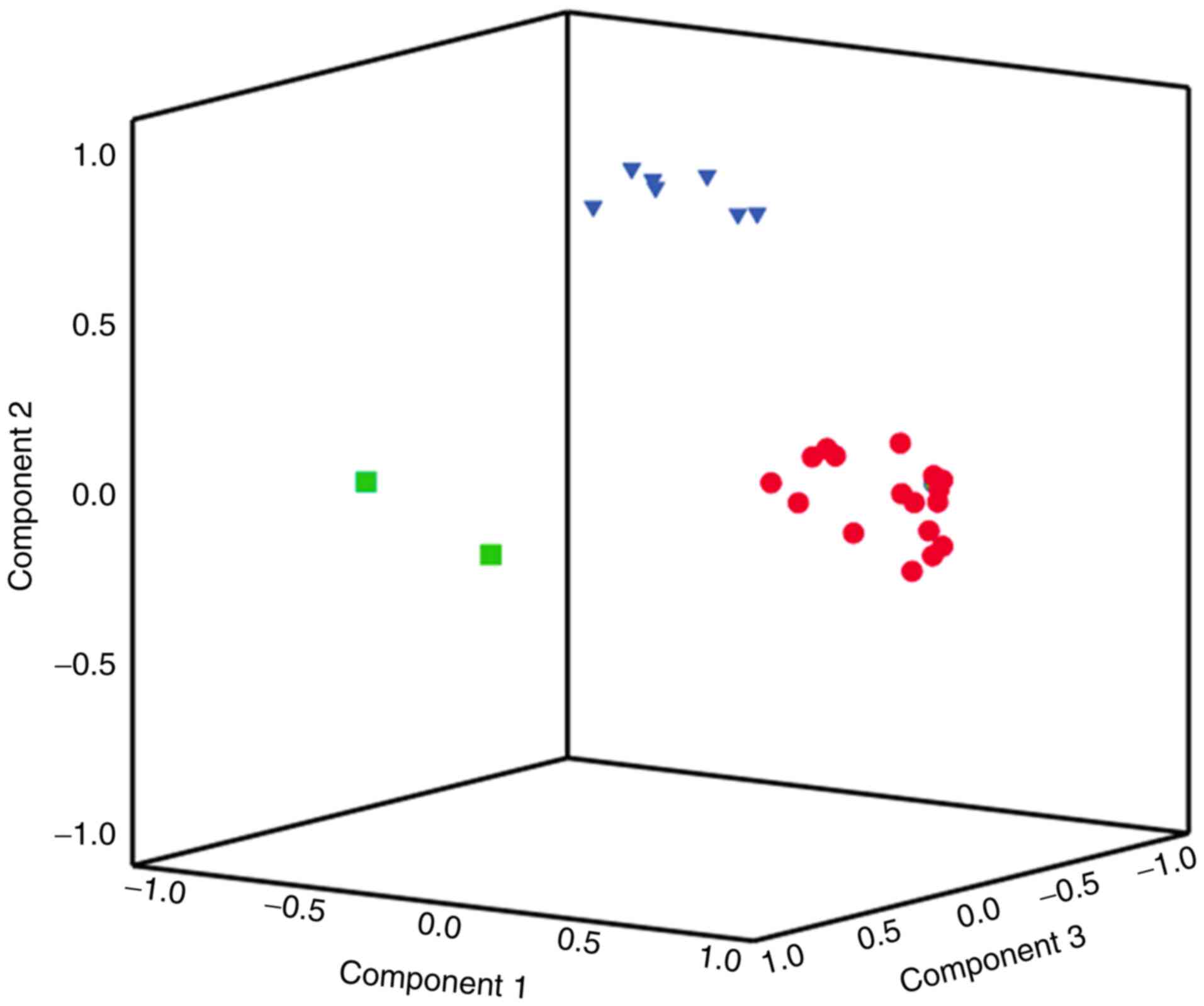
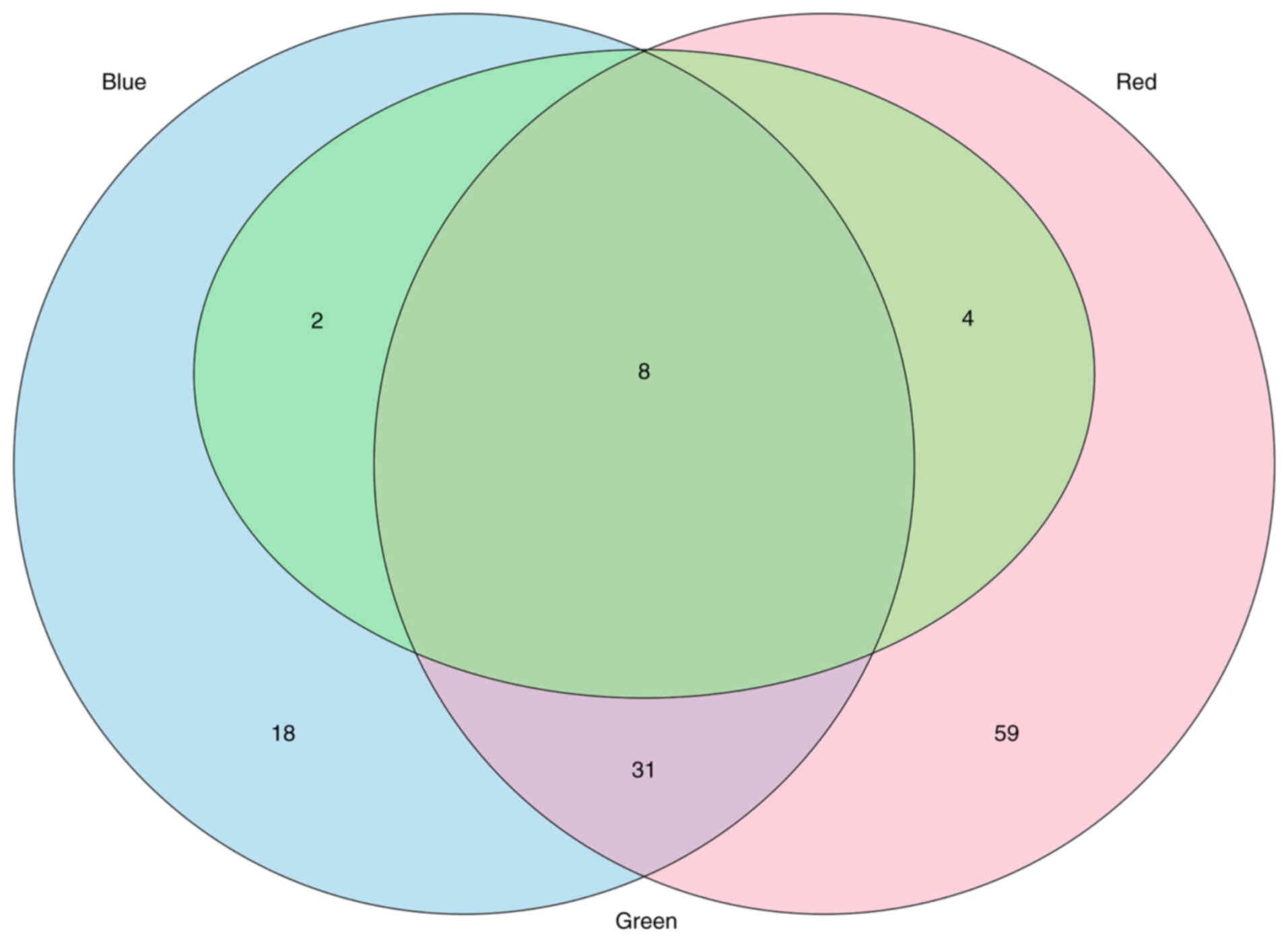
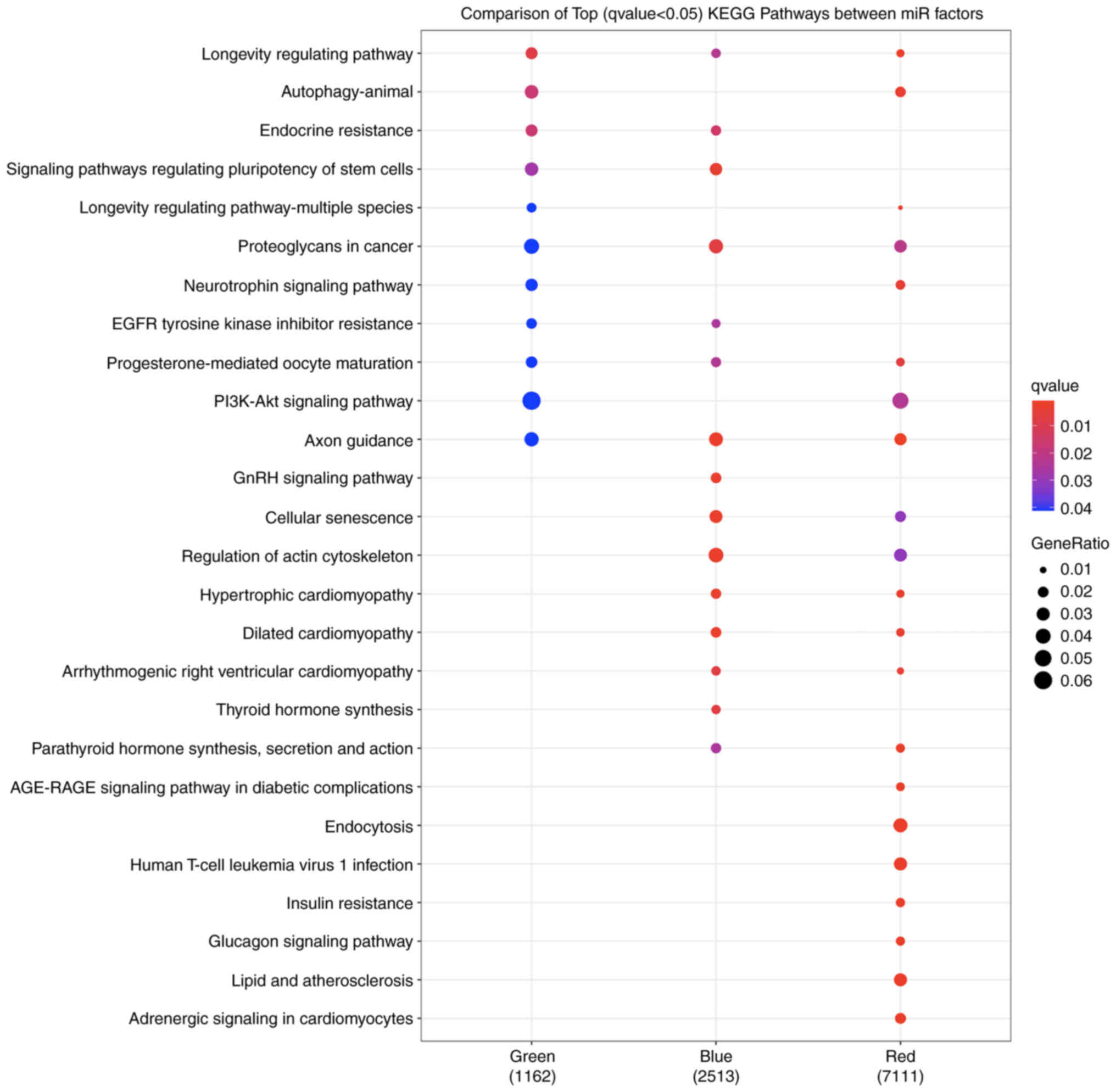
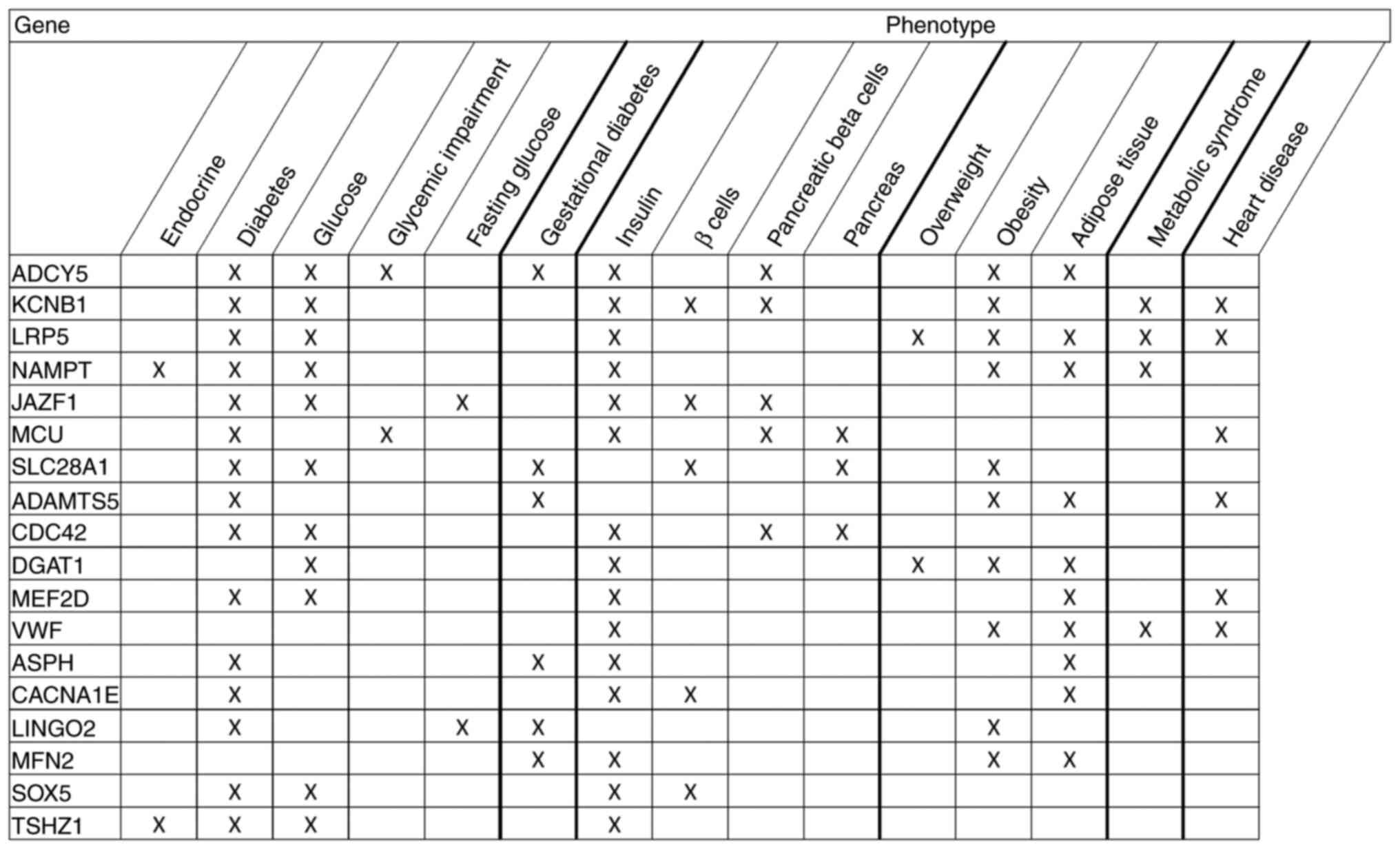
|
1
|
National Diabetes Statistics Report, .
Centers for Diseaes Control and Prevention, U.S. Department of
Health and Human Services; Atlanta, GA: 2017
|
|
2
|
Manolio TA, Collins FS, Cox NJ, Goldstein
DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR,
Chakravarti A, et al: Finding the missing heritability of complex
diseases. Nature. 461:747–753. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xue A, Wu Y, Zhu Z, Zhang F, Kemper KE,
Zheng Z, Yengo L, Lloyd-Jones LR, Sidorenko J, Wu Y, et al:
Genome-wide association analyses identify 143 risk variants and
putative regulatory mechanisms for type 2 diabetes. Nat Commun.
9:29412018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bagchi D: Nutritional and therapeutic
interventions for diabetes and metabolic syndrome. Elsevier;
Waltham, MA: 2018
|
|
5
|
Ahlqvist E, Storm P, Käräjämäki A,
Martinell M, Dorkhan M, Carlsson A, Vikman P, Prasad RB, Aly DM,
Almgren P, et al: Novel subgroups of adult-onset diabetes and their
association with outcomes: A data-driven cluster analysis of six
variables. Lancet Diabetes Endocrinol. 6:361–369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zou X, Zhou X, Zhu Z and Ji L: Novel
subgroups of patients with adult-onset diabetes in Chinese and US
populations. Lancet Diabetes Endocrinol. 7:9–11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Anjana RM, Baskar V, Nair ATN, Jebarani S,
Siddiqui MK, Pradeepa R, Unnikrishnan R, Palmer C, Pearson E and
Mohan V: Novel subgroups of type 2 diabetes and their association
with microvascular outcomes in an Asian Indian population: A
data-driven cluster analysis: The INSPIRED study. BMJ Open Diabetes
Res Care. 8:e0015062020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Herder C, Maalmi H, Strassburger K,
Zaharia OP, Ratter JM, Karusheva Y, Elhadad MA, Bódis K, Bongaerts
BWC, Rathmann W, et al: Differences in biomarkers of inflammation
between novel subgroups of recent-onset diabetes. Diabetes.
70:1198–1208. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hill-Briggs F, Adler NE, Berkowitz SA,
Chin MH, Gary-Webb TL, Navas-Acien A, Thornton PL and Haire-Joshu
D: Social determinants of health and diabetes: A scientific review.
Diabetes Care. 44:258–279. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Flowers E, Won GY and Fukuoka Y: MicroRNAs
associated with exercise and diet: A systematic review. Physiol
Genomics. 47:1–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Parr EB, Camera DM, Burke LM, Phillips SM,
Coffey VG and Hawley JA: Circulating microRNA responses between
‘high’ and ‘low’ responders to a 16-Wk diet and exercise weight
loss intervention. PLoS One. 11:e01525452016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vaishya S, Sarwade RD and Seshadri V:
MicroRNA, proteins, and metabolites as novel biomarkers for
prediabetes, diabetes, and related complications. Front Endocrinol
(Lausanne). 9:1802018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Flowers E, Allen IE, Kanaya AM and
Aouizerat BE: Circulating microRNAs predict glycemic improvement
and response to a behavioral intervention. Biomark Res. 9:652021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Flowers E, Aouizerat BE, Abbasi F,
Lamendola C, Grove KM, Fukuoka Y and Reaven GM: Circulating
microRNA-320a and microRNA-486 predict thiazolidinedione response:
Moving towards precision health for diabetes prevention.
Metabolism. 64:1051–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Flowers E, Gadgil M, Aouizerat BE and
Kanaya AM: Circulating micrornas associated with glycemic
impairment and progression in Asian Indians. Biomark Res. 3:1–8.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Luo J, Zhang H and Lu J: MicroRNAs
in the same clusters evolve to coordinately regulate functionally
related genes. Mol Biol Evol. 33:2232–2247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu H and Leung SW: Identification of
microRNA biomarkers in type 2 diabetes: A meta-analysis of
controlled profiling studies. Diabetologia. 58:900–911. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mononen N, Lyytikäinen LP, Seppälä I,
Mishra PP, Juonala M, Waldenberger M, Klopp N, Illig T, Leiviskä J,
Loo BM, et al: Whole blood microRNA levels associate with glycemic
status and correlate with target mRNAs in pathways important to
type 2 diabetes. Sci Rep. 9:88872019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanaya AM, Araneta MR, Pawlowsky SB,
Barrett-Connor E, Grady D, Vittinghoff E, Schembri M, Chang A,
Carrion-Petersen ML, Coggins T, et al: Restorative yoga and
metabolic risk factors: the practicing restorative yoga vs.
stretching for the metabolic syndrome (PRYSMS) randomized trial. J
Diabetes Complications. 28:406–412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alberti KG, Eckel RH, Grundy SM, Zimmet
PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith
SC Jr, et al: Harmonizing the metabolic syndrome: A joint interim
statement of the international diabetes federation task force on
epidemiology and prevention; national heart, lung, and blood
institute; American heart association; world heart federation;
international atherosclerosis society; and international
association for the study of obesity. Circulation. 120:1640–1645.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Friedewald WT, Levy RI and Fredrickson DS:
Estimation of the concentration of low-density lipoprotein
cholesterol in plasma, without use of the preparative
ultracentrifuge. Clin Chem. 18:499–502. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dweep H, Sticht C, Pandey P and Gretz N:
MiRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statistical Society. Series B
(Methodological). 57:289–300. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Common Metabolic Diseases Knowledge
Portal. https://hugeamp.org/phenotype.html?phenotype=T2DMay
26–2021
|
|
27
|
Sell H, Habich C and Eckel J: Adaptive
immunity in obesity and insulin resistance. Nat Rev Endocrinol.
8:709–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiang W, Zhang B, Lv F, Feng G, Chen L,
Yang F, Zhang K, Cao C, Wang P and Chu M: The potential regulatory
mechanisms of the gonadotropin-releasing hormone in gonadotropin
transcriptions identified with bioinformatics analyses. Reprod Biol
Endocrinol. 15:462017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Johnson JL: Diabetes control in thyroid
disease. Diabetes Spectrum. 19:148–153. 2006. View Article : Google Scholar
|
|
30
|
Dupuis J, Langenberg C, Prokopenko I,
Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji
N, Gloyn AL, et al: New genetic loci implicated in fasting glucose
homeostasis and their impact on type 2 diabetes risk. Nat Genet.
42:105–116. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Esteghamati A, Alamdari A, Zandieh A,
Elahi S, Khalilzadeh O, Nakhjavani M and Meysamie A: Serum visfatin
is associated with type 2 diabetes mellitus independent of insulin
resistance and obesity. Diabetes Res Clin Pract. 91:154–158. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Y, Wang J, Kang R, Dong J, Zhang Y, Liu
F, Yan Y, Zhu R, Xia L, Peng X, et al: Association of KCNB1
polymorphisms with lipid metabolisms and insulin resistance: A
case-control design of population-based cross-sectional study in
Chinese Han population. Lipids Health Dis. 14:1122015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Raum JC, Soleimanpour SA, Groff DN, Coré
N, Fasano L, Garratt AN, Dai C, Powers AC and Stoffers DA: Tshz1
regulates pancreatic β-cell maturation. Diabetes. 64:2905–2914.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fogarty MP, Panhuis TM, Vadlamudi S,
Buchkovich ML and Mohlke KL: Allele-specific transcriptional
activity at type 2 diabetes-associated single nucleotide
polymorphisms in regions of pancreatic islet open chromatin at the
JAZF1 locus. Diabetes. 62:1756–1762. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Adabi E, Omidfar A, Farahani NA, Faghihi
F, Asghar Malek Hosseini SA, Maghbooli Z and Shirvani A: The
association of LRP5 (rs556442) polymorphism with body composition
and obesity in postmenopausal women. Diabetes Metab Syndr.
13:2381–2385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Wang J, Zhang M, Wang G, Shen Y,
Wu D, Wang C, Li L, Ren Y, Wang B, et al: Association of type 2
diabetes mellitus with the interaction between low-density
lipoprotein receptor-related protein 5 (LRP5) polymorphisms and
overweight and obesity in rural Chinese adults. J Diabetes.
9:994–1002. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Perry JR, Voight BF, Yengo L, Amin N,
Dupuis J, Ganser M, Grallert H, Navarro P, Li M, Qi L, et al:
Stratifying type 2 diabetes cases by BMI identifies genetic risk
variants in LAMA1 and enrichment for risk variants in lean compared
to obese cases. PLoS Genet. 8:e10027412012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Manning AK, Hivert MF, Scott RA, Grimsby
JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko
I, et al: A genome-wide approach accounting for body mass index
identifies genetic variants influencing fasting glycemic traits and
insulin resistance. Nat Genet. 44:659–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gene (Internet), . National Library of
Medicine (US). National Center for Biotechnology Information;
Bethesda, MD: 1988
|
|
40
|
Chang AM and Halter JB: Aging and insulin
secretion. Am J Physiol Endocrinol Metab. 284:E7–E12. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Newgard CB and Pessin JE: Recent progress
in metabolic signaling pathways regulating aging and life span. J
Gerontol A Biol Sci Med Sci. 69 (Suppl 1):S21–S27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Corezola do Amaral ME, Kravets V, Dwulet
JM, Farnsworth NL, Piscopio R, Schleicher WE, Miranda JG and
Benninger RK: Caloric restriction recovers impaired β-cell-β-cell
gap junction coupling, calcium oscillation coordination, and
insulin secretion in prediabetic mice. Am J Physiol Endocrinol
Metab. 319:E709–E720. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He XY, Zhao XL, Gu Q, Shen JP, Hu Y and Hu
RM: Calorie restriction from a young age preserves the functions of
pancreatic β cells in aging rats. Tohoku J Exp Med. 227:245–252.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stienstra R, Haim Y, Riahi Y, Netea M,
Rudich A and Leibowitz G: Autophagy in adipose tissue and the beta
cell: Implications for obesity and diabetes. Diabetologia.
57:1505–1516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
American Diabetes Association. 10.
Cardiovascular disease, risk management, . Standards of medical
care in diabetes-2021. Diabetes Care. 44 (Suppl 1):S125–S150. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Condrat CE, Thompson DC, Barbu MG, Bugnar
OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM and Voinea SC: miRNAs
as biomarkers in disease: Latest findings regarding their role in
diagnosis and prognosis. Cells. 9:2762020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|