Introduction
Obesity and type 2 diabetes mellitus (T2DM) have
become diseases that endanger the health of individuals all over
the world, especially in China. In 2018, 10% of the global
population was obese and China ranks first in the world in terms of
obesity (1). T2DM was declared a
global public health emergency as early as 2015, and in 2017, the
number of T2DM patients reached 425 million; China also has many
patients with T2DM, and 90% of these patients have obesity combined
with T2DM (2). Obesity is closely
associated with T2DM. The incidence rate of obesity combined with
T2DM increases year by year (3).
Compared with traditional drug therapy and diet regulation,
surgical weight loss surgery improves the remission rate of T2DM in
obese patients, reduces the weight of obese patients and has fewer
postoperative complications (4).
Due to the rapid development of metabolic surgery, weight loss
surgery has become the main treatment strategy for obesity and T2DM
(5).
Patients with T2DM often suffer from a high glucose
and high fat environment, leading to damage of myocardial cells,
podocytes and other tissue cells, which affects cell proliferation,
morphology and apoptosis (6–8).
Long non-coding (lnc)RNAs have transcripts of >200 nt and are
widely distributed in the nucleus and cytoplasm. They cannot encode
proteins, but can regulate gene expression (9). It has been found that lncRNAs are
widely involved in physiological and pathological processes,
serving an important role in the occurrence and development of a
variety of tumors (10). A study
showed that lncRNA transcribed ultraconserved element 338 was
overexpressed in lung cancer and its expression might be associated
with the prognosis of lung cancer (11). Another study indicated that lncRNA
nuclear enriched abundant transcript 1 activated Wnt signaling to
promote colorectal cancer progression and metastasis (12). In addition to tumor diseases,
lncRNAs also serve a regulatory role in a number of other diseases,
including metabolic diseases. It has been found that lncRNA
glycolysis-associated lncRNA of colorectal cancer (GLCC1) affects
carbohydrate metabolism by regulating c-Myc and further promoting
cell proliferation (13). Another
study found that lncRNA breast cancer anti-estrogen resistance 4
(BCAR4) coordinates Hippo and Hedgehog signaling to enhance the
transcription of glycolysis activators HK2 and PFKFB3, then affects
glycolysis via the YAP-breast cancer anti-estrogen resistance
4-glycolysis axis (14).
Therefore, it was hypothesized that an lncRNA may also be involved
in the regulation of T2DM, serving a role in mediating high glucose
and high fat-induced alterations in cells.
lncRNA taurine upregulated gene 1 (TUG1) was
initially identified and confirmed in the related research of
retinal development (15). TUG1
displays abnormal expression and function in bladder cancer, glioma
and other tumors (16,17). A previous study showed that lncRNA
TUG1 expression was obviously upregulated in patients with CHD, and
metformin activated the AMPK/mTOR pathway by regulating lncRNA TUG1
to promote autophagy and inhibit atherosclerosis (18). Based on the aforementioned
studies, the present study detected alterations in lncRNA TUG1
expression levels in blood samples from patients with T2DM before
and after sleeve gastrectomy (SG), and detected the effect of
lncRNA TUG1 on the viability of intestinal epithelial cells under
high glucose and high fat conditions, as well as the possible
signaling pathway involved in these regulatory roles.
Materials and methods
Cell culture and treatment
The human normal intestinal epithelial cell line
(HIEC-6) and human colorectal adenocarcinoma epithelial cell line
(SW480) were obtained from American Type Culture Collection. Cells
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C with 5% CO2. The cell injury model was induced
using DMEM supplemented with 50 mmol/l glucose and 500 mmol/l
saturated free fatty acid (palmitate; Sigma-Aldrich; Merck KGaA)
for 48 h at 37°C. The control group was treated with 30 mmol/l
mannitol for 48 h at 37°C. lncRNA TUG1 was overexpressed using a
TUG1 lentiviral vector (LV; LV-TUG1) and a lentiviral negative
control (NC; LV-NC) obtained from Shanghai GeneChem Co., Ltd. After
cell counting, cells (1.5×105 cells/well) were
inoculated into a 6-well plate and divided into the LV-NC and
LV-TUG1 groups, followed by culture in an incubator at 37°C. After
12 h of cell inoculation and the number of cells reached
2×105 in each well, the empty vector (LV-NC) and the
lentiviral vector (Shanghai GeneChem Co., Ltd., China)
overexpressing lncRNA TUG1 (LV-TUG1) were transfected into the
LV-NC and LV-TUG1 groups, respectively, followed by culture in an
incubator at 37°C. After 48 h post-transfection, cells were
collected by trypsin digestion and TUG1 expression was detected by
reverse transcription-quantitative PCR (RT-qPCR) to verify the
transfection efficiency of the lentiviral vector.
T2DM patients
A total of 50 T2DM patients (26 female patients and
24 male patients; age range, 32–71 years; average age, 45.7±4.6
years) were selected in the present study. Patients in the study
were diagnosed with T2DM and treated at the First Affiliated
Hospital of Jiamusi University (Jiamusi, China) between March 2017
and March 2019. The following inclusion criteria were used: i)
Presented with T2DM for the first time with no complications; ii)
follow-up for >1 year; and iii) underwent SG after the diagnosis
of T2DM. The basis for selecting patients was polyuria, fatigue,
weight loss or polydipsia, 2 h post-load glucose ≥11.1 mmol/l
following 75 g, oral glucose uptake and random plasma glucose ≥11.1
mmol/l. All patients has 10 ml peripheral blood sample from venous
blood at the elbow and the 1 week prior to SG, 6 months after
surgery and 12 months after surgery. All blood samples were stored
at −80°C after adding anticoagulants. The present study was
approved by the Clinical Research Ethics Committee of the First
Affiliated Hospital of Jiamusi University (approval no. 2019018)
and all the patients signed informed consent.
RT-qPCR
Total RNA was extracted from HIEC-6 cells, SW480
cells and blood samples using TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.), then reverse transcribed into cDNA
using the PrimeScript RT reagent kit with gDNA Eraser (Takara Bio,
Inc.) according to the manufacturer's protocol. qPCR was performed
using SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Inc.). The
thermocycling conditions were: 1 min 10 sec at 95°C and then 39
cycles of 12 sec at 95°C and 30 sec at 59.5°C. The relative
expression of mRNA was normalized to the internal reference gene
β-actin and calculated using 2−ΔΔCt method (19). The primers used for qPCR were
synthesized by Guangzhou RiboBio Co., Ltd. The primer sequences
were as follows: TUG1 forward, 5′-CTGGACCTGGAACCCCAAAG-3′ and
reverse, 5′-GGTAGTGCTTGCTCAGTCGT-3′; AMPK forward,
5′-GGTAGTGCTTGCTCAGTCGT-3′ and reverse, 5′-GGTAGTGCTTGCTCAGTCGT-3′;
SIRT1 forward, 5′-AAGATGACGTCTTATCCTCT-3′ and reverse,
5′-GCTTCATTAATTGCCTCTTG-3′; UCP2 forward, 5′-GCTGGTGGTGGTCGGAGAT-3′
and reverse, 5′-TGAAGTGGCAAGGGAGGT−3′; β-actin forward,
5′-AAGATGACGTCTTATCCTCT-3′ and reverse,
5′-GCTTCATTAATTGCCTCTTG-3′.
Cell counting kit-8 (CCK-8) assay
HIEC-6 and SW480 cells challenged with high glucose
and high fat and/or transfected with LV-TUG1 were seeded into a
96-well plate at 1×104 in each well and incubated for 12
h. Cell viability was analyzed using the CCK-8 kit (Dojindo
Molecular Technologies, Inc.) according to the manufacturer's
protocol. CCK-8 (10 µl) were added to each well for incubation at
37°C with 5% CO2 for 1 h. A microplate reader at 450 nm
wavelength was used for detecting optical density values.
TUNEL assay
To detect cell apoptosis, a TUNEL detection kit
(Roche Diagnostics) was used according to the manufacturer's
protocol. Treated cells were cultured overnight and then washing
twice with PBS and collected on the slide, fixed with 4%
paraformaldehyde for 15 min at 25°C and permeabilized in 0.25%
Triton X-100 for 20 min. The cells were incubated in terminal
deoxynucleotidyl transferase (TdT) reaction cocktail for 45 min at
37°C, followed by treatment with Click-iT reaction cocktail. The
nucleus of HIEC-6 and SW480 were stained with DAPI (0.5 µg/ml) at
room temperature for 15 min and then observed under a fluorescence
microscope (five fields were selected at a magnification of
×40).
LDH assay
Treated cells were seeded in 96-well plates
(4×103 cells/well) and incubated for 48 h at 37°C. Cell
damage was detected by the LDH Cytotoxicity Assay kit (Beyotime
Institute of Biotechnology). Briefly, LDH release regent (150 µl)
was added to the 96-well plate following removal of the
supernatant. The cells were incubated at 37°C for 1 h with 5%
CO2. The absorbance at 490 nm was detected with
Microplate Reader (Bio-Rad Laboratories, Inc.).
Flow cytometry assay
Flow cytometry was performed to further assess the
apoptotic rate. HIEC-6 or SW480 cells were plated into 6-well
plates (6×105 cells/well). At 48 h treatment with high
glucose and high fat-containing medium, pre-cooled PBS was used to
wash cells three times. Subsequently, the cells were analyzed using
the Annexin V-APC Apoptosis Detection kit (Beyotime Institute of
Biotechnology) following the manufacturer's protocol. The apoptotic
rate including early apoptosis and late apoptosis was assessed
using a flow cytometer (FACSCanto II; BD Biosciences) by measuring
the percentage of Annexin V+ and PI− cells.
The data was analyzed using FlowJo software (version 7.2.4; FlowJo
LLC).
Western blot analysis
Cells on the walls of the culture dish were washed
three times with PBS. Subsequently, total proteins were harvested
with RIPA buffer (Beyotime Institute of Biotechnology) and
quantified using a bicinchoninic acid assay (Beyotime Institute of
Biotechnology). Proteins (30 µg/lane) were separated via 10%
SDS-PAGE and transferred onto PVDF membranes. Following blocking
with 5% skimmed milk for 1 h at room temperature, the membranes
were incubated overnight at 4°C with the following primary
antibodies: Anti-SIRT1 (cat. no. ab189494; Abcam; 1:1,000),
anti-Bcl-2 (cat. no. ab32124; Abcam; 1:1,000), anti-AMPK (cat. no.
ab32047; Abcam; 1:1,000), anti-UCP2 (cat. no. ab97931; Abcam;
1:1,000) and anti-β-actin (cat. no. ab8226; Abcam; 1:8,000). The
membranes were incubated with horseradish peroxidase-conjugated
second antibody (goat anti-rabbit IgG, 1:2,000, goat anti-mouse
IgG, 1:2,000, TransGen Biotech Co., Ltd.) for 1 h at room
temperature. The proteins were determined by immunoblotting
analysis using an ECL immunoblotting kit (Millipore, Sigma). Each
protein expression was normalized to β-actin. Densitometry was
performed using ImageJ software (version 1.38X; National Institutes
of Health).
ELISA
Briefly, cells were inoculated (2×105
cells/well) into a 6-well plate and cultured overnight with 1
ml/well DMEM supplemented with 10% FBS at 37°C. After 24 h of
culture, cells were divided into several groups and treated with
DMEM containing concentrations of high glucose and high fat without
FBS. The IL-1β (cat. no. P1305), IL-6 (cat. no. P1330), IL-8 (cat.
no. P1640) and IL-10 (cat. no. P1528) levels in culture medium were
determined using ELISA kits (Beyotime Institute of Biotechnology)
according to the manufacturer's protocols.
Statistical analysis
Statistical analysis was performed using a paired
t-test or one-way ANOVA followed by Tukey's post hoc test with SPSS
17.0 statistical software (SPSS, Inc.). Each experiment was
performed in triplicate. Data are presented as the mean ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference.
Results
The expression of lncRNA TUG1, AMPK
and SIRT1 is affected by SG
Peripheral blood samples were obtained from T2DM
patients at 1 week before surgery, 6 months after surgery and 12
months after surgery. RT-qPCR was used to measure the mRNA
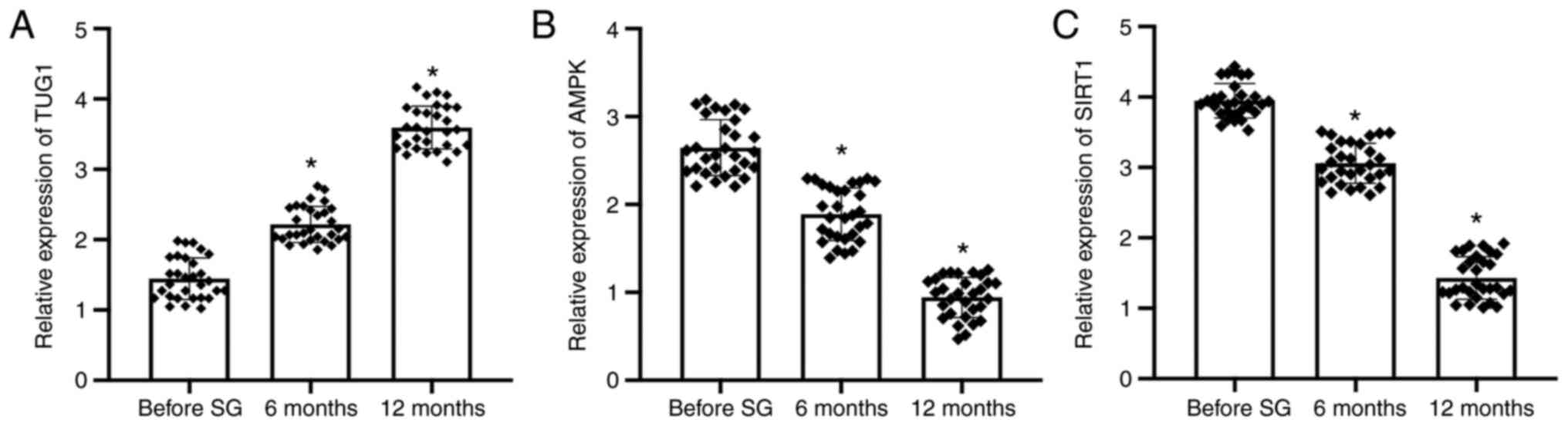
expression levels of TUG1, SIRT1 and AMPK. The results confirmed
that following SG, the expression of lncRNA TUG1 was significantly
increased, reaching a maximum at 12 months post-SG surgery,
compared with that of the before SG samples (Fig. 1A). The expression levels of AMPK
and SIRT1 were significantly downregulated after SG compared with
those before SG, reaching the lowest levels at 12 months among the
three group (Fig. 1B and C).
High glucose and high fat induce a
high apoptotic rate and low viability
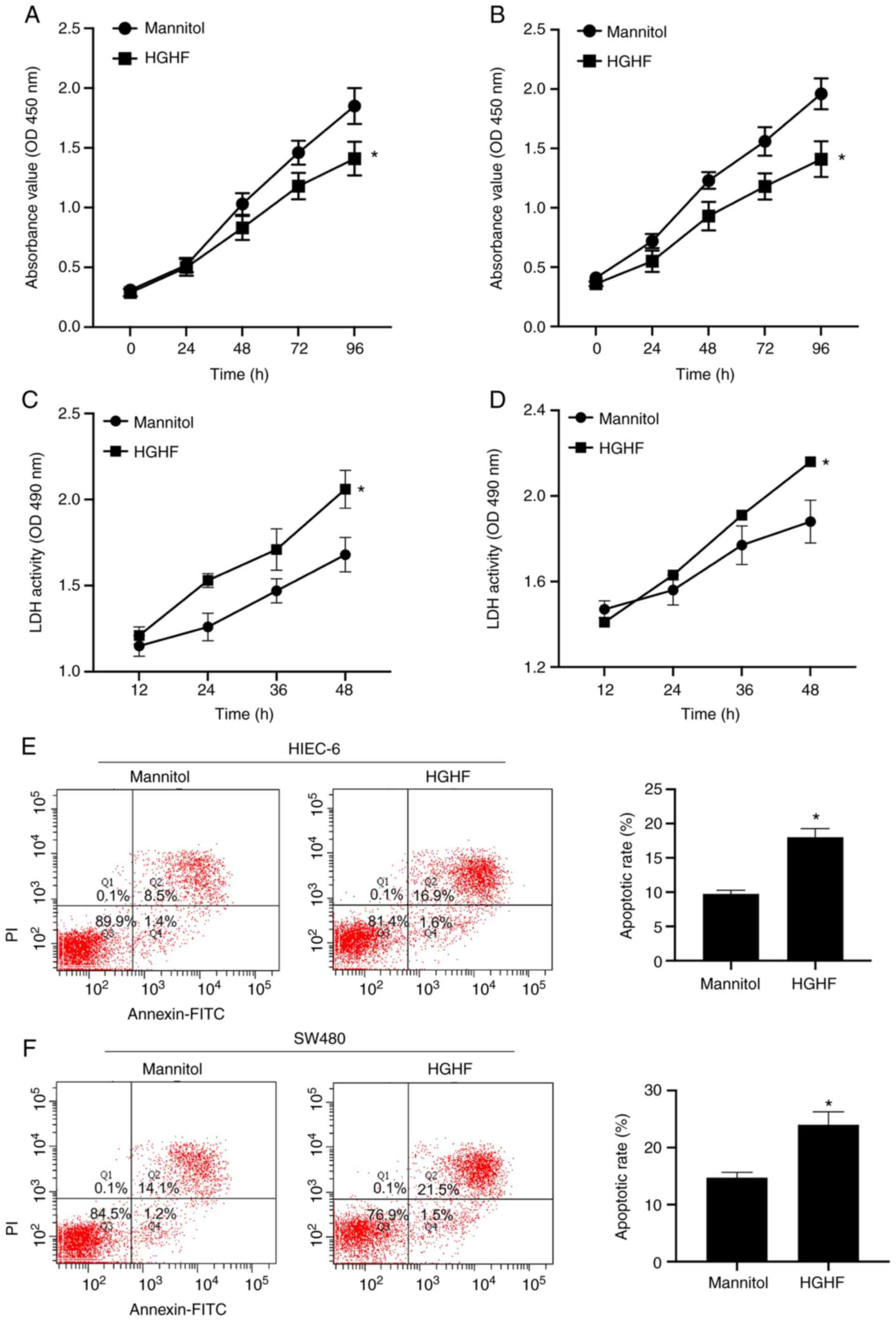
The effects of high glucose and high fat on the
apoptosis and viability of HIEC-6 and SW480 cells were explored.
The CCK-8 assay was performed to measure cell viability, and the
apoptotic rate was detected by performing lactate dehydrogenase
(LDH) cytotoxicity and flow cytometry assays. As expected, compared
with the mannitol group, under high glucose and high fat
conditions, the viability of HIEC-6 and SW480 cells was
significantly inhibited (Fig. 2A and
B), the release of LDH was significantly increased (Fig. 2C and D) and the apoptotic rate was
significantly increased (Fig. 2E and
F).
High glucose and high fat induce
downregulation of lncRNA TUG1 and UCP2, and upregulation of AMPK
and SIRT1
Following the detection of alterations in the
apoptosis and viability of HIEC-6 and SW480 cells, it was
hypothesized that the expression of lncRNA TUG1 and glucose
metabolism-associated proteins was also affected. The RT-qPCR
results confirmed that lncRNA TUG1 and UCP2 were significantly
downregulated (Fig. 3A and B),
whereas AMPK and SIRT1 were significantly upregulated under high
glucose and high fat conditions compared with those in the mannitol
group (Fig. 3C and D). The
western blotting results showed markedly lower protein expression
levels of UCP2 and Bcl-2, and higher protein expression levels of
SIRT1 and AMPK following high glucose and high fat treatment
compared with those following mannitol treatment (Fig. 3E).
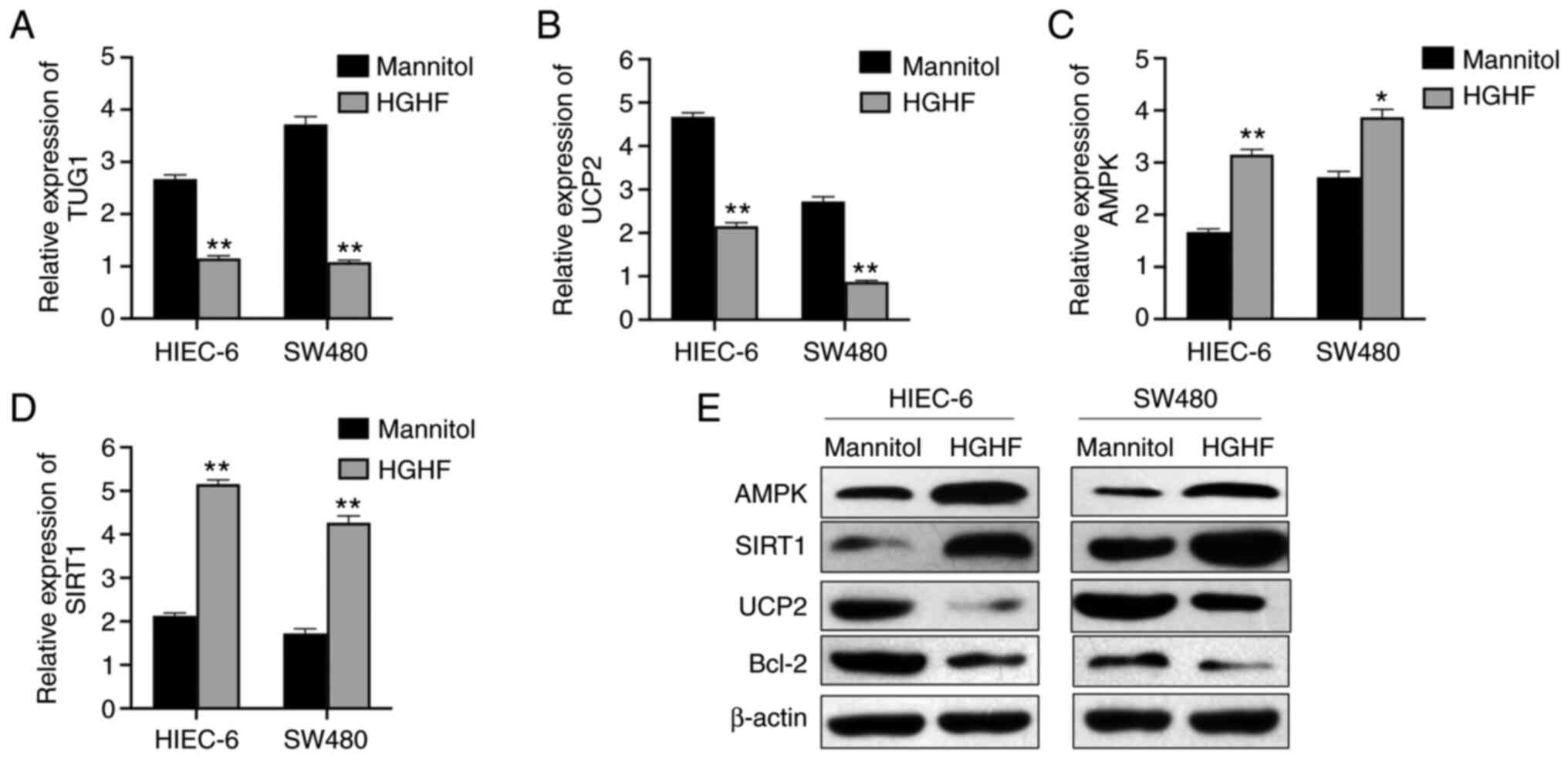 | Figure 3.HGHF inhibits the expression of
lncRNA TUG1 and UCP2, and promotes the expression of AMPK and
SIRT1. (A) Expression of lncRNA TUG1 in HIEC-6 and SW480 cells as
measured by reverse transcription-quantitative PCR. HGHF decreased
the expression of (B) UCP2, and increased the expression of (C)
AMPK and (D) SIRT1. (E) HGHF-mediated alterations to the protein
expression levels of AMPK, SIRT1, UCP2 and Bcl-2 were determined by
western blotting. Data are presented as the mean ± standard
deviation. *P<0.05 and **P<0.01 vs. mannitol. HGHF, high
glucose and high fat; lncRNA, long non-coding RNA; TUG1,
taurine-upregulated gene 1; UCP2, uncoupling protein 2; AMPK,
AMP-activated protein kinase; SIRT1, Sirtuin 1. |
High glucose and high fat alter the
cytokine release from HIEC-6 and SW480
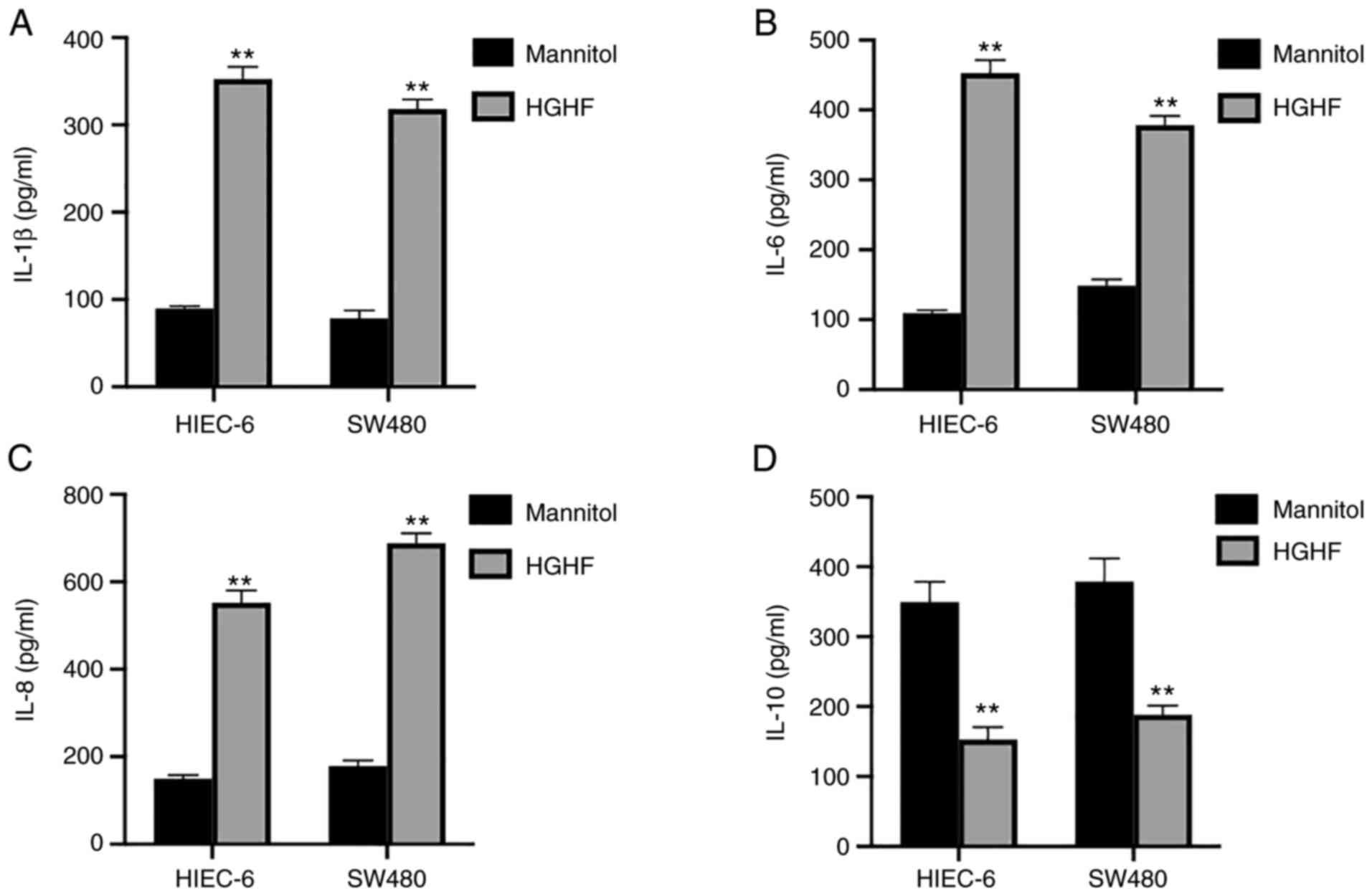
To study the release of cytokines associated with
inflammation by HIEC-6 and SW480 cells under high glucose and high
fat conditions, ELISAs were performed. The results showed a
significant increase in the secretion of IL-1β (Fig. 4A), IL-6 (Fig. 4B) and IL-8 (Fig. 4C), which could promote the
inflammatory reaction, but a significant decrease in the secretion
of IL-10 (Fig. 4D) under high
glucose and high fat conditions compared with those in the mannitol
group.
lncRNA TUG1 overexpression alleviates
the effects of high glucose and high fat on cell viability
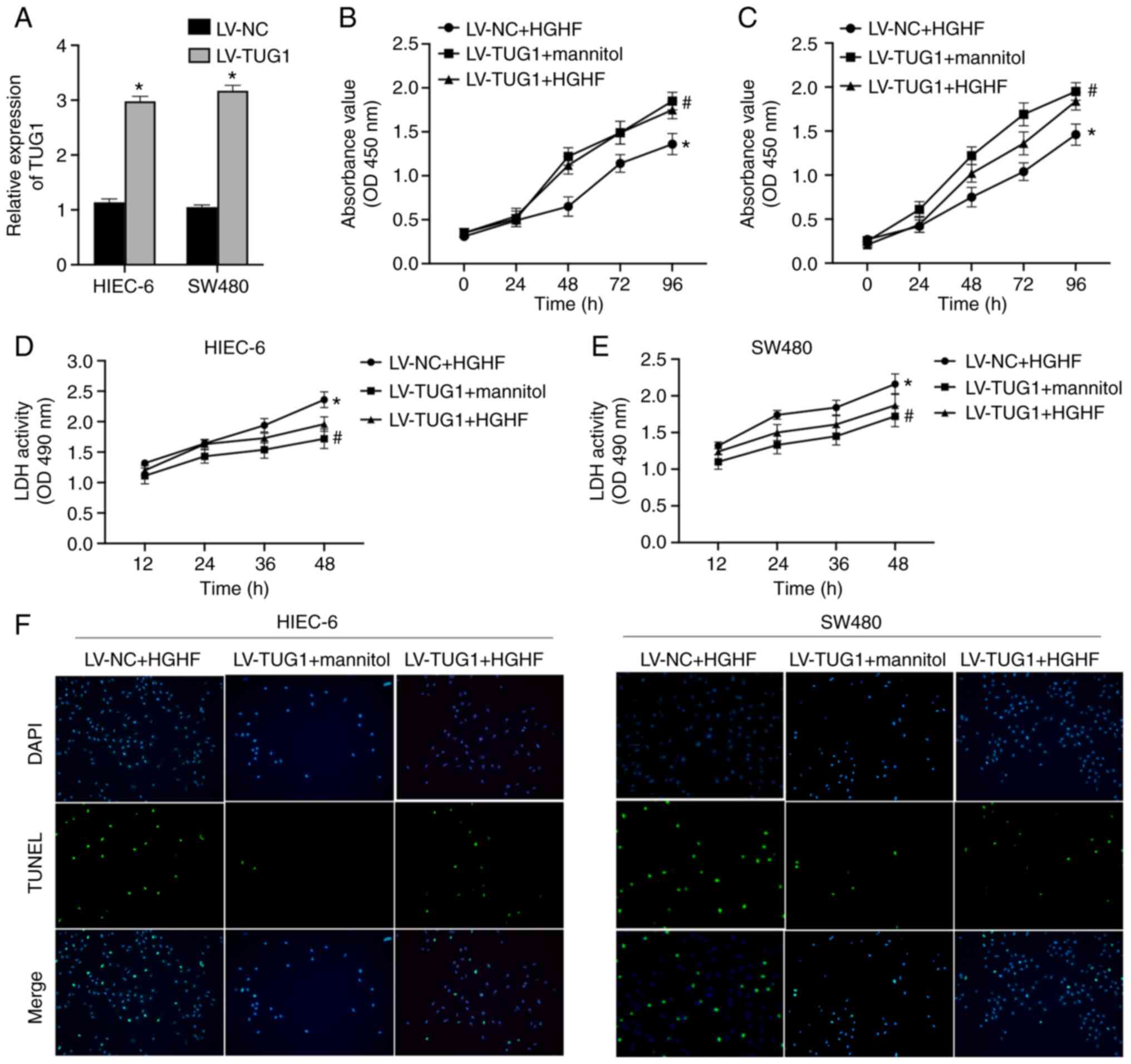
To study the effect of lncRNA TUG1 and high glucose
and high fat on intestinal epithelial cells, the expression of TUG1
in HIEC-6 and SW480 cells was overexpressed using LV-TUG1. TUG1
overexpression was confirmed by RT-qPCR (Fig. 5A). The treatment groups were as
follows: i) LV-NC + high glucose and high fat; ii) LV-TUG1 + high
glucose and high fat; and iii) LV-TUG1 + mannitol. TUG1
overexpression significantly enhanced the viability of HIEC-6 and
SW480 cells to alleviate high glucose and high fat-induced
inhibition of cell viability (Fig. 5B
and C). In addition, high glucose and high fat-mediated effects
on the secretion of LDH were also ameliorated by TUG1
overexpression (Fig. 5D and E).
It was also demonstrated that TUG1 markedly inhibited high glucose
and high fat-induced increases in the apoptotic rate of HIEC-6 and
SW480 cells (Fig. 5F).
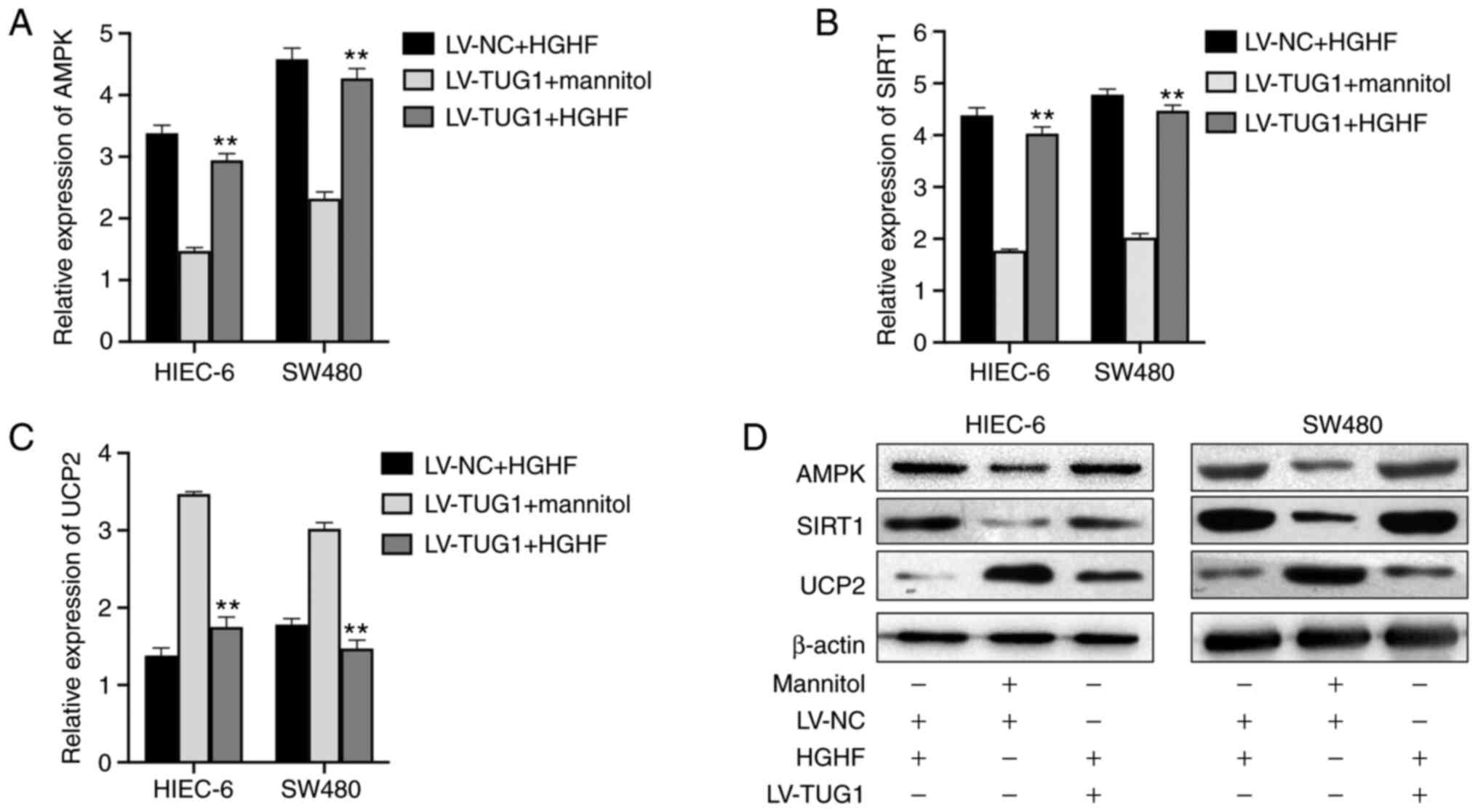
TUG1 reverses high glucose and high
fat-mediated alterations to the expression levels of AMPK, SIRT1
and UCP2
The gene and protein expression levels in HIEC-6 and
SW480 cells were evaluated by RT-qPCR and western blotting,
respectively, following treatment with LV-TUG1 and/or high glucose
and high fat. At the mRNA level, the high expression of AMPK
(Fig. 6A) and SIRT1 (Fig. 6B), and the low expression of UCP2
(Fig. 6C) induced by HGHF was
partially reversed by LV-TUG1 in HIEC-6 and SW480 cells, the
results of LV-TUG1+ Mannitol also confirmed that high glucose and
high fat inhibited the changes of expression caused by TUG1. The
western blotting results revealed that the increase in UCP2
expression, and decrease in AMPK and SIRT1 expression caused by
high glucose and high fat was partly reversed by TUG1
overexpression in HIEC-6 and SW480 cells (Fig. 6D).
Discussion
Obesity is an important problem threatening human
health. Excessive obesity causes great stress on human bones,
organs and systems, and leads to a variety of chronic diseases,
including hypertension and diabetes (20). A previous study (21) found that diabetic patients are
also more likely to be obese, which further aggravates the disease.
In addition, under the effect of obesity and hyperglycemia,
patients are more likely to present with coronary heart disease,
stroke, hyperlipidemia and other complications, increasing the risk
of disease and mortality (21).
Therefore, for T2DM patients with obesity, effective treatment
strategies should be employed as soon as possible to intervene and
control the weight and blood glucose of patients. SG has a
significant effect on the treatment of metabolic syndrome
associated with obesity, as well as a positive effect on the
complications of T2DM, hypertension and dyslipidemia, and the
remission rate of diabetes following SG is as ≤62% (22). However, the underlying molecular
mechanisms are not completely understood.
lncRNAs are a type of non-coding RNA that widely
exist in plasma, serum and organ tissues. lncRNAs serve an
important role in the process of body proliferation and
development, immune regulation, cell proliferation, migration,
signal transduction, autophagy and inflammation (23–26). A total of 55 differentially
expressed lncRNAs were screened from the peripheral blood of six
patients with T2DM and 60 healthy subjects by microarray analysis.
The top three most differentially expressed lncRNAs were verified
again in 60 patients with T2DM and 60 healthy individuals. It was
found that these three lncRNAs are associated with fasting plasma
glucose (FPG) and hemoglobin A1c (HbA1c) (27). In addition, a variety of lncRNAs,
including metastasis associated in lung denocarcinoma transcript 1
(MALAT1), maternally expressed gene 3 (MEG3), growth
arrest-specific transcript 5 (GAS5), neighbor of BRCA1 gene 2
(NBR2), cyclin-dependent kinase inhibitor 2B antisense RNA
1/antisense non-coding RNA in the INK4 locus (CDKN2BAS1/ANRIL) were
identified in the peripheral blood mononuclear cells of patients
with T2DM and were positively associated with blood glucose control
(28). A previous study found
that lncRNAs are involved in the regulation of insulin synthesis
and secretion, liver gluconeogenesis and lipid metabolism (29), and adipose tissue glucose uptake
through multi-level gene regulation (30), ultimately affecting blood glucose
in the human body (31). A
previous study found that carbohydrate responsive element binding
protein (ChREBP) can coordinate glucose homeostasis by regulating
lncRNA TUG1 transcription in the podocytes in response to increased
glucose levels, which also indicates that TUG1 is closely
associated with glucose metabolism (32). A study suggests that TUG1 restores
high glucose and high fat-treated endothelial progenitor cells
function by regulating microRNA (miR)-29c-3p/platelet-derived
growth factor-BB (PDGF-BB)/Wnt signaling (33). In addition, the role of lncRNA
TUG1 in diabetes also includes inhibiting diastolic dysfunction of
diabetic cardiomyopathy by regulating miR-499-5p (34). The aforementioned results
indicated that lncRNA TUG1 is closely associated with T2DM, which
is of great significance in regulating the dynamic balance of blood
glucose in the body. Therefore, the present study hypothesized that
SG may affect the expression level of lncRNA TUG1 in the body,
which may in turn affect glucose uptake and metabolism, and
ultimately alleviate the injury of intestinal epithelial cells
induced by high glucose and high fat.
SIRT1 interacts with a number of target proteins
involved in metabolism, inflammation, genomic stability and
apoptosis. SIRT1 changes the catalytic activity of proteins or
serves as an epigenetic signal to change the stability of proteins
by removing the acetyl groups of these target proteins (35). The metabolic regulation of SIRT1
includes regulating gluconeogenesis, increasing fatty acid
oxidation, decreasing fat production, increasing insulin secretion
and regulating autophagy to prolong life (36). SIRT1 activators have been proposed
to prevent and counteract metabolic age-related diseases, such as
T2DM (37). AMPK is a key
regulator of cell energy homeostasis, which regulates cell
metabolism through the ratio of AMP/ATP; when the ratio of AMP/ATP
decreases, the expression of AMPK increases (38). SIRT1 can deacetylate and activate
liver kinase B1 (LKB1) and the activated LKB1 can phosphorylate and
activate AMPK (39). A study
found that AMPK/SIRT1 can participate in the regulation of glucose
metabolism pathway by lncRNA CDKN2B antisense RNA 1, thus affecting
cell viability (40). SIRT1 is
able to directly bind the UCP2 promoter, repressing its
transcription and affecting blood glucose by regulating β-cells
(41). The present study
hypothesized that lncRNA TUG1 promoted glucose metabolism through
the AMPK/SIRT1 pathway following SG, thus affecting the blood
glucose level of T2DM patients.
The present study found that high levels of TUG1
were associated with SG in T2DM patients. However, in a high
glucose and high fat environment, the expression of TUG1 and the
viability of HIEC-6 and SW480 cells was inhibited, whereas
apoptosis was promoted. These results suggested that SG surgery may
affect blood glucose by altering the expression of lncRNAs in cells
and further regulating the downstream genes. In addition, the
results of RT-qPCR of blood samples obtained from T2DM patients
confirmed that SIRT1 and AMPK expression decreased following SG
surgery. Under high glucose and high fat conditions, the trends in
expression, cell viability and apoptosis displayed an opposite
tendency. Following high glucose and high fat treatment combined
with TUG1 overexpression, TUG1 alleviated high glucose and high
fat-induced alterations in cell viability and the expression levels
of AMPK, SIRT1 and UCP2. In the present study, the research on
TUG1-related pathways was not detailed enough and more possible
pathways, including glucose metabolism and fat metabolism, need to
be explored further. The clinical implications of the present study
needs further investigation. In conclusion, these results suggested
that AMPK/SIRT1/UCP2 may be one of the pathways altered by SG
through modulation of the expression of lncRNA TUG1, which may
result in the control of blood glucose in T2DM patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific Research of
Heilongjiang Health and Family Planning Commission (grant no.
2019-294).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WW, XW, YW, SL and DS primarily designed and
performed the study. DS, SG, HT and WW analyzed the data. WW and XW
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Clinical
Research Ethics Committee of the First Affiliated Hospital of
Jiamusi University (approval no. 2019018). All patients signed
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang Y, Xue H, Sun M, Zhu X, Zhao L and
Yang Y: Prevention and control of obesity in China. Lancet Glob
Health. 9:e1166–e1167. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jia W, Weng J, Zhu D, Ji L, Lu J, Zhou Z,
Zou D, Guo L, Ji Q, Chen L, et al: Standards of medical care for
type 2 diabetes in China 2019. Diabetes Metab Res Rev.
35:e31582019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ortega MA, Fraile-Martínez O, Naya I,
García-Honduvilla N, Álvarez-Mon M, Buján J, Asúnsolo Á and de la
Torre B: Type 2 diabetes mellitus associated with obesity
(diabesity). The central role of gut microbiota and its
translational applications. Nutrients. 12:27492020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsilingiris D, Koliaki C and Kokkinos A:
Remission of type 2 diabetes mellitus after bariatric surgery: Fact
or fiction? Int J Environ Res Public Health. 16:31712019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng ZY, Sun J and Kang WM: History,
recent advancements, and prospects in bariatric/metabolic surgery.
Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 40:581–590. 2018.(In Chinese).
PubMed/NCBI
|
|
6
|
Macauley M, Percival K, Thelwall PE,
Hollingsworth KG and Taylor R: Altered volume, morphology and
composition of the pancreas in type 2 diabetes. PLoS One.
10:e01268252015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fan M, Jiang H, Zhang Y, Ma Y, Li L and Wu
J: Liraglutide enhances autophagy and promotes pancreatic β cell
proliferation to ameliorate type 2 diabetes in high-fat-fed and
streptozotocin-treated mice. Med Sci Monit. 24:2310–2316. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu H, Wang Q, Sun Q, Qin Y, Han A, Cao Y,
Yang Q, Yang P, Lu J, Liu Q and Xiang Q: In type 2 diabetes induced
by cigarette smoking, activation of p38 MAPK is involved in
pancreatic β-cell apoptosis. Environ Sci Pollut Res Int.
25:9817–9827. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
St Laurent G, Wahlestedt C and Kapranov P:
The landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng WX, Koirala P and Mo YY:
lncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang YX, Yuan J, Gao ZM and Zhang ZG:
lncRNA TUC338 promotes invasion of lung cancer by activating MAPK
pathway. Eur Rev Med Pharmacol Sci. 22:443–449. 2018.PubMed/NCBI
|
|
12
|
Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan
C, Xu M, Sun H, Liu C, Wei P and Du X: The lncRNA NEAT1 activates
Wnt/β-catenin signaling and promotes colorectal cancer progression
via interacting with DDX5. J Hematol Oncol. 11:1132018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X,
Tian X, Guo F, Liang Q, Liu Q, et al: lncRNA GLCC1 promotes
colorectal carcinogenesis and glucose metabolism by stabilizing
c-Myc. Nat Commun. 10:34992019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng X, Han H, Liu GP, Ma YX, Pan RL,
Sang LJ, Li RH, Yang LJ, Marks JR, Wang W and Lin A: lncRNA wires
up Hippo and Hedgehog signaling to reprogramme glucose metabolism.
EMBO J. 36:3325–3335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu G, Zhou H, Yao W, Meng L and Lang B:
lncRNA TUG1 promotes cisplatin resistance by regulating CCND2 via
epigenetically silencing miR-194-5p in bladder cancer. Mol Ther
Nucleic Acids. 16:257–271. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li J, An G, Zhang M and Ma Q: Long
non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells.
Biochem Biophys Res Commun. 477:743–748. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
You G, Long X, Song F, Huang J, Tian M,
Xiao Y, Deng S and Wu Q: Metformin activates the AMPK-mTOR pathway
by modulating lncRNA TUG1 to induce autophagy and inhibit
atherosclerosis. Drug Des Devel Ther. 14:457–468. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arocho A, Chen B, Ladanyi M and Pan Q:
Validation of the 2-DeltaDeltaCt calculation as an alternate method
of data analysis for quantitative PCR of BCR-ABL P210 transcripts.
Diagn Mol Pathol. 15:56–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Umemura A, Sasaki A, Nitta H, Baba S, Ando
T, Kajiwara T and Ishigaki Y: Pancreas volume reduction and
metabolic effects in Japanese patients with severe obesity
following laparoscopic sleeve gastrectomy. Endocr J. 64:487–498.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li YX, Fang DH and Liu TX: Laparoscopic
sleeve gastrectomy combined with single-anastomosis
duodenal-jejunal bypass in the treatment of type 2 diabetes
mellitus of patients with body mass index higher than 27.5 kg/m2
but lower than 32.5 kg/m2. Medicine (Baltimore).
97:e115372018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramos AC, Bastos EL, Ramos MG, Bertin NT,
Galvão TD, de Lucena RT and Campos JM: Medium-term follow-up
results with laparoscopic sleeve gastrectomy. Arq Bras Cir Dig. 28
Suppl 1:S61–S64. 2015.(In English, Portuguese).s. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murillo-Maldonado JM and Riesgo-Escovar
JR: The various and shared roles of lncRNAs during development. Dev
Dyn. 248:1059–1069. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
25
|
Xiong H, Ni Z, He J, Jiang S, Li X, He J,
Gong W, Zheng L, Chen S, Li B, et al: lncRNA HULC triggers
autophagy via stabilizing Sirt1 and attenuates the chemosensitivity
of HCC cells. Oncogene. 36:3528–3540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun Y, Zhong L, He X, Wang S, Lai Y, Wu W,
Song H, Chen Y, Yang Y, Liao W, et al: lncRNA H19 promotes vascular
inflammation and abdominal aortic aneurysm formation by functioning
as a competing endogenous RNA. J Mol Cell Cardiol. 131:66–81. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Chang X, Zhang P, Fan L, Zhou T
and Sun K: Aberrant expression of long non-coding RNAs in newly
diagnosed type 2 diabetes indicates potential roles in chronic
inflammation and insulin resistance. Cell Physiol Biochem.
43:2367–2378. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sathishkumar C, Prabu P, Mohan V and
Balasubramanyam M: Linking a role of lncRNAs (long non-coding RNAs)
with insulin resistance, accelerated senescence, and inflammation
in patients with type 2 diabetes. Hum Genomics. 12:412018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen J, Ke S, Zhong L, Wu J, Tseng A,
Morpurgo B, Golovko A, Wang G, Cai JJ, Ma X, et al: Long noncoding
RNA MALAT1 regulates generation of reactive oxygen species and the
insulin responses in male mice. Biochem Pharmacol. 152:94–103.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu X, Li H, Wu Y, Zhou J, Yang G, Wang W,
Kang D and Ye S: CREB-upregulated lncRNA MEG3 promotes hepatic
gluconeogenesis by regulating miR-302a-3p-CRTC2 axis. J Cell
Biochem. 120:4192–4202. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu S, Sheng L, Miao H, Saunders TL,
MacDougald OA, Koenig RJ and Xu B: SRA gene knockout protects
against diet-induced obesity and improves glucose tolerance. J Biol
Chem. 289:13000–13009. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Long J, Galvan DL, Mise K, Kanwar YS, Li
L, Poungavrin N, Overbeek PA, Chang BH and Danesh FR: Role for
carbohydrate response element-binding protein (ChREBP) in high
glucose-mediated repression of long noncoding RNA Tug1. J Biol
Chem. 295:15840–15852. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Y, Zhi K, Han S, Li X, Li M, Lian W,
Zhang H and Zhang X: TUG1 enhances high glucose-impaired
endothelial progenitor cell function via miR-29c-3p/PDGF-BB/Wnt
signaling. Stem Cell Res Ther. 11:4412020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao L, Li W and Zhao H: Inhibition of
long non-coding RNA TUG1 protects against diabetic cardiomyopathy
induced diastolic dysfunction by regulating miR-499-5p. Am J Transl
Res. 12:718–730. 2020.PubMed/NCBI
|
|
35
|
Rajendran R, Garva R, Krstic-Demonacos M
and Demonacos C: Sirtuins: Molecular traffic lights in the
crossroad of oxidative stress, chromatin remodeling, and
transcription. J Biomed Biotechnol. 2011:3682762011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
D'Onofrio N, Servillo L and Balestrieri
ML: SIRT1 and SIRT6 signaling pathways in cardiovascular disease
protection. Antioxid Redox Signal. 28:711–732. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kitada M, Ogura Y, Monno I and Koya D:
Sirtuins and type 2 diabetes: Role in inflammation, oxidative
stress, and mitochondrial function. Front Endocrinol (Lausanne).
10:1872019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Herzig S and Shaw RJ: AMPK: Guardian of
metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol.
19:121–135. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bai B, Man AW, Yang K, Guo Y, Xu C, Tse
HF, Han W, Bloksgaard M, De Mey JG, Vanhoutte PM, et al:
Endothelial SIRT1 prevents adverse arterial remodeling by
facilitating HERC2-mediated degradation of acetylated LKB1.
Oncotarget. 7:39065–39081. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun LY, Li XJ, Sun YM, Huang W, Fang K,
Han C, Chen ZH, Luo XQ, Chen YQ and Wang WT: lncRNA ANRIL regulates
AML development through modulating the glucose metabolism pathway
of AdipoR1/AMPK/SIRT1. Mol Cancer. 17:1272018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bordone L, Motta MC, Picard F, Robinson A,
Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A,
et al: Sirt1 regulates insulin secretion by repressing UCP2 in
pancreatic beta cells. PLoS Biol. 4:e312006. View Article : Google Scholar : PubMed/NCBI
|