Introduction
Liver cancer is the fifth most frequent fatal
malignancy in the United States (1), but ranks second in China (2) due to the high risk of factors, such
as hepatitis B virus (HBV) or hepatitis C virus (HCV) infection,
fatty liver disease and alcohol-related cirrhosis. In total 5-15%
of patients are suitable for surgical removal, which is only for
early-stage patients (3). The
prognosis for unresectable stage liver cancer is very poor, with a
median survival time of 6-20 months and <5% for the 5-year
survival rate (4). Therefore,
discovering prognostic or therapeutic biomarkers is urgently
required for patients with liver cancer.
Long non-coding RNAs (lncRNAs) are defined as
non-coding RNAs >200 nucleotides (5). Although lncRNAs undergo similar
processing as mRNAs, including splicing, capping, polyadenylation
and editing, they lack significant open reading frames (5). Increasing evidence suggests that
lncRNAs participate in every aspect of the life cycle of a gene,
including transcription, splicing, RNA decay and translation
(6). Cytoplasmic lncRNAs can
regulate gene expression via diverse mechanisms, either by sponging
microRNAs (miRNAs/miRs) or altering mRNA stability (7).
To date, a total of 74 deregulated liver
cancer-associated lncRNAs have been reported, with 52 upregulated
lncRNAs exhibiting oncogenic properties and 22 downregulated
lncRNAs exhibiting tumor-suppressive properties (8). Therefore, these insights reveal that
novel lncRNAs may be potential biomarkers and enable the design of
precision therapy for liver cancer. Previously, it was reported
that HBV infection inhibited the expression of LINC00238, which was
found to be significantly downregulated in HBV-positive liver
tissues, HBV-expressing cell lines and HBV transient-expressing
cells (9). Furthermore,
overexpression of LINC00238 could suppress HBV replication
(9). Chronic HBV infection is a
major risk factor for liver cancer, and notably, in China,
HBV-related liver cancer accounts for ~85% of liver cancer cases
due to the high prevalence of HBV infection (10). However, to the best of our
knowledge, the role of LINC00238 in liver cancer progression is
still unclear.
Gene Expression Profiling Interactive Analysis
(GEPIA) is a web-based tool that can deliver fast and customizable
functionalities based on The Cancer Genome Atlas (TCGA) and The
Genotype-Tissue Expression data (11). Researchers can perform
comprehensive expression analyses through GEPIA by customizable
functions, including differential expression analysis, profiling
plotting, correlation analysis, patient survival analysis, similar
gene detection and dimensionality reduction analysis (11). The chick embryo chorioallantoic
membrane (CAM) assay is an alternative in vivo experimental
model suitable for cancer studies (12). Compared with the mouse xenograft
model, the CAM is a low cost method that is naturally
immune-incompetent, favorable to tumor grafting, and enables the
ability of perform neovascularization and invasion assays to
evaluate the ability of cells to be tumorigenic and invade and
metastasize into the embryo (13). It is widely used for the
investigation of multiple steps of tumor progression, metastatic
behavior and molecular deregulated pathways, including the
evaluation of stem cell activity in breast cancer (13), sarcoma (14,15), lymphoma (16), liver cancer (17) and melanoma (18).
In the present study, data mining was performed by
GEPIA analysis of lncRNAs for liver hepatocellular carcinoma [LIHC
(TCGA-LIHC)], which led to the detection of the expression profile
of LINC00238 in liver cancer, and identified it as a tumor
suppressor. Mechanistically, LINC00238 acted as a molecular sponge
to adsorb miR-522, resulting in the reversal of the inhibitory
effects on two downstream targets, secreted frizzled related
protein 2 (SFRP2) and dickkopf1 (DKK1). Therefore, the functions of
a novel lncRNA that regulated the malignant phenotype of liver
cancer was described.
Materials and methods
GEPIA analysis of TCGA and Gene
Expression Omnibus (GEO) data
GEPIA (11)
(http://gepia.cancer-pku.cn) was used to
analyze the aberrantly expressed lncRNAs by differential expression
analysis of TCGA-LIHC. The downregulated lncRNAs among the
differentially expressed genes (DEGs) with log2 fold
change (FC) cutoff=1 and q-value cutoff=0.01 were selected. Next,
Kaplan-Meier survival analysis was performed followed by a log-rank
test for these downregulated lncRNAs to obtain lncRNAs associated
with the overall survival of patients after surgery by the median
expression level of each lncRNA in LIHC (P<0.1, two-tailed test
P<0.1 means one-tailed test P<0.05 because this study was
only concerned about one direction, not both).
Cell line and cell culture
Two liver cancer cell lines (HepG2 and Huh7) and
239T cells were obtained from the National Infrastructure of Cell
Line Resource. Cell lines were confirmed to be free of mycoplasma
contamination by PCR and three short tandem repeat (STR) loci.
Cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with penicillin (50 U/ml), streptomycin (50
g/ml) and 10% heat-inactivated fetal bovine serum (FBS, Thermo
Fisher Scientific Inc.) in a humidified 5% CO2
atmosphere at 37°C.
In vivo tumor growth and
metastasis
A modified chick embryo CAM assay was used to assess
tumor growth and metastatic characteristics. Briefly, a total of 35
10-day-old SPF white leghorn chicken embryo eggs (Beijing Vital
River Laboratory Animal Technology Co., Ltd.) were randomized into
groups (n=5/group). A square window was opened in the shell under
aseptic conditions after sterilization with 75% ethanol. A total of
5×106 cells in 50 µl PBS (including Huh7 control cells,
LINC00238 overexpression Huh7 cells, scramble HepG2 cells,
LINC00238 shRNA HepG2 cells, LINC00238 overexpression Huh7 cells
transfected with NC and miR-522 mimics) were labeled with CM-DiI
(red fluorescent dye) in 5% glucose for 15 min at 37°C and
inoculated onto each CAM. Eggs were resealed with sterilized tape
and returned to a humidified 37°C incubator for an additional 7
days (before hatching out, normally chicken eggs would hatch in 21
days). The tumors that grew on each CAM of the 18-day-old chicken
embryos were dissected and weighed. Then, the chicken embryos were
sacrificed by cervical dislocation on day 18 after the incubation
period began. The fresh lungs were isolated, flattened by two
slides and evaluated under a fluorescence microscope (Leica
Microsystems, Inc.) to track the distant metastatic tumor loci
(with red fluorescence) (19).
RNA extraction and reverse
transcription-quantitative (RT-q) PCR analysis
Total RNA was isolated from control or LINC00238
overexpression Huh7 cells, scramble or LINC00238-shRNA HepG2 cells,
NC or miR-522 transfected Huh7 or LINC00238 overexpression Huh7
cells by using a miRNeasy mini kit (Qiagen, Inc.). Separation and
purification of cytoplasmic and nuclear RNA from HepG2 and Huh7
cells was performed using a Cytoplasmic & Nuclear RNA
Purification kit (Norgen Biotek Corp.). lncLocator
(csbio.sjtu.edu.cn/bioinf/lncLocator/) was used to predict the
lncRNA subcellular location. First strand cDNA was synthesized from
5 µg RNA using random primers and Moloney Murine Leukemia Virus
reverse transcriptase (M-MLV RT; Invitrogen; Thermo Fisher
Scientific, Inc.). For miRNA detection, 100 ng of RNA was first
added to polyA tails by polyA polymerase (NEB), and then cDNA was
synthesized by OligodT-Adaptor and M-MLV RT according to the
manufacturer's protocol. qPCR was performed using SYBR Green PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) on
an ABI 7500 System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were initial denaturation at
95°C for 5 min, followed by 45 amplification cycles of 95°C for 10
sec, 60°C for 20 sec, 72°C for 42 sec and a final extension 1 min
at 72°C. The gene expression level was calculated by the
2−ΔΔCq method (20),
where ΔCq=Cq (gene)-Cq (GAPDH). For lncRNA and miRNA expression,
GAPDH and U6 were used as internal reference genes, respectively.
qPCR data are represented as the mean ± SD from three independent
experiments. Sequences for primers are listed in Table SI. The primers for amplification
of LINC00238 were designed in the common region of three isotypes
of V1, V3 and V4.
Plasmid construction and cell
transfection
LINC00238 (NR_024338.3, isotype V1) was subcloned
into a pcDNA3.1 (+) (Invitrogen; Thermo Fisher Scientific, Inc.)
expression vector. A total of 10 µg of blank vector or LINC00238
plasmids were transfected into 5×106 Huh7 cells for 48 h
and selected with 500 mg/ml G418 solution for 1 week to obtain
control group and stable LINC00238 overexpression group maintained
in 250 mg/ml G418 solution. A total of 5 µg of 3rd lentivirus RNAi
shuttle vector (plenti6-U6, originally from Invitrogen plenti6
backbone) containing the sequence for scramble control or for the
short hairpin RNA (shRNA) of LINC00238 was transfected into
1×106 293FT cells together with the 9 µg packaging
plasmids containing PLP1, PLP2 and pLP/VSVG to generate
lentiviruses for 48 h at 37°C to collect lentiviral particles in
the supernatant by centrifuge at 1,000 × g for 5 min. Lentivirus at
an MOI of 20 were added into HepG2 cells seeded in 6-well plates
(at a density of 5×104 cells/well), screened with 1
µg/ml blasticidine S and maintained in 0.5 µg/ml blasticidine S
solution. All plasmids were confirmed by sequencing. Targeting
sequences for shRNAs are listed in Table SI. For miRNA overexpression, 10
pmol/ml double-stranded miR-522 mimics
(5′-aaaaugguucccuuuagagugu-3′) and negative control (micrON mimic
NC #22) oligonucleotides were purchased from Guangzhou Ruibio Co.,
Ltd., and transfected into LINC00238 overexpression
1×106 Huh7 cells by using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. At 24 h post-transfection, subsequent
experiments were performed.
Cell viability and plate colony
formation assays
A total of 1×103 cells/well, including
control or LINC00238 overexpression Huh7 cells, scramble or
LINC00238-shRNA HepG2 cells, NC or miR-522 transfected Huh7 or
LINC00238 overexpression Huh7 cells, were seeded into 96-well
plates. Cell viability was evaluated by a Cell Counting Kit-8
(CCK-8; Dojindo Molecular Technologies, Inc.). Briefly, 10 µl CCK-8
reagents were incubated with cells for 1 h, and then the absorbance
at 450 nm was measured with a microplate reader. For the plate
colony formation assay, 500 cells/well, including control or
LINC00238 overexpression Huh7 cells, scramble or LINC00238-shRNA
HepG2 cells, NC or miR-522 transfected Huh7 or LINC00238
overexpression Huh7 cells, were cultured in 6-well plates for 1
week. Colonies were fixed in 4% formaldehyde for 5 min at room
temperature and stained with 0.1% crystal violet for 10 min at room
temperature. Colonies numbers were counted by Image J software.
Cell migration and invasion
assays
Control or LINC00238 overexpression Huh7 cells,
scramble or LINC00238-shRNA HepG2 cells, NC or miR-522 transfected
Huh7 or LINC00238 overexpression Huh7 cells (1×104)
pretreated with 10 µg/ml mitomycin-C (Sigma-Aldrich; Merck KGaA)
for 1 h at 37°C were added to the upper chamber of a Transwell
plate (for invasion assay, Transwell membrane were precoated with
50 µl 1:40 diluted Matrigel for 1 h at 37°C) with an 8.0-µm pore
polycarbonate membrane insert in 100 µl RPMI-1640 containing 1%
FBS. A total of 500 µl RPMI-1640 with 10% FBS was added to the
lower chamber as a chemoattractant. After 24 h at 37°C in the cell
incubator, cells were fixed for 5 min with 4% formaldehyde at room
temperature and stained with 1% crystal violet for 1 min at room
temperature. After removing the cells inside the membrane, the
number of migrated and invasive cells was imaged in four randomly
selected microscopic fields under light microscope (Leica).
Immunoblotting
Cells or tumor tissues were lysed with
radioimmunoprecipitation assay (RIPA) buffer containing a protease
inhibitor cocktail (Roche Diagnostics). The protein concentration
was assessed with a BCA protein assay kit (Bio-Rad Laboratories,
Inc.). Equal amounts of protein for each sample (30 µg) were
separated on 10% SDS-PAGE gels and subsequently transferred to PVDF
membranes (Millipore Sigma). The membranes were blocked for 1 h in
PBS containing 5% non-fat milk at room temperature and then
incubated overnight with SFRP2 (cat. no. #4687, Cell Signaling
Technology, Inc.), DKK1 (cat. no. ab109416, Abcam) and GAPDH (cat.
no. #5174, Cell Signaling Technology, Inc.) antibodies (1:5,000) at
4°C. The membranes were then incubated with HRP-conjugated
secondary antibodies (1:50,000, #111-005-003, Jackson
ImmunoResearch) at room temperature for 1 h and visualized with ECL
detection reagents (Millipore Sigma). Images were captured by AI600
version 1.2.0 (GE Healthcare) on an Amersham Imager 600 (GE
Healthcare).
RNA pull-down
For the in vivo pull-down assay,
LINC00238-6×MS2bs plasmids in the pcDNA3.1 backbone (Invitrogen;
Thermo Fisher Scientific, Inc.) containing six repeat MS2-binding
site RNA sequences and MS2 expression plasmids with Flag tags in
the pcDNA3.1 backbone (Invitrogen; Thermo Fisher Scientific, Inc.)
were co-transfected into 1×106 293T cells. After 48 h
transfection cells were lysed by 500 µl NP40 cell lysis buffer
(Thermo Fisher Scientific, Inc.), then 500 µl lysate were incubated
with 50 µl Anti-FLAG® M2 Magnetic Beads (# M8823,
Sigma-Aldrich; Merck KGaA) overnight at 4°C. After washing with PBS
three times using magnetic separation rack at room temperature, the
pull-down product was eluted by 100 µl 3X FLAG® peptide.
Biotin-based RNA pull-down assays were carried out according to the
protocol (21). The pull-down
RNAs were isolated, purified and cDNA was synthesized, followed by
analysis via qPCR as aforementioned.
Reporter gene assay
Predicted consequential pairing of target region
‘CCAUUU’ and miR-522 were searched by Gene Runner software version
6.5.52 Beta (Frank Buquicchio and Michael Spruyt). The wild-type
and mutant LINC00238 were incorporated into the pGL3-control vector
(Promega Corporation) via the In-Fusion HD Cloning kit (Takara
Bio). To determine the relative luciferase activity,
1×104 HEK 293 cells were seeded into 24-well plates and
co-transfected with 500 ng pGL3-control containing LINC00238
wild-type or mutant, 26 ng pRL-TK plasmid (Promega Corporation)
expressing Renilla luciferase, and 20 pmol miR-522 mimics or
NC using Lipofectamine 2000 (n=4). Firefly and Renilla
luciferase activity in the cell lysates was measured 24 h after
transfection using a dual-luciferase reporter assay kit (Promega
Corporation). Firefly luciferase activity was normalized to that of
Renilla luciferase for each sample.
Statistical analysis
Experiments were performed in triplicate. Data were
analyzed with SPSS 26 (IBM Corp.) and GraphPad Prism 8 (GraphPad
Software, Inc.). The continuous variables with normal distribution
and equal variance (F test) between/within the groups are expressed
as the mean ± SD, and the statistical significance was determined
using an unpaired two-tailed Student's t-test between two groups,
one-way ANOVA followed by a Bonferroni post hoc test for multiple
comparisons and two-way ANOVA for cell viability analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of the downregulated
lncRNAs in liver cancer tissues
A total of 15 downregulated lncRNAs were screened
out in liver cancer tissues by GEPIA (FC>2,
Padj<0.01, Table
SII). Next, Kaplan-Meier survival analysis suggested that five
lncRNAs (LINC01554, LINC01093, LINC01018, LINC01370 and LINC00238)
were associated with the overall survival of patients after surgery
by the median expression level of each lncRNA as cut-off value
(Log-rank test P<0.1, Table
SII; Fig. 1D for LINC00238
and Fig. S1 for the remaining
lncRNAs). LINC01554 (22,23), LINC01093 (24,25) and LINC01018 (26,27) have been reported to have roles in
liver cancer. LINC00238 has been reported to be associated with HBV
infection (9), which accounts for
~50% of liver cancer cases worldwide (28). Recently, LINC00238 was also
reported as a tumor suppressor in liver cancer (29). Although Jiang reported that
LINC00238 inhibited liver cancer progression by activating the
TMEM106C-mediated apoptosis pathway through in vitro assays,
further experiments are still required, including in vivo
experiments on the subcellular location and other mechanisms
involved (29). Thus, LINC00238
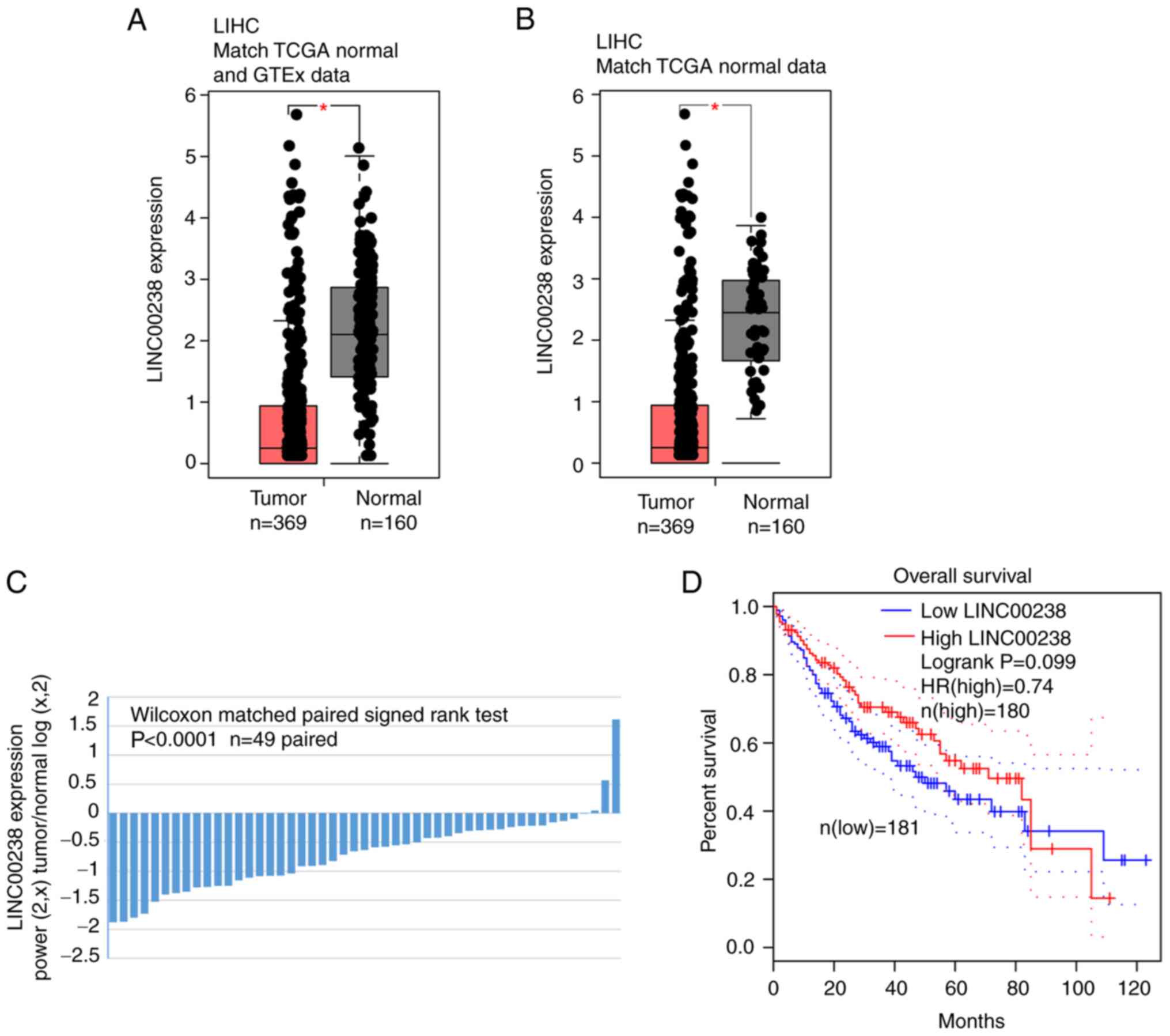
was selected for further study. As shown in Fig. 1A and B, the expression of
LINC00238 was significantly decreased in liver cancer tissues
compared with normal tissues according to the GEPIA website. In
addition, as indicated in Fig.
1C, LINC00238 was downregulated in 91.8% (45/49, P<0.0001)
of liver cancer tissues by paired Student's t-test analysis of
matched and normal tissues from TCGA dataset. K-M survival curves
indicated that patients with high LINC00238 expression showed
longer overall survival times (median survival 80 months vs. 50
months; hazard ratio, 0.74; P=0.099; Fig. 1D).
LINC00238 overexpression impairs the
liver cancer malignant phenotype in vitro
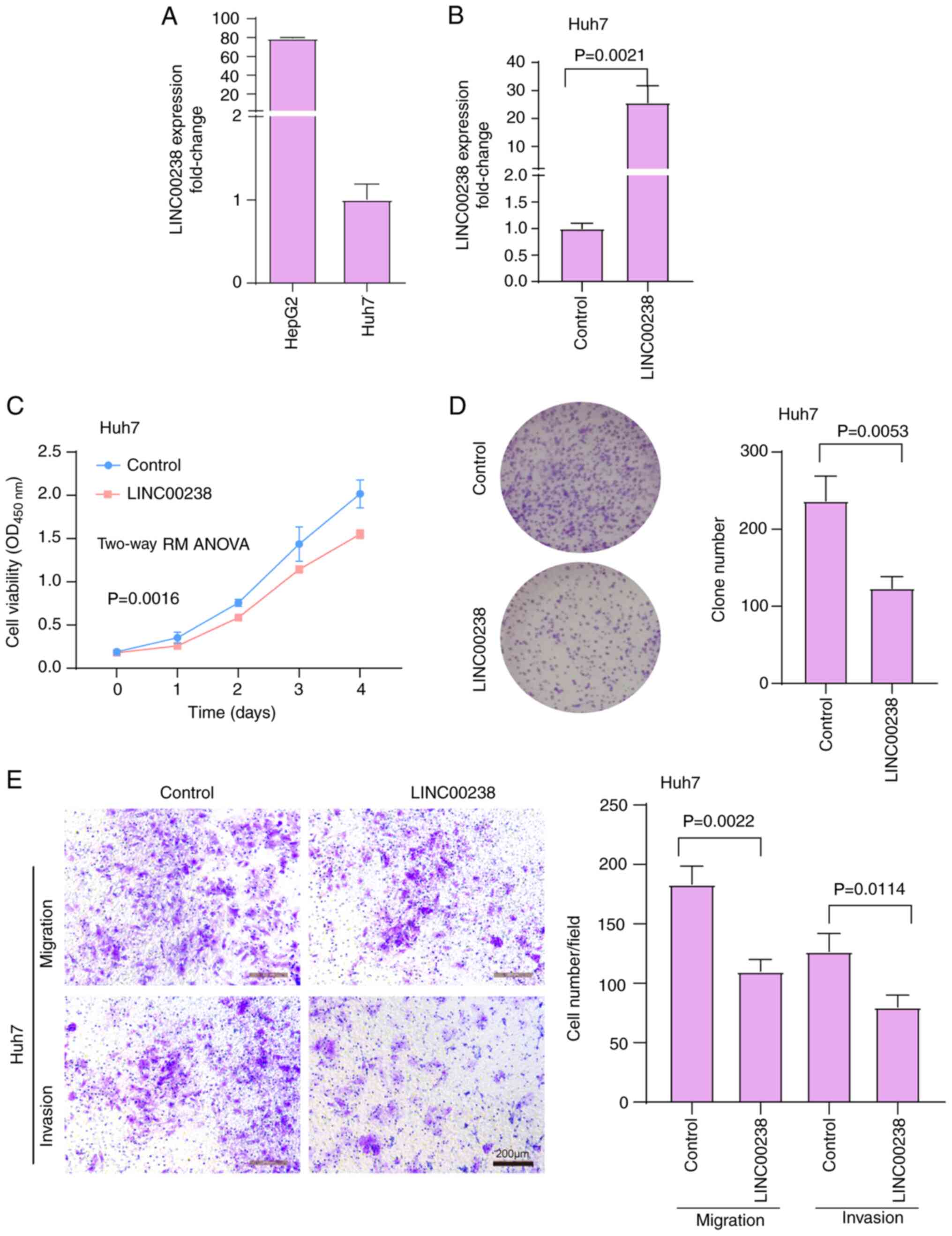
LINC00238 expression in human liver cancer cells,
including HepG2 and Huh7 cells, was then quantified. The relative
expression of LINC00238 in Huh7 cells was lower than that in HepG2
cells (Fig. 2A). Huh7 cells
(relatively low LINC00238 expression) were chosen for the
overexpression experiments and HepG2 cells (relatively high
LINC00238 expression) were chosen for the knockdown experiments.
Compared with control cells, LINC00238 expression was increased by
24-fold after overexpression, as detected by qPCR (Fig. 2B). Overexpression of LINC00238
reduced Huh7 cell viability (Fig.
2C) and colony formation (~47.9% inhibition; Fig. 2D). In addition, overexpression of
LINC00238 significantly decreased the migratory and invasive
abilities of Huh7 cells, as determined by Transwell assays (~40.0%
inhibition for cell migration and 36.9% inhibition for cell
invasion; Fig. 2E). These in
vitro findings suggested that LINC00238 might act as a tumor
suppressor in liver cancer.
Knockdown of LINC00238 promotes the
liver cancer malignant phenotype in vitro
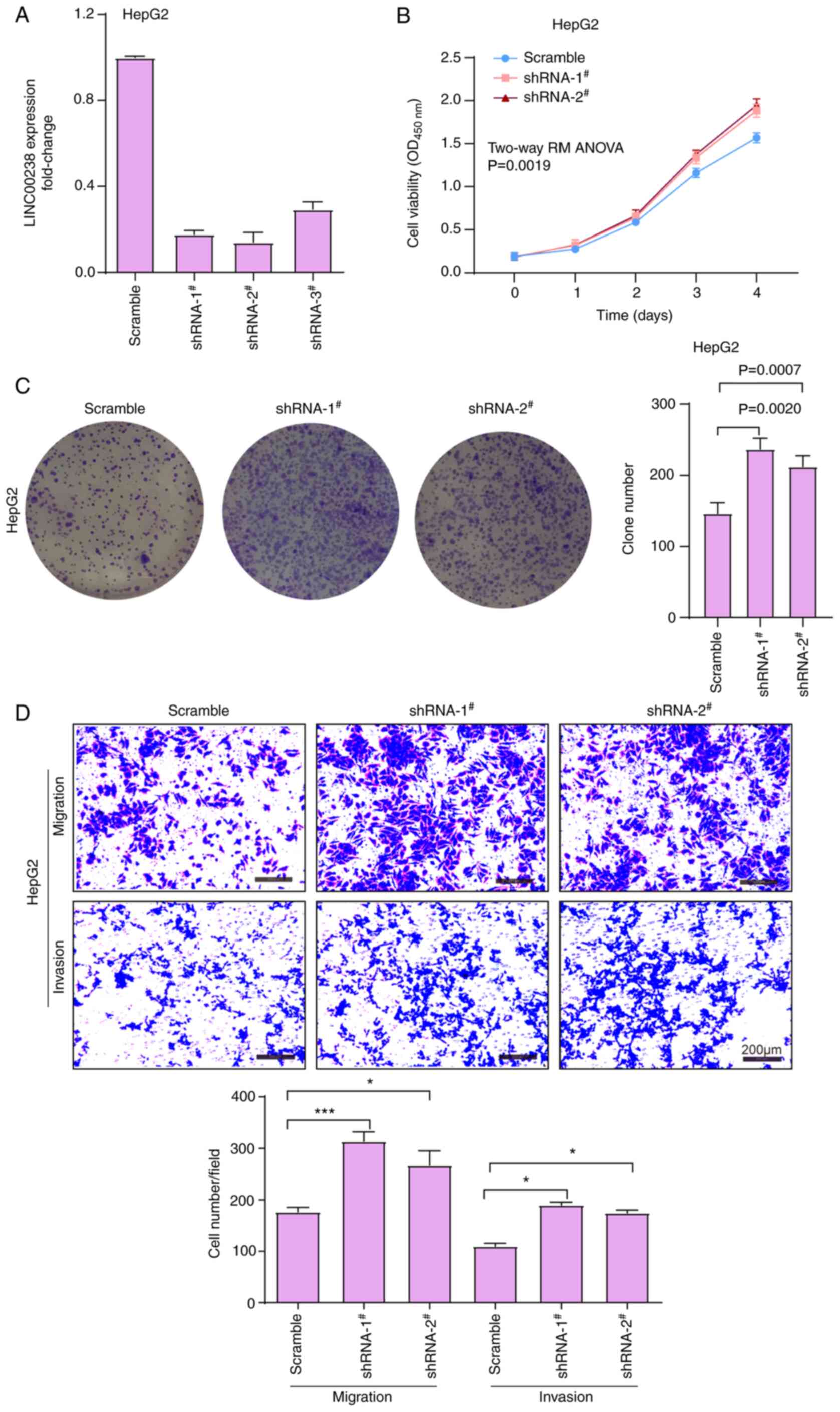
The expression of LINC00238 was knocked down in
HepG2 cells using three shRNAs targeting different sites. The three
shRNAs had 82, 88 and 72% knockdown efficiency, respectively
(Fig. 3A), and two silencers,
shRNA1 and shRNA2, were chosen in subsequent experiments. The CCK-8
and plate colony formation assay results demonstrated that
LINC00238 depletion promoted the viability (Fig. 3B) and colony formation ability of
HepG2 cells (Fig. 3C).
Furthermore, the Transwell assay results showed that the knockdown
of LINC00238 significantly enhanced the migratory and invasive
abilities of Hep2 cells compared with the scramble control group
(Fig. 3D).
LINC00238 functions as a tumor
suppressor in vivo
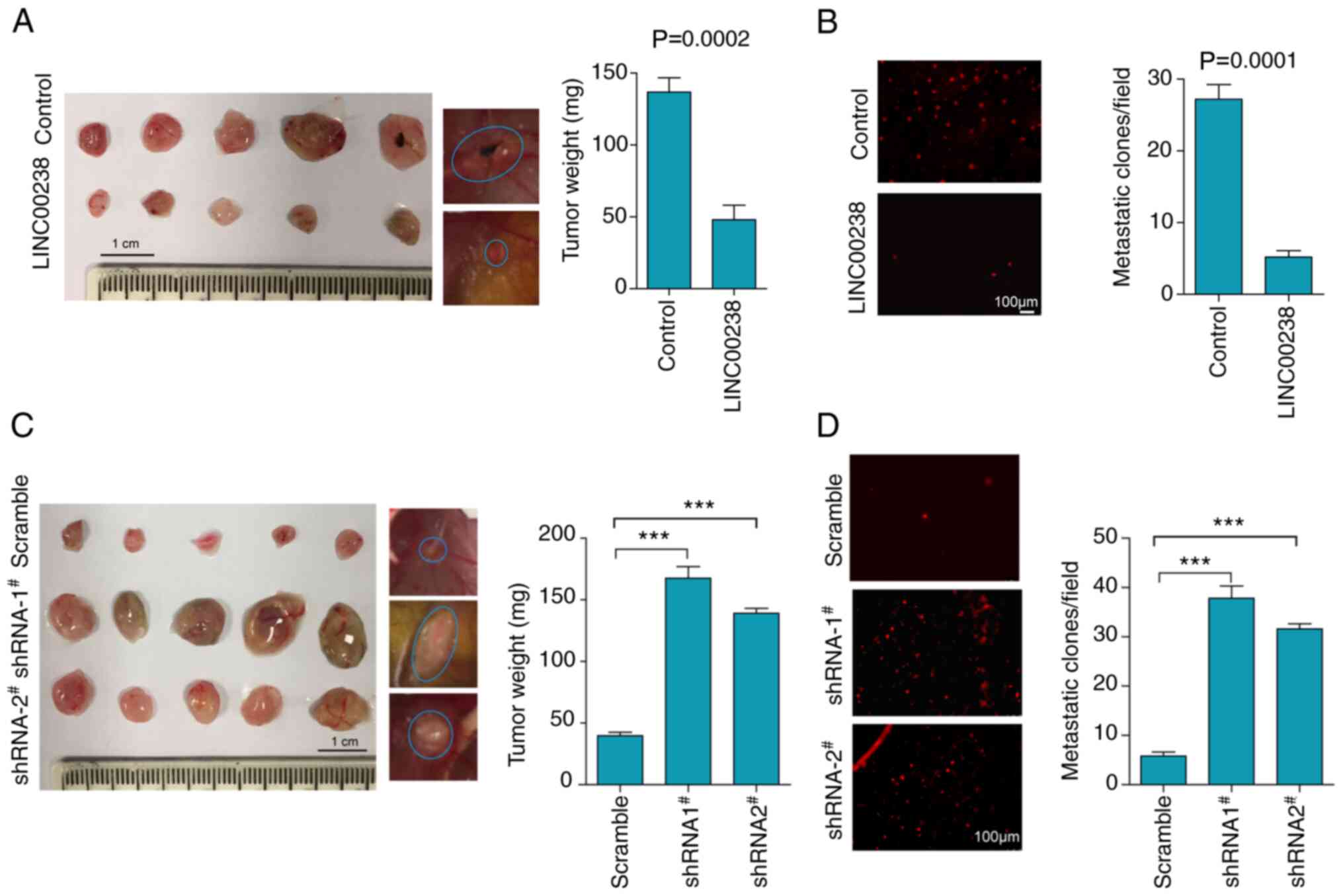
To further confirm the suppressive effect of
LINC00238 in liver cancer in vivo, a modified chick embryo
CAM assay was performed to assess tumor growth and metastasis.
Control and LINC00238-overexpressing Huh7 cells labeled with CM-DiI
(red fluorescent dye) were inoculated onto CAM to monitor tumor
growth and distant metastasis to lung tissues of chick embryos.
Compared with the control cells, the overexpression of LINC00238
resulted in a decrease in tumor weight by 64.8% (Fig. 4A). Moreover, the number of
metastatic tumor colonies, which represented metastatic ability to
lung tissues, was decreased by 80.9% in cells after LINC00238
overexpression (Fig. 4B).
Consistently, knockdown of LINC00238 expression resulted in a
significant increase in not only the tumor weight, but also the
extent of metastasis to the lungs (Fig. 4C and D) by CAM assay. Notably,
metastatic nodules in lung tissues of the LINC00238 shRNA groups
were significantly more abundant than those of the scramble group
(Fig. 4D). Hence, the reciprocal
effects of LINC00238 overexpression and knockdown both in
vitro and in vivo supported the idea that LINC00238
acted as a tumor suppressor in liver cancer.
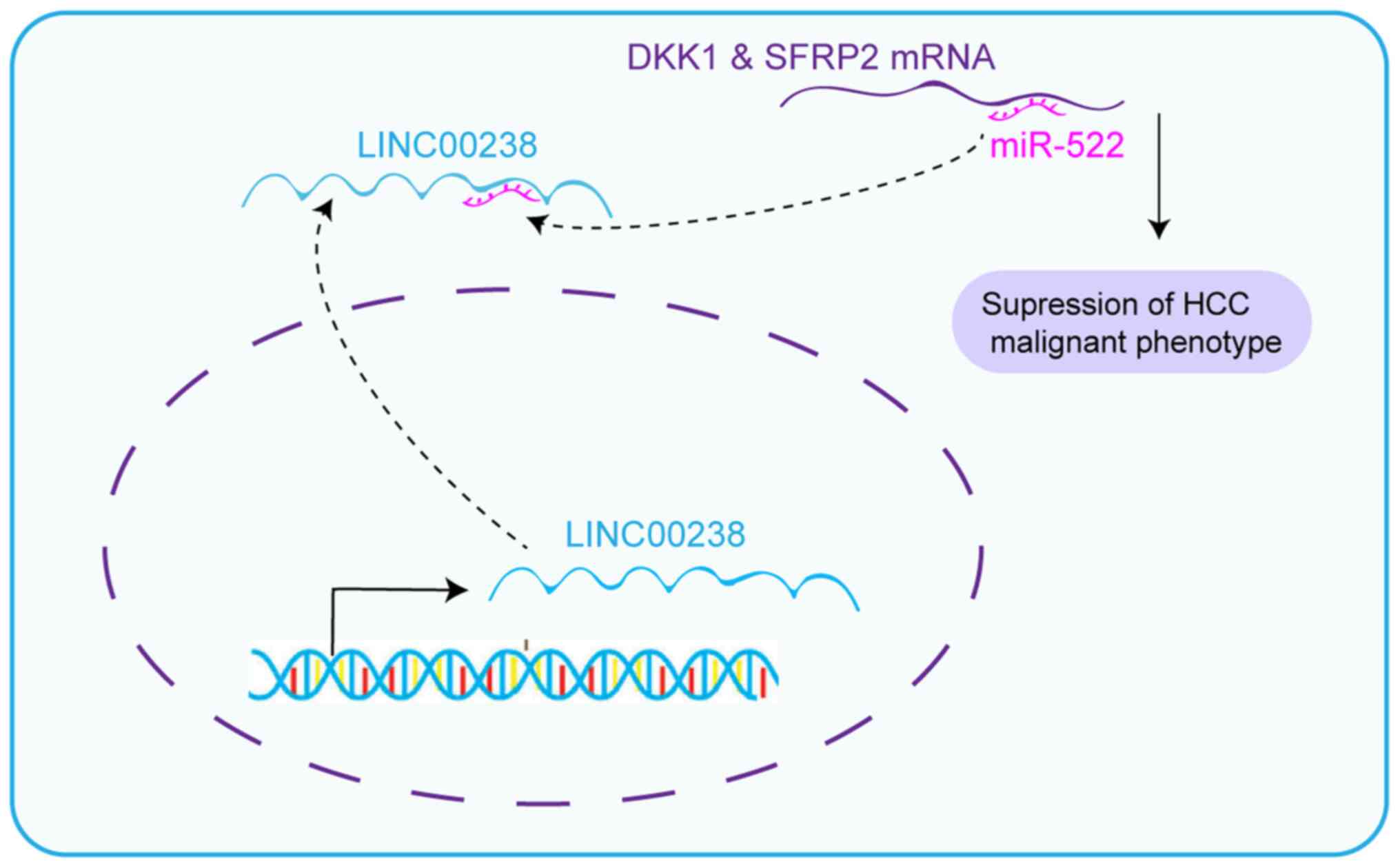
LINC00238 sponges miR-522
Cytoplasm lncRNAs can regulate cell phenotype by
sponging miRNAs (7). The
prediction by lncLocator (30,31)
(csbio.sjtu.edu.cn/bioinf/lncLocator/) predicted that the cytoplasm
locations score of 0.73, nucleus locations score of 0.03, ribosome
locations score of 0.04, cytosol locations score of 0.15 and
exosome locations score of 0.05, suggesting that LINC00238 was
predominantly located in the cytoplasm. Consistently, nuclear
cytoplasmic separation experiments confirmed that LINC00238 was
distributed mainly in the cytoplasm (Fig. 5A), which suggested that LINC00238
may sponge miRNAs. Gene Runner software predicted that the target
regions ‘CCAUUU’ of miR-522 were complementary with three isotypes
of LINC00238 (Fig. 5C).
Furthermore, the expression of miR-522 was decreased in
LINC00238-overexpressing Huh7 cells, but increased in LINC00238
knockdown HepG2 cells (Fig. 5B).
Then, RNA pull-down experiments suggested that the expression of
miR-522 was enriched ~8.3 times by LINC00238 compared with the
control group (Fig. 5D). In
addition, the expression of LINC00238 in the bio-miR-522 group was
~5.8 times that of the bio-NC group (Fig. 5E). Wild-type and mutant versions
of LINC00238 downstream of the luciferase gene were constructed
using the PGL3-control vector (Fig.
5F). Using a dual-luciferase reporter assay, overexpression of
miR-522 was associated with a decrease in the luciferase activity
of the LINC00238 wild-type vector, but had no effect on the mutant
vector (Fig. 5G). It was reported
previously that miR-522 contributes to the proliferation of liver
cancer cells by targeting two Wnt signaling inhibitors, DKK1 and
SFRP2 (32). Therefore, it was
next investigated whether LINC00238 could suppress the inhibitory
effect of miR-522 on these target genes. Consistent with this
hypothesis, SFRP2 and DKK1 were increased in Huh7 cell lines after
overexpression of LINC00238, but decreased in HepG2 cells after
knockdown of LINC00238 (Fig. 5H).
Taken together, LINC00238 may regulate the malignant phenotype in
liver cancer by sponging miR-522, which leads to release of the
inhibition of downstream targets of miR-522.
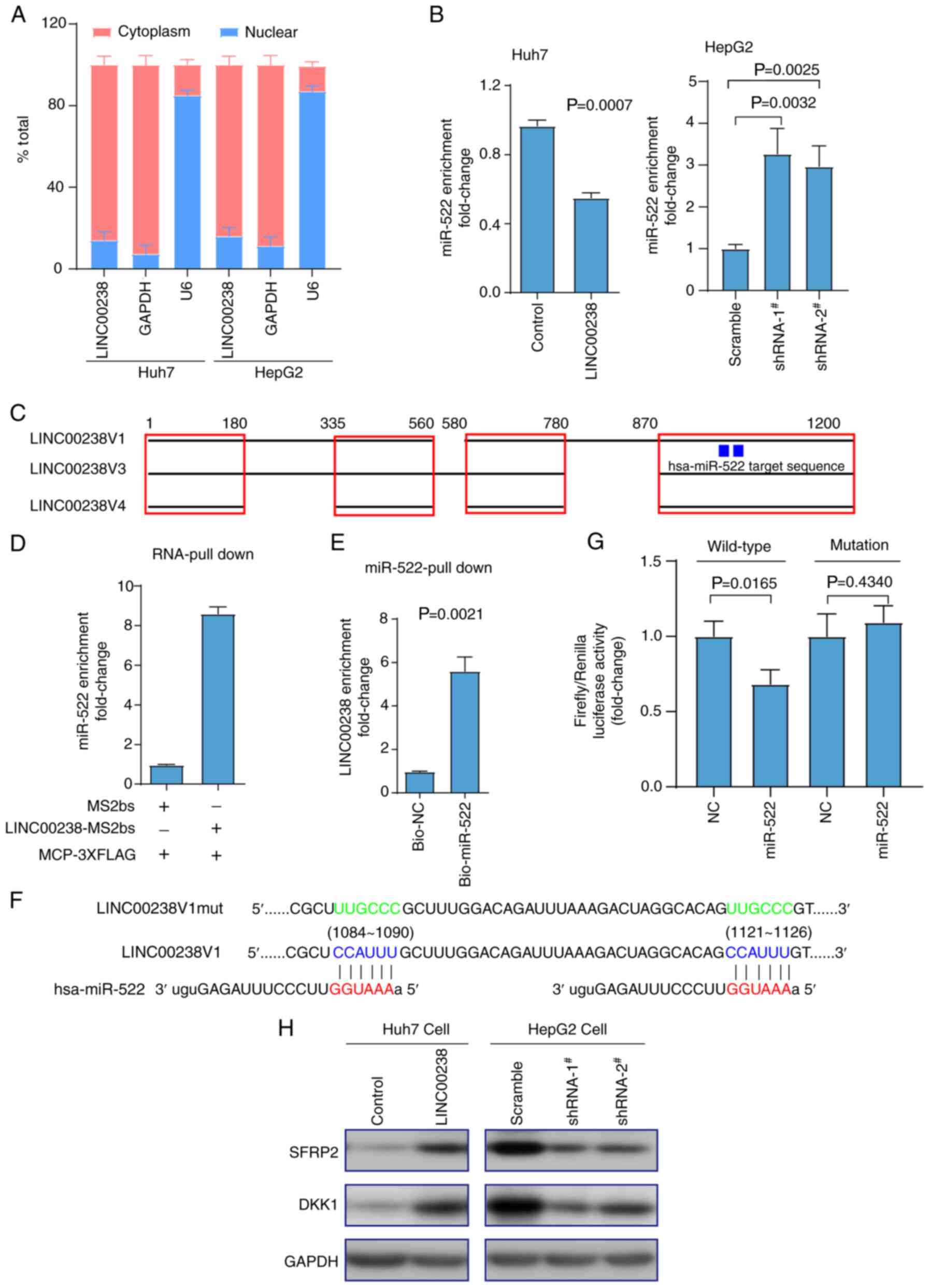 | Figure 5.Cytoplasmic LINC00238 sponges
miR-522. (A) The bar graphs show that LINC00238 was predominantly
localized in cytoplasmic Huh7 and HepG2 cells, as calculated via
reverse transcription-quantitative PCR. Cytoplasmic control (GAPDH)
and nuclear control (U6) were determined in their expected
localization. (B) The expression of miR-522 was downregulated in
LINC00238-overexpressing Huh7 cells, but upregulated in
LINC00238-knockdown HepG2 cells. (C) Schematic model indicating
that three variants of LINC00238 contain the target region of
miR-522. (D,E) In vivo and in vitro RNA pull-down
assays indicated the interaction between LINC00238 and miR-522. (F)
Complementary pairing and corresponding mutation between the
sequences of miR-522 and LINC00238. (G) Compared with NC, the
miR-522 mimic inhibited the relative fluorescence activity of the
LINC00238 wild-type PGL3-control plasmids, but not the mutant type.
(H) Western blotting detected that alterations in two known miR-522
targets (SFRP2 and DKK1) were upregulated in
LINC00238-overexpressing Huh7 cells and downregulated in
LINC00238-knockdown HepG2 cells. miR, microRNA; NC, negative
control; SFRP2, secreted frizzled related protein 2; DKK1,
dickkopf1; shRNA, short hairpin RNA. |
Overexpression of miR-522 partially
reverses the suppressive effects of LINC00238
To corroborate the role of miR-522, a
gain-of-function experiment in Huh7-LINC00238 cells was performed
by using miR-522 mimics (Fig.
6A). The expression of miR-522 was enhanced 163-fold and
186-fold after transfection with mimics in Huh7 and Huh7-LINC00238
cells, respectively. Consistent with our prediction and a previous
report (32), the overexpression
of miR-522 partially reversed the inhibitory effects of LINC00238
on Huh7 cell viability (Fig. 6B),
plate colony formation ability (Fig.
6C), migratory and invasive abilities (Fig. 6D), and tumor growth (Fig. 6E) and metastasis to the lungs
(Fig. 6F) in the CAM assay. These
results confirmed that LINC00238 functioned as a tumor suppressor
by sequestering miR-522 at least partially.
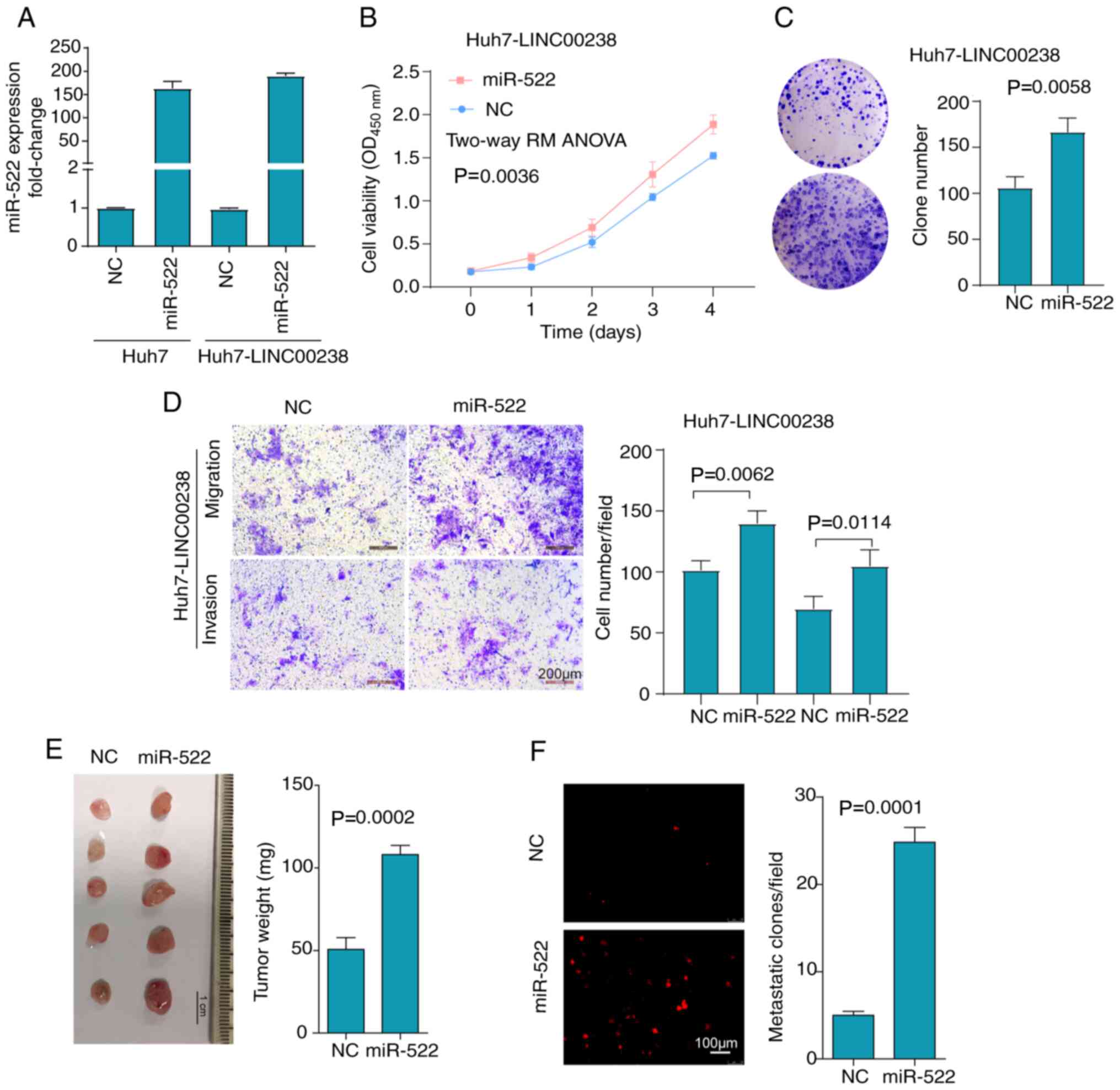 | Figure 6.Overexpression of miR-522 partially
reverses the suppressive effects of LINC00238. (A) Huh7 cells and
LINC00238-overexpressing Huh7 cells were transfected with miR-522
mimics or NC. miR-522 expression was detected via reverse
transcription-quantitative PCR. Overexpression of miR-522 in
LINC00238-overexpressing Huh7 cells partially reversed the
inhibitory effect of LINC00238 on not only (B) cell viability, as
determined by Cell Counting Kit-8 assays, (C) cell colony
formation, (D) and cell migratory and invasive abilities, as
detected by Transwell assays, but also (E) tumor growth and (F)
lung metastasis, as determined by the chorioallantoic membrane
model. Scale bar, 200 µm. miR, microRNA; NC, negative control;
SFRP2, secreted frizzled related protein 2; DKK1, dickkopf1. |
Discussion
In the present study, a suppressive function for
LINC00238 in the regulation of the malignant phenotype of liver
cancer was proposed. These results suggested that LINC00238
expression was decreased in liver cancer tissues and served as a
competing endogenous RNA (ceRNA) that inhibited tumor cell growth
and migration via sponging miR-522, which relieved the inhibition
of two tumor suppressor targets of miR-522, SFRP2 and DKK1
(Fig. 7).
In the present study, DEGs in liver cancer and
normal tissues were assessed by using a web server for cancer and
normal gene expression profiling and interactive analyses, GEPIA
(11). Among these DEGs, 15
downregulated known lncRNAs were screened out by survival curve
analysis, and LINC00238 was selected for further study. Log-rank
test supplied by GEPIA was used for the analysis of survival
curves. Although there are often obvious violations of the
proportional hazard rates due to late-stage crossover, the log-rank
test is still used in 70% of studies (33). Thus, whether the results of the
survival curve for lncRNAs where there is late-stage crossover are
significant in the current study need further confirmation either
by restricting the analyzed period of time to exclude this late
crossover event or using a weighted test, such as Renyi or
Cramer-von Mises (33). LINC00238
has been reported to be significantly downregulated in liver
tissues and liver cells after HBV infection (9). Notably, chronic HBV infection
accounts for ~50% of liver cancer cases worldwide (28). Recently, LINC00238 has been
reported in liver cancer as a tumor suppressor by activating the
apoptosis pathway (29). However,
further experiments, including in vivo assays, are still
required to support the suppressive functional role of LINC00238,
and determine its subcellular localization and underlying
mechanisms. In the current study, consistent with a previous report
(29), the in vitro
experiments validated LINC00238 as a tumor suppressor. In addition,
its tumor suppressor role was further validated by in vivo
experiments and a novel mechanism as a ceRNA via the sponging
miR-522 was reported. Although LINC00238 was confirmed as a
predictor of clinical prognosis from the databank, further studies
are needed to confirm LINC00238 as a clinical predictor from our
own cohort of patients. A considerable number of studies have
examined whether lncRNAs, either as oncogenes or suppressor genes,
function as ceRNAs in liver cancer. For example, HOXD-AS1, as an
oncogene, sponges regulatory miR-130a-3p to enhance the expression
of the transcription factor SOX4, thus resulting in the promotion
of liver cancer metastasis (34).
LncRNA CASC2, a novel tumor suppressor, exerts antimetastatic
effects through the miR-367/FBXW7 axis in liver cancer cells
(35). Previous work suggests
that lncRNAs in the cytoplasm function as molecular sponges or
modulate mRNA stability (6). In
the present study, LINC00238 was identified to be located in the
cytoplasm and considered to be a ceRNA that regulates liver cancer
progression. The RNA binding immunoprecipitation assay suggested
that LINC00238 had the potential to interact with miR-522. Dual
luciferase reporter and biotin-miR-522 pull-down assays also
revealed that LINC00238 was a target of miR-522. It is commonly
known that miR-522 is an oncogene in a number of cancers, including
liver (36) and lung (37) cancer. SFRP2 and DKK1, two known
Wnt signaling inhibitors involved in HCV-induced multistep
hepatocarcinogenesis (38), have
already been identified as two direct targets of miR-522 in liver
cancer cells (32). In the
current study, although it was demonstrated that LINC00238 sponged
miR-522, and that the expression of miR-522 was decreased in
LINC00238-overexpression Huh7 cells and increased in
LINC00238-knockdown HepG2 cells, further study to determine the
associations between the expression levels of LINC00238 and miR-522
in patients with liver cancer are required.
In summary, it was speculated that the suppressive
effects of LINC00238 on the liver cancer malignant phenotype may be
due to miR-522-mediated regulation of SFRP2 and DKK1. The ceRNA
regulatory network (LINC00238/miR-522/SFRP2 and DKK1) shed light on
the mechanisms of lncRNA regulation of liver cancer development. It
is worth noting that LINC00238 may also regulate liver cancer
progression through other mechanisms, such as the regulation of
mRNA stability.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HQ and CH designed and performed the research, wrote
the manuscript and confirm the authenticity of all the raw data. QW
analyzed the data from The Cancer Genome Atlas. JW, XT and WX
analyzed the data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
TCGA
|
The Cancer Genome Atlas
|
|
LIHC
|
liver hepatocellular carcinoma
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
CCK-8
|
Cell Counting Kit-8
|
|
SFRP2
|
secreted frizzled related protein
2
|
|
DKK1
|
dickkopf1
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anwanwan D, Singh SK, Singh S, Saikam V
and Singh R: Challenges in liver cancer and possible treatment
approaches. Biochim Biophys Acta Rev Cancer. 1873:1883142020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mikhail S, Cosgrove D and Zeidan A:
Hepatocellular carcinoma: Systemic therapies and future
perspectives. Expert Rev Anticancer Ther. 14:1205–1218. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lim LJ, Wong SYS, Huang F, Lim S, Chong
SS, Ooi LL, Kon OL and Lee CG: Roles and regulation of long
noncoding RNAs in hepatocellular carcinoma. Cancer Res.
79:5131–5139. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qin H, Zhenzhen Z and Quanbo L: Effect of
long intergenic non-coding RNA 238 on HBV replication and
expression in vitro. J Third Military Med Univ. 40:1942–1947.
2018.
|
|
10
|
Gao Q, Zhu H, Dong L, Shi W, Chen R, Song
Z, Huang C, Li J, Dong X, Zhou Y, et al: Integrated Proteogenomic
characterization of HBV-related hepatocellular carcinoma. Cell.
179:561–577.e22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ribatti D: The Chick Embryo
Chorioallantoic Membrane in the Study of Angiogenesis and
Metastasis. Springer; Netherlands: 2010, View Article : Google Scholar
|
|
13
|
Pinto MT, Ribeiro AS, Conde I, Carvalho R
and Paredes J: The chick chorioallantoic membrane model: A new in
vivo tool to evaluate breast cancer stem cell activity. Int J Mol
Sci. 22:3342020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zabielska K, Lechowski R, Król M,
Pawłowski KM, Motyl T, Dolka I and Zbikowski A: Derivation of
feline vaccine-associated fibrosarcoma cell line and its growth on
chick embryo chorioallantoic membrane-a new in vivo model for
veterinary oncological studies. Vet Res Commun. 36:227–233. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cimpean AM, Lalosevic D, Lalosevic V,
Banovic P, Raica M and Mederle OA: Disodium Cromolyn and
Anti-podoplanin antibodies strongly inhibit growth of BHK
21/C13-derived Fibrosarcoma in a chick Embryo chorioallantoic
membrane model. In Vivo. 32:791–798. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Klingenberg M, Becker J, Eberth S, Kube D
and Wilting J: The chick chorioallantoic membrane as an in vivo
xenograft model for Burkitt lymphoma. BMC Cancer. 14:3392014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li M, Pathak RR, Lopez-Rivera E, Friedman
SL, Aguirre-Ghiso JA and Sikora AG: The In Ovo chick
chorioallantoic membrane (CAM) assay as an efficient Xenograft
model of hepatocellular carcinoma. J Vis Exp. 524112015.doi:
10.3791/52411. PubMed/NCBI
|
|
18
|
Avram S, Coricovac DE, Pavel IZ, Pinzaru
I, Ghiulai R, Baderca F, Soica C, Muntean D, Branisteanu DE,
Spandidos DA, et al: Standardization of A375 human melanoma models
on chicken embryo chorioallantoic membrane and Balb/c nude mice.
Oncol Rep. 38:89–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wilson SM and Chambers AF: Experimental
metastasis assays in the chick embryo. Curr Protoc Cell Biol.
Chapter 19: Unit 19.6. 2004.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dash S, Balasubramaniam M, Dash C and
Pandhare J: Biotin-based Pulldown assay to validate mRNA targets of
cellular miRNAs. J Vis Exp. 577862018.doi: 10.3791/57786.
PubMed/NCBI
|
|
22
|
Zheng YL, Li L, Jia YX, Zhang BZ, Li JC,
Zhu YH, Li MQ, He JZ, Zeng TT, Ban XJ, et al: LINC01554-mediated
glucose metabolism reprogramming suppresses tumorigenicity in
hepatocellular carcinoma via downregulating PKM2 expression and
inhibiting Akt/mTOR signaling pathway. Theranostics. 9:796–810.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding Y, Sun Z, Zhang S, Chen Y, Zhou B, Li
G, Sun Q, Zhou D, Ge Y, Yan S and Wang W: Down-regulation of Long
Non-coding RNA LINC01554 in hepatocellular cancer and its clinical
significance. J Cancer. 11:3369–3374. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He J, Zuo Q, Hu B, Jin H, Wang C, Cheng Z,
Deng X, Yang C, Ruan H, Yu C, et al: A novel, liver-specific long
noncoding RNA LINC01093 suppresses HCC progression by interaction
with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett.
450:98–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng Y, Yu K, Huang C, Liu L, Zhao H, Huo
M and Zhang J: Integrated bioinformatics analysis reveals role of
the LINC01093/miR-96-5p/ZFAND5/NF-κB signaling axis in
hepatocellular carcinoma. Exp Ther Med. 18:3853–3860.
2019.PubMed/NCBI
|
|
26
|
Wang S, Xu M, Sun Z, Yu X, Deng Y and
Chang H: LINC01018 confers a novel tumor suppressor role in
hepatocellular carcinoma through sponging microRNA-182-5p. Am J
Physiol Gastrointest Liver Physiol. 317:G116–G126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wen Q, Wang D, Yang Y, Chen X, Pan X, Han
Q, Deng Y, Li X, Chen X, Yan J and Zhou J: Competing endogenous RNA
screening based on long noncoding RNA-messenger RNA co-expression
profile in Hepatitis B virus-associated hepatocarcinogenesis. J
Tradit Chin Med. 37:510–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie Y: Hepatitis B Virus-associated
hepatocellular carcinoma. Adv Exp Med Biol. 1018:11–21. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang C, Li F, Yang M, Duan J, Lai J, Sun
S and Fan S: LINC00238 inhibits hepatic carcinoma progression by
activating TMEM106C-mediated apoptosis pathway. Mol Med Rep.
24:7572021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin Y, Pan X and Shen HB: lncLocator 2.0:
A cell-line-specific subcellular localization predictor for long
non-coding RNAs with interpretable deep learning. Bioinformatics.
37:2308–2316. 2021. View Article : Google Scholar
|
|
31
|
Cao Z, Pan X, Yang Y, Huang Y and Shen HB:
The lncLocator: A subcellular localization predictor for long
non-coding RNAs based on a stacked ensemble classifier.
Bioinformatics. 34:2185–2194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang H, Yu C, Chen M, Li Z, Tian S, Jiang
J and Sun C: miR-522 contributes to cell proliferation of
hepatocellular carcinoma by targeting DKK1 and SFRP2. Tumour Biol.
37:11321–11329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang
C, Dou C, Xu M, Liu Q and Tu K: Long non-coding RNA CASC2
suppresses epithelial-mesenchymal transition of hepatocellular
carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer.
16:1232017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi YH, Qi BB, Liu XB and Ding HM:
Upregulation of miR-522 is associated with poor outcome of
hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 20:3194–3198.
2016.PubMed/NCBI
|
|
37
|
Zhang T, Hu Y, Ju J, Hou L, Li Z, Xiao D,
Li Y, Yao J, Wang C, Zhang Y and Zhang L: Downregulation of miR-522
suppresses proliferation and metastasis of non-small cell lung
cancer cells by directly targeting DENN/MADD domain containing 2D.
Sci Rep. 6:193462016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Umer M, Qureshi SA, Hashmi ZY, Raza A,
Ahmad J, Rahman M and Iqbal M: Promoter hypermethylation of Wnt
pathway inhibitors in hepatitis C virus-induced multistep
hepatocarcinogenesis. Virol J. 11:1172014. View Article : Google Scholar : PubMed/NCBI
|