Introduction
Bone is made up of an organic protein matrix, and is
continuously remodeled in a dynamic process maintained by
osteoblasts and osteoclasts. During this process, osteoblasts and
osteoclasts are important for normal bone metabolism, and
imbalances in the relative activities of these cell types can cause
various pathological conditions, including rheumatoid arthritis and
osteoporosis (1,2). Osteoclasts are multinucleated cells
that are formed by the fusion of mononuclear macrophages derived
from hematopoietic stem cells (3). Macrophage colony-stimulating factor
(M-CSF) and the receptor activator of NF-κB ligand (RANKL) are
important factors that induce the differentiation of osteoclast
precursors into mature osteoclasts (4). A previous study evaluated regulation
of various genes during osteoclastogenesis (5); however, the mechanism underlying
osteoclast differentiation has not been clearly elucidated.
Vega et al (6) proposed that the
RANK/RANKL/osteoprotegerin (OPG) system is a vital signal during
osteoclast differentiation. In addition, a previous study (7) demonstrated that the RANK/RANKL/OPG
system may trigger a series of signaling molecules during
osteoclast differentiation, and the calcium signaling pathway has
been identified as essential during osteoclast differentiation. The
activation of calcium signaling can induce osteoclast
differentiation through Ca2+ oscillations (8). The P2X7 receptor (P2X7R), an ion
channel receptor, constitutes a calcium-permeable cationic channel
(9). It has previously been
reported that P2X7 receptor activation has a vital role in
physiological and pathological reactions, including autophagy and
metabolic responses (10). A
previous study (11) focused on
the role of the P2X7 receptor in skeletal diseases, and loss of
function polymorphisms in the P2X7 receptor have been shown to be
associated with bone loss and increased fracture risk. In addition,
the activation of the P2X7 receptor is vital in the formation and
apoptosis of osteoclasts (12).
These studies suggested that P2X7 may have an important role in
bone disease, and could act as a potential therapeutic target for
medicinal development. Therefore, elucidating its role during
osteoclast differentiation may have critical implications for
therapeutic strategies in bone disease.
Autophagy is a process associated with the
degradation of long-lived proteins and organelles, which has an
important role in maintaining cell homeostasis. Notably, autophagy
has been shown to be activated during osteoclastogenesis (13). A series of autophagy-related
proteins are involved in polarization of osteoclasts, including
autophagy-related 5 (Atg5), Atg7 and LC3 (14). In addition, Beclin-1 serves a
crucial role in osteoclastogenesis. Li and Bai (15) reported that the level of autophagy
was associated with the activation state of P2X7R and P2X7R
activation could affect a series of signals to regulate
autophagy.
Based on previous findings, it was hypothesized that
P2X7 may regulate autophagy and calcium signaling, and thereby
modulate osteoclast differentiation. In addition, it was
hypothesized that RANKL may affect P2X7R activation to modify
osteoclast function by regulating calcium signaling. Therefore, the
present study aimed to investigate the effects of P2X7 on
osteoclast differentiation.
Materials and methods
Animals
In the present study, all mice were provided by
Yangzhou University. BALB/c mice (age, 4 weeks; weight, 10 g) were
given free access to food and water and were sacrificed by cervical
dislocation. A total of 20 mice were used to isolate primary cells.
The health and behavior of mice were monitored every day; no mice
died prior to sacrifice and all mice were maintained in specific
pathogen-free animal housing for 1 month at 20–25°C and 40–70%
humidity under a 12/12-h light/dark cycle.
Cell culture
The 4-week-old BALB/c mice were euthanized and
primary bone marrow cells were isolated from mice femurs; the
marrow was aspirated from the marrow cavity using a syringe to
collect the cells. Cells were maintained in an α-MEM (Gibco; Thermo
Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) for 2 days in a humidified
atmosphere containing 5% CO2 at 37°C. After 2 days, the
suspended cells were collected and cultured in the presence of
M-CSF (30 ng/ml; R&D Systems, Inc.) and RANKL (60 ng/ml;
R&D Systems, Inc.) for 4 days. When the percentage of
osteoclast confluence reached ~60%, they were used for subsequent
experiments. The medium was replaced every 2 days and the cells
used for osteoclastogenesis were from passage one. In addition,
BMMs overexpressing P2X7 were cultured in the presence of M-CSF and
RANKL for 12 h; after 12 h, cells were treated treatment with 2 µM
FK506 (cat. no. HY-13756; MedChemExpress) or 5 mM 3-MA (cat. no.
19312; MedChemExpress) for 12 h in a humidified atmosphere
containing 5% CO2 at 37°C. BMMs were cultured in the
presence of M-CSF as a control.
Tartrate-resistant acid phosphatase
(TRAP) staining
To identify osteoclasts, the cells were washed with
1X PBS, and then fixed in 4% paraformaldehyde for 10 min at room
temperature. The cells were stained using the TRAP staining kit
(MilliporeSigma) according to the manufacturer's instructions. The
TRAP-positive cells were observed under a light microscope (Leica
Microsystems GmbH), and multinucleated (≥3 nuclei) cells were
counted as osteoclasts. The number of mature osteoclasts was
counted in three randomly selected fields of view.
Resorption activity
BMMs (1×106/ml) were seeded in 48-well
plates with 120–140-µm bovine cortical bone slices (IDS Nordic) in
the presence of M-CSF. BMMs were transduced with shRNA-P2X7
adenovirus and HBLV-m-P2Rx7-3×fag-2s Green-Puro for 12 h. RANKL was
then added. After 3 days, the cells were washed with 1X PBS, fixed
in 2.5% glutaraldehyde solution overnight at 4°C, dehydrated using
an alcohol gradient and dried. To observe their morphology, the
specimens were coated with gold using an SCD 500 sputter-coater
(Leica Microsystems GmbH) and examined using a GeminiSEM 300
field-emission environmental scanning electron microscope (Carl
Zeiss). Bone resorption area was calculated by ImageJ 1.48
(National Institutes of Health).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total cellular RNA was extracted from mature
osteoclasts using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Subsequently, RNA was reverse transcribed into complementary (c)DNA
using the HiScript qRT SuperMix kit according to the manufacturers
protocol. (Vazyme Biotech Co., Ltd.). cDNA templates were then
amplified using a ChamQ SYBR q-PCR Master Mix kit according to the
manufacturers protocol. (Vazyme Biotech Co., Ltd.). Thermocycling
conditions were as follows: Initial denaturation for 2 min at 95°C,
followed by 40 cycles of denaturation at 95°C for 15 sec, annealing
at 60°C for 1 min and final extension for 15 sec at 95°C, 15 sec at
60°C and 15 sec at 95°C. The expression levels of target genes
[P2X7, c-fos, NFATc1 and TRAP] were normalized to the reference
gene GAPDH. The 2−ΔΔCq method was applied to calculate
relative gene expression; 2−ΔΔCq =
2−(Cq,target-
Cq,GAPDH)experiment
group-(Cq,target-Cq,GAPDH)control
group (16). All of the
qPCR reactions were performed in triplicate and the primers used
for qPCR are shown in Table I.
The primers were obtained from Shenzhen Huada Gene Technology Co.,
Ltd.
 | Table I.Nucleotide sequences of primers used
for reverse transcription-quantitative polymerase chain
reaction. |
Table I.
Nucleotide sequences of primers used
for reverse transcription-quantitative polymerase chain
reaction.
| Gene name | Annealing
temperature (°C) | Forward primer
sequence | Reverse primer
sequence |
|---|
| GAPDH | 60 |
5-TCAAGAAGGTGGTGAAGCAG-3' |
5-AGTGGGAGTTGCTGTTGAAGT-3' |
| NFATc1 | 58 |
5-CTCGAAAGACAGCACTGGAGCAT-3′ |
5-CGGCTGCCTTCCGTCTCATAG-3' |
| TRAP | 62 |
5-CTGGAGTGCACGATGCCAGCGACA-3' |
5-TCCGTGCTCGGCGATGGACCAGA-3' |
| c-fos | 64 |
5-GGAGAATCCGAAGGGAACGG-3′ |
5-GCAATCTCAGTCTGCAACGC-3' |
| CAII | 52 |
5-GGGGATACAGCAAGCACAAC-3' |
5-GACTGCCGGTCTCCATTG-3' |
| CK | 52 |
5-CGAAAAGAGCCTAGCGAACA-3' |
5-TGGGTAGCAGCAGAAACTTG-3' |
| MMP-9 | 52 |
5-ACGACATAGACGGCATCCA-3′ |
5-GCTGTGGTTCAGTTGTGGTG-3′ |
| P2X7 | 60 |
5′-AGATCGTGGAGAATGGAGTG-3′ |
5′-TTCTCGTGGTGTAGTTGTGG-3′ |
Western blot analysis
Western blot analysis was performed to detect
protein expression levels. Total protein was isolated from mature
osteoclasts using radioimmunoprecipitation assay buffer (Beyotime
Institute of Biotechnology). Equal amounts of protein (25 µg/lane)
were separated by SDS-PAGE on 10% gels and transferred onto PVDF
membranes. After blocking with 5% non-fat skim milk at room
temperature for 2 h, the membranes were incubated at 4°C overnight
with primary antibodies against P2X7 (cat. no. NBP1-20180; 1:1,000;
Novus Biologicals, LLC), LC3 (cat. no. L7543; 1:1,000;
MilliporeSigma), TRAP (cat. no. ab238033; 1:1,000), CK (cat. no.
ab37259; 1:1,000), CAII (cat. no. ab182611; 1:1,000), MMP-9 (cat.
no. ab76003; 1:1,000) (all from Abcam), NFATc1 (cat. no. 8032;
1:1,000), c-fos (cat. no. 2250; 1:1,000), calcineurin (cat. no.
2614; 1:1,000), Beclin-1 (cat. no. 3495; 1:1,000) and β-actin (cat.
no 4970; 1:1,000; all from Cell Signaling Technology, Inc.).
Subsequently, membranes were incubated for 2 h at room temperature
with horseradish peroxidase-conjugated secondary antibodies (cat.
nos. 7074 and 7076; 1:1,000; Cell Signaling Technology, Inc.). The
proteins were visualized using an ECL kit (cat. no. P2300; New Cell
& Molecular Biotech) and the data were analyzed using ImageJ
software; the protein expression levels were calculated according
to the gray value of the bands.
Cell transduction
A P2X7 short hairpin RNA (shRNA) knockdown construct
(pHBAd-U6-Scarmble-GFP) and P2X7 overexpression system were
constructed by Hanheng Biology. Empty vector lentivirus was used as
negative control. Knockdown and overexpression were performed using
shRNA-P2X7 adenovirus and HBLV-m-P2Rx7-3×fag-2s Green-Puro,
respectively. BMMs were grown in α-MEM supplemented with 10% fetal
bovine serum at 37°C and 5% CO2. An adenoviral vector
containing shRNA-P2X7 and HBLV-m-P2Rx7-3×fag-2s Green-Puro (MOI,
1–3) were introduced into BMMs in the presence of M-CSF (30 ng/ml);
after 24 h, transduction efficiency was observed using fluorescence
microscopy and silencing and overexpression efficiency were
determined by western blotting. The sequences of the shRNA-P2X7 and
non-targeting negative control (NC)-shRNA were as follows: NC-shRNA
sense,
5′-AATTCGTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGACACGTTCGCAGAATTTTTTG-3′
and antisense,
5′-GATCCAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGAATTACGTGACACGTTCGGAGAACT-3′
and shRNA-P2X7 sense,
5′-AATTCGACGAAGTTAGGACACAGCATCTTTGTTCAAGAGACAAAGATGCTGTGTCCTAACTTCGTTTTTTTG-3′
and antisense,
5′-GATCCAAAAAAACGAAGTTAGGACACAGCATCTTTGTctcttgaaCAAAGATGCTGTGTCCTAACTTCGTCG-3′.
Immunofluorescence staining
BMMs on glass coverslips were incubated in the
presence of M-CSF and RANKL, and cells were transduced with
adenovirus-mediated shRNA-P2X7 and NC-shRNA. For SP2X7
overexpression, cells were transduced with HBLV-m-P2Rx7-3×fag-2s
Green-Puro and NC. When the confluence of osteoclasts reached ~60%,
they were used for the subsequent experiment. Osteoclasts were
fixed with 4% paraformaldehyde for 20 min at room temperature,
washed in PBS twice (5 min/wash), permeabilized with 0.5% Triton
X-100 for 30 min at room temperature, washed in PBS twice (5
min/wash) and blocked with 5% FBS for 30 min at room temperature.
Osteoclasts were then incubated overnight at 4°C with an anti-LC3
primary antibody (cat. no. L7543; 1:200; Sigma-Aldrich; Merck
KGaA). Subsequently, the coverslips were washed and incubated for 2
h at room temperature with Alexa Fluor® 488-conjugated
secondary antibody (cat. no. A0428; 1:200; Beyotime Institute of
Biotechnology), followed by two further washed with PBS (5
min/wash). Cells were then observed under a confocal microscope. In
the present study, mature osteoclasts were chosen for confluence
analysis. Fluorescence intensity was analyzed using ImageJ.
Intracellular Ca2+
measurement
BMMs were seeded onto confocal
microscope-specialized cover glass. When the confluence of
osteoclasts reached ~60%, they were used for the subsequent
experiment. Osteoclasts were incubated with 1 µM Fluo3-AM (Beijing
Solarbio Science & Technology Co., Ltd.) for 30 min at 5%
CO2 and 37°C. The medium was then replaced and
intracellular Ca2+ concentrations were observed using a
confocal microscope. Fluorescence images were captured and analyzed
using ImageJ.
Statistical analysis
Each experiment was repeated at least three times.
All of experimental data were analyzed using SPSS 21.0 (IBM Corp.).
An independent samples Student's t-test was used for two-sample
comparisons with data exhibiting a normal distribution. Multiple
groups were compared using one-way analysis of variance followed by
Tukey's or Tamhane's test. P<0.05 was considered to indicate a
statistically significant difference.
Results
P2X7R expression and Ca2+
levels increase during osteoclast differentiation
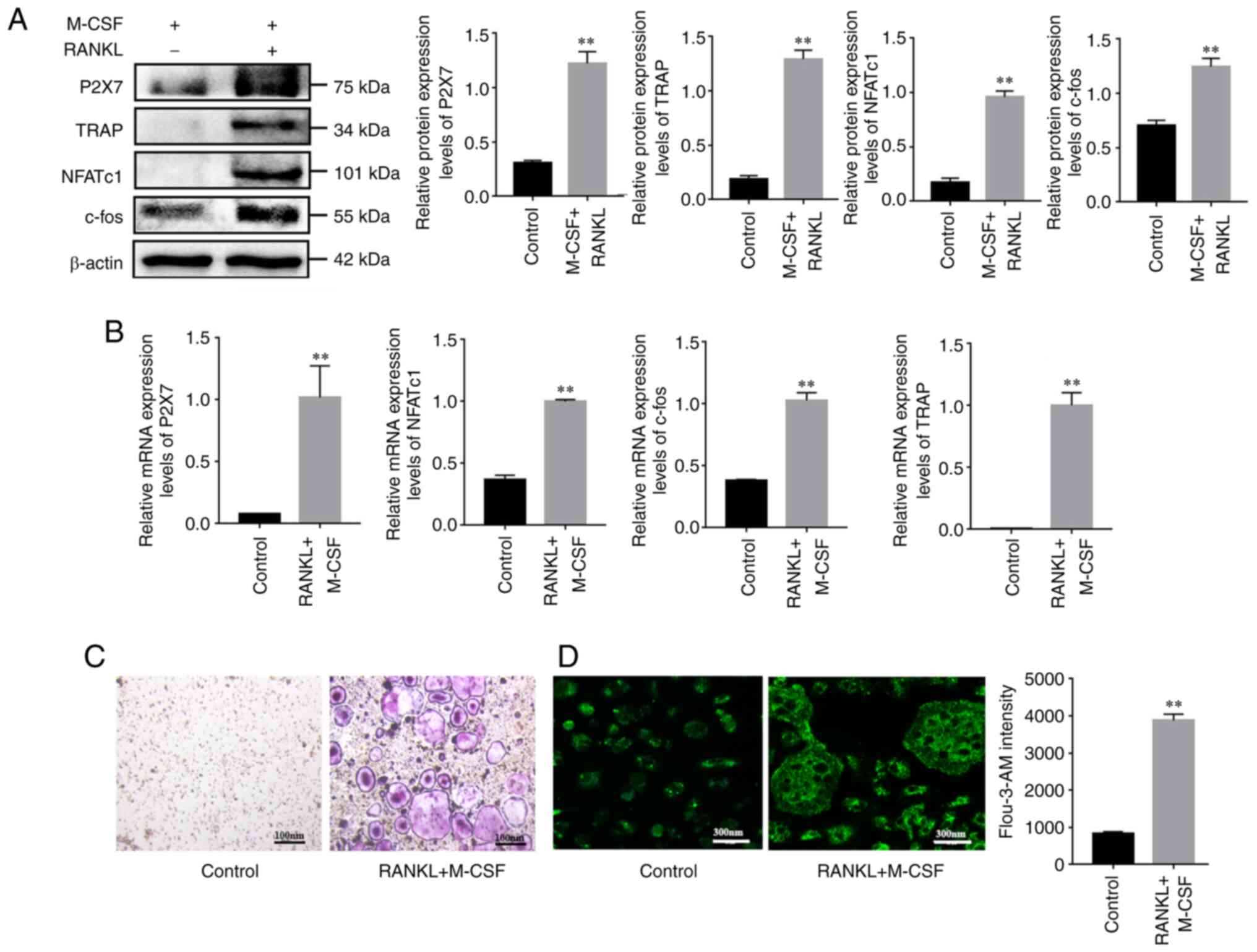
The results of the present study demonstrated that
the expression levels of osteoclastogenesis key genes were
significantly increased at the mRNA and protein levels following
M-CSF and RANKL stimulation compared with in the control cells
(M-CSF stimulation; Fig. 1A and
B). Furthermore, the number of TRAP-positive osteoclasts was
markedly increased after stimulating BMMs with M-CSF and RANKL for
4 days (Fig. 1C). To determine
whether P2X7R was involved in regulating osteoclast differentiation
by changing intracellular Ca2+ influx, the present study
measured the mRNA and protein expression levels of P2X7, as well as
Ca2+ concentration, during osteoclast differentiation.
The results suggested that the expression levels of P2X7R were
significantly upregulated in mature osteoclasts compared with those
in the control group (Fig. 1A and
B). In addition, Ca2+ concentration was
significantly increased in mature osteoclasts compared with that in
the control group (Fig. 1D).
Knockdown of P2X7R inhibits osteoclast
bone resorption, Ca2+ signaling and autophagy during
osteoclast differentiation
Based on the aforementioned results, the present
study knocked down P2X7R expression using adenovirus-mediated
transduction of BMMs with P2X7-shRNA, and induced overexpression of
P2X7R in BMMs using HBLV-m-P2Rx7-3×fag-2s Green-Puro. Knockdown and
overexpression efficiencies were confirmed by western blot analysis
(Fig. 2A and B). As expected,
P2X7R knockdown significantly reduced the number of TRAP-positive
osteoclasts (Fig. 2C) and
inhibited bone resorption in the presence of RANKL and M-CSF
(Fig. 2D. In addition, P2X7R
knockdown in the presence of M-CSF and RANKL significantly
downregulated the protein expression levels of osteoclast key
genes, including TRAP, CK, CAII, MMP-9, NFATc1 and c-fos compared
with in the NC group (Fig. 2A).
By contrast, P2X7 overexpression exerted the opposite effects
(Fig. 2B-D). These results
indicated that the absence of P2X7R may inhibit osteoclast
differentiation.
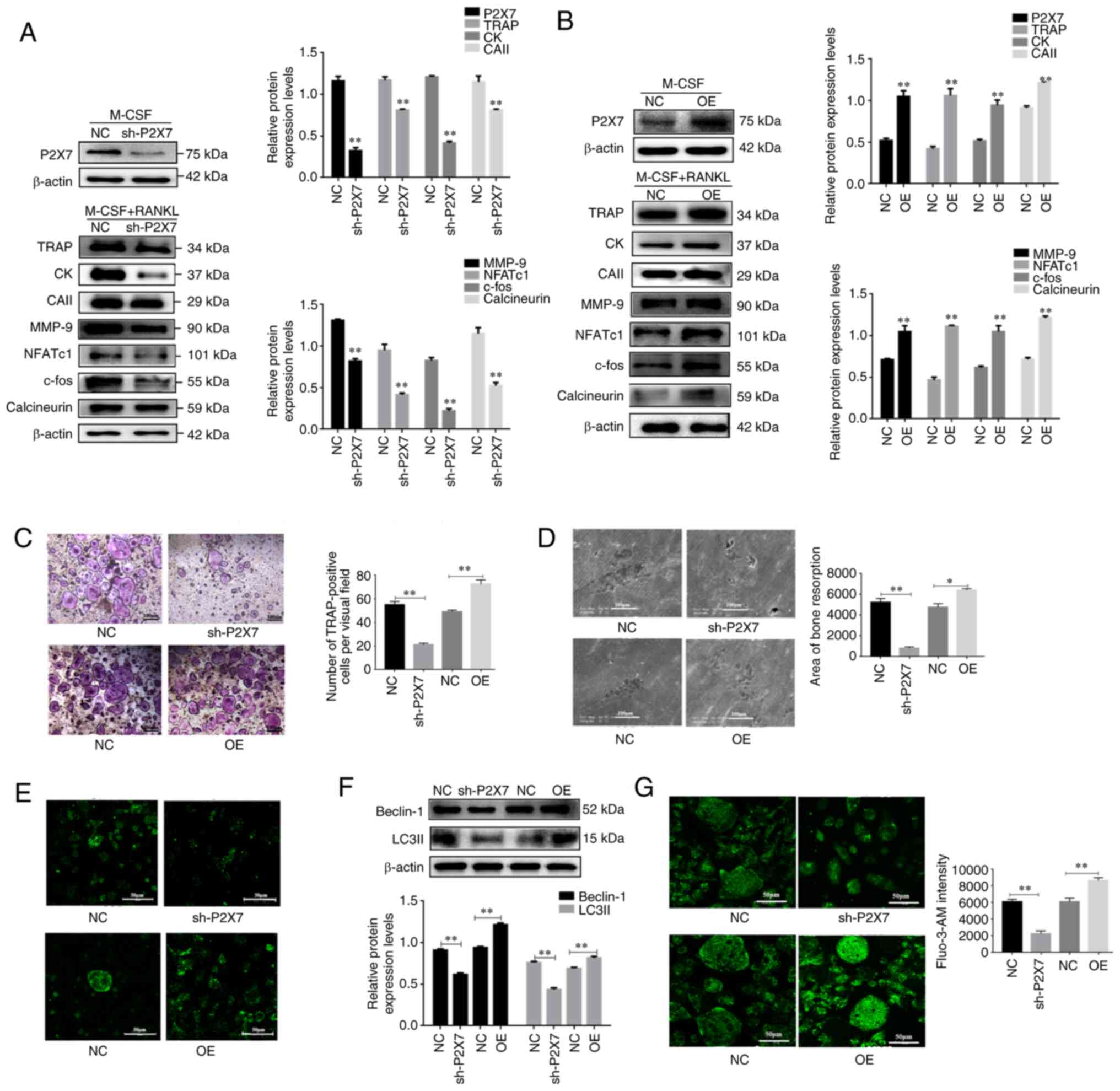 | Figure 2.Effects of P2X7 overexpression on
osteoclast differentiation, Ca2+/calcineurin/NFATc1
signaling and autophagy. Bone marrow mononuclear macrophages were
transduced with sh-P2X7 or transduced with HBLV-m-P2Rx7-3×fag-2s
Green-Puro in the presence of M-CSF with or without RANKL. (A and
B) Western blot analysis of the protein expression levels of P2X7,
TRAP, CK, CAII, MMP-9, NFATc1, c-fos and calcineurin. (C and D)
TRAP-positive multinucleated cells were counted as osteoclasts and
scanning electron microscopy was used to observe bone resorption
lacuna. (E and F) Western blot analysis of the protein expression
levels of Beclin-1 and LC3II, and immunofluorescence analysis of
LC3 fluorescent puncta. (G) Fluo3-AM staining was performed to
detect intracellular Ca2+ concentration. Data are
presented as the mean ± SD from three independent experiments.
*P<0.05, **P<0.01 vs. NC group or as indicated. CAII,
carbonic anhydrase II; CK, cathepsin K; M-CSF, macrophage
colony-stimulating factor; MMP-9, matrix metalloproteinase-9; NC,
negative control; NFATc1, nuclear factor of activated T cells c1;
OE, overexpression; RANKL, receptor activator of NF-κB ligand; sh,
short hairpin; TRAP, tartrate-resistant acid phosphatase. |
Ca2+ signaling is an essential axis of
osteoclast differentiation. Expanding on the aforementioned
observations of the role of Ca2+ signaling during
osteoclastogenesis, the present study revealed that knockdown of
P2X7R significantly inhibited the expression levels of calcineurin,
whereas P2X7R overexpression upregulated the expression of
calcineurin (Fig. 2A and B).
Moreover, knockdown of P2X7R suppressed Ca2+
concentration in M-CSF and RANKL-stimulated BMMs (Fig. 2G). By contrast, P2X7R
overexpression exerted the opposite effects (Fig. 2G). These results suggested that
the absence of P2X7R suppressed Ca2+ signaling during
osteoclast differentiation.
The present study also investigated the effects of
P2X7R on autophagy. Immunofluorescence analysis revealed that
knockdown of P2X7R led to a reduction in LC3 puncta in mature
osteoclasts, whereas P2X7R overexpression increased LC3
immunofluorescence puncta (Fig.
2E). Furthermore, knockdown of P2X7R significantly reduced the
protein expression levels of Beclin-1 and LC3II, as determined by
western blotting (Fig. 2F). Taken
together, these results suggested that knockdown of P2X7 inhibited
autophagy during osteoclast differentiation.
Inhibition of autophagy and
calcineurin attenuates P2X7R overexpression-induced osteoclast
differentiation
To explore whether
Ca2+/calcineurin/NFATc1 signaling was involved in P2X7
overexpression-induced osteoclast differentiation and autophagy
activation, P2X7R overexpression-induced BMMs were treated with
FK506 (a calcineurin inhibitor) in the presence of M-CSF and RANKL.
The results confirmed that FK506 significantly inhibited P2X7R
overexpression-induced osteoclast differentiation in the presence
of M-CSF and RANKL, and decreased the protein expression levels of
NFATc1, calcineurin and TRAP (Fig. 3A
and B). In addition, FK506 significantly attenuated P2X7R
overexpression-induced activation of autophagy, as indicated by a
decrease in the protein expression levels of Beclin-1 and LC3II
(Fig. 3B). These results
indicated that Ca2+/calcineurin/NFATc1 signaling may be
involved in P2X7 overexpression-induced osteoclast differentiation
and activation of autophagy.
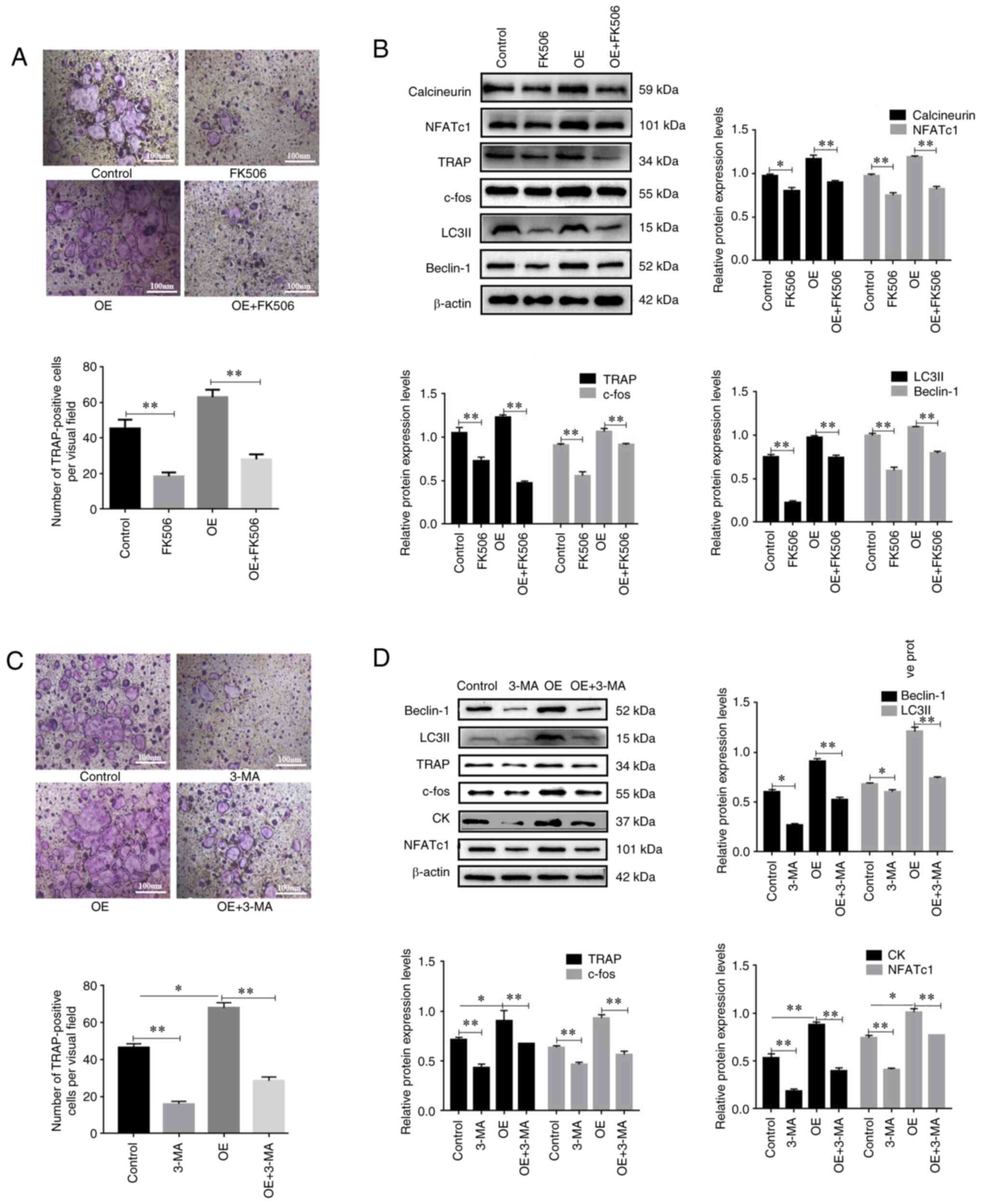 | Figure 3.Calcineurin/NFATc1 signaling and
autophagy are involved in P2X7R overexpression-induced osteoclast
differentiation. Bone marrow mononuclear macrophages were cultured
with M-CSF and RANKL. Cells were transduced with
HBLV-m-P2Rx7-3×fag-2s Green-Puro in the presence of M-CSF and
RANKL, followed by treatment with FK506 (2 µM) for 12 h. (A)
TRAP-positive multinucleated cells were counted as osteoclasts. (B)
Western blot analysis of the protein expression levels of
calcineurin, NFATc1, TRAP, c-fos, Beclin-1 and LC3II. Cells were
transduced with HBLV-m-P2Rx7-3×fag-2s Green-Puro in the presence of
M-CSF and RANKL, followed by treatment with 3-MA (5 mM) for 12 h.
(C) TRAP-positive multinucleated cells were counted as osteoclasts.
(D) Western blot analysis of the protein expression levels of
NFATc1, c-fos, TRAP, CK, Beclin-1 and LC3II. Data are presented as
the mean ± SD from three independent experiments. *P<0.05,
**P<0.01 vs. NC group or as indicated. CK, cathepsin K; M-CSF,
macrophage colony-stimulating factor; NFATc1, nuclear factor of
activated T cells c1; OE, overexpression; RANKL, receptor activator
of NF-κB ligand; sh, short hairpin; TRAP, tartrate-resistant acid
phosphatase. |
The present study also investigated whether
autophagy was involved in P2X7R overexpression-induced osteoclast
differentiation. The present results showed that 3-MA (an autophagy
inhibitor) significantly inhibited the P2X7R overexpression-induced
expression levels of Beclin-1 and LC3II (Fig. 3D). Notably, it was revealed that
P2X7R overexpression significantly increased the number of
TRAP-positive osteoclasts (Fig.
3C). Furthermore, P2X7R overexpression increased the protein
expression levels of osteoclast key genes (Fig. 3D). 3-MA reversed the effect of OE
on TRAP cells and protein expression (Fig. 3D). These results indicated that
autophagy may be involved in P2X7 overexpression-induced osteoclast
differentiation.
Discussion
The present study revealed that P2X7R expression was
significantly increased at both the mRNA and protein levels in
mature osteoclasts, indicating a potential role for P2X7R during
osteoclast differentiation. Accordingly, P2X7R knockdown
significantly reduced the number of TRAP-positive osteoclasts and
the expression levels of osteoclastogenesis-related genes in the
presence of M-CSF and RANKL, whereas P2X7R overexpression exerted
the opposite effects. These results suggest that P2X7R may be
involved in osteoclast differentiation, and that absence of P2X7R
could inhibit osteoclast differentiation.
Given the complexity of cell differentiation, the
role of P2X7R during osteoclast differentiation is unclear.
However, the present study revealed the effects of P2X7R on
osteoclast differentiation, which contrasted with the results of
some previous studies. Wang et al (17) examined the effect of P2X7R on
osteoclast formation and bone loss using a mouse model of
osteoporosis. The results of this previous study showed that loss
of P2X7R increased estrogen-deficient bone loss, likely due to
increased activity of osteoclasts in the absence of estrogen. In
this previous study, osteoclast precursors were isolated from
P2RX7−/− mice, which generated more mature osteoclasts,
but the resorption activity of osteoclasts was significantly
reduced. This would suggest that the increased number of
osteoclasts observed in vivo may well be a compensatory
mechanism for their reduced activity. Notably, osteoclasts from
P2RX7−/− mice exhibited an inhibition in osteoclast
resorption function. By contrast, Agrawal et al (18) revealed that a P2X7R-specific
antagonist significantly inhibited osteoclast formation and bone
resorption. Gartland et al (19) also demonstrated that blockade of
P2X7R inhibited osteoclast formation in vitro. However, in a
previous study, P2X7R-deficient mice displayed no overt skeletal
problems, and were able to form multinucleated cells (20). It was hypothesized that other
regulatory factors may have a compensatory effect, thus short-term
loss of P2X7R would not affect osteoclast function. These findings
indicated that P2X7R may exert specific effects on osteoclast
formation, which require further exploration in vivo and
vitro.
In the present study, the absence of P2X7R inhibited
osteoclast formation and bone resorption in BMMs, and how P2X7R
affects osteoclastogenesis was examined. NFATc1 has an essential
role in osteoclastogenesis. In response to activation of
Ca2+/calcineurin signaling, NFATc1 is a key modulator,
which activates transcription of a series of key osteoclast genes
by translocating to the nucleus (21). NFATc1 is not only required but
also sufficient for osteoclastogenesis, as its overexpression in
osteoclast precursors has been shown to induce osteoclast
differentiation in the absence of RANKL (22). Ca2+/calcineurin/NFATc1
signaling serves an important role in osteoclast differentiation,
and can accelerate osteoclast differentiation through inducing an
increase in intracellular Ca2+ concentration (23). P2X7 directly regulates
differentiation and function of osteoclasts by mediating
intracellular Ca2+ influx. P2X7-mediated Ca2+
influx during osteoclast differentiation is critical when ATP or
BzATP concentration is relatively high; a previous study revealed
that ATP can stimulate the formation and resorption activity of
osteoclasts by activating purinergic signaling (24). P2X7-mediated Ca2+
influx affects intracellular Ca2+ levels, ensuring
NFATc1-mediated gene transcription, and thereby regulating
osteoclast differentiation; however, it remains to be determined as
to whether P2X7 is involved in ATP-induced osteoclast
differentiation. In the present study, knockdown of P2X7R was
associated with reduced intracellular Ca2+
concentration, reduced NFATc1 activity, and reduced osteoclast
differentiation and bone resorption. Therefore, based on these
observations, the present study explored whether
Ca2+/calcineurin/NFATc1 signaling was involved in
P2X7-mediated osteoclast differentiation. The results revealed that
knockdown of P2X7 significantly inhibited
Ca2+/calcineurin/NFATc1 signaling during osteoclast
differentiation. By contrast, P2X7R overexpression increased
Ca2+ concentration and activated calcineurin/NFATc1
signaling. Notably, inhibition of calcineurin abrogated the effect
of P2X7R overexpression on osteoclast differentiation.
Autophagy is the chief machinery for bulk
degradation of superfluous or aberrant cytoplasmic components.
Dysregulated autophagy has been involved in the development of
several diseases. Zhang et al (25) reported that autophagy was
associated with the formation and development of osteoporosis. This
previous study indicated that autophagy may have an essential role
in modulating bone metabolism and in disease. Although increasing
evidence has indicated that OPG may inhibit osteoclast
differentiation by promoting autophagy (26,27), other studies have identified a
potential mechanism for autophagy in promoting osteoclast
differentiation (28,29). Autophagy-related proteins,
including Beclin-1and Atg7, have been shown to increase in the
osteoclasts of patients with rheumatoid arthritis, leading to bone
destruction (30). A previous
study suggested that autophagy activation promoted osteoclast
formation and differentiation. By contrast, 3-MA (an autophagy
inhibitor) interrupted TRPV4 overexpression-induced
osteoclastogenesis in a previous study; these results demonstrated
that the absence of TRPV4 inhibited osteoclast differentiation
(31). The present study revealed
that knockdown of P2X7R reduced the expression levels of
autophagy-related proteins (LC3II and Beclin-1) during osteoclast
differentiation, whereas P2X7R overexpression exerted the opposite
effects. Taken together, interrupting autophagy may affect
osteoclast formation and differentiation. Notably, a previous study
suggested that the overactivation of P2X7R may increase calcium
influx, which may activate a series of downstream signaling
(32). In general, autophagy may
serve a dual role in regulating osteoclast differentiation and cell
death. The present study also revealed that inhibition of
calcineurin by FK506 abrogated P2X7R overexpression-induced
autophagy. Moreover, inhibition of autophagy attenuated P2X7R
overexpression-induced osteoclast differentiation. It is well known
that bone homeostasis is maintained by the communication between
osteoblasts, osteoclasts and osteocytes. Autophagy is a complex
process, which exerts dual roles in other bone cells. Another study
demonstrated that overactivation of autophagy promoted osteoblast
apoptosis and bone loss (33);
however, further studies are required to confirm the relationship
between autophagy dysfunction, and bone loss and osteoclast
differentiation.
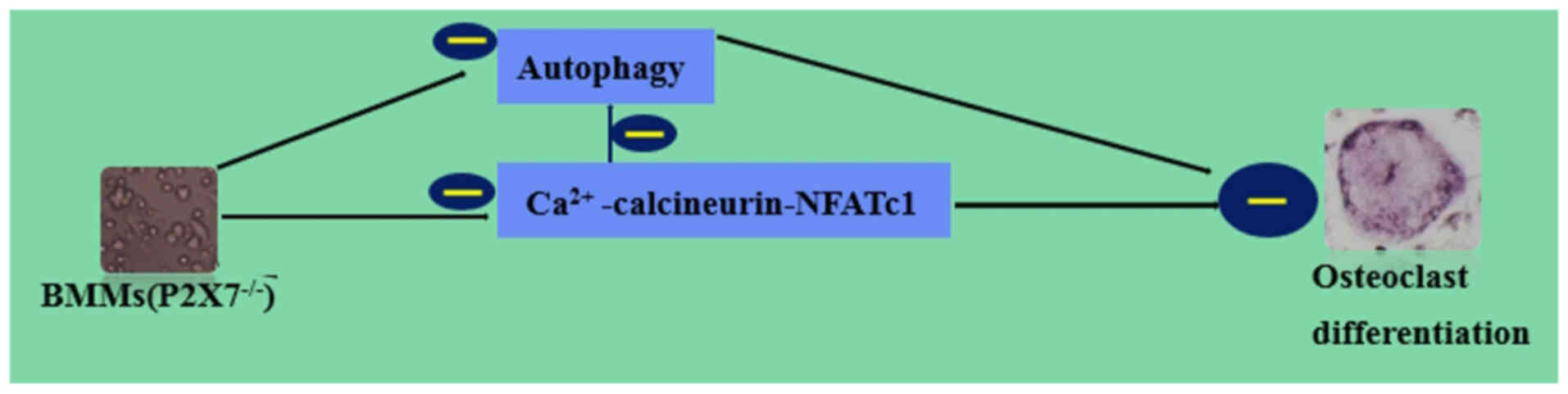
In conclusion, the results of the present study
indicated that knockdown of P2X7R suppressed osteoclast
differentiation by inhibiting autophagy through the
Ca2+/calcineurin/NFATc1 signaling pathway (Fig. 4). However, the relationship
between the absence of P2X7 receptor and the progression of
osteoporosis in mice remains elusive; further study of knockout of
P2X7 receptor in a mouse model should be performed to determine the
association between osteoporosis and P2X7R. The absence of P2X7R
affects osteoclast formation, thereforeP2X7R may be a useful
therapeutic target for metabolic bone disease.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National Nature
Science Foundation of China (grant nos. 31702304, 31672620,
31872533 and 31872534), the Natural Science Foundation of Jiangsu
Province (grant no. BK20150447), the Natural Science Foundation
Research Grants of Jiangsu Province (grant no. BK20181452), the
China Postdoctoral Science Foundation (grant no. 2017M611932), and
the Priority Academic Program Development of Jiangsu Higher
Education Institutions (PAPD) and the Graduate Innovation Project
of Jiangsu Province.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZPL and HYZ conceived and designed the study. YGM
and RD performed the cell transduction and culture. YGM and RLS
performed the western blotting and immunofluorescence. YGM and HZ
performed the TRAP staining and bone resorption. YGM, RD and HYZ
analyzed the data. YGM, RD and HZ organized the data. YGM and RD
wrote the manuscript. ZPL and HYZ proofread the manuscript and
confirm the authenticity of all the raw data. All authors
contributed to the article, and read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal experiments and procedures were approved
by the Animal Care and Use Committee of Yangzhou University, China
[approval number: SYXK (Su) 2016–0020].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Atg5
|
autophagy-related 5
|
|
CK
|
cathepsin K
|
|
CAII
|
carbonic anhydrase II
|
|
MMP-9
|
matrix metalloproteinase-9
|
|
NC
|
negative control
|
|
OPG
|
osteoprotegerin
|
|
OE
|
overexpression
|
|
TRAP
|
tartrate-resistant acid
phosphatase
|
|
NFATc1
|
nuclear factor of activated T cells
c1
|
References
|
1
|
Murata K, Fang C, Terao C, Giannopoulou
EG, Lee YJ, Lee MJ, Mun SH, Bae S, Qiao Y, Yuan R, et al:
Hypoxia-sensitive COMMD1 integrates signaling and cellular
metabolism in human macrophages and suppresses osteoclastogenesis.
Immunity. 47:66–79.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alves CH, Farrell E, Vis M, Colin EM and
Lubberts E: Animal models of bone loss in inflammatory arthritis:
From cytokines in the bench to novel treatments for bone loss in
the bedside-a comprehensive review. Clin Rev Allergy Immunol.
51:27–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al Faraj A, Shaik AS and Alnafea M:
Intrapulmonary administration of bone-marrow derived M1/M2
macrophages to enhance the resolution of LPS-induced lung
inflammation: Noninvasive monitoring using free-breathing MR and CT
imaging protocols. BMC Med Imaging. 15:162015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakashima T, Hayashi M, Fukunaga T, Kurata
K, Oh-Hora M, Feng JQ, Bonewald LF, Kodama T, Wutz A, Wagner EF, et
al: Evidence for osteocyte regulation of bone homeostasis through
RANKL expression. Nat Med. 17:1231–1234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang XL, Liu C, Shi XM, Cheng YT, Zhou Q,
Li JP and Liao J: Zoledronic acid inhibits osteoclastogenesis and
bone resorptive function by suppressing RANKL-mediated NF-κB and
JNK and their downstream signaling pathways. Mol Med Rep.
25:592022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vega D, Maalouf NM and Sakhaee K: Clinical
review #: The role of receptor activator of nuclear factor-kappaB
(RANK)/RANK ligand/osteoprotegerin: Clinical implications. J Clin
Endocrinol Metab. 92:4514–4521. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou S, Yuan X, Liu Q, Zhang X, Pan X,
Zang L and Xu L: BAPTA-AM, an intracellular calcium chelator,
inhibits RANKL-induced bone marrow macrophages differentiation
through MEK/ERK, p38 MAPK and Akt, but not JNK pathways. Cytokine.
52:210–214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin H, Yao L, Chen K, Liu Y, Wang Q, Wang
Z, Liu Q, Cao Z, Kenny J, Tickner J, et al: Evodiamine inhibits
RANKL-induced osteoclastogenesis and prevents ovariectomy-induced
bone loss in mice. J Cell Mol Med. 23:522–534. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Surprenant A, Rassendren F, Kawashima E,
North RA and Buell G: The cytolytic P2Z receptor for extracellular
ATP identified as a P2X receptor (P2X7). Science. 272:735–748.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Orioli E, De Marchi E, Giuliani AL and
Adinolfi E: P2X7 receptor orchestrates multiple signalling pathways
triggering inflammation, autophagy and metabolic/trophic responses.
Curr Med Chem. 24:2261–2275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jørgensen NR, Syberg S and Ellegaard M:
The role of P2X receptors in bone biology. Curr Med Chem.
22:902–914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gartland A, Buckley KA, Bowler WB and
Gallagher JA: Blockade of the pore-forming P2X7 receptor inhibits
formation of multinucleated human osteoclasts in vitro. Calcif
Tissue Int. 73:361–369. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ke D, Fu X, Xue Y, Wu H, Zhang Y, Chen X
and Hou J: IL-17A regulates the autophagic activity of osteoclast
precursors through RANKL-JNK1 signaling during osteoclastogenesis
in vitro. Biochem Biophys Res Commun. 497:890–896. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
DeSelm CJ, Miller BC, Zou W, Beatty WL,
van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL and
Virgin HW: Autophagy proteins regulate the secretory component of
osteoclastic bone resorption. Dev Cell. 21:966–974. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Huang Z and Bai L: The P2X7 receptor
in osteoarthritis. Front Cell Dev Biol. 9:6283302021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang N, Agrawal A, Jørgensen NR and
Gartland A: P2X7 receptor regulates osteoclast function and bone
loss in a mouse model of osteoporosis. Sci Rep. 8:35072018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agrawal A, Buckley KA, Bowers K, Furber M,
Gallagher JA and Gartland A: The effects of P2X7 receptor
antagonists on the formation and function of human osteoclasts in
vitro. Purinergic Signal. 6:307–315. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gartland A, Buckley KA, Hipskind RA, Perry
MJ, Tobias JH, Buell G, Chessell I, Bowler WB and Gallagher JA:
Multinucleated osteoclast formation in vivo and vitro by P2X7
receptor-deficient mice. Crit Rev Eukaryot Gene Expr. 23:243–253.
2003.PubMed/NCBI
|
|
20
|
Ke HZ, Qi H, Weidema AF, Zhang Q,
Panupinthu N, Crawford DT, Grasser WA, Paralkar VM, Li M, Audoly
LP, et al: Deletion of the P2X7 nucleotide receptor reveals its
regulatory roles in bone formation and resorption. Mol Endocrinol.
17:1356–1367. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fujita K, Iwasaki M, Ochi H, Fukuda T, Ma
C, Miyamoto T, Takitani K, Negishi-Koga T, Sunamura S, Kodama T, et
al: Vitamin E decreases bone mass by stimulating osteoclast fusion.
Nat Med. 18:589–594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takayanagi H, Kim S, Koga T, Nishina H,
Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, et al:
Induction and activation of the transcription factor NFATc1 (NFAT2)
integrate RANKL signaling in terminal differentiation of
osteoclasts. Dev Cell. 3:889–901. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Xu H, Han Z, Chen P, Yu Q, Lei Y,
Li Z, Zhao M and Tian J: Pulsed electromagnetic field inhibits
RANKL-dependent osteoclastic differentiation in RAW264.7 cells
through the Ca2+-calcineurin-NFATc1 signaling pathway.
Biochem Biophys Res Commun. 482:289–295. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morrison MS, Turin L, King BF, Burnstock G
and Arnett TR: ATP is a potent stimulator of the activation and
formation of rodent osteoclasts. J Physiol. 511:495–500. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Guo YF, Liu YZ, Liu YJ, Xiong DH,
Liu XG, Wang L, Yang TL, Lei SF, Guo Y, et al: Pathway-based
genome-wide association analysis identified the importance of
regulation-of-autophagy pathway for ultradistal radius BMD. J Bone
Miner Res. 25:1572–1580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tong X, Min W, Li S, Chen M, Song R, Bian
J, Gu J and Liu Z: Beclin 1 positively regulates
osteoprotegerin-induced inhibition of osteoclastogenesis by
increasing autophagy in vitro. Differentiation. 121:35–43. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji Li, Sun Z, Lin Y, Yan Y, Yan H, Jing B
and Han Z: Syndecan 4 contributes to osteoclast differentiation
induced by RANKL through enhancing autophagy. Int Immunopharmacol.
91:1072752021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tong X, Gu J, Song R, Wang D, Sun Z, Sui
C, Zhang C, Liu X, Bian J and Liu Z: Osteoprotegerin inhibit
osteoclast differentiation and bone resorption by enhancing
autophagy via AMPK/mTOR/p70S6K signaling pathway in vitro. J Cell
Biochem. Sep 6–2018.(Epub ahead of print). doi:
10.1002/jcb.27468.
|
|
29
|
Chu B, Chen S, Zheng X, Ye J, Cheng X,
Zhang L, Guo D, Wang P, Hong D and Hong Z: Nepetin inhibits
osteoclastogenesis by inhibiting RANKL-induced activation of NF-κB
and MAPK signalling pathway, and autophagy. J Cell Mol Med.
24:14366–14380. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cejka D, Hayer S, Niederreiter B, Sieghart
W, Fuereder T, Zwerina J and Schett G: Mammalian target of
rapamycin signaling is crucial for joint destruction in
experimental arthritis and is activated in osteoclasts from
patients with rheumatoid arthritis. Arthritis Rheum. 62:2294–2302.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao B, Dai X and Wang W: Knockdown of
TRPV4 suppresses osteoclast differentiation and osteoporosis by
inhibiting autophagy through Ca2+ -calcineurin-NFATc1
pathway. J Cell Physiol. 234:6831–6841. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sekar P, Huang DY, Hsieh SL, Chang SF and
Lin WW: AMPK-dependent and independent actions of P2X7 in
regulation of mitochondrial and lysosomal functions in microglia.
Cell Commun Signal. 16:832018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ran D, Ma Y, Liu W, Luo T, Zheng J, Wang
D, Song R, Zhao H, Zou H, Gu J, et al: TGF-β-activated kinase 1
(TAK1) mediates cadmium-induced autophagy in osteoblasts via the
AMPK/mTORC1/ULK1 pathway. Toxicology. 442:1525382020. View Article : Google Scholar : PubMed/NCBI
|