and activates
downstream pathways. In this process, the phosphorylation of Y490
and Y785 in the ICD involved in the activation of the Raf/MEK/ERK
and PLCγ-PKC signalling pathways, respectively. To characterize the
receptor expression, GFP was also fused to the receptor. AuLOV,
light-oxygen-voltage-sensing domain of aureochrome1 from Vaucheria
frigida; CRY, cryptochrome 2; ECD, extracellular domain; GFP, green
fluorescent protein; ICD, intracellular domain; NGF, nerve growth
factor; p, phosphorylation; PLC, phosphoinositide phospholipase
C-γ; PKC, protein kinase C; TM, transmembrane; Trk, tropomyosin
receptor kinase; CIB1, calcium and integrin-binding 1. |
Following recent technological developments in
proteomics and functional genomics, research on RTK signalling
pathways has shifted from a study on isolated components in the
ligand-receptor interaction to a broader understanding of the
signalling pathway as an integrated system, including signalling
proteins and the membrane environment. Such a system-wide analysis
provided an unprecedented ‘panoramic’ view of tyrosine
phosphorylation events (8).
The molecular weight range of the Trk receptor
family is 140-145 kDa, and they all have glycosyl modifications.
Neurotrophins are the most common ligands for Trk receptors, and
they serve crucial roles in nervous system function (9). TrkA was the first member of the Trk
receptor family to be discovered (5). During the discovery process, a fusion
of tropomyosin to the kinase domain of TrkA was also identified,
which was shown to be oncogenic and, therefore, TrkA was named
tropomyosin kinase receptor A (8,9). A
subsequent study isolated and identified two additional members of
the family, TrkB and TrkC (10).
Notably, all of these Trk genes are expressed in the nervous system
(9,10). In addition, their ligands are
neurotrophins, and each receptor has a preference for a specific
neurotrophin; for example, nerve growth factor (NGF) activates TrkA
(10), brain-derived growth factor
(BDNF) and neurotrophin (NT)-4/5 activates TrkB (11), and NT-3 activates TrkC. In a
specific cellular environment, NT-3 can also activate both TrkA and
TrkB (12).
However, alteration of ligand specificity of these
receptors has been noted. For example, differential splicing of the
Trk receptor genes results in the synthesis of proteins that have
functionally essential differences in their extracellular domains
(13). There is a fundamental
difference in the function of receptors owing to splicing variants
of the receptor genes. These splicing events are mainly divided
into two categories: i) Splicing at the extracellular domain
(13), such as the different
isoforms of TrkA mentioned above; and ii) splicing at the kinase
domain, such as the differential splicing found in TrkC that
produces an isoform with a modified kinase domain with altered
substrate specificity. Other examples are the isoforms of TrkB and
TrkC that lack the kinase domain [as reviewed in (1)].
The most well-studied of Trk receptor is TrkA, owing
to its association with NGF, which was the first discovered ligand
of RTK receptor family. Before TrkA was identified (14,15),
NGF had been extensively studied and was reported to exhibit a
broad spectrum of neuronal functions, including neuronal survival,
growth, differentiation, synapse formation and synaptic plasticity
(16,17). In the adult nervous system, NGF
supports the maintenance and repair of nerves. Defects in NGF
signalling are associated with a variety of neurodegenerative
diseases. The currently accepted mechanism of TrkA activation is
that the binding of NGF to the extracellular domain of the receptor
results in phosphorylation of specific tyrosine residues in the
phosphokinase region of the cytoplasmic domain (Fig. 1A) (1,5). These
negatively charged phosphate groups then recruit signalling
molecules and activate multiple signalling pathways, with Y490 and
Y785 phosphorylation in the ICD involved in the activation of the
Raf/MEK/ERK and PLCγ-PKC signalling pathways being the most well
characterized (18) (Fig. 1E). After TrkA binds to NGF, the
receptor complex together with their signalling molecule is
internalized into cells either by the classical clathrin-mediated
endocytosis (19,20) or the pincher-mediated macrophage
action (21). The fate of the
receptor complex is less well understood, although previous studies
have shown it may propagate the survival and differentiation
signals through retrograde transport along the axon (22,23).
Therefore, comprehensive elucidation of the TrkA signalling should
enable a better understanding of its function and life cycle.
Although there have been numerous studies on Trk
signalling using TrkA as a model system, the lack of a method to
accurately activate Trk receptor greatly limits our understanding
of TrkA signalling and its dynamics, which further hinders the
development of related treatment strategies. There is a lack of
in-depth analysis in the following three aspects: First, although
the identities of the downstream signalling effectors are known, it
is extremely difficult to determine which signalling molecules are
involved in transporting the TrkA signalling complex into the cell.
Second, since the signalling of TrkA is dynamically regulated in
time and space, how does the difference in cell membrane caused by
subcellular environment affect signal specificity? Third, the
intracellular kinase domain of TrkA consists of multiple
phosphorylation sites, but how the phosphorylation of these sites
regulates different signalling proteins and activates a specific
pathway through coordination with each other is unclear. These
problems all require the development of a proper tool to activate
TrkA receptor with more precise spatial and temporal controls than
their ligands relying on relatively slow diffusion and binding. The
emergence of optogenetics, which is based on natural
light-sensitive proteins, provides a new strategy for controlling
protein function with high spatial and temporal resolution
(24–26). To date, two types of light-sensitive
proteins, the light-oxygen-voltage-sensing (LOV) domain and the
cryptochrome 2 (CRY2), have been tested on TrkA receptors,
providing new molecular tools to study intracellular signalling of
this important family of receptors. The present study proposed a
signal transduction model for light-induced receptor activation.
This model is based on analysis of recently engineered
light-induced Trk receptors reported to date and comparisons to the
activation mechanism of natural receptors. The careful delineation
of the dimerization mechanisms fine-tuning activation will guide
future design for a specific cellular output using precise
light.
Materials and methods
Analysis of structures
Crystal structures of TrkA and insulin receptor
kinase (IRK) were chosen for analysis. Crystal structures were
downloaded from the Research Collaboratory for Structural
Bioinformatics Protein Data Bank (PDB; www.rcsb.org)
for the inactive TrkA receptor kinase domain (PDB: 4GT5), the IRK
in the inactive (PDB: 1IRK) and phosphorylated/activated (PDB:
1IR3) state. PyMOL software (version 1.8) was used to display
structures.
Analysis of experimental data
Functional assay readouts were extracted from
previously published data (24–26),
including ERK translation [also termed as ERK expression (26)], neurite growth and neuronal cell
survival rate. Neurite growth assay (often referred as neurite
outgrowth) quantifies the process wherein developing neurons
produce new projections as they grow in response to guidance cues.
The readouts were measured before and after light treatment
conditions. The neurite growth and cell survival rate data were
normalized using ratio of the number of surviving cells/total
number of cells. Functional outputs are represented by the
activities ratio, (readout obtained + light)/(readout
obtained-light), measured in three types of assays: ERK
translocation, neurite growth and neuron survival rate (24–26).
Data are presented as the mean ± SD, n ≥3.
Statistical analysis
Statistical analyses were performed using
StatPlus:mac software version 7 (AnalystSoft, Inc.). ANOVA followed
by Tukey post-hoc test for pairwise multiple comparison procedure
was performed. P<0.05 was considered to indicate a statistically
significant difference.
Results and Discussion
TrkA activation by light with spatial
and temporal precision
Over the past five years, several Trk proteins have
been engineered to respond to light (summarized in Fig. 1B-E and Table I). The first design was demonstrated
in 2014 by Chang et al (24), who inserted the light-sensitive
protein photolyase homology region (PHR) of CRY2 into the
full-length Trk receptor sequence and also combined mCerulean (a
cyan fluorescent protein) at the end of the PHR as the fluorophore
(Fig. 1B). CRY2, together with
CRY1, belongs to a class of flavoproteins found in plants and
animals that respond to blue light. They regulate circadian rhythms
and may also function in the sensing of magnetic fields in several
species. To validate the functional expression of the
light-sensitive Trks, intracellular ERK, Ca2+ indicator
R-GECO1, AKT and EGFR inhibitor in signal activity were compared
before and after light stimulations. Translocation ERK expression
from the cytosol to nucleus was also measured by using fluoresce
intensity and analysed by the nucleus/cytosol intensity ratio
before and after light treatment. Dynamic axon movement was
recorded by live video dark 400 sec and light 370 sec. These
functional outputs were summarized in Table II. Although this system was
successful in controlling ERK activation and dynamic movement of
the receptors along neurites using light, there was high basal
activity presumably due to the presence of ligand-banding domain in
the Trk receptors.
 | Table I.Various light-sensitive Trk
receptors, design, light stimulation and pathway activation
studied. |
Table I.
Various light-sensitive Trk
receptors, design, light stimulation and pathway activation
studied.
|
|
|
|
|
| Excitation
wavelength |
|
|---|
|
|
|
|
|
|
|
|
|---|
| Author, year | Light-sensitive
TrkA | MW, kDa | Light module | Linker length (aa
sequence) | Light stimulation,
nm (power) | Fluorescence
imaging (nm) | Pathways | (Refs.) |
|---|
| Chang KY et al,
2014 | TrkB-PHR-mCit | 143 | PHR was inserted
into full length Trk receptor at C-terminus | 6 (DPPVAT) | 488 (0.5 sec
pulses; intensity, 6.5 µW) | mCit (514),
excitation laser | ERK; AKT;
Ca2+ | (24) |
| Duan L et al,
2018 | opto-full-TrkA | 169 | CRY2 was inserted
into full length TrkA | 2 (GS) | 488 (0.2 sec pulse;
intensity, 9.7 W/cm) | GFP (472/30) and
mCh (560/40) excitation filters | AKT; ERK;
PIK3K | (25) |
| Duan L et al,
2018 | opto-iTrkA | ~120 | CRY2 was inserted
into iTrkA at C-terminus | 3 (GSS) | 488 (0.2 sec pulse;
intensity, 9.7 W/cm) | GFP (472/30) and
mCh (560/40) excitation filters | AKT; ERK;
PIK3K | (25) |
| Duan L et al,
2018 | opto-Lyn-iTrkA | ~120 | CRY2 was inserted
into iTrkA at C-terminus | 15
(MGCIKSKRKDNLNDD) | 488 (0.2 sec pulse;
intensity, 9.7 W/cm) | GFP (472/30) and
mCh (560/40) excitation filters | AKT; ERK;
PIK3K | (25) |
| Duan L et al,
2018 | opto-iTrkA +
CAAX | ~120 | CRY2 was inserted
into iTrkA at N-terminus | n/a | 488 (0.2 sec pulse;
intensity, 9.7 W/cm) | GFP (472/30) and
mCh (560/40) excitation filters | AKT; ERK;
PIK3K | (25) |
| Khamo JS et al,
2019 |
Lyn-iTrkA-AuLOV | 83 | AuLOV was inserted
into iTrkA at C-term | 15
(MGCIKSKRKDNLNDD) | 488 (single pulse,
6.5 µW) 488 (single pulse; 6.5 µW) | GFP: Excitation
filter (472/30), dichroic mirror (495), emission filter
(520/35) | AKT; ERK; PKC; EGFR
inhibitor | (26) |
 | Table II.Functional output of light-sensitive
Trk receptors. |
Table II.
Functional output of light-sensitive
Trk receptors.
|
|
| Functional
outputa |
|---|
|
|
|
|
|---|
| Light-sensitive
Trk | Propagation
direction | ERK
translocation | Neurite growth | Neuron survival
rateb |
|---|
| TrkB-PHR-mCit | Bottom-to-top | 1.5 | 7 | n/a |
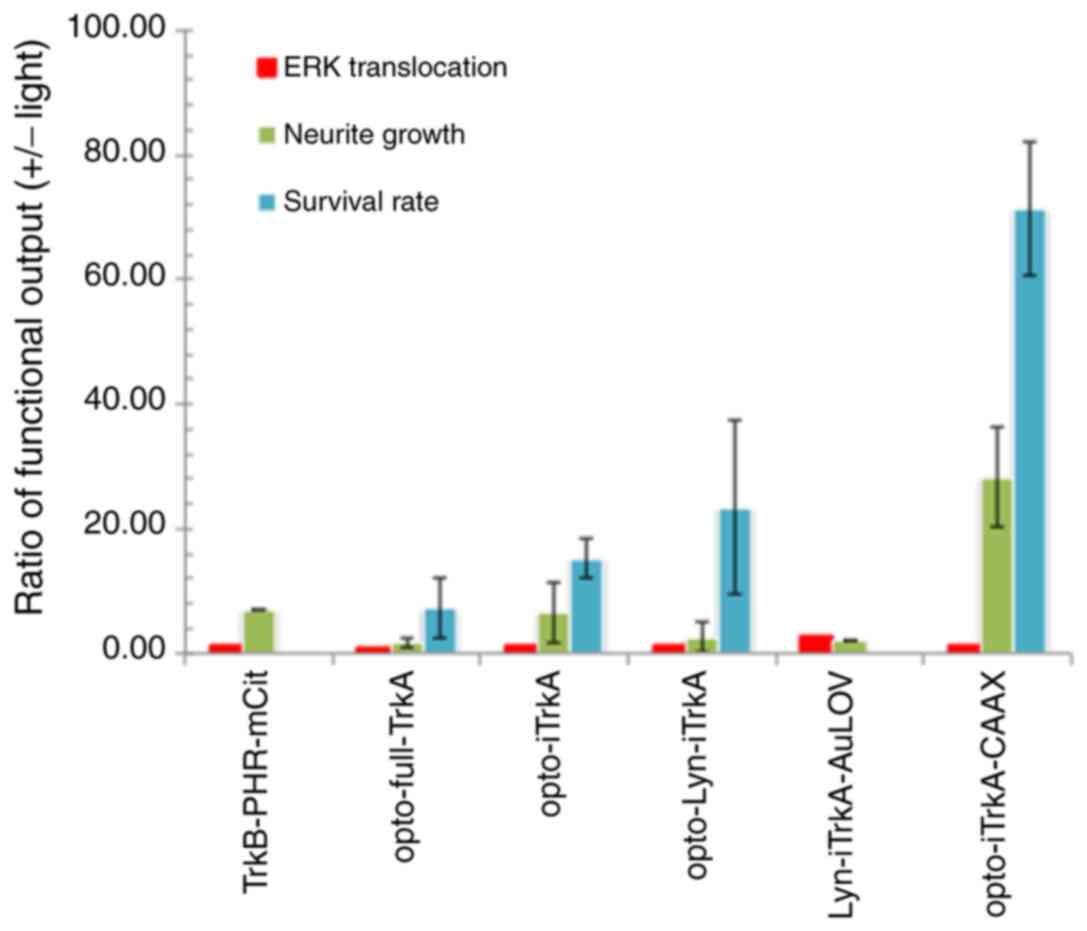
| opto-full-TrkA | Bottom-to-top | 1.1 | 1 | 4.11±2.3 |
| opto-iTrkA | Bottom-to-top | 1.25 | 5.48 | 12.72±3.4 |
| opto-Lyn-iTrkA | Bottom-to-top | 1.5 | 1.69 | 27.14±14 |
|
Lyn-iTrkA-AuLOV | Bottom-to-top | 3 | 2 | n/a |
|
opto-iTrkA-CAAX | Top-to-bottom | 1.5 | 35.88 | 73±30 |
In 2018, Duan et al introduced Arabidopsis
thaliana CRY2 into intracellular TrkA (iTrkA) (25), a truncated receptor without the
ligand-binding domain (Fig. 1C and
D), which significantly reduced receptors' basal activity
without light stimulation. Altogether, Duan et al made four
different designs (Table I), i)
opto-full-TrkA, ii) opto-iTrkA, iii) opto-Lyn-iTrkA, and iv)
opto-iTrkA+CAAX. The first construct has a similar design as Chang
et al (24), which has a
fusion of CRY motif into the full-length receptor. The second
construct has the ligand-binding domain removed, whereas the third
and fourth construct have an N-terminal membrane anchor (Lyn motif
or CAAX motif) to localize the receptors onto the plasma membrane.
Among these four different constructs, functional output before and
after light activation was evaluated; these include ERK
translocation, neurite growth and neuron survival rate. Even though
all constructs demonstrated light-induced activities, the
opto-iTrkA+CAAX design outperformed the other three constructs by
demonstrating higher contrasts after light stimulation (Table II).
This approach was subsequently adapted to another
light-sensitive protein containing the LOV domain (Fig. 1E; Tables
I and II), which is a protein
module present in many plants, microalgae, fungi and bacteria
(25). This naturally occurring
protein enables cells to sense environmental conditions and to
control phototropism. Compared with CRY2 (~67 kDa), the LOV domain
is a small, light-sensing module (~12-19 kDa). In this construct,
Khamo et al (26) fused LOV
domain of the aureochrome1 protein from the yellow-green alga
Vaucheria frigida (AuLOV) with iTrkA. AuLOV responds to blue
light and is excited at 488 nm, the same wavelength as for CRY2.
Following the design by Duan et al (25), Khamo et al (26) used the Lyn motif to position the
receptor in the plasma membrane (Fig.
1E). The resultant light-sensitive Lyn-iTrkA-AuLOV was 83 kDa
the smallest in size among all designs. Khamo et al
(26)demonstrated that activation
of optogenetic TrkA significantly promoted cell differentiation.
The present study summarized and analysed all the light-induced
activities of the different constructs created and characterized
from the three independent laboratories (Fig. 2; Table
II). There is a general trend among the various functional
assays performed that the neuron survival rate test has the highest
sensitivity toward light stimulation. Furthermore, the construct
(opto-iTrkA-CAAX) with a top-to-bottom design has the highest light
sensitivity, manifested by the neuron survival rate assay (ANOVA;
F=34.83; P=0.00006). Finally, various flexible linkers (Table I) inserted into the TrkA did not
play significant roles in dictating the light-sensitivities of the
receptors.
Light modules lend support to the
dimerization model
The light-dependent alterations of the TrkA
receptors analysed above, allow us to reach new conclusions
regarding possible activation mechanisms in TrkA signalling when
combined with structural information. In contrast with the
extensive characterization of kinase domains of EGFR and insulin
receptors (27–30), very little is known for the TrkA
receptor. To date, there is only one isolated kinase domain of TrkA
available (PDB: 4GT5), in which three of the tyrosines (Y670, Y674,
Y675) could be located in the activation loop (highlighted in
orange in Fig. 3A). This structure
is considered to represent the inactive state because none of the
tyrosine residues are phosphorylated. This makes it possible to
analyse TrkA in the inactive state. In this structure, Y670
situates on a slightly more structured α-helix compared with the
Y674 and Y675 sites, which are both in the non-structured loop
region. Inspired by the publication of Cunningham and Greene
(27), which proposed TrkA kinase
3D structural models based on the insulin receptor IRK, the present
study compared the available X-ray crystal structure of inactive
TrkA kinase domain with the inactivated (PDB: 1IRK) (Fig. 3B) (28) and the activated (PDB: 1IR3)
(Fig. 3C) (30) kinase domain of IRK. To the best of
our knowledge, such TrkA structural comparison with IRK was not yet
available. This analysis, for the first time, demonstrated that
these domains both have the three activation loop phosphotyrosines
(Y1158, Y1162, and Y1163) that are the direct homologous sequence
of those in TrkA (Fig. 3). It was
noticed that in IRK, the activation loop had undergone a
significant conformational switch from inactivated to the activated
state, exposing Y1158 (the equivalent of Y670 in TrkA) to the
protein exterior in an entirely solvent-exposed environment,
whereas the Y1162 (the equivalent of Y674 in TrkA) and Y1163 (the
equivalent of Y675 in TrkA) remain relatively unchanged. Because
the conservation of TrkA activation within the family of RTK,
therefore, we hypothesized that the activation loop of TrkA may
adopt conformational changes similar to what has been observed in
IRK, which means that such change in the activation loop may
promote dimerization of the kinase domain. A threshold model was
proposed by Zinkle and Mohammadi (8) to explain RTK activation. In their
model, they differentiate transit dimers and potent dimers. When
RTK forms transit dimers, they are unable to phosphorylate
kinetically disadvantaged sites, whereas RTK forms potent a dimer
which enables specific downstream signals and influence the
cellular response.
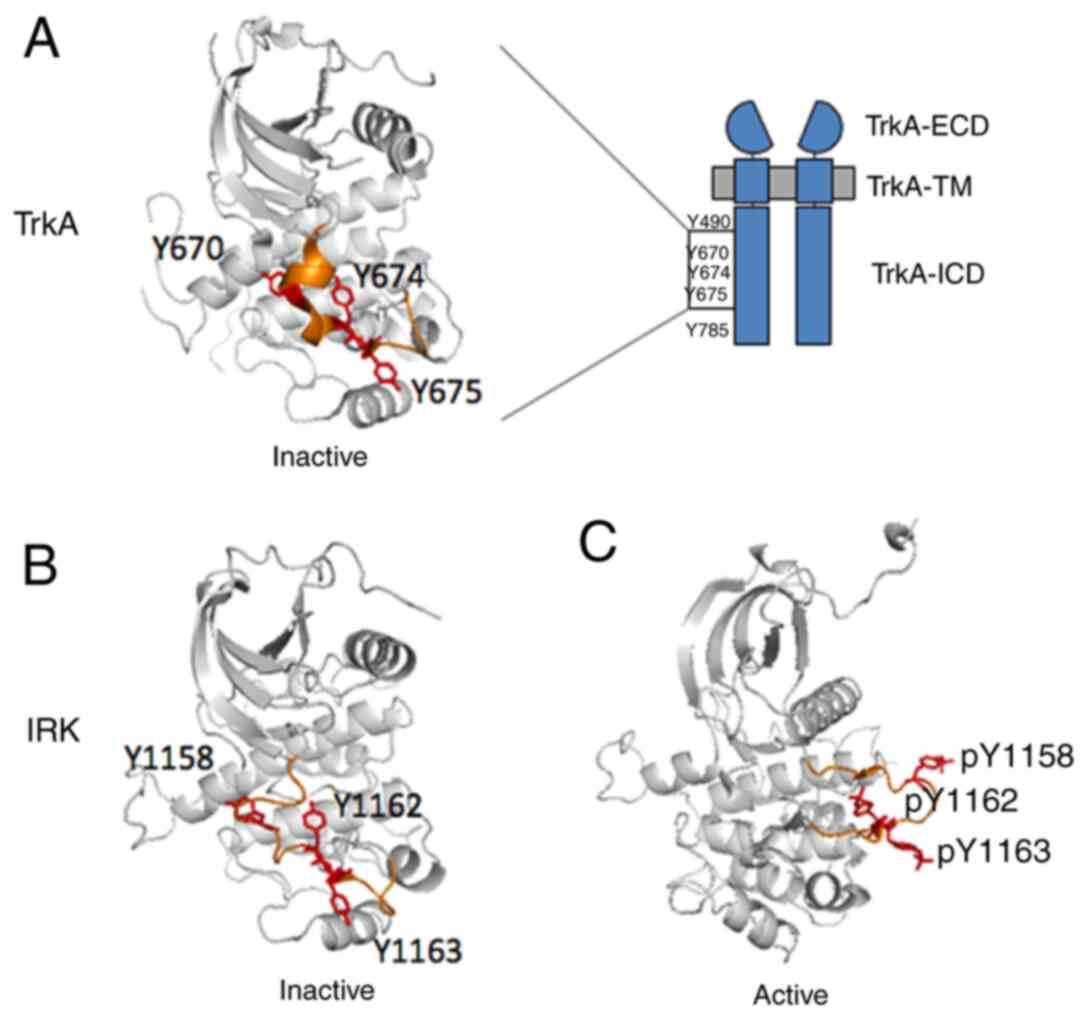 | Figure 3.Crystal structures of TrkA and IRK
tyrosine kinase domains. (A) Crystal structure of the TrkA tyrosine
kinase domain (PDB: 4GT5) in the inactive state (37). The activation loop is coloured
orange, with Y670, Y674 and Y675 residues coloured red. Crystal
structures of the IRK in (B) the inactive (PDB: 1IRK) (28) and (C) in the activated (PDB: 1IR3)
state (30). IRK crystal structures
were adapted to depict graphically the TrkA tyrosine kinase domain
in (A). The activation loop is coloured orange, with the homologous
Y1158, Y1162, Y1163 residues coloured red. ECD, extracellular
domain; ICD, intracellular domain; IRK, insulin receptor kinase; p,
phosphorylation; TM, transmembrane; Trk, tropomyosin receptor
kinase. |
Based on above observations and comparisons, we
propose a new light-induced activation model (Fig. 4) inspired by the photoreceptor
rhodopsin, a G-protein-coupled receptor (31) and the ligand activated GPCR
beta-adrenergic receptors (32).
This model predicts that the unliganded receptor, which is defined
as the R° state that can undergo transitions through the
stimulation of light, to the R* state, which is defined as the
active state. In this model, at least one intermediate state R' is
included, which is before the binding of the signalling molecules.
R° is stabilized by an unphosphorylated kinase domain, and
intracellular substrates stabilize R* through the interactions with
the phosphorylated kinase domain. Mechanistically, this may be
explained by considering that the R° receptor (either un-liganded
or un-induced by light) exists in a monomeric state, whereas R*
exists as oligomeric (dimer) state. Moreover, R° may undergo
spontaneous transitions to the R' state, explaining the high basal
activity observed for some TrkA constructs when the extracellular
ligand-binding domain was removed. As shown in Fig. 3, binding of a ligand is similar to
the light-induced activation that promotes kinase domain
dimerization to occur sequentially, resulting in a series of
conformational states that are intermediates (Rlig and
Rop. Subscript ‘lig’ refers to the ligand induced
activation and ‘op’ refers to optogenetic activation.). Here, it is
necessary to point out a factor that has been overlooked between
the Rlig and Rop state, which is the
direction of dimerization induction. For the ligand induction, the
driving forces always come from the top of the receptors (induced
from the extracellular side). However, for the light induction, the
driving forces may come either from the top or bottom of the
receptor, depending where the light-dependent module was fused
into. From the experimental analyses, aforementioned, results from
light-engineered TrkA receptors indicate that dimerization promoted
from top-to-bottom direction are more efficient than bottom-to-top
direction, as compared among four different TrkA CRY2 fusion
constructs (especially between CAAX-CRY2-mCh-iTrkA vs.
Lyn-iTrkA-CRY2; Fig. 2; Table II). Following the initial
dimerization of the light-sensitive protein module (PHR, CRY2 or
AuLOV), the kinase domain may undergo rearrangement and
conformation changes (Fig. 3) that
lead to autophosphorylation and dimerization. Each phosphorylation
on the kinase domain stabilizes the dimer and depending on its
conformation preferably recruits an intracellular substrate until
the receptor has been stabilized by interactive proteins to reach
the active R* state. Any agents that can stabilize the R° state can
reverse the process and dephosphorylate the activate R' state of
the receptor. Conversely, some ligands may promote stronger dimers
than other ligands, and some RTK mutants may stabilize unique
conformational states having a lower affinity for the intracellular
interactive proteins. Techniques that rely on the genetic code
expansion (33), which allows the
site-specific incorporation of unnatural amino acids into proteins,
can potentially enable smaller modifications of the protein.
Recently, we have successfully demonstrated the feasibility of
controlling TrkA activity using genetic code expansion technique
(34). Another important
alternative strategy to reengineering kinase activity is through
chemical genetics. Several recent reviews have summarized topics
related to engineering at the gene, protein and ligand level
(35–37), as well as the eukaryotic model
organisms (38) in which the
technique can be implemented. These strategies also have not yet
been applied to the RTK receptors. The exploitation of these
alternative approaches to engineer light-sensitive RTKs will
certainly contribute to the future studies of RTK receptors and
associated therapeutic strategies.
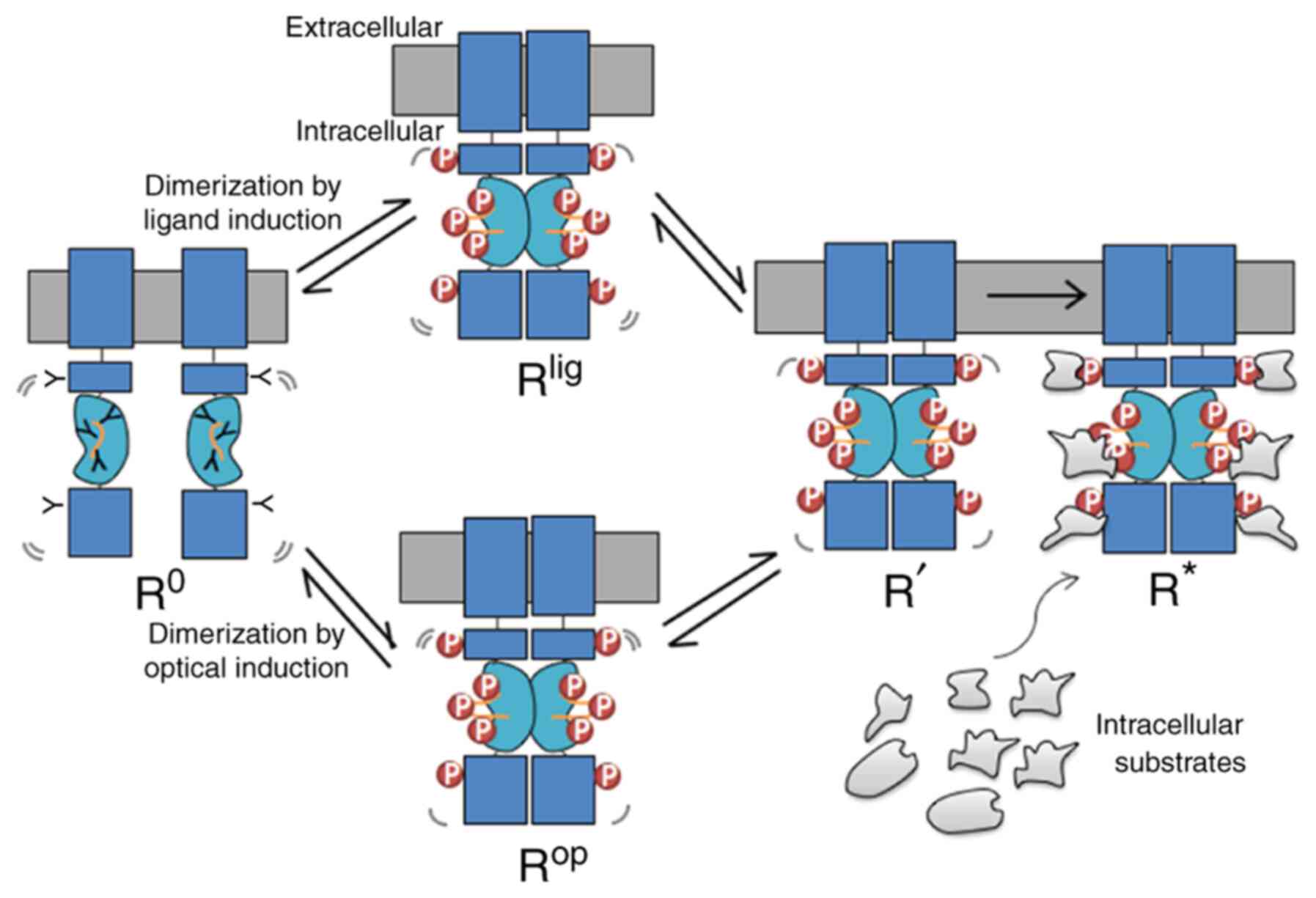 | Figure 4.Light modules lend support to the
dimerization model. RTKs exist in the monomeric ground state, R°.
After binding to a ligand, or stimulated by the light, RTK
dimerizes and reaches to the Rlig or the Rop
state, respectively, with the intracellular kinase domains entering
into a proper proximity such that tyrosines residues in the
intracellular domain and activation loop (orange lines) are
phosphorylated. Such kinase activation leads to the intermediate R'
state, which subsequently creates docking sites for the recruitment
of distinct intracellular in the activated R* state. Lig, ligand;
op, optogenetic; p, phosphorylation; RTK, receptor tyrosine
kinase. |
Finally, it was noted that the use of
light-sensitive TrkA has thus been restricted to cells in tissue
culture. However, optogenetics can also been applied in
vivo. In the future, it may be possible to apply these the most
robust light-sensitive TrkA in a living animal and to determine the
nature of the tissue specific functions of TrkA in various cell
types and organs. The major challenge associated with these
experiments would be achieving effective photoactivation; for
example, by using a transparent animal such as zebrafish or by
surgically implanting a light emitter.
Acknowledgements
The authors thank Professor Michael Schumacher
(INSERM U1195) for valuable suggestions.
Funding
Financial supports were made available by Sorbonne University
start-up fund INRA S18LRPV022 and CNRS PEPS (grant: Exomod) to
S.Y.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SY monitored the project progression, data analysis,
interpretation, writing and editing the manuscript. WZ, LL, ZF and
SZ contributed to data analysis. SY and LL prepared the initial
draft of the manuscript; ZF and SY revised and finalized the
manuscript. All authors reviewed, read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lemmon MA and Schlessinger J: Cell
signalling by receptor tyrosine kinases. Cell. 141:1117–1134. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sangwan V and Park M: Receptor tyrosine
kinases: Role in cancer progression. Curr Oncol. 13:191–193. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Robinson DR, Wu YM and Lin SF: The protein
tyrosine kinase family of the human genome. Oncogene. 19:5548–5557.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fraser J, Cabodevilla AG, Simpson J and
Gammoh N: Interplay of autophagy, receptor tyrosine kinase
signalling and endocytic trafficking. Essays Biochem. 61:597–607.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hubbard SR: Juxtamembrane autoinhibition
in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 5:464–471.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pawson T, Gish GD and Nash P: SH2 domains,
interaction modules and cellular wiring. Trends Cell Biol.
11:504–511. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren S, Yang G, He Y, Wang Y, Li Y and Chen
Z: The conservation pattern of short linear motifs is highly
correlated with the function of interacting protein domains. BMC
Genomics. 9:4522008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zinkle A and Mohammadi M: A threshold
model for receptor tyrosine kinase signaling specificity and cell
fate determination. F1000Res. 7:8722018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ip NY, Stitt TN, Tapley P, Klein R, Glass
DJ, Fandl J, Greene LA, Barbacid M and Yancopoulos GD: Similarities
and differences in the way neurotrophins interact with the Trk
receptors in neuronal and nonneuronal cells. Neuron. 10:137–149.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brodeur GM, Nakagawara A, Yamashiro DJ,
Ikegaki N, Liu XG, Azar CG, Lee CP and Evans AE: Expression of
TrkA, TrkB and TrkC in human neuroblastomas. J Neurooncol.
31:49–55. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marshall J, Szmydynger-Chodobska J,
Rioult-Pedotti MS, Lau K, Chin AT, Kotla SKR, Tiwari RK, Parang K,
Threlkeld SW and Chodobski A: TrkB-enhancer facilitates functional
recovery after traumatic brain injury. Sci Rep. 7:109952017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou XF and Rush RA: Functional roles of
neurotrophin 3 in the developing and mature sympathetic nervous
system. Mol Neurobiol. 13:185–197. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Segal RA: Selectivity in neurotrophin
signalling: Theme and variations. Annu Rev Neurosci. 26:299–330.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaplan DR, Martin-Zanca D and Parada LF:
Tyrosine phosphorylation and tyrosine kinase activity of the trk
proto-oncogene product induced by NGF. Nature. 350:158–160. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Klein R, Conway D, Parada LF and Barbacid
M: The trkB tyrosine protein kinase gene codes for a second
neurogenic receptor that lacks the catalytic kinase domain. Cell.
61:647–656. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Moheban DB, Conway BR,
Bhattacharyya A and Segal RA: Cell surface Trk receptors mediate
NGF-induced survival while internalized receptors regulate
NGF-induced differentiation. J Neurosci. 20:5671–5678. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ninan I: Synaptic regulation of affective
behaviors; role of BDNF. Neuropharmacology. 76:684–695. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Biarc J, Chalkley RJ, Burlingame AL and
Bradshaw RA: Dissecting the roles of tyrosines 490 and 785 of TrkA
protein in the induction of downstream protein phosphorylation
using chimeric receptors. J Biol Chem. 288:16606–16618. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Beattie EC, Howe CL, Wilde A, Brodsky FM
and Mobley WC: NGF signals through TrkA to increase clathrin at the
plasma membrane and enhance clathrin-mediated membrane trafficking.
J Neurosci. 20:7325–7333. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Howe CL, Valletta JS, Rusnak AS and Mobley
WC: NGF signaling from clathrin-coated vesicles: Evidence that
signaling endosomes serve as a platform for the Ras-MAPK pathway.
Neuron. 32:801–814. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shao Y, Akmentin W, Toledo-Aral JJ,
Rosenbaum J, Valdez G, Cabot JB, Hilbush BS and Halegoua S:
Pincher, a pinocytic chaperone for nerve growth factor/TrkA
signaling endosomes. J Cell Biol. 157:679–691. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Riccio A, Pierchala BA, Ciarallo CL and
Ginty DD: An NGF-TrkA-mediated retrograde signal to transcription
factor CREB in sympathetic neurons. Science. 227:1097–1100. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bhattacharyya A, Watson FL, Bradlee TA,
Pomeroy SL, Stiles CD and Segal RA: Trk receptors function as rapid
retrograde signal carriers in the adult nervous system. J Neurosci.
17:7007–7016. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang KY, Woo D, Jung H, Lee S, Kim S, Won
J, Kyung T, Park H, Kim N, Yang HW, et al: Light-inducible receptor
tyrosine kinases that regulate neurotrophin signalling. Nat Commun.
5:40572014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Duan L, Hope JM, Guo S, Ong Q, François A,
Kaplan L, Scherrer G and Cui B: Optical activation of TrkA
signaling. ACS Synth Biol. 7:1685–1693. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khamo JS, Krishnamurthy VV, Chen Q, Diao J
and Zhang K: Optogenetic delineation of receptor tyrosine kinase
subcircuits in PC12 cell differentiation. Cell Chem Biol.
26:400–410.e3. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cunningham ME and Greene LA: A
function-structure model for NGF-activated TRK. EMBO J.
17:7282–7293. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hubbard SR, Wei L, Ellis L and Hendrickson
WA: Crystal structure of the tyrosine kinase domain of the human
insulin receptor. Nature. 372:746–754. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Artim SC, Mendrola JM and Lemmon MA:
Assessing the range of kinase autoinhibition mechanisms in the
insulin receptor family. Biochem J. 448:213–220. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hubbard SR: Crystal structure of the
activated insulin receptor tyrosine kinase in complex with peptide
substrate and ATP analog. EMBO J. 16:5572–5581. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fahmy K, Siebert F and Sakmar TP:
Photoactivated state of rhodopsin and how it can form. Biophys
Chem. 56:171–178. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gether G and Kobilka BK: G protein-coupled
receptors. II. Mechanism of agonist activation. J Biol Chem.
273:17979–17982. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Y, Lu L and Ye S: Genetic code
expansion and optoproteomics. Yale J Biol Med. 90:599–610.
2017.PubMed/NCBI
|
|
34
|
Zhao S, Shi J, Yu G, Li D, Wang M, Yuan C,
Zhou H, Parizadeh A, Li Z, Guan MX and Ye S: Photosensitive
tyrosine analogues unravel site-dependent phosphorylation in TrkA
initiated MAPK/ERK signaling. Commun Biol. 3:7062020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Leopold AV, Chernov KG and Verkhusha VV:
Optogenetically controlled protein kinases for regulation of
cellular signaling. Chem Soc Rev. 47:2454–2484. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Islam K: The bump-and-hole tactic:
Expanding the scope of chemical genetics. Cell Chem Biol.
25:1171–1184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McCormick JW, Pincus D, Resnekov O and
Reynolds KA: Strategies for engineering and rewiring kinase
regulation. Trends Biochem Sci. 45:259–271. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ryu J and Park SH: Simple synthetic
protein scaffolds can create adjustable artificial MAPK circuits in
yeast and mammalian cells. Sci Signal. 383:ra662015.PubMed/NCBI
|