Introduction
The increasing rates of morbidity and mortality in
patients with ischemic heart disease (IHD) pose a serious threat to
human health (1,2). The timely and effective restoration
of coronary blood flow is an effective strategy for the treatment
of IHD. However, the process of restoration of blood flow
inevitably induces an additional type of damage, namely myocardial
ischemia-reperfusion injury (MIRI) (3). Thus, the treatment of IHD can result
in sequelae, and it is of significance to further study the
relevant mechanisms underlying MIRI to identify effective targets
and strategies for alleviating or preventing MIRI.
Long non-coding RNAs (lncRNAs) are non-coding RNAs
that are >200 nucleotides in length (4). Recent studies have shown that lncRNA
small nucleolar RNA host gene 1 (SNHG1) is involved in the
occurrence and development of a variety of diseases (5,6).
lncRNA SNHG1 can enhance cell proliferation, migration and invasion
in cervical cancer (7). lncRNA
SNHG1 also suppresses gastric cancer cell proliferation and
promotes apoptosis via the Notch1 signaling pathway (8). Moreover, lncRNA SNHG1 displays a
protective effect on the myocardium. For example, lncRNA SNHG1 can
serve a protective role in cardiomyocyte hypertrophy by targeting
the microRNA (miRNA/miR)-15a-5p/high mobility group AT-hook 1 axis
(6). lncRNA SNHG1 also protects
AC16 cells against adriamycin-induced toxicity by regulating the
miR-195/Bcl-2 axis (9). Moreover,
Liang et al (10)
demonstrated that upregulated lncRNA SNHG1 expression promoted the
proliferation, migration and angiogenesis of vascular endothelial
cells, and reduced the injury to these cells after
hypoxia/reoxygenation (H/R) induction. However, to the best of our
knowledge, no relevant studies on lncRNA SNHG1 in MIRI have been
reported.
lncRNAs can function as molecular sponges to adsorb
miRNAs, competitively binding with miRNAs internally, thus exerting
their biological effects via downregulated inhibition of the target
genes of these miRNAs, effectively increasing the activity of the
target gene, which is termed the lncRNA/miRNA/mRNA pathway
(11). A previous study has shown
that inhibition of miR-450b-5p significantly alleviates H/R injury
and improves liver function in mice by targeting crystallin αB
(CRYAB) (12). However, whether
miR-450b-5p serves a role in MIRI is not completely understood.
IGF1 serves an important role in MIRI. For example, it has been
shown that IGF1 serves a protective role in MIRI model rats by
activating the PI3K/Akt signaling pathway (13). Overexpression of suppressor of
cytokine signaling 2 inhibited IGF1 expression via the Janus kinase
1/STAT signaling pathway, thereby exacerbating MIRI in type 2
diabetes (14). Moreover, lncRNA
SNHG1 was involved in sorafenib resistance by activating the Akt
signaling pathway (15). lncRNA
SNHG1 can inhibit neuronal apoptosis in rats with cerebral
infarction via the PI3K/Akt signaling pathway (16). However, to the best of our
knowledge, the specific role of the lncRNA SNHG1/miR-450b-5p/IGF1
axis in H/R-induced MIRI has not been previously reported.
In the present study, the effects of the lncRNA
SNHG1/miR-450b-5p/IGF1 axis on H/R-induced myocardial apoptosis and
oxidative stress levels were investigated and the underlying
mechanisms were also assessed. The results of the present study may
provide a theoretical basis for future clinical studies and
highlight potential targets for the treatment of MIRI.
Materials and methods
Cell culture and establishment of the
H/R model
AC16 cells (American Type Culture Collection) were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) in a humidified incubator with 5% CO2 at 37°C. For
establishment of the H/R model, the cell culture atmosphere was
N2 (94%), O2 (1%) and CO2 (5%) to
simulate hypoxia for 6 h, followed by 12-h reoxygenation under
normal atmospheric culture conditions as previously described
(17).
Database
ENCORI database (http://starbase.sysu.edu.cn/index.php) was used to
predict the interactions among lncRNA SNHG1, miR-450b-5p and IGF1
(Figs. S1 and S2).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. RNA was reverse
transcribed into cDNA using a TIANScript II RT Kit (Tiangen Biotech
Co., Ltd.) according to the manufacturer's protocol. Subsequently,
qPCR was performed using a RealMastcrMix (SYBR-Green) kit (Tiangen
Biotech Co., Ltd.). The following thermocycling conditions were
used for qPCR: 95°C for 2 min; 40 cycles at 95°C for 20 sec, 60°C
for 15 sec and 72°C for 30 sec. The sequences of the primers
(obtained from Sangon Biotech) used for qPCR were as follows:
lncRNA SNHG1 forward, 5′-AGGCTGAAGTTACAGGT-3′ and reverse,
5′-TTGGCTCCCAGTGTCTT-3′; miR-450b-5p forward,
5′-GCTTTTGCAATATGTTCCG-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′;
IGF1 forward, 5′-CCCAGAAGGAAGTACATTTG-3′ and reverse,
5′-GTTTAACAGGTAACTCGTGC-3′; U6 forward,
5′-AGAGAAGATTAGCATGGCCCCTG-3′ and reverse,
5′-ATCCAGTGCAGGGTCCGAGG-3′; and GAPDH forward,
5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-GTGAGGGAGATGCTCAGTGT-3′.
miRNA and mRNA expression levels were quantified using the
2−∆∆Cq method (18)
and normalized to the internal reference genes U6 and GAPDH,
respectively.
Cell transfection
A SNHG1 overexpression plasmid (Oe-SNHG1;
pBluescript vector) and the empty vector [Oe-negative control
(NC)], miR-450b-5p mimic (forward, 5′-UUUUGCAGUAUGUUCCUGAAUA-3′ and
reverse 5′-UUCAGGAACAUACUGCAAAAUU-3′; lentiviral vector pLVTHM),
mimic NC (forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′; lentiviral vector pLVTHM), IGF1 short
hairpin (sh)RNA lentiviral particles (shRNA-IGF1; shRNA-IGF1#1,
5′-GAAGAATTGTGAAAGTTTA-3′ and shRNA-IGF2#1,
5′-GCTAGAGTGTCATAATAAA-3′) and non-targeting NC shRNA lentiviral
particles (shRNA-NC; 5′-TTCTCCGAACGTGTCACGT-3′) were all purchased
from Genecopoeia, Inc. Cells (1×105 cells/well) were
transfected with 20 nM overexpression plasmid, miRNA mimic, shRNA
or the corresponding NC using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h
according to the manufacturer's protocol. At 48 h
post-transfection, RT-qPCR and western blotting were used to detect
transfection efficiency.
MTT
AC16 cells were seeded (8×103 cells/well)
into 96-well plates. At 48 h post-transfection, the medium was
removed and 10 µl MTT (Sigma-Aldrich; Merck KGaA) was added to each
well for 4 h. Subsequently, DMSO was added. The optical density was
measured at a wavelength of 570 nm using a microplate analyzer.
TUNEL assay
Apoptosis was measured using a TUNEL staining kit
(Beyotime Institute of Biotechnology). Transfected cells were fixed
using 4% paraformaldehyde for 1 h at 4°C and then 0.1% Triton X-100
was added at room temperature for 5 min. Subsequently, cells were
washed with PBS and incubated with TUNEL reagent for 1 h at 37°C.
Subsequently, 50 µl DAB color development was performed for 10 min
at 15°C according to the manufacturer's instructions. The cells
were stained with DAPI for 10 min at room temperature in the dark.
Stained cells were observed under a glass coverslip with PBS using
a fluorescent microscope (magnification, ×200; Carl Zeiss AG).
ImageJ software (version 1.8.0; National Institutes of Health) was
used to count the total cells and the TUNEL+ cells. The
number of apoptotic cells was calculated to be the average number
of positive cells out of the total number of cells in six fields of
view per slide.
Western blotting
Total protein was isolated from cells using RIPA
lysis buffer (Thermo Fisher Scientific, Inc.) and then quantified
using a BCA kit (Bio-Rad Laboratories, Inc.). Proteins (30 µg) were
separated via 12% SDS-PAGE and transferred to PVDF membranes
(Bio-Rad Laboratories, Inc.). After blocking with 5% skimmed milk
at room temperature for 1.5 h, the membranes were incubated at 4°C
overnight with primary antibodies targeted against: Bcl-2 (1:1,000;
cat. no. ab32124; Abcam), Bax (1:1,000; cat. no. ab182733; Abcam),
cytochrome c (1:1,000; cat. no. ab133504; Abcam), cleaved
caspase 3 (1:1,000; cat. no. ab32042; Abcam), IGF1 (1:1,000; cat.
no. ab134140; Abcam), phosphorylated (p)-PI3K (1:1,000; cat. no.
ab278545; Abcam), p-Akt (1:1,000; cat. no. ab8805; Abcam), PI3K
(1:1,000; cat. no. ab191606; Abcam), Akt (1:1,000; cat. no. ab8805;
Abcam) and GAPDH (1:1,000; cat. no. ab8245; Abcam). Subsequently,
the membranes were probed with horseradish peroxidase-conjugated
secondary antibodies (1:5,000; cat. no. ab150113 or ab9482; Abcam)
for 2 h at room temperature. Signals were visualized using Super
Signal ECL (Thermo Fisher Scientific, Inc.) and semi-quantified
using ImageJ software (version 1.46; National Institutes of
Health).
Detection of malondialdehyde (MDA),
superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px)
The level of SOD was detected using the SOD Assay
Kit (cat. no. A001-3-2; Nanjing Jiancheng Bioengineering Institute)
according to the manufacturer's protocol. MDA concentrations were
determined using an ELISA kit (cat. no. A003-1-2; Nanjing Jiancheng
Bioengineering Institute) according to the manufacturer's protocol.
All above were used in accordance with the manufacturer's protocol.
GSH-Px enzyme activity was measured using a GSH-Px Assay Kit (cat.
no. A005-1-2, Nanjing Jiancheng Bioengineering Institute) according
to a manufacturer's protocol.
Luciferase reporter assay
The amplified PCR products of the 3′ untranslated
regions of SNHG1 and IGF1 were ligated into the miRNA target
expression vector pmirGLO-dual-luciferase (Promega Corporation). A
QuickChange II XL site-directed mutagenesis kit (Agilent
Technologies, Inc.) was used to generate the SNHG1 (SNHG1 MUT) and
IGF (IGF1 MUT) mutants. Cells were co-transfected with miR-450b-5p
mimic or miR-NC and SNHG1 MUT, SNHG1 wild-type (WT), IGF1 WT or
IGF1 MUT using Lipofectamine 2000 reagent, all at a final
concentration of 100 nmol/l. At 48 h post-transfection, the
luciferase activities were assayed using a dual-luciferase reporter
gene assay system (Promega Corporation) according to the
manufacturer's protocol. Renilla luciferase activity was
used for normalization and results were recorded using a GloMax 96
Microplate Luminometer (Promega Corporation).
Statistical analysis
Data are presented as the mean ± standard deviation.
Comparisons among multiple groups were analyzed using one-way ANOVA
followed by Tukey's post hoc test. All statistical analyses were
performed using SPSS software (version 22.0; IBM Corp.). P<0.05
was considered to indicate a statistically significant difference.
Each experiment was repeated at least three times.
Results
lncRNA SNHG1 overexpression inhibits
H/R-induced apoptosis and oxidative stress in AC16 cells
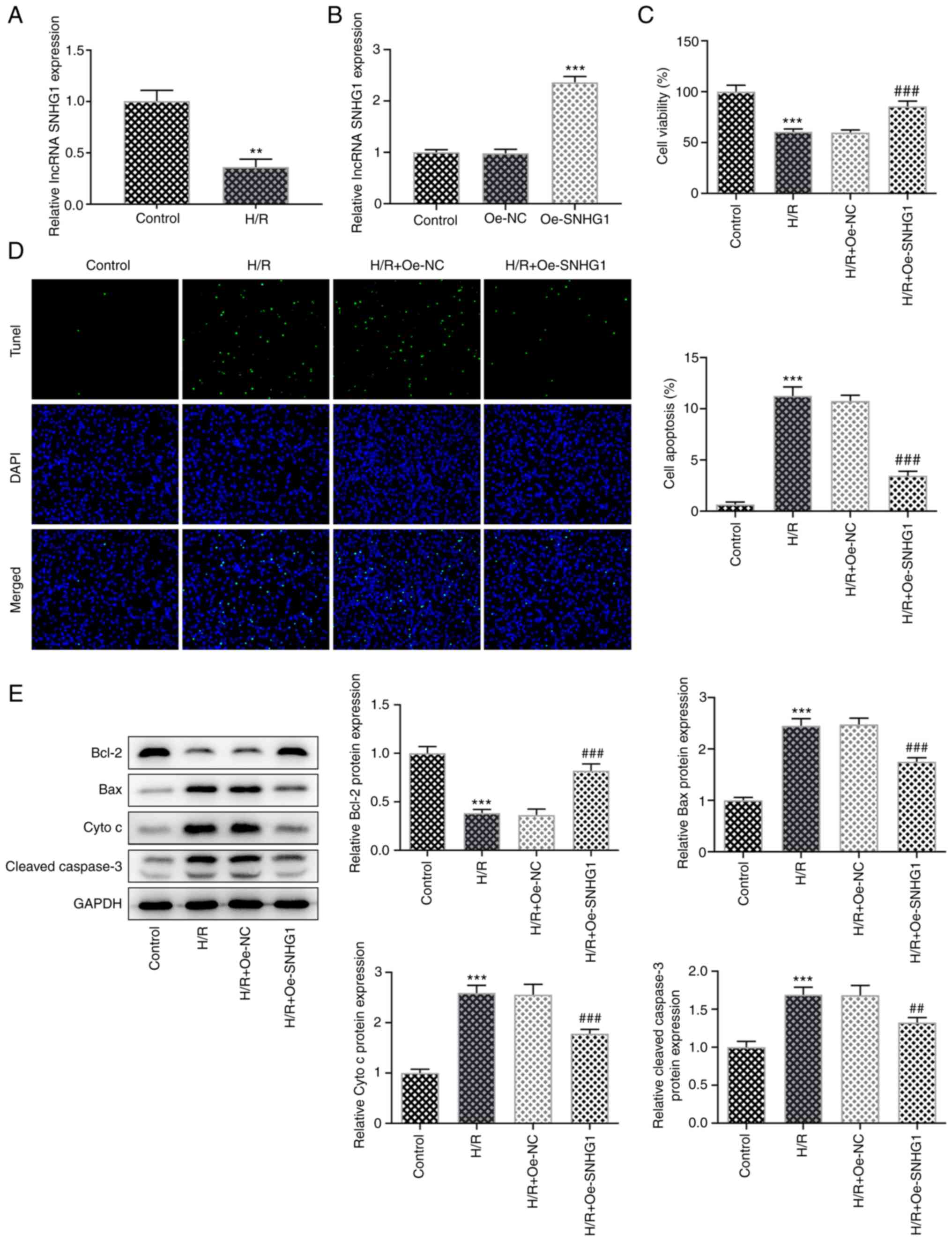
After AC16 cells were induced by H/R, the expression
levels of lncRNA SNHG1 were detected using RT-qPCR. The results
showed that lncRNA SNHG1 expression in the H/R group was
significantly decreased compared with that in the control group
(Fig. 1A). Subsequently, lncRNA
SNHG1 was overexpressed in cells (Fig. 1B). The MTT assay showed that cell
viability was significantly decreased following H/R induction
compared with that in the control group. SNHG1 overexpression
significantly increased cell viability compared with that in the
H/R + Oe-NC group (Fig. 1C). Cell
apoptosis was detected using a TUNEL assay and western blotting.
Compared with the control group, apoptosis was significantly
increased following H/R induction, which was accompanied by
significantly increased expression levels of Bax, cytochrome
c and cleaved caspase 3 and significantly decreased
expression levels of Bcl-2. Compared with the H/R + Oe-NC group,
apoptosis was significantly decreased in the H/R + Oe-SNHG1 group,
which was accompanied by significantly decreased expression levels
of Bax, cytochrome c and cleaved caspase 3 and significantly
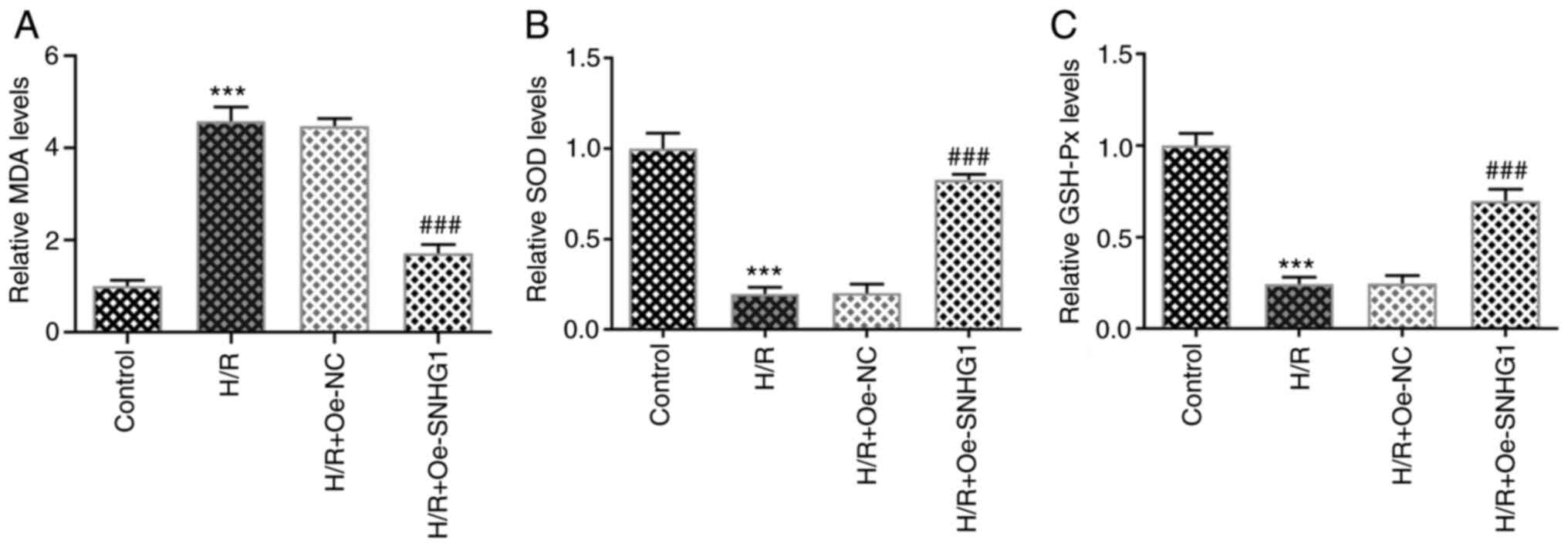
increased expression levels of Bcl-2 (Fig. 1D and E). The levels of MDA, SOD
and GSH-Px were detected using ELISAs. The results demonstrated
that the level of MDA was significantly increased, whereas the
levels of SOD and GSH-Px were significantly decreased in the H/R
group compared with those in the control group. However, SNHG1
overexpression significantly inhibited the effects of H/R induction
on the levels of oxidative stress-related indicators (Fig. 2A-C). The aforementioned results
indicated that lncRNA SNHG1 overexpression inhibited H/R-induced
apoptosis and oxidative stress levels in AC16 cells.
lncRNA SNHG1 overexpression increases
the expression of IGF1 by sponging miR-450b-5p
The ENCORI database predicted the presence of
binding sites between lncRNA SNHG1 and miR-450b-5p, as well as
between miR-450b-5p and IGF1 (Fig. 3A
and B). In addition, compared with that in the control group,
the expression of miR-450b-5p was significantly increased in
H/R-induced cells, whereas the expression of IGF1 was significantly
decreased (Fig. 3C and D). Cell
transfection was used to overexpress miR-450b-5p, which was
confirmed via RT-qPCR (Fig. 3E).
Subsequently, the luciferase reporter gene assay demonstrated that
in SNHG1 WT, the luciferase activity significantly decreased when
transfected with the miR-450b-5p mimic compared with the mimic NC
group. However, no significant change in luciferase activity was
exhibited in the SNHG1 MUT group. In the IGF1 WT group, luciferase
activity was significantly decreased following transfection with
the miR-450b-5p mimic compared with the mimic NC group. However, no
significant change in luciferase activity was exhibited following
miR-450b-5p mimic transfection in the IGF1 MUT group. These results
verified the targeted binding between lncRNA SNHG1 and miR-450b-5p
and between miR-450b-5p and IGF1 (Fig. 3F). In addition, following lncRNA
SNHG1 overexpression, the expression of miR-450b-5p in H/R-induced
cells was significantly decreased, whereas the expression of IGF1
was significantly increased. The expression of IGF1 in the H/R +
miR-450b-5p mimic group was significantly increased compared with
that in the H/R + mimic NC group (Fig. 3G-I). These results indicated that
lncRNA SNHG1 overexpression upregulated IGF1 expression by sponging
miR-450b-5p.
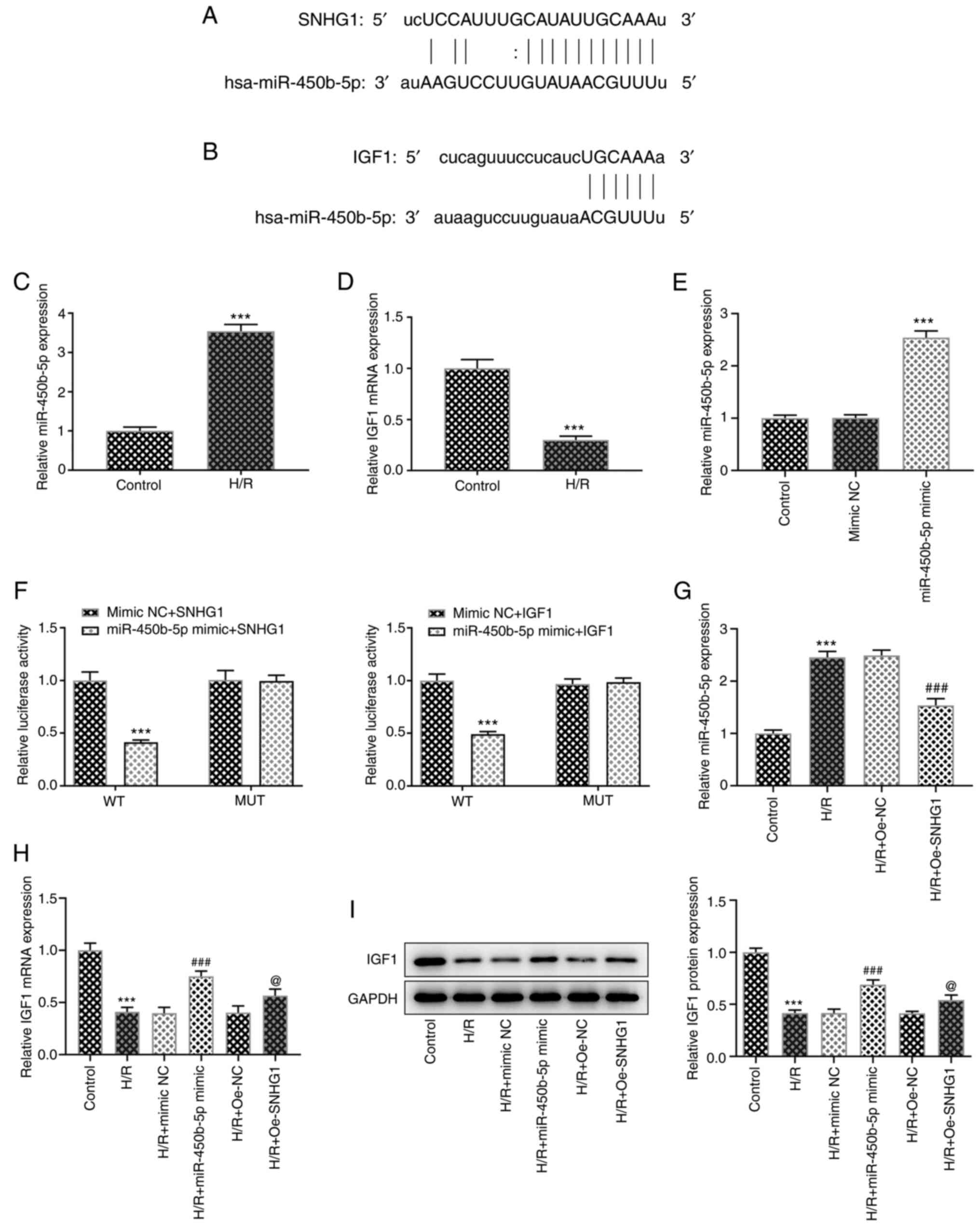 | Figure 3.lncRNA SNHG1 overexpression
upregulates IGF1 expression by sponge adsorption of miR-450b-5p.
Binding sides of (A) lncRNA SNHG1 and miR-450b-5p, and (B)
miR-450b-5p and IGF1. (C) RT-qPCR was performed to detect the
expression levels of (C) miR-450b-5p and (D) IGF1. ***P<0.001
vs. control. (E) RT-qPCR was performed to assess the transfection
efficiency of miR-450b-5p mimic. ***P<0.001 vs. mimic NC. (F)
Luciferase reporter assays confirmed the binding sides between
lncRNA SNHG1 and miR-450b-5p, and between miR-450b-5p and IGF1.
***P<0.001 vs. mimic NC + IGF1 or mimic NC + SNHG1. (G) RT-qPCR
was performed to assess the effect of lncRNA SNHG1 overexpression
on miR-450b-5p expression. ***P<0.001 vs. control;
###P<0.001 vs. H/R + Oe-NC. (H) RT-qPCR and (I)
western blotting were performed to assess the effect of lncRNA
SNHG1 and miR-450b-5p overexpression on IGF1 expression.
***P<0.001 vs. control; ###P<0.001 vs. H/R + mimic
NC; @P<0.05 vs. H/R + Oe-NC. lncRNA, long non-coding
RNA; SNHG1, small nucleolar RNA host gene 1; IGF1, insulin-like
growth factor 1; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; H/R,
hypoxia/reoxygenation; Oe, overexpression; WT, wild-type; MUT,
mutant. |
lncRNA SNHG1/miR-450b-5p/IGF1 axis
regulates PI3K/Akt signaling and affects apoptosis and oxidative
stress levels of H/R-induced AC16 cells
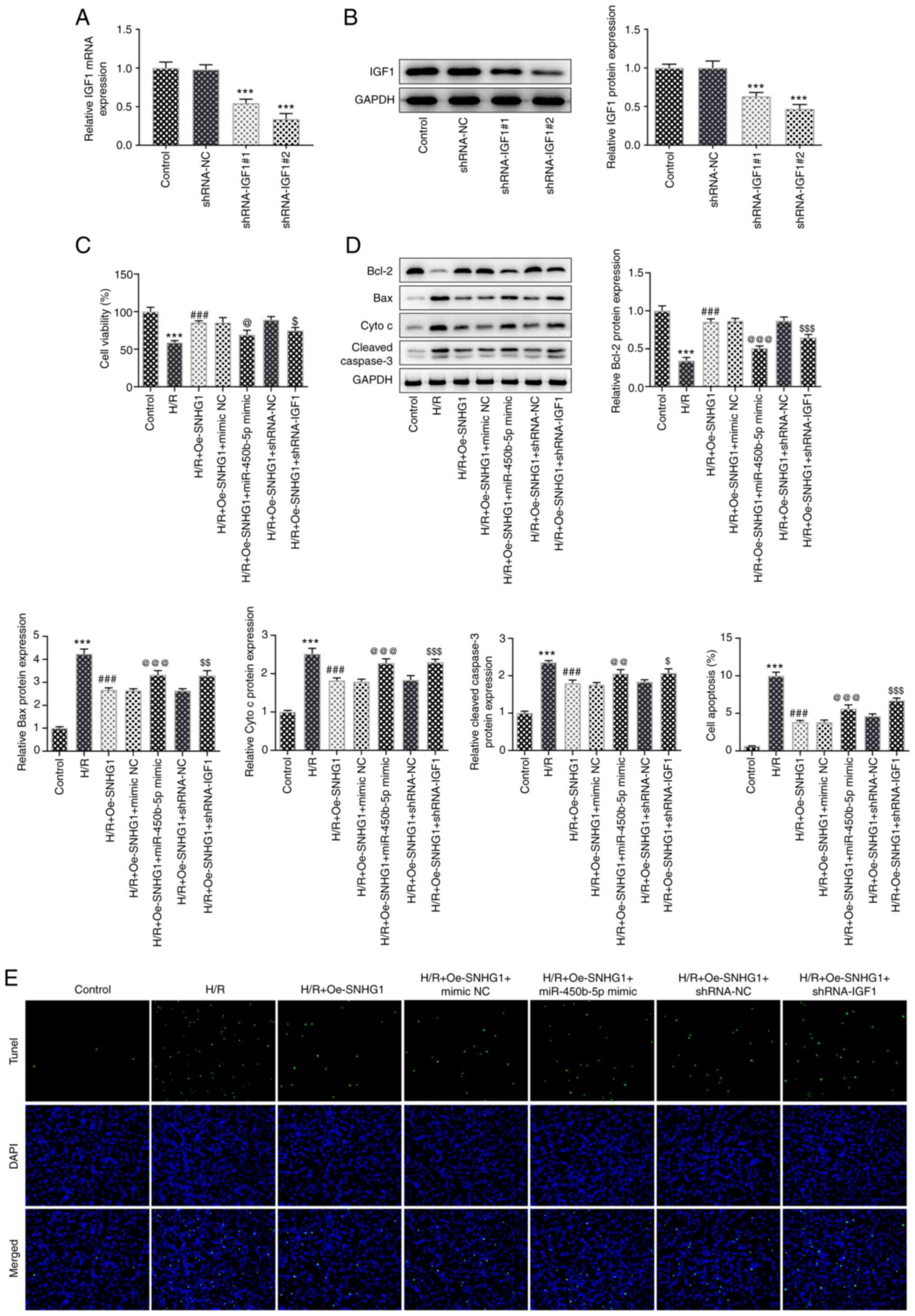
IGF1 expression was knocked down using shRNAs and
successful knockdown was confirmed via RT-qPCR (Fig. 4A). shRNA-IGF1#2 was selected for
subsequent experiments as it displayed the optimum knockdown
efficiency. Cells were divided into the following groups: i)
Control; ii) H/R; iii) H/R + Oe-SNHG1; iv) H/R + Oe-SNHG1 + mimic
NC; v) H/R + Oe-SNHG1 + miR-450b-5p mimic; vi) H/R + Oe-SNHG1 +
shRNA-NC; and vii) H/R + Oe-SNHG1 + shRNA-IGF1. The MTT and TUNEL
assay results demonstrated that cell viability was significantly
decreased and apoptosis was significantly increased in the H/R +
Oe-SNHG1 + shRNA-IGF1 group compared with that in the H/R +
Oe-SNHG1 + mimic NC group. This was accompanied by a significant
increase in the expression levels of the apoptosis-related proteins
Bax, cytochrome c and cleaved caspase 3, whereas Bcl-2
expression was significantly decreased. Compared with that in the
H/R + Oe-SNHG1 + shRNA-NC group, cell viability was significantly
decreased and apoptosis was significantly increased in the H/R +
Oe-SNHG1 + shRNA-IGF1 group, which was accompanied by increased
Bax, cytochrome c and cleaved caspase 3 expression levels
and decreased Bcl-2 expression levels (Fig. 4A-D). These results indicated that
miR-450b-5p overexpression and IGF1 knockdown could reverse the
effects of SNHG1 overexpression on cell viability and apoptosis.
Subsequently, the levels of oxidative stress-related indicators
were detected. Compared with those in the H/R + Oe-SNHG1 + mimic NC
group, SOD and GSH-Px levels were significantly decreased and MDA
levels were significantly increased in the H/R + Oe-SNHG1 +
miR-450b-5p mimic group. Compared with those in the H/R + Oe-SNHG1
+ shRNA-NC group, the levels of SOD and GSH-Px were significantly
decreased and the levels of MDA were significantly increased in the
H/R + Oe-SNHG1 + shRNA-IGF1 group (Fig. 5A-C).
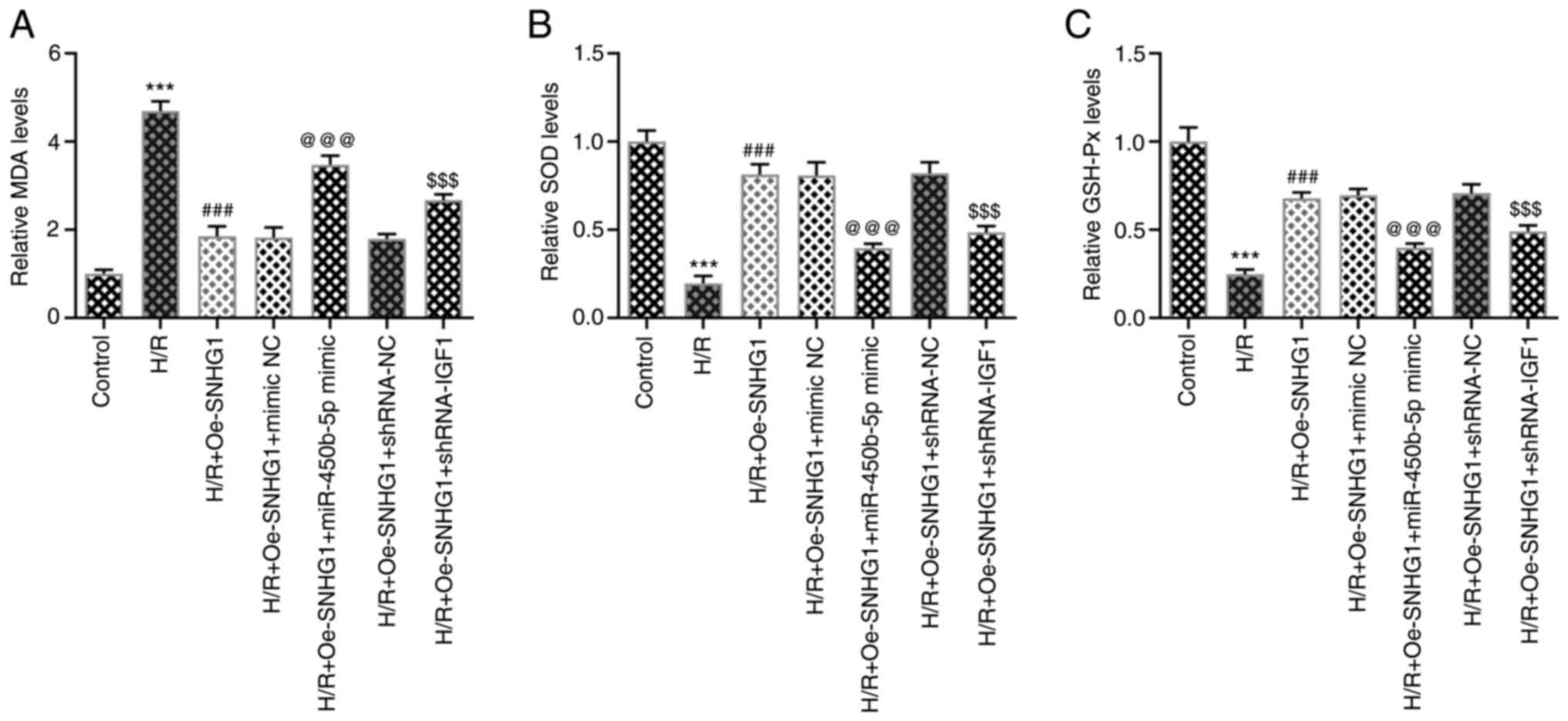 | Figure 5.Long non-coding RNA
SNHG1/miR-450b-5p/IGF1 axis affects oxidative stress levels of
H/R-induced AC16 cells. (A) MDA, (B) SOD and (C) GSH-Px were
detected using commercial kits. ***P<0.001 vs. control;
###P<0.001 vs. H/R; @@@P<0.001 vs. H/R + Oe-SNHG1 + mimic NC;
$$$P<0.001 vs. H/R + Oe-SNHG1 + shRNA-NC. SNHG1,
small nucleolar RNA host gene 1; miR, microRNA; IGF1, insulin-like
growth factor 1; H/R, hypoxia/reoxygenation; MDA, malondialdehyde;
SOD, superoxide dismutase; GSH-Px, glutathione peroxidase; Oe,
overexpression; NC, negative control; shRNA, short hairpin RNA. |
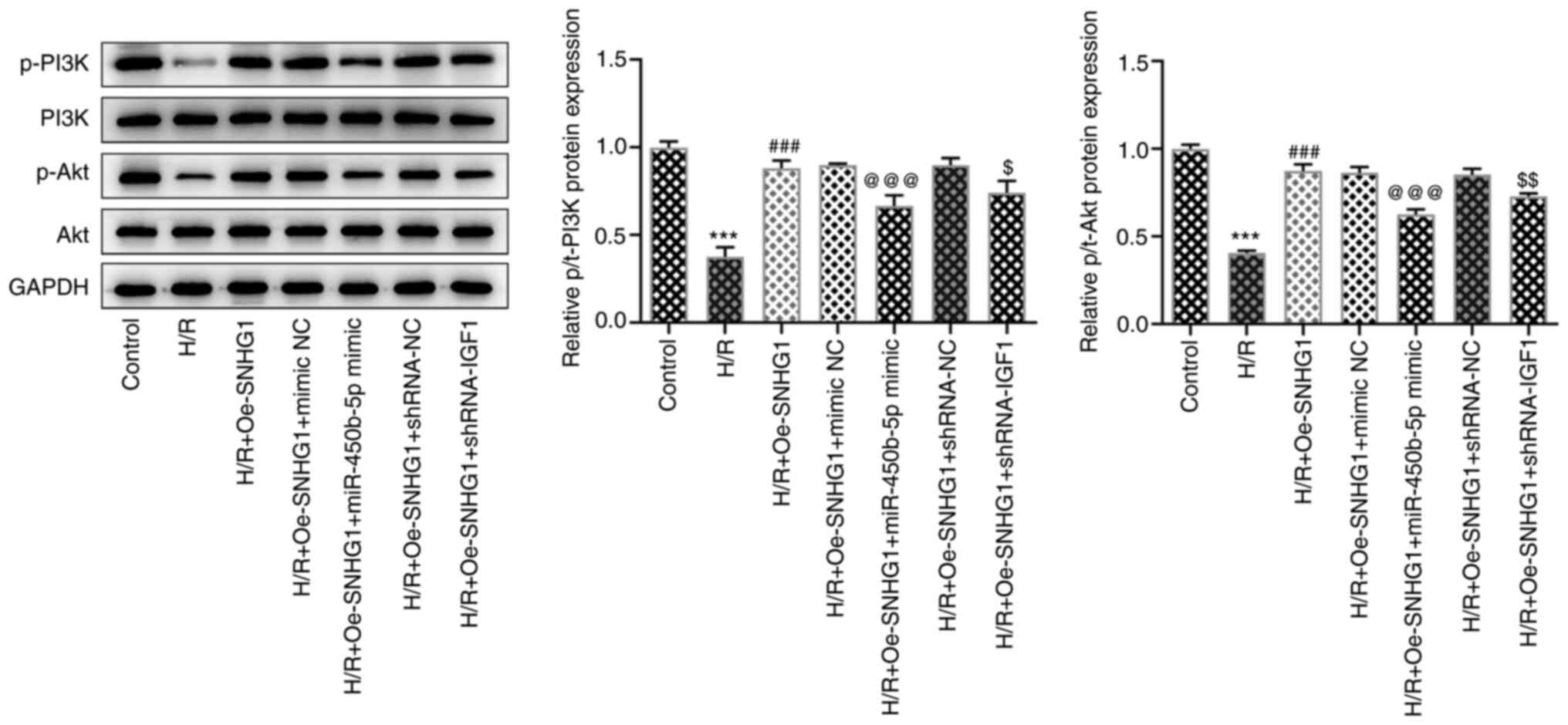
Additionally, the expression levels of PI3K/Akt
signaling pathway-related proteins were dysregulated. Compared with
that in the control group, the expression of p-PI3K and p-AKT was
significantly decreased following H/R induction, whereas SNHG1
overexpression resulted in significant upregulation of p-PI3K and
p-Akt expression levels. miR-450b-5p overexpression or IGF1
knockdown significantly inhibited the effects of SNHG1
overexpression on the PI3K/Akt signaling pathway (Fig. 6).
Discussion
In the present study, the role of the lncRNA
SNHG1/miR-450b-5p/IGF1 axis in H/R injury was assessed. Oxidative
stress serves a significant disruptive role in the state of
ischemia and reperfusion, which lead to cell apoptosis and
ultimately result in MIRI (19).
In the present study, the results demonstrated that the lncRNA
SNHG1/miR-450b-5p/IGF1 axis regulated the PI3K/Akt signaling
pathway to affect H/R-induced apoptosis and oxidative stress levels
in AC16 cells, thus suggesting a regulatory role in MIRI.
lncRNA SNHG1 downregulation reduced apoptosis,
oxidative stress and inflammation in a model of Parkinson's disease
via inhibition of the miR-125b-5p/MAPK1 axis (20). In H/R-induced endothelial cells,
the expression of lncRNA SNHG1 was significantly decreased, and as
an endogenous competitive RNA, lncRNA SNHG1 can alleviate
H/R-induced injury of vascular endothelial cells via the
hypoxia-inducible factor-1α/VEGF signaling pathway (10). In the present study, lncRNA SNHG1
expression levels were significantly decreased in H/R-induced AC16
cells. In addition, lncRNA SNHG1 has been shown to serve a
protective effect on the myocardium. Zhang et al (21) demonstrated that hydrogen peroxide
treatment could significantly inhibit the activity and promote the
apoptosis of cardiomyocytes, whereas SNHG1 overexpression could
significantly inhibit hydrogen peroxide-induced damage to
cardiomyocytes. Another study reported that SNHG1 overexpression
could significantly reduce the myocardial hypertrophy induced by
deoxyadrenaline (6). The results
of the present study demonstrated that lncRNA SNHG1 overexpression
significantly inhibited apoptosis and decreased oxidative stress
levels in H/R-induced AC16 cells, which may serve a therapeutic
role in the treatment of MIRI.
In the present study, the ENCORI database predicted
that lncRNA SNHG1 could bind to miR-450b-5p and miR-450b-5p could
bind to IGF1. These interactions were verified using luciferase
reporter assays. miR-450b-5p was reported to be a latent biomarker
in transient ischemic attack and liver self-healing plasmodium
malaria (22,23). Additionally, it has been shown
that the expression of miR-450b-5p decreased in mice with hepatic
ischemia/reperfusion injury, and miR-450b-5p inhibition
significantly alleviates hepatic ischemia/reperfusion injury and
improves liver function in mice by targeting CRYAB (12). In the present study, miR-450b-5p
expression levels were significantly increased in H/R-induced AC16
cells. Following myocardial ischemia reperfusion, miR-29a and
lethal-7 can affect cell apoptosis by regulating IGF1 (24). miR-320 downregulation inhibited
myocardial apoptosis and protected against myocardial
ischemia-reperfusion injury by targeting IGF1 (25). Degradation of IGF1 by mouse mast
cell protease 4 promoted cell death and adverse cardiac remodeling
after myocardial infarction (26). In the present study, IGF1
expression levels were significantly decreased in H/R-induced AC16
cells. lncRNA SNHG1 overexpression significantly downregulated the
expression of miR-450b-5p, resulting in upregulated IGF1 expression
to inhibit apoptosis and decrease the levels of oxidative stress in
H/R-induced AC16 cells.
PI3K/Akt is a classical signaling pathway that
serves an important role in MIRI (27,28). In the present study, the
expression levels of p-PI3K and p-Akt were downregulated after H/R
induction. In addition, IGF1 has been reported to serve a
protective role on MIRI model rats by activating the PI3K/Akt
signaling pathway (13). These
results indicated that IGF1 regulated the PI3K/Akt signaling
pathway. In the present study, the results demonstrated that the
lncRNA SNHG1/miR-450b-5p/IGF1 axis regulated the PI3K/Akt signaling
pathway and affected H/R-induced apoptosis and oxidative stress
levels in AC16 cells. The results of the present study should be
verified with the use of PI3K/Akt signaling pathway inhibitors or
activators in future studies.
The present study had a number of limitations.
Firstly, the study only used one cell line (AC16), which is not
reliable enough to explain the mechanism. Future experiments should
investigate the results of the present study in additional cell
lines. Moreover, the results were not verified by performing animal
experiments, thus further investigations are required to verify the
conclusions of the present study.
In conclusion, the present study demonstrated that
the activation of the PI3K/Akt signaling pathway via the lncRNA
SNHG1/miR-450b-5p/IGF1 axis inhibited the apoptosis and oxidative
stress levels of H/R-induced AC16 cells. These results provided a
theoretical basis for the mechanistic study of MIRI.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
LH and JZ contributed to the conception and design
of the present study, analyzed and interpreted the data, and
critically revised the manuscript for important intellectual
content. QY and PZ contributed to designing the study, analyzed the
data, and drafted and revised the manuscript. All authors read and
approved the final manuscript. LH and JZ confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Severino P, D'Amato A, Pucci M, Infusino
F, Adamo F, Birtolo LI, Netti L, Montefusco G, Chimenti C, Lavalle
C, et al: Ischemic heart disease pathophysiology paradigms
overview: From plaque activation to microvascular dysfunction. Int
J Mol Sci. 21:81182020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kaski JC, Crea F, Gersh BJ and Camici PG:
Reappraisal of ischemic heart disease. Circulation. 138:1463–1480.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Frank A, Bonney M, Bonney S, Weitzel L,
Koeppen M and Eckle T: Myocardial ischemia reperfusion injury: From
basic science to clinical bedside. Semin Cardiothorac Vasc Anesth.
16:123–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X
and Tai S: LncRNA HOXA-AS2 and its molecular mechanisms in human
cancer. Clin Chim Acta. 485:229–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thin KZ, Tu JC and Raveendran S: Long
non-coding SNHG1 in cancer. Clin Chim Acta. 494:38–47. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yan SM, Li H, Shu Q, Wu WJ, Luo XM and Lu
L: LncRNA SNHG1 exerts a protective role in cardiomyocytes
hypertrophy via targeting miR-15a-5p/HMGA1 axis. Cell Biol Int.
44:1009–1019. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G
and Song H: LncRNA SNHG1 enhances cell proliferation, migration,
and invasion in cervical cancer. Biochem Cell Biol. 96:38–43. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Z and Wang H: lncRNA SNHG1
suppresses gastric cancer cell proliferation and promotes apoptosis
via Notch1 pathway. J BUON. 25:302–307. 2020.PubMed/NCBI
|
|
9
|
Chen S, Wang J and Zhou Y: Long non-coding
RNA SNHG1 protects human AC16 cardiomyocytes from doxorubicin
toxicity by regulating miR-195/Bcl-2 axis. Biosci Rep. Jul
25–2019.(Epub ahead of print). doi: 10.1042/BSR20191050. View Article : Google Scholar
|
|
10
|
Liang S, Ren K, Li B, Li F, Liang Z, Hu J,
Xu B and Zhang A: LncRNA SNHG1 alleviates
hypoxia-reoxygenation-induced vascular endothelial cell injury as a
competing endogenous RNA through the HIF-1α/VEGF signal pathway.
Mol Cell Biochem. 465:1–11. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Y: The novel regulatory role of
lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med.
22:5768–5775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang Z, Mou T, Luo Y, Pu X, Pu J, Wan L,
Gong J, Yang H, Liu Y, Li Z, et al: Inhibition of miR-450b-5p
ameliorates hepatic ischemia/reperfusion injury via targeting
CRYAB. Cell Death Dis. 11:4552020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liao Y, Li H, Pi Y, Li Z and Jin S:
Cardioprotective effect of IGF-1 against myocardial
ischemia/reperfusion injury through activation of PI3K/Akt pathway
in rats in vivo. J Int Med Res. 47:3886–3897. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sheng M, Huang Z, Pan L, Yu M, Yi C, Teng
L, He L, Gu C, Xu C and Li J: SOCS2 exacerbates myocardial injury
induced by ischemia/reperfusion in diabetic mice and H9c2 cells
through inhibiting the JAK-STAT-IGF-1 pathway. Life Sci.
188:101–109. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li W, Dong X, He C, Tan G, Li Z, Zhai B,
Feng J, Jiang X, Liu C, Jiang H and Sun X: LncRNA SNHG1 contributes
to sorafenib resistance by activating the Akt pathway and is
positively regulated by miR-21 in hepatocellular carcinoma cells. J
Exp Clin Cancer Res. 38:1832019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen J, Zhang W, Wu YQ, Chen H and Zhao
JF: LncRNA SNHG1 inhibits neuronal apoptosis in cerebral infarction
rats through PI3K/Akt signaling pathway. Eur Rev Med Pharmacol Sci.
23:5366–5373. 2019.PubMed/NCBI
|
|
17
|
Tang M, Pan H, Zheng Z, Guo Y, Peng J,
Yang J, Luo Y, He J, Yan S, Wang P, et al: Prostaglandin E1
protects cardiomyocytes against hypoxia-reperfusion induced injury
via the miR-21-5p/FASLG axis. Biosci Rep. 39:BSR201905972019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eltzschig HK and Eckle T: Ischemia and
reperfusion-from mechanism to translation. Nat Med. 17:1391–1401.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao X, Tan Z, Jia M, Zhou X, Wu K, Ding Y
and Li W: Long Noncoding RNA SNHG1 knockdown ameliorates apoptosis,
oxidative stress and inflammation in models of Parkinson's disease
by inhibiting the miR-125b-5p/MAPK1 Axis. Neuropsychiatr Dis Treat.
17:1153–1163. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang N, Meng X, Mei L, Hu J, Zhao C and
Chen W: The long non-coding RNA SNHG1 attenuates cell apoptosis by
regulating miR-195 and BCL2-Like protein 2 in human cardiomyocytes.
Cell Physiol Biochem. 50:1029–1040. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Delic D, Dkhil M, Al-Quraishy S and
Wunderlich F: Hepatic miRNA expression reprogrammed by Plasmodium
chabaudi malaria. Parasitol Res. 108:1111–1121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ohuchi K, Watanabe M, Hirasawa N,
Tsurufuji S, Ozeki T and Fujiki H: Inhibition by gossypol of tumor
promoter-induced arachidonic acid metabolism in rat peritoneal
macrophages. Biochim Biophys Acta. 971:85–91. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Niu X, Hu J, Xing H, Sun M, Wang
J, Jian Q and Yang H: After myocardial ischemia-reperfusion,
miR-29a, and Let7 could affect apoptosis through regulating IGF-1.
Biomed Res Int. 2015:2454122015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song CL, Liu B, Diao HY, Shi YF, Zhang JC,
Li YX, Liu N, Yu YP, Wang G, Wang JP and Li Q: Down-regulation of
microRNA-320 suppresses cardiomyocyte apoptosis and protects
against myocardial ischemia and reperfusion injury by targeting
IGF-1. Oncotarget. 7:39740–39757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tejada T, Tan L, Torres RA, Calvert JW,
Lambert JP, Zaidi M, Husain M, Berce MD, Naib H, Pejler G, et al:
IGF-1 degradation by mouse mast cell protease 4 promotes cell death
and adverse cardiac remodeling days after a myocardial infarction.
Proc Natl Acad Sci USA. 113:6949–6954. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arslan F, Lai RC, Smeets MB, Akeroyd L,
Choo A, Aguor EN, Timmers L, van Rijen HV, Doevendans PA,
Pasterkamp G, et al: Mesenchymal stem cell-derived exosomes
increase ATP levels, decrease oxidative stress and activate
PI3K/Akt pathway to enhance myocardial viability and prevent
adverse remodeling after myocardial ischemia/reperfusion injury.
Stem Cell Res. 10:301–312. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen E, Chen C, Niu Z, Gan L, Wang Q, Li
M, Cai X, Gao R, Katakam S, Chen H, et al: Poly(I:C)
preconditioning protects the heart against myocardial
ischemia/reperfusion injury through TLR3/PI3K/Akt-dependent
pathway. Signal Transduct Target Ther. 5:2162020. View Article : Google Scholar : PubMed/NCBI
|