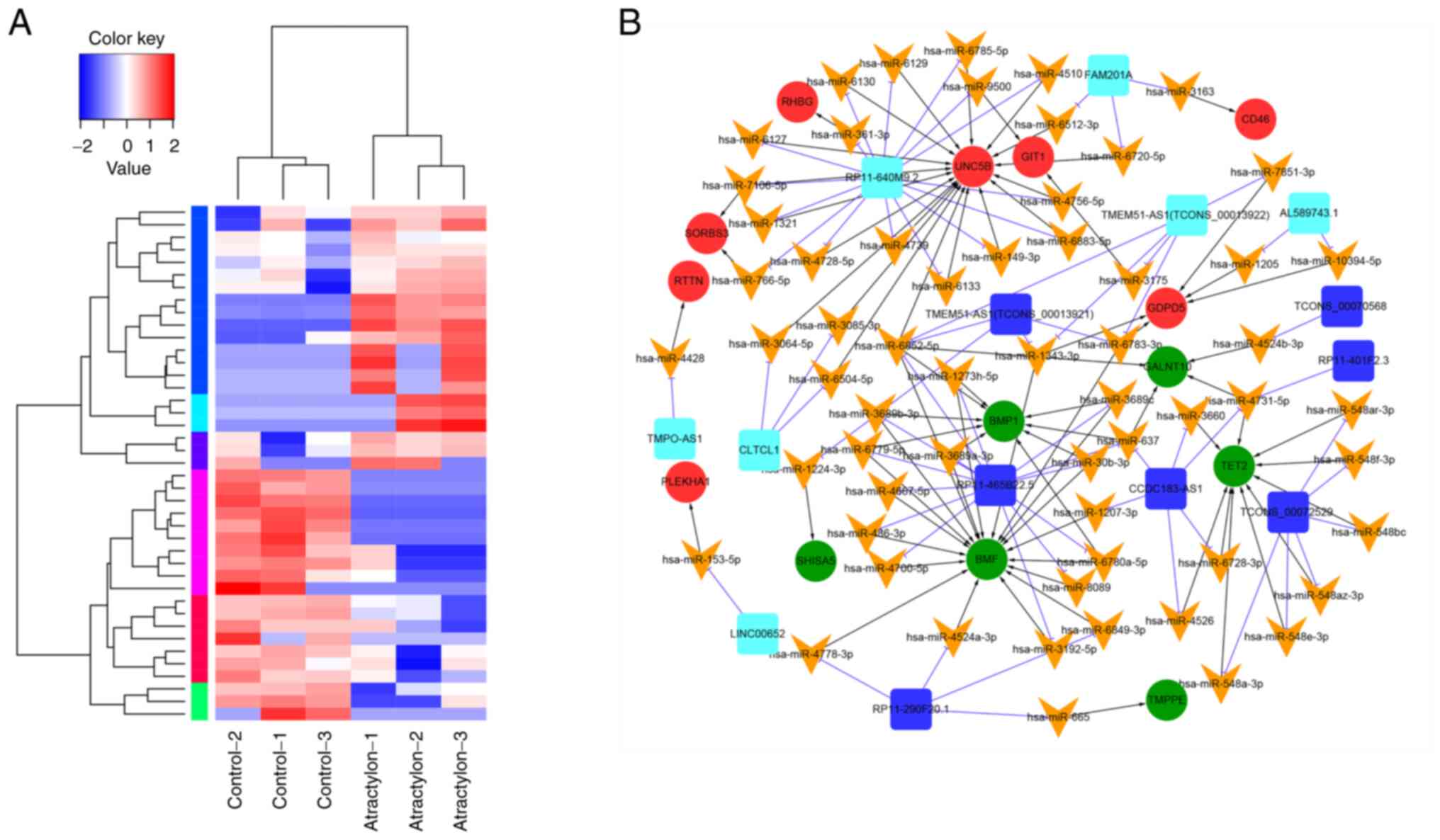
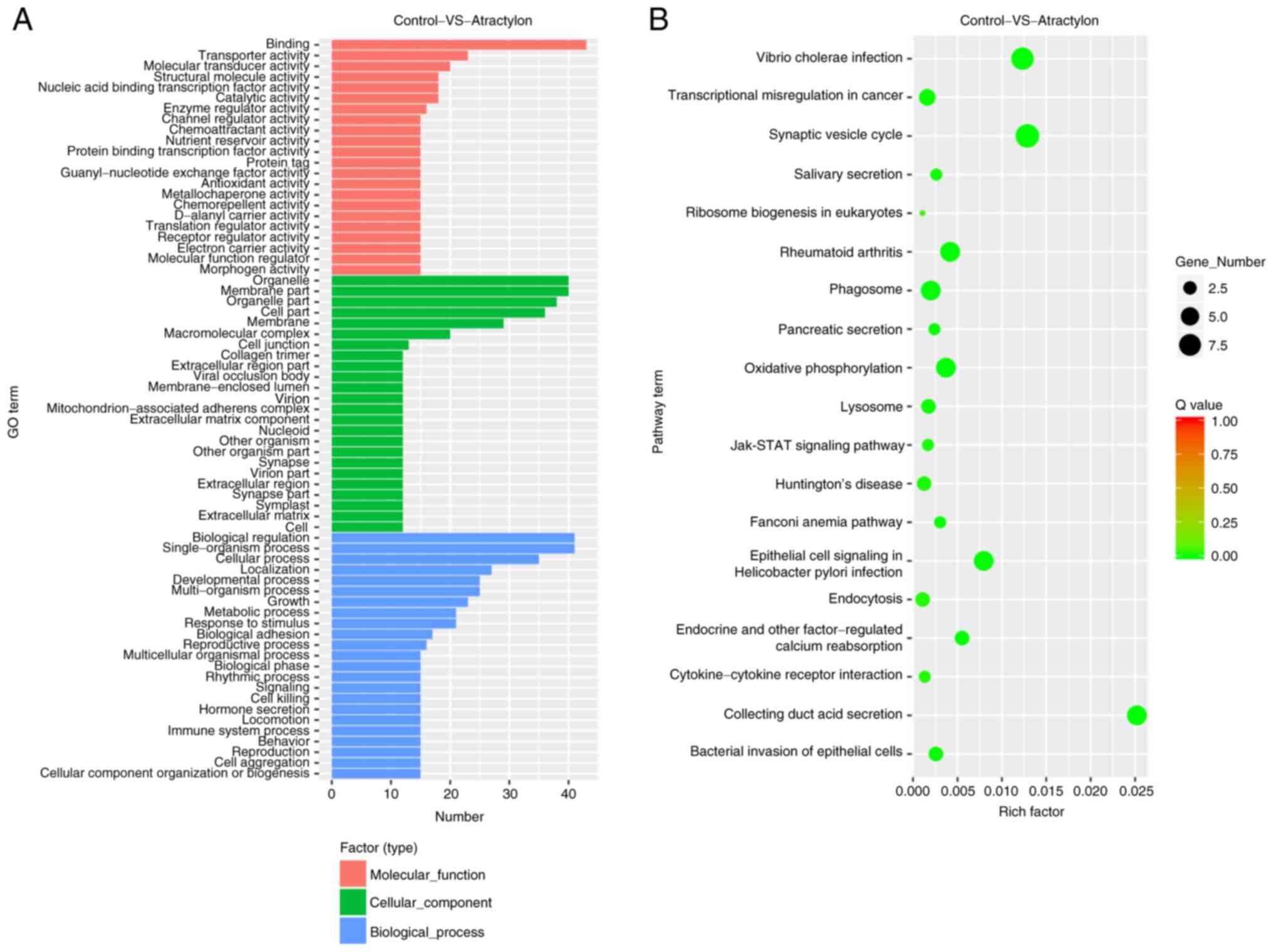
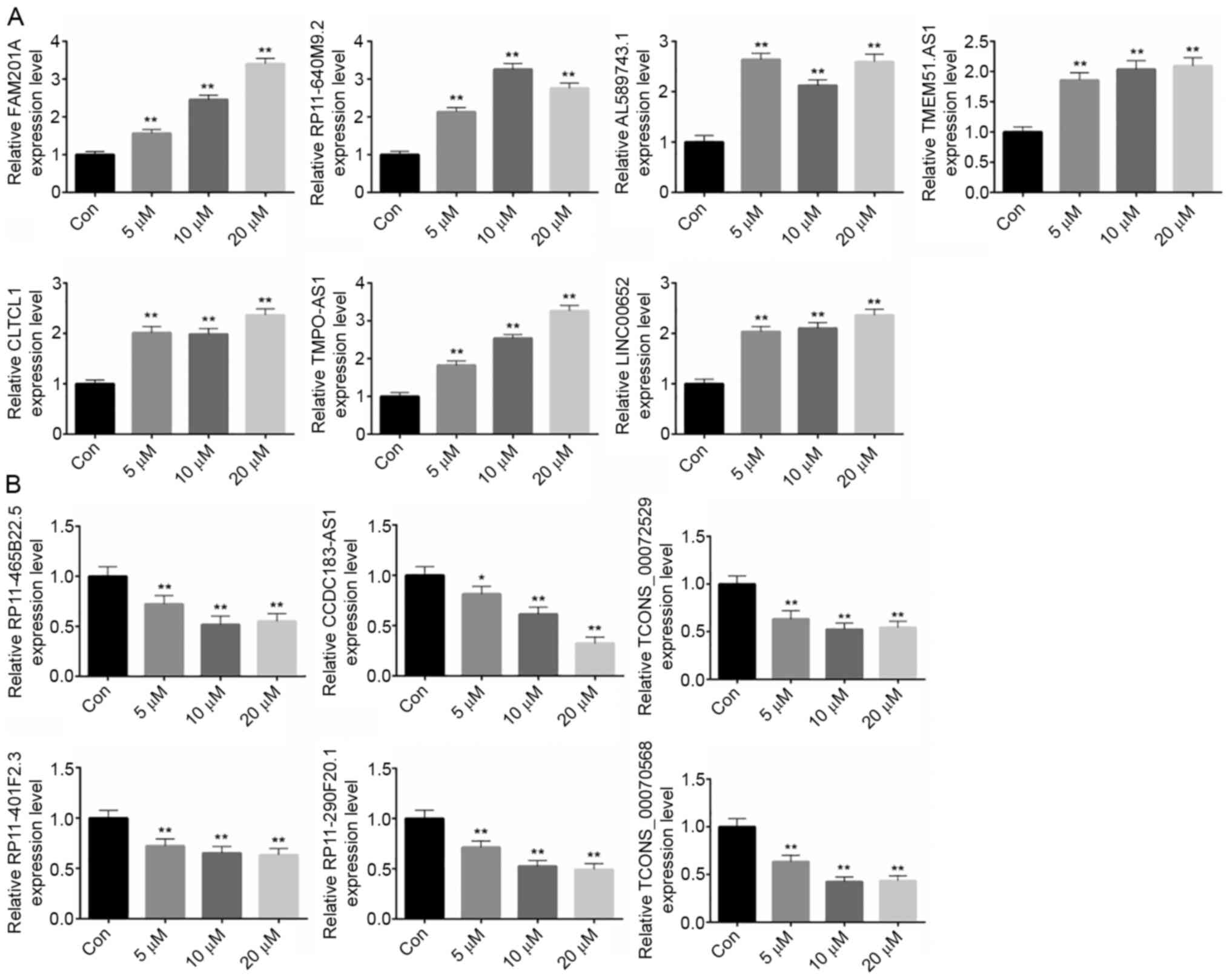
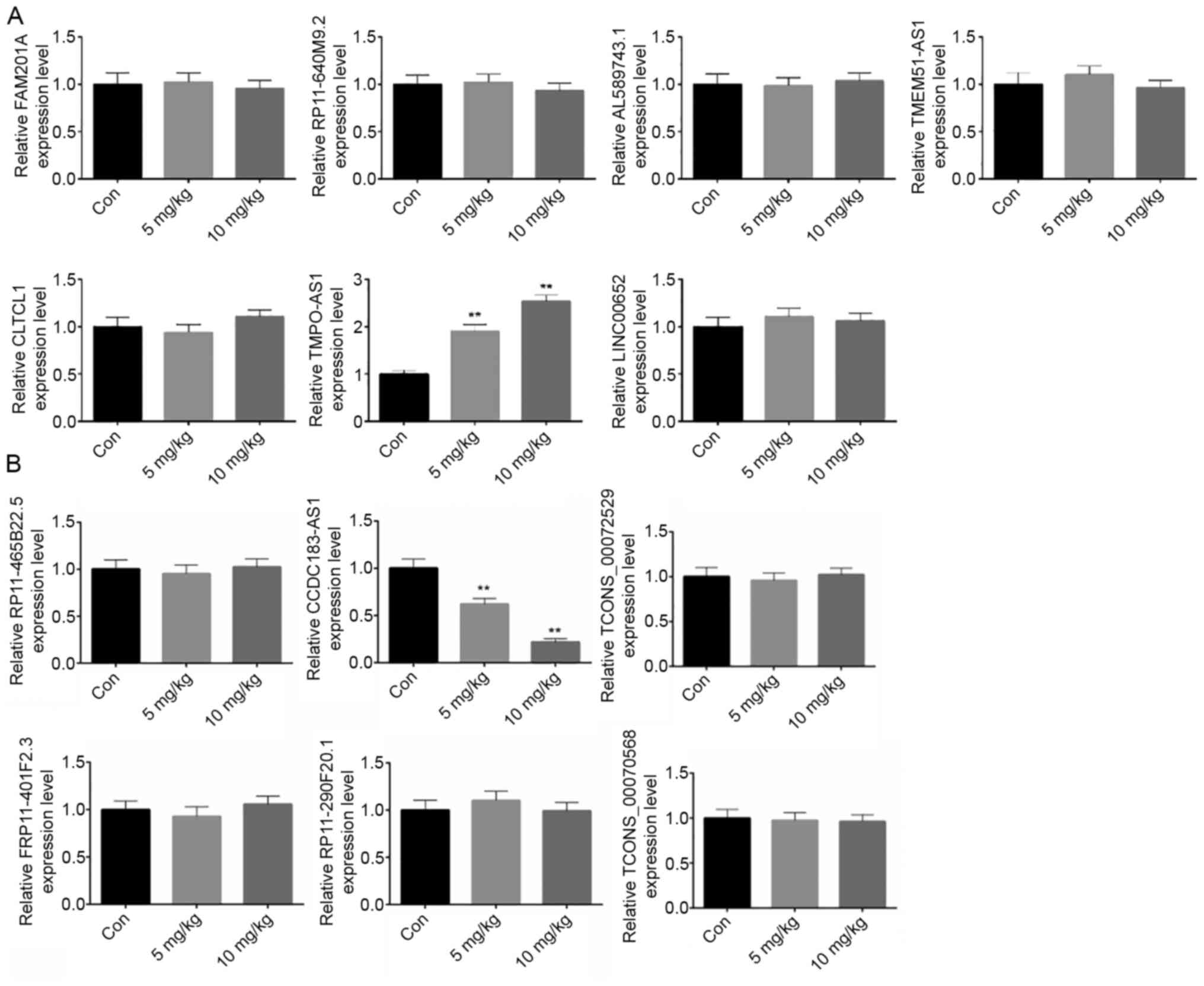
|
1
|
An L, Zeng HM, Zheng RS, Zhang SW, Sun KX,
Zou XN, Chen R, Wang SM, Gu XY, Wei WW and He J: Liver cancer
epidemiology in china, 2015. Zhonghua Zhong Liu Za Zhi. 41:721–727.
2019.(In Chinese). PubMed/NCBI
|
|
2
|
Ding C, Fu X, Zhou Y, Liu X, Wu J, Huang
C, Deng M, Li Y, Li L and Yang S: Disease burden of liver cancer in
China from 1997 to 2016: An observational study based on the global
burden of diseases. BMJ Open. 9:e0256132019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fukui N, Golabi P, Otgonsuren M, de Avila
L, Bush H and Younossi ZM: Hospice care in medicare patients with
primary liver cancer: The impact on resource utilisation and
mortality. Aliment Pharmacol Ther. 47:680–688. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
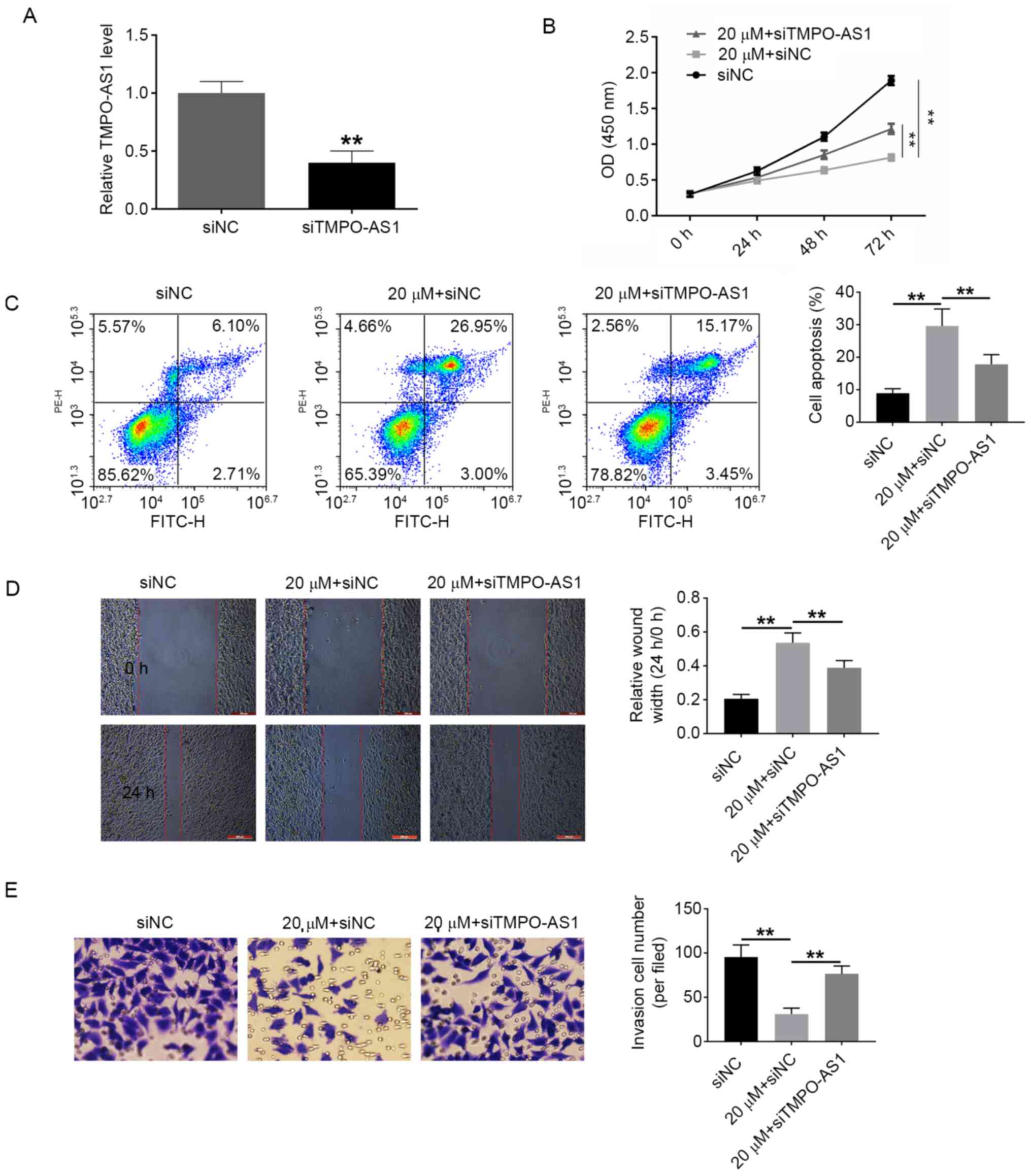
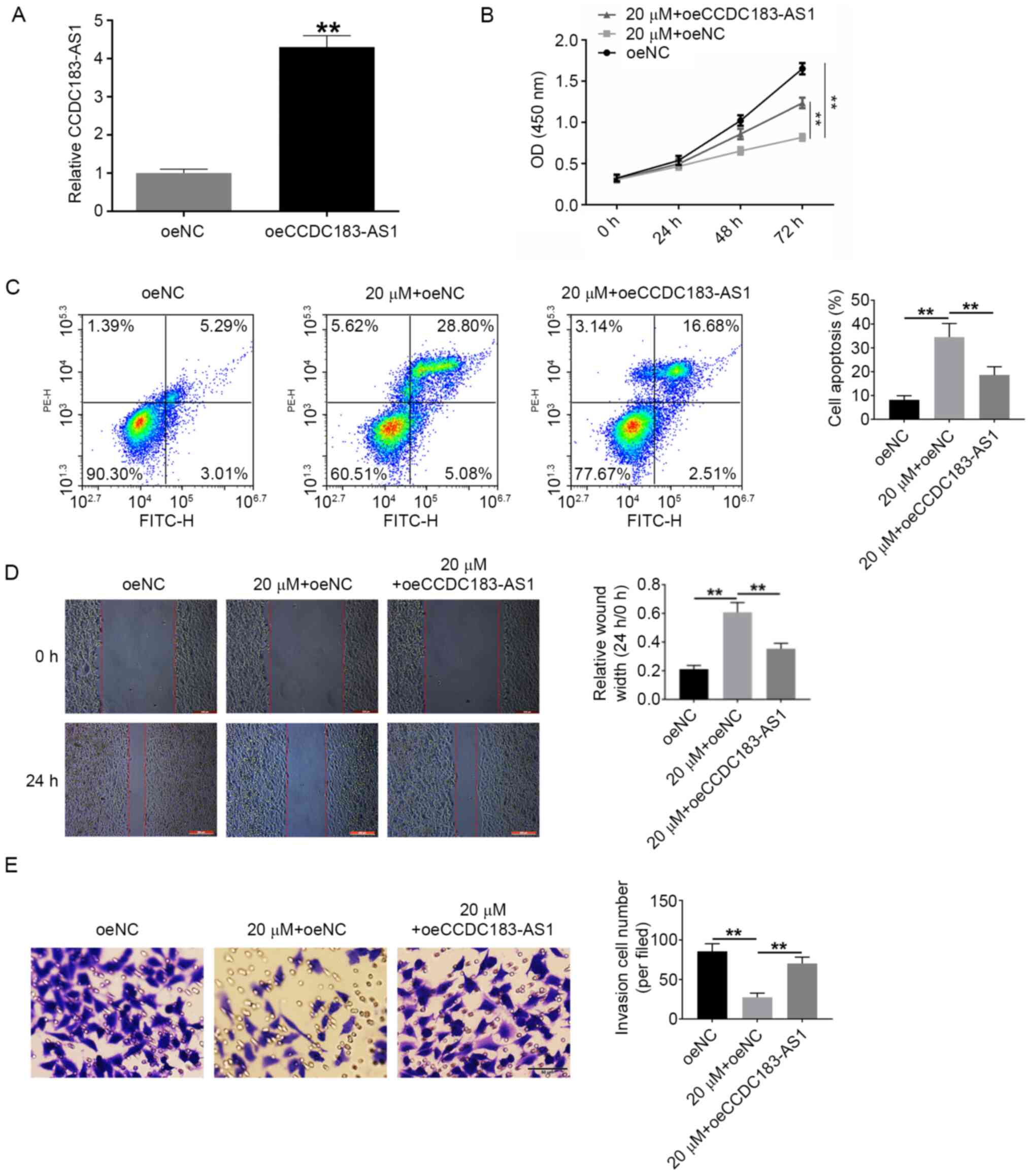
Fisher RA, Maluf DG, Wolfe L, Williams B,
Cotterell A, Stravitz RT, Heuman D and Posner M: Is hepatic
transplantation justified for primary liver cancer? J Surg Oncol.
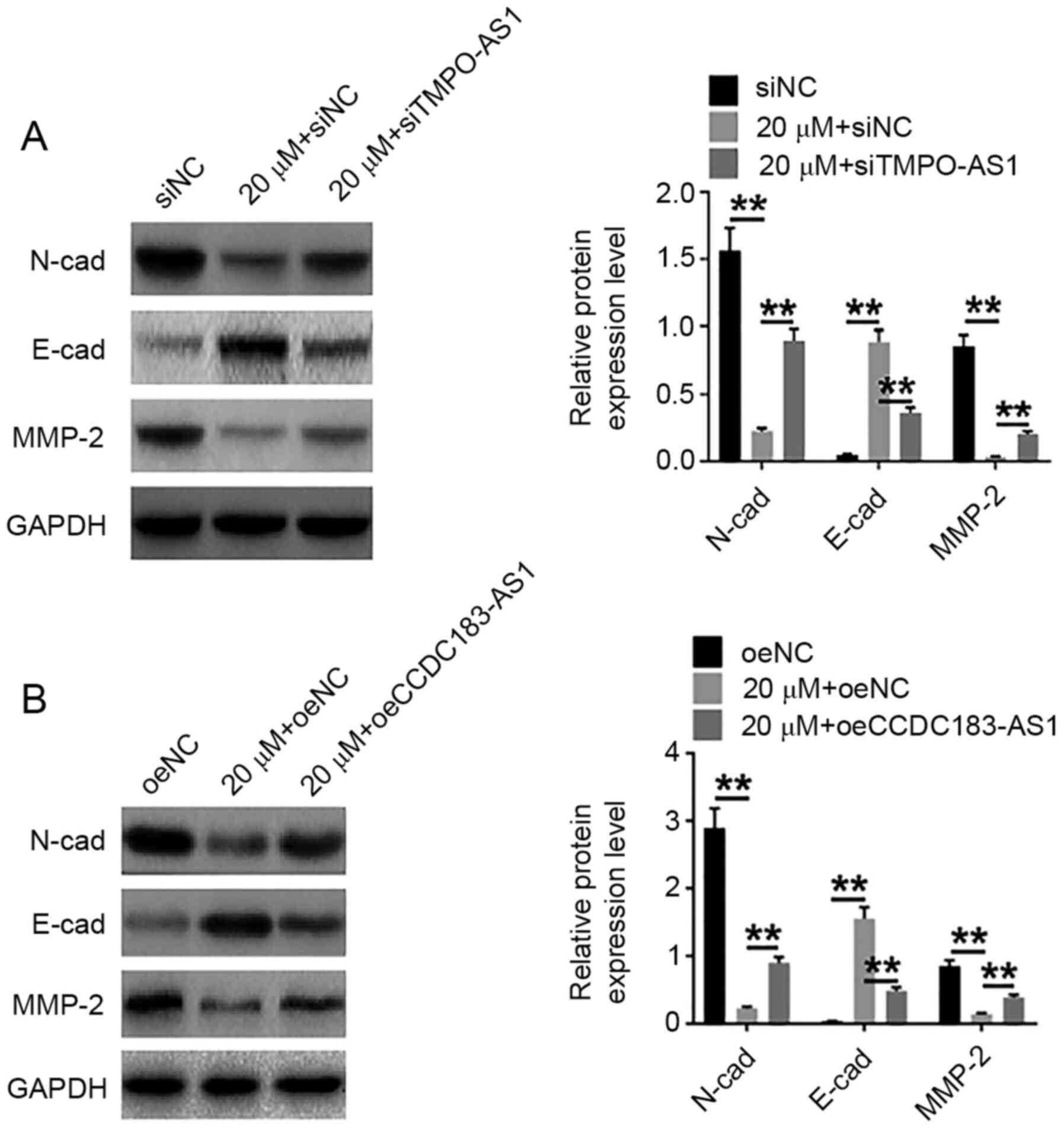
95:674–679. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan YX, Chen JC, Fang AP, Wang XH, Chen
JB, Wang JC, He W, Fu YZ, Xu L, Chen MS, et al: A nomogram
predicting the recurrence of hepatocellular carcinoma in patients
after laparoscopic hepatectomy. Cancer Commun (Lond). 39:552019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Wang X, Ma M, et al: Liver
transplantation for non-hepatocellular carcinoma malignant liver
tumors: A single center case study. Pract J Organ Transplant.
7:44–47. 2019.
|
|
7
|
Kim HK, Yun YK and Ahn YJ: Toxicity of
atractylon and atractylenolide III identified in atractylodes ovata
rhizome to dermatophagoides farinae and dermatophagoides
pteronyssinus. J Agric Food Chem. 55:6027–6031. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Makówka A, Olejniczak-Fortak M and Nowicki
M: A comparison of the antihypertensive and anti-inflammatory
effects of aliskiren and ramipril add-on therapy in peritoneal
dialysis patients-a pilot open label study. Kidney Blood Press Res.
36:18–25. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen LG, Jan YS, Tsai PW, Norimoto H,
Michihara S, Murayama C and Wang CC: Anti-inflammatory and
antinociceptive constituents of atractylodes japonica koidzumi. J
Agric Food Chem. 64:2254–2262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hwang JM, Tseng TH, Hsieh YS, Chou FP,
Wang CJ and Chu CY: Inhibitory effect of atractylon on tert-butyl
hydroperoxide induced DNA damage and hepatic toxicity in rat
hepatocytes. Arch Toxicol. 70:640–644. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tin MM, Cho CH, Chan K, James AE and Ko
JK: Astragalus saponins induce growth inhibition and apoptosis in
human colon cancer cells and tumor xenograft. Carcinogenesis.
28:1347–1355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tao Y, Zhan S, Wang Y, Zhou G, Liang H,
Chen X and Shen H: Baicalin, the major component of traditional
Chinese medicine Scutellaria baicalensis induces colon cancer cell
apoptosis through inhibition of oncomiRNAs. Sci Rep. 8:144772018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng X, Su X, Chen X, Zhao H, Bo C, Xu J,
Bai H and Ning K: Biological ingredient analysis of traditional
Chinese medicine preparation based on high-throughput sequencing:
The story for Liuwei Dihuang Wan. Sci Rep. 4:51472014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nichols AC, Black M, Yoo J, Pinto N,
Fernandes A, Haibe-Kains B, Boutros PC and Barrett JW: Exploiting
high-throughput cell line drug screening studies to identify
candidate therapeutic agents in head and neck cancer. BMC Pharmacol
Toxicol. 15:662014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng Y, Chen T, Yang X, Xue J and Chen J:
Atractylon induces apoptosis and suppresses metastasis in hepatic
cancer cells and inhibits growth in vivo. Cancer Manag Res.
11:5883–5894. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mane VP, Heuer MA, Hillyer P, Navarro MB
and Rabin RL: Systematic method for determining an ideal
housekeeping gene for real-time PCR analysis. J Biomol Tech.
19:342–347. 2008.PubMed/NCBI
|
|
17
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng Y, Geng L, Wang K, Sun J, Xu W, Gong
S and Zhu Y: Long noncoding RNA expression signatures of colon
cancer based on the ceRNA network and their prognostic value. Dis
Markers. 2019:76367572019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao C and He C: Preparative isolation and
purification of atractylon and atractylenolide III from the Chinese
medicinal plant Atractylodes macrocephala by high-speed
counter-current chromatography. J Sep Sci. 29:1630–1636. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kiso Y, Tohkin M and Hikino H:
Antihepatotoxic principles of atractylodes rhizomes. J Nat Prod.
46:651–654. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui XB, Shan CX, Wen HM, Wei L and Hao W:
UFLC/Q-TOF-MS based analysis on material base of atractylodis
macrocephalae rhizoma stir-fried with wheat bran. Zhongguo Zhong
Yao Za Zhi. 38:1929–1933. 2013.(In Chinese). PubMed/NCBI
|
|
22
|
Cheng Y, Mai JY, Hou TL, Ping J and Chen
JJ: Antiviral activities of atractylon from atractylodis rhizoma.
Mol Med Rep. 14:3704–3710. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shou D, Dai S, Zhang J, Li H and Yu Z:
Simultaneous determination of atractylenolide III, atractylenolide
I and atractylon in artactylodis macrocephala using microbore
liquid chromatography. Se Pu. 26:637–639. 2008.(In Chinese).
PubMed/NCBI
|
|
24
|
Satoh K, Nagai F, Ushiyama K and Kano I:
Specific inhibition of Na+,K(+)-ATPase activity by atractylon, a
major component of byaku-jutsu, by interaction with enzyme in the
E2 state. Biochem Pharmacol. 51:339–343. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang DN, Wei JM, Zhu ZM, Wen HX and Guo
LY: Experimental study the on relationship between inhibiting the
ubiquitin-proteasome pathway and drug sensitivity in human liver
carcinoma cell. Chin J Hepatobiliary Surg. 9:634–636. 2006.(In
Chinese).
|
|
26
|
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z
and Zhang LW: Identification of key long non-coding RNAs as
competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma.
Eur Rev Med Pharmacol Sci. 20:2285–2295. 2016.PubMed/NCBI
|
|
27
|
Mu X, Wu H, Liu J, Hu X, Wu H, Chen L, Liu
W, Luo S and Zhao Y: Long noncoding RNA TMPO-AS1 promotes lung
adenocarcinoma progression and is negatively regulated by
miR-383-5p. Biomed Pharmacother. 125:1099892020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang W, Su X, Yan W, Kong Z, Wang D,
Huang Y, Zhai Q, Zhang X, Wu H, Li Y, et al: Overexpression of
AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and
poor prognosis in prostate cancer. Prostate. 78:1248–1261. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qin Z, Zheng X and Fang Y: Long noncoding
RNA TMPO-AS1 promotes progression of non-small cell lung cancer
through regulating its natural antisense transcript TMPO. Biochem
Biophys Res Commun. 516:486–493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Simsir A, Fetsch P, Mehta D, Zakowski M
and Abati A: E-cadherin, N-cadherin, and calretinin in pleural
effusions: The good, the bad, the worthless. Diagn Cytopathol.
20:125–130. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Andersen H, Mejlvang J, Mahmood S, Gromova
I, Gromov P, Lukanidin E, Kriajevska M, Mellon JK and Tulchinsky E:
Immediate and delayed effects of E-cadherin inhibition on gene
regulation and cell motility in human epidermoid carcinoma cells.
Mol Cell Biol. 25:9138–9150. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin CJ, Lin CY, Wu JC, Huang SH, Wang SM
and Chen GH: 357 Macrophage activation promotes invasive properties
of hepatocellular carcinoma cells through destablization of the
E-cadherin/B-catenin complex. J Hepatol. 44 (Suppl 2):S1362006.
View Article : Google Scholar
|