Introduction
Diabetic retinopathy (DR) is a severe microvascular
complication of diabetes and is also the main cause of blindness in
the adult population. Due to the increasing incidence of diabetes,
the number of patients suffering from DR is continuously increasing
(1). Blindness caused by DR may
lead to a reduced quality of life and increased medical burden on
both patients and society (2,3).
Special gene targeting therapy strategies have been investigated in
previous years (4).
The retinal pigment epithelial (RPE) cells form the
outer blood retinal barrier (BRB). In diabetic animal models,
increased leakage from the outer BRB (5) and disruptions of the RPE layer have
been observed (6), indicating
that RPE damage is involved in the pathogenesis of DR (7,8).
Accumulating data have indicated that oxidative stress (8,9)
inflammation (10) and apoptosis
(11,12) are all involved in RPE damage
caused by high glucose (HG) treatment, contributing to the
breakdown of the BRB and RPE dysfunction observed in DR.
Long non-coding RNAs (lncRNAs) can be expressed in
RPE cells under certain stimuli (13,14) and participate in RPE cell damage
in DR (12). lncRNA
metastasis-associated lung adenocarcinoma transcript 1 (lncRNA
MALAT1) is a highly conserved lncRNA originally found in tumors
(15). Studies conducted in
previous years have demonstrated that increased MALAT1 expression
may be involved in the development of DR (16,17), and participates in promoting
inflammation (18), apoptosis and
oxidative stress (19,20) in other cell types. However,
systematic evaluation of the effect of MALAT1 on RPE cells has
rarely been performed.
Therefore, the present study aimed to determine the
effect of MALAT1 on human RPE cells in a HG environment and to
explore the role of MALAT1 in DR.
Materials and methods
Cell culture and small interfering RNA
(siRNA)-mediated interference
ARPE-19 cells, which were derived from human retinal
pigment epithelium, were commercially obtained from Procell Life
Science & Technology Co., Ltd. (cat. no. CL-0026) and
maintained in six-well plates containing DMEM/F12 (cat. no.
SH30022.01; HyClone; Cytiva) with 100 U/ml penicillin plus 100
µg/ml streptomycin (cat. no. 0503; Sciencell) and 10% fetal bovine
serum (Every Green, cat. no. 11011-8611; Tianhang Biotech,
Hangzhou) in a tissue-culturing bioincubator with 95% O2
and 5% CO2 at 37°C. Cells were passaged approximately
once or twice every week.
siRNA targeting MALAT1 (si-MALAT1) and scrambled
siRNA (si-Scrambled) were purchased from Sangon Biotech Co., Ltd.
ARPE-19 cells were transfected using Lipofectamine® 2000
Reagent (cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific,
Inc.) for 4 h at a final siRNA concentration of 200 nmol/l in a
tissue-culturing bio-incubator with 95% O2 and 5%
CO2 at 37°C. At 4 h after transfection, the transfected
cells were incubated with HG (25 mM D-glucose; cat. no. G8270;
Millipore Sigma) at 37°C for 48 h (21) after the medium was replaced with
fresh culture medium. ARPE-19 cells exposed to normal glucose were
set as controls (control; 5.5 mM D-glucose; cat. no. G8270;
Millipore Sigma). The oligonucleotides used were as follows:
siMALAT1 sense, 5′-AGGUAAAGCUUGAGAAGAUTT-3′ and antisense,
5′-AUCUUCUCAAGCUUUACCUTT-3′; and control siScrambled sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′ (22). Cells were harvested and subjected
to further measurements.
Cell apoptosis assay
In order to evaluate whether MALAT1 can cause cell
apoptosis, flow cytometry was used to assess the early + late
apoptosis ratio. ARPE-19 cells were collected, and cell suspensions
were prepared. Subsequently, 100 µl cell suspension
(1×105 cells) was added to a tube. Cells were stained at
room temperature with 5 µl Annexin V-FITC and then 5 µl PI (Annexin
V-FITC/PI Apoptosis Detection kit; cat. no. CA1020; Beijing
Solarbio Science & Technology Co., Ltd.), and incubated at room
temperature in dark for 10 min and 5 min separately, and apoptotic
cells were separated and analyzed via flow cytometry by its
equipped Mfa32 software (Cytomics FC500 flow cytometer; both
Beckman Coulter, Inc.).
Oxidative stress detection
Since HG may promote oxidative stress in cells,
cellular reactive oxygen species (ROS) were detected using a
Reactive Oxygen Species Assay kit (cat. no. E004; Nanjing Jiancheng
Bioengineering Institute) according to the manufacturer's protocol.
Briefly, cells were harvested and adjusted to 1×106
cells/ml. The cells were resuspended in 0.5 ml diluted
dichloro-dihydro-fluorescein diacetate (DCFH-DA) and mixed well.
Analysis was carried out on a flow cytometer immediately after
incubation for 20 min at 37°C. ROS in cells were identified by flow
cytometry (Cytomics FC500; Beckman Coulter, Inc.) in the FITC
channel and data was analyzed by its equipped Mfa32 software.
Superoxide dismutase (SOD) and
malondialdehyde (MDA) measurement
Cellular MDA content and SOD activity were detected
using an MDA Assay kit (cat. no. A003-1) and a SOD Assay kit (cat.
no. A001-1; both from Nanjing Jiancheng Bioengineering Institute),
respectively, according to the manufacturer's protocols.
RNA isolation and PCR
Total RNA was extracted from cultured cells using
TRIzol® reagent (cat. no. 15596026; Invitrogen; Thermo
Fisher Scientific, Inc.). RNA quality and concentration were
assessed by UV spectroscopy at 260 and 280 nm.
cDNA was synthesized using a HiFiScript gDNA Removal
cDNA Synthesis kit (cat. no. CW2582S; CoWin Biosciences) according
to the manufacturer's protocol. In brief, the RT reaction mixture
was incubated at 42°C for 15 min, then incubated at 85°C for 5 min.
Primers (Table I) were designed
and synthesized by Sangon Biotech Co., Ltd. Quantitative PCR was
performed using MonAmp™ SYBR Green qPCR Mix (cat. no.
RN04005M; Monad Biotech Co., Ltd.) according to standard
thermocycler conditions. Relative gene expression at the mRNA level
was calculated from Cq values using the 2−ΔΔCq method
(23). The thermocycling
conditions were as follows: 95°C for 5 min; 40 cycles at 95°C for
10 sec, 58°C for 30 sec and 72°C for 30 sec. The expression of each
gene was normalized to that of β-actin.
 | Table I.Reverse transcription-quantitative
PCR primer sequences. |
Table I.
Reverse transcription-quantitative
PCR primer sequences.
| Gene name | Primer sequences
(5′→3′) |
|---|
| MALAT1 | F:
TACCTAACCAGGCATAACA |
|
| R:
GTAGACCAACTAAGCGAAT |
| TNF-α | F:
CGAGTGACAAGCCTGTAGCC |
|
| R:
TGAAGAGGACCTGGGAGTAGAT |
| MCP-1 | F:
CTTCTGTGCCTGCTGCTC |
|
| R:
TGCTGCTGGTGATTCTTCT |
| ICAM-1 | F:
GCAAGAAGATAGCCAACCAA |
|
| R:
TGCCAGTTCCACCCGTTC |
| VEGF | F:
CCCACTGAGGAGTCCAACA |
|
| R:
CAAATGCTTTCTCCGCTCT |
| β-actin | F:
AAGGCCAACCGCGAGAA |
|
| R:
ATGGGGGAGGGCATACC |
Protein isolation and western
blotting
Total protein was extracted from cultured cells
using RIPA cell lysis buffer (cat. no. R0020; Beijing Solarbio
Science & Technology Co., Ltd.). Protein concentrations were
measured by spectrophotometer using absorbance at 280 nm (model
22331; Eppendorf AG). Total protein (40 µg/lane) was
electrophoresed on 10% SDS-polyacrylamide gels and transferred onto
PVDF membranes (cat. no. IPVH00010; Millipore; Merck KGaA) by
electroblotting. The PVDF membranes were blocked for 1 h in 5%
nonfat milk at room temperature before incubation with primary
antibodies, including rabbit anti-TNF-α (cat. no. bs-0078R;
poly-antibody; rabbit-anti-human; 1:600; BIOSS), monocyte
chemotactic protein 1 (MCP-1; cat. no. PAA087HU01; poly-antibody;
rabbit-anti-human; 1:1,000; Wuhan USCN Business Co., Ltd.),
intercellular cell adhesion molecule 1 (ICAM-1; cat. no. bs-4615R;
poly-antibody; rabbit-anti-human; 1:800; BIOSS), vascular
endothelial growth factor (VEGF; cat. no. bs-0279R; poly-antibody;
rabbit-anti-human; 1:600; BIOSS) and anti-β-actin antibodies (cat.
no. TA-09; poly-antibody; mouse-anti-human; 1:1,000; OriGene
Technologies, Inc.) overnight at 4°C. The membranes were further
incubated with anti-rabbit IgG-HRP secondary antibody (cat. no.
ZB2301; poly-antibody; goat-anti-rabbit; 1:3,000; OriGene
Technologies, Inc.) at room temperature for 1 h. The blots were
developed using ECL (cat. no. sc-2048; Santa Cruz Biotechnology,
Inc.). Tanon Gis software (ver. 4.00; Tanon Science and Technology
Co., Ltd.) was used for semi-quantification of protein expression
and data were normalized to that of β-actin.
Statistical analysis
Experiments were repeated 3 times. Data are
presented as the mean ± standard deviation. Statistical analyses
among groups were performed by one-way analysis of variance
followed by Tukey's multiple comparison test. P<0.05 was
considered to indicate a statistically significant difference.
Analyses were performed using GraphPad Prism 9.0.0 for MacOS
(GraphPad Software, Inc.).
Results
HG increases MALAT1 expression in
ARPE-19 cells
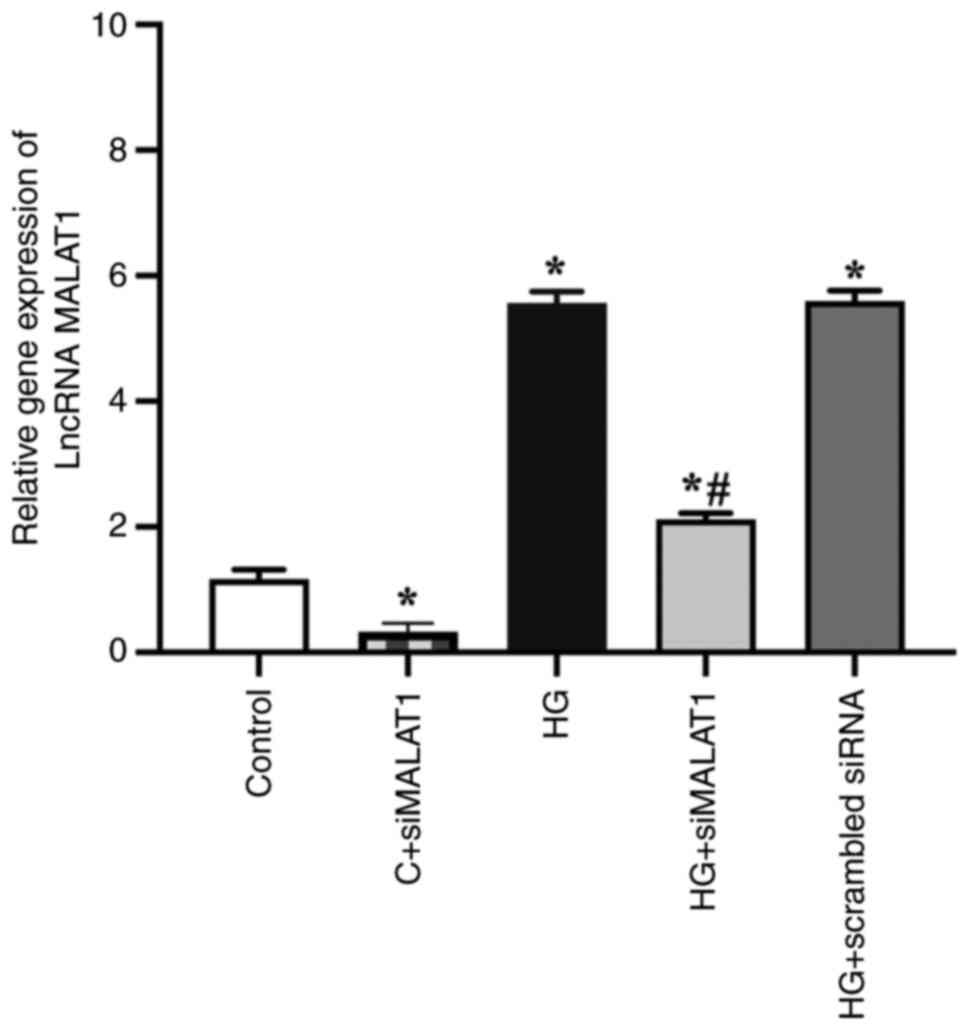
The results revealed that the expression of MALAT1
was successfully knocked down by siMALAT1 transfection.
Significantly increased MALAT1 gene expression was found in the
HG-treated ARPE-19 cells compared with the control cells (Fig. 1; P<0.05), indicating that HG
treatment increased MALAT1 expression in ARPE-19 cells. This
expression was strongly impaired by siMALAT1 transfection (Fig. 1; P<0.05).
lncRNA MALAT1 knockdown decreases
HG-induced oxidative stress in ARPE-19 cells
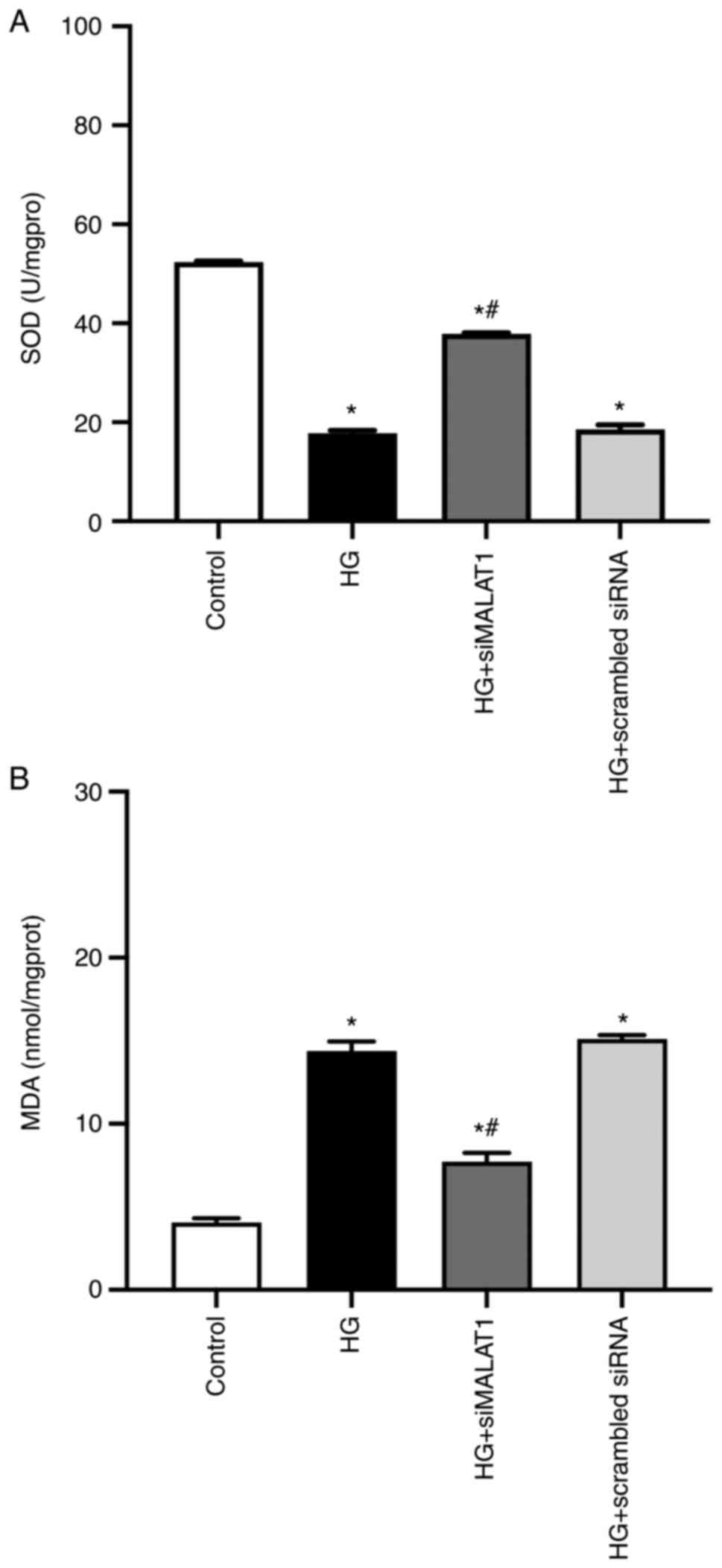
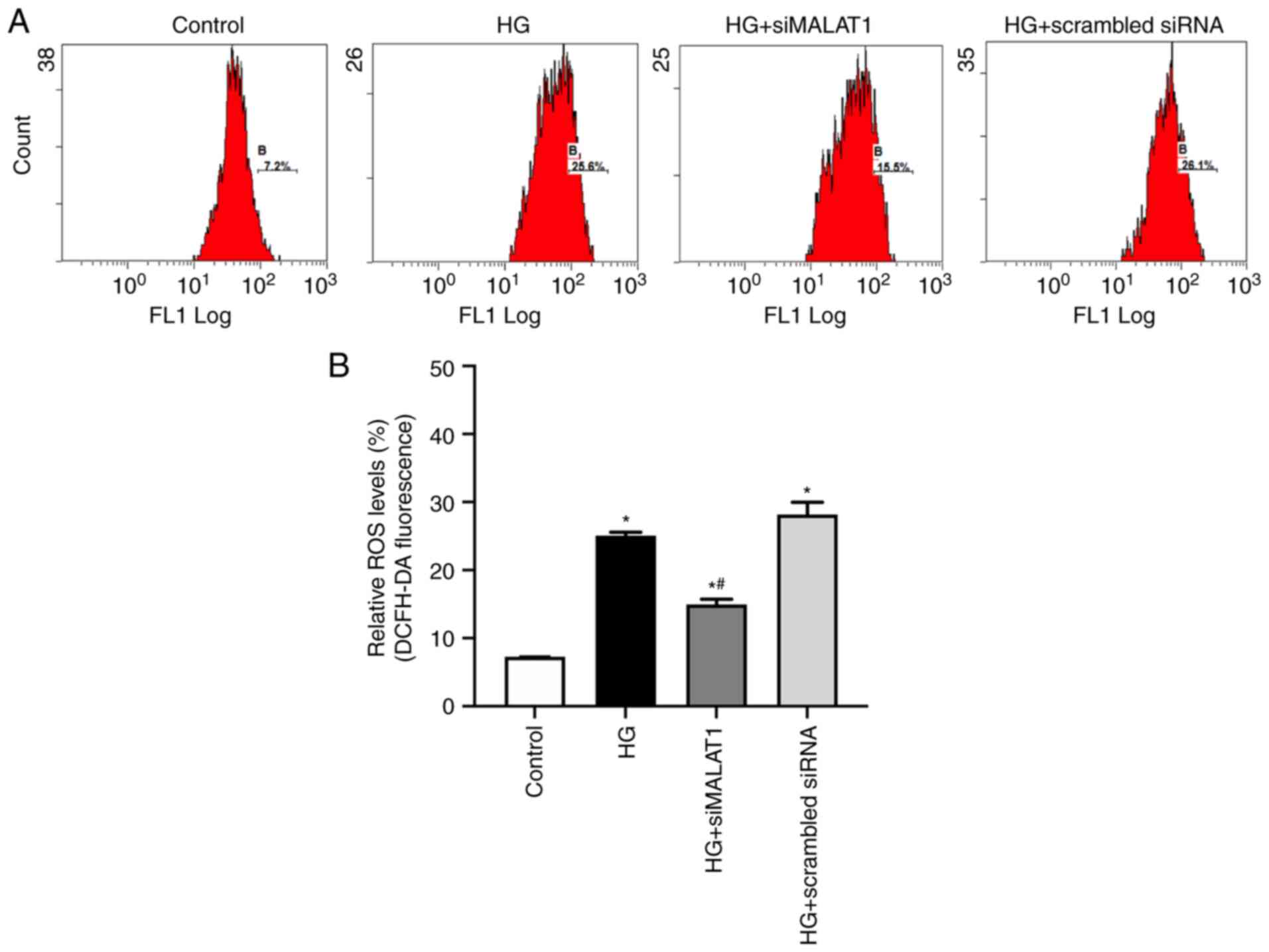
Increased cellular MDA levels and decreased SOD
activity may lead to increased ROS production (24). In the present study, compared with
the control, HG treatment led to significantly increased MDA
content and reduced SOD activity (Fig. 2A and B; both P<0.05) and
significantly increased ROS levels in ARPE-19 cells (Fig. 3A and B; P<0.05), indicating
that HG treatment led to increased ARPE-19 cellular oxidative
stress by elevating MDA content while inhibiting SOD activities.
All these effects achieved by HG could be partly reversed by MALAT1
knockdown (Fig. 2A and B; both
P<0.05), which resulted in reduced ROS levels (Fig. 3A and B; P<0.05), indicating
that increased MALAT1 may be involved in the increased retinal
endothelial oxidative stress caused by HG. Inhibiting MALAT1 may
alleviate ROS overload by rebalancing the cellular effects of MDA
and SOD.
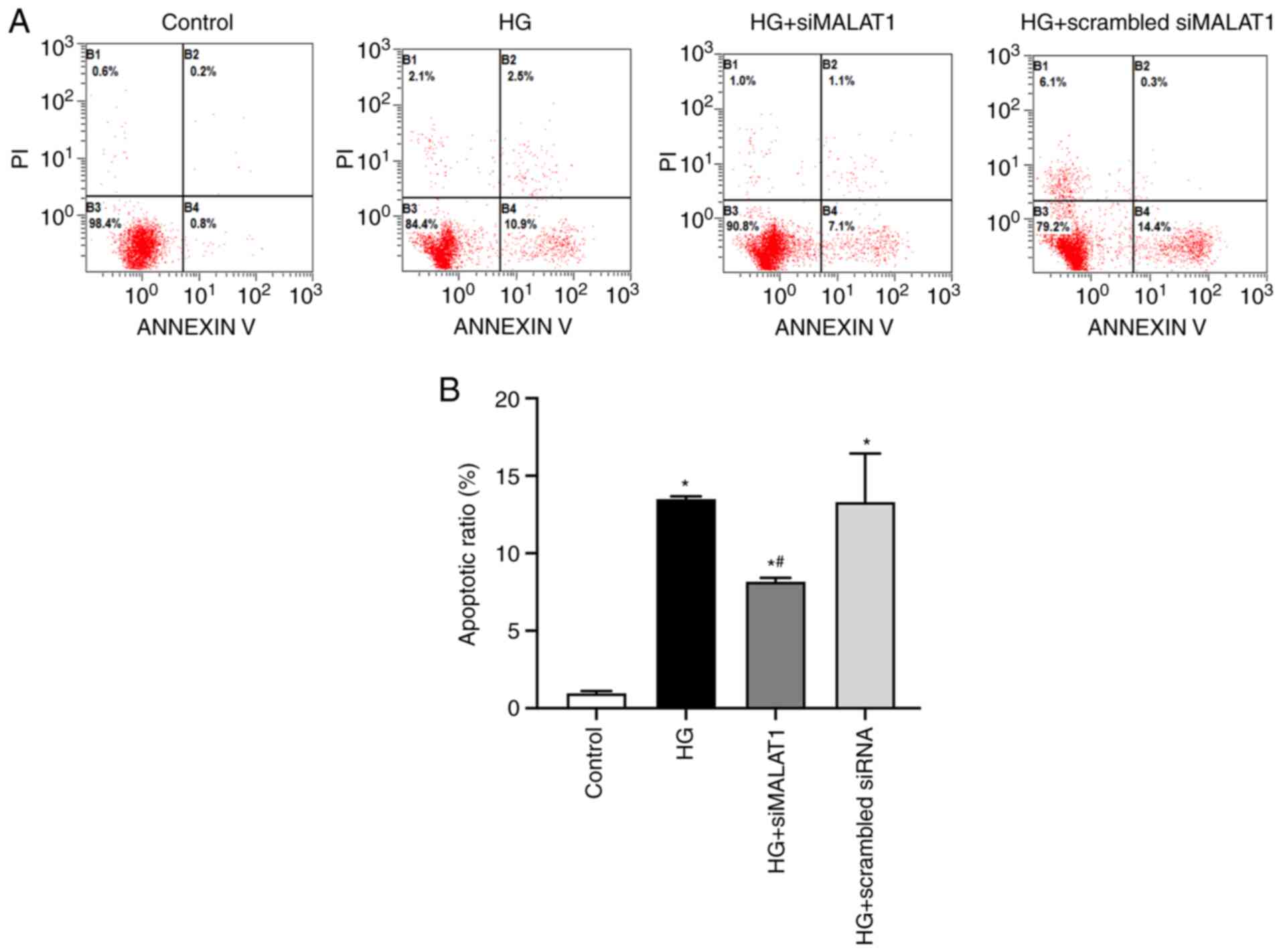
MALAT1 knockdown reduces ARPE-19 cell
apoptosis induced by HG treatment
In the present study, HG treatment was associated
with significantly increased apoptosis of ARPE-19 cells compared
with that of cells under normal glucose conditions (Fig. 4A and B; P<0.05) indicating that
HG treatment may cause ARPE-19 cell damage partly by increasing
apoptosis, and this effect could be substantially inhibited by
MALAT1 knockdown (Fig. 4A and B;
P<0.05). Therefore, elevated MALAT1 expression may be involved
in HG-induced ARPE-19 cell apoptosis.
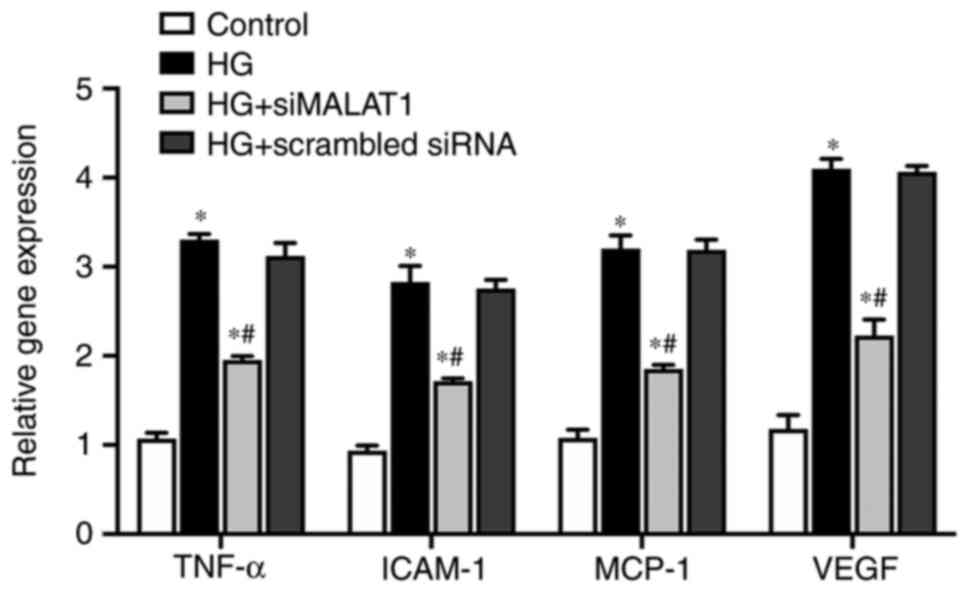
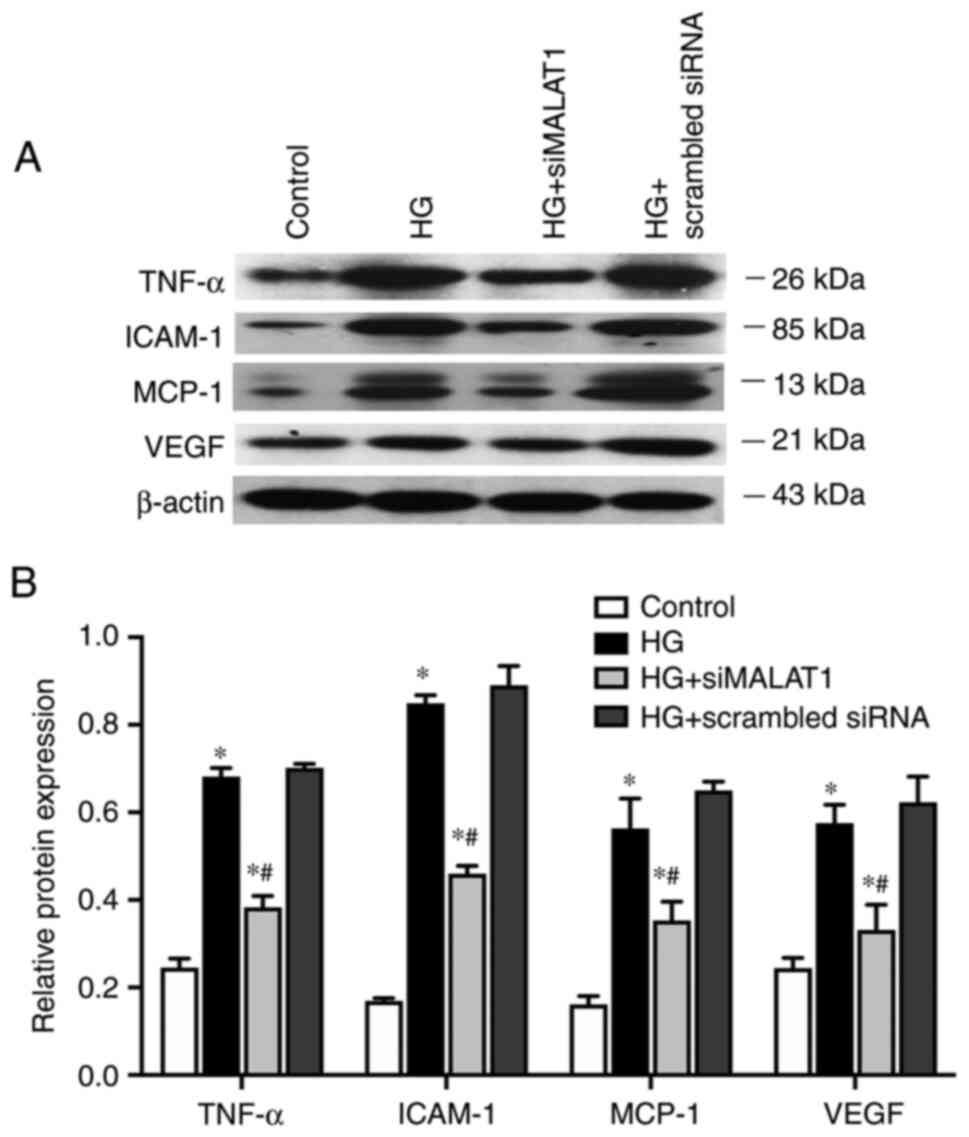
MALAT1 knockdown attenuates
inflammation induced by HG
In the present study, compared with those in the
control cells, the expression levels of genes involved in
inflammation of ARPE-19 cells, including the inflammatory cytokine
TNF-α, the endothelial adhesion molecule ICAM-1 and the immunogenic
cytokine MCP-1, were significantly increased under HG treatment at
both the mRNA (Fig. 5; all
P<0.05) and protein levels (Fig.
6A and B; all P<0.05).
VEGF, which is considered both an inflammatory and
angiogenic factor, was also upregulated by HG treatment at both the
mRNA (Fig. 5; P<0.05) and
protein levels (Fig. 6A and B;
P<0.05). The expression levels of all the genes elevated by HG
were largely impaired by MALAT1 knockdown (all P<0.05; Figs. 5 and 6). Therefore, HG treatment may promote
the ARPE-19 cellular inflammatory response by upregulating the
expression levels of inflammation-related genes, which may partly
be due to activating MALAT1 expression. Increased MALAT1 expression
may be detrimental in APRE-19 cells by promoting the downstream
inflammatory response.
Discussion
In recent years, an increasing number of studies
have indicated that structural and functional disorders in the RPE
are involved in the pathogenesis of DR (14,21,25), and that numerous lncRNAs are
involved in this process (12,26–28). lncRNA MALAT1, which was first
identified in lung carcinoma cells (29), performs multiple functions as a
stress response gene that can be differently expressed under some
stress, such as HG (30). MALAT1
has been found to be closely associated with a number of diabetic
complications (31). MALAT1
expression has been reported to be increased in the retinas of
diabetic animal models (32,33) and in endothelial cells of the
retina under HG treatment, which may contribute to the occurrence
of DR (16,17,34). It has also been suggested that
MALAT1 can be expressed in RPE cells (35) and elevated MALAT1 expression in
RPE cells is considered to be involved in the pathogenesis of
proliferative retinal disease (36,37).
The present study revealed that HG may stimulate
MALAT1 expression in ARPE-19 cells. However, at present, the
mechanisms underlying HG stimulation of MALAT1 in RPE cells remain
largely unknown. Gong et al (19) demonstrated that HG may upregulate
MALAT1 expression via SP1 binding to MALAT1 promoter regions in
human lens epithelial cells. Whether similar mechanisms also exist
in RPE cells still needs to be further explored.
Oxidative stress is a key contributor to the
pathogenesis of DR (38). Data
from the present study demonstrated that MALAT1 was involved in
HG-induced oxidative stress in ARPE-19 cells by increasing MDA
levels while reducing antioxidant SOD activity. It has been
previously reported that increased expression of genes, including
microRNA-34a (9) and NLR family
pyrin domain containing 3 (39),
and impairment of the nuclear factor-erythroid factor 2-related
factor 2 signaling pathway (21)
in RPE cells are all involved in oxidative stress caused by HG, and
these genes have been demonstrated to be the downstream target
genes of MALAT1 in other cells types (17,40,41).
HG treatment may induce RPE cell apoptosis, which
has been illustrated previously (42,43). However, the concentration of HG
mentioned in those studies varied from 25 mM (21,42) to 50 mM (43). In the present study, ARPE-19 cell
apoptosis could be induced at 25 mM glucose, which was consistent
with previous studies. MALAT1 has been found to be involved in
HG-induced apoptosis in cartilage endplate cells and lens
epithelial cells by targeting the p38 MAPK signaling pathway
(19,20), while the p38 MAPK signaling
pathway has been found to participate in HG-induced RPE cell
apoptosis (42). Therefore,
MALAT1 may be a pivotal mediator in HG-induced RPE cell
apoptosis.
MALAT1 has been found to be a pro-inflammatory
factor and may regulate glucose-induced inflammatory action in
cells (34,44). In the present study, MALAT1
knockdown could substantially blunt the effect of HG on the
expression of genes involved in inflammation, indicating that
MALAT1 may be involved in the inflammatory response in RPE cells
caused by HG. Similar results in RPE cells have rarely been
reported previously. MALAT1 may target some inflammatory pathways,
such as the NF-κB signaling pathway (45), which is an important mediator in
the inflammatory response in RPE cells (46,47).
The RPE may express and secret VEGF (48), and this procedure can be
stimulated by HG (49), hypoxia
(50) and oxidative stress
(51). In the present study,
increased VEGF gene expression induced by HG was also impaired by
MALAT1 knockdown. MALAT1 may elevate VEGF expression in RPE cells
directly (52) or indirectly by
increasing oxidative stress.
There are still some limitations in the present
study. First, the number of dead cells were found to be
unexpectedly higher in the HG group and could also be alleviated by
MALAT1 knockdown, thus, there may be certain mechanisms underlying
MALAT1 modulating cell death which were not originally designed in
the present study. Second, since this is a preliminary study
concerning the role of MALAT1 in ARPE-19 cells, detailed signaling
pathways and relative intervention studies were insufficient.
In conclusion, MALAT1 was involved in ARPE-19 cell
damage caused by HG by prompting oxidative stress, the inflammatory
response and apoptosis. Targeting MALAT1 may be a promising
therapeutic strategy for DR treatment. However, detailed mechanisms
underlying the effects of MALAT1 on RPE cells still need to be
further explored in both in vitro and in vivo
studies.
Acknowledgements
The authors would like to thank Professor Huijie Ma
(Department of Physiology, Hebei Medical University, Shijiazhuang,
China) for technical help and discussion of the results obtained in
the experiments.
Funding
The present study was supported by Projects of the Medical
Science Research of Health Commission of Hebei Province, China
(grant nos. 20210725 and 20210513). The funders had no role in the
study design, data collection and analysis, decision to publish or
preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JM and YL conceived and designed the study. XJ, YW
and PZ performed the experiments, and wrote, reviewed and revised
the manuscript. YZ, HM, XJ and YL were involved in the analysis and
interpretation of data, and performed the statistical analysis. XJ
and YL confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DR
|
diabetic retinopathy
|
|
SOD
|
superoxide dismutase
|
|
MDA
|
malondialdehyde
|
|
ROS
|
reactive oxygen species
|
|
MCP-1
|
monocyte chemotactic protein 1
|
|
ICAM-1
|
intercellular cell adhesion molecule
1
|
|
VEGF
|
vascular endothelial growth factor
|
|
lncRNA MALAT1
|
long non-coding RNA metastasis
associated lung adenocarcinoma transcript 1
|
References
|
1
|
Wong TY, Cheung CM, Larsen M, Sharma S and
Simó R: Diabetic retinopathy. Nat Rev Dis Primers. 2:160122016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saaddine JB, Honeycutt AA, Narayan KM,
Zhang X, Klein R and Boyle JP: Projection of diabetic retinopathy
and other major eye diseases among people with diabetes mellitus:
United States, 2005-2050. Arch Ophthalmol. 126:1740–1747. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mazhar K, Varma R, Choudhury F,
McKean-Cowdin R, Shtir CJ and Azen SP; Los Angeles Latino Eye Study
Group, : Severity of diabetic retinopathy and health-related
quality of life: The Los Angeles latino eye study. Ophthalmology.
118:649–655. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amadio M, Pascale A, Cupri S, Pignatello
R, Osera C, D'Agata V, D'Amico AG, Leggio GM, Ruozi B, Govoni S, et
al: Nanosystems based on siRNA silencing HuR expression counteract
diabetic retinopathy in rat. Pharmacol Res. 111:713–720. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu HZ and Le YZ: Significance of outer
blood-retina barrier breakdown in diabetes and ischemia. Invest
Ophthalmol Vis Sci. 52:2160–2164. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tarchick MJ, Bassiri P, Rohwer RM and
Samuels IS: Early functional and morphologic abnormalities in the
diabetic nyxnob mouse retina. Invest Ophthalmol Vis Sci.
57:3496–3508. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simó R, Villarroel M, Corraliza L,
Hernández C and Garcia-Ramírez M: The retinal pigment epithelium:
Something more than a constituent of the blood-retinal
barrier-implications for the pathogenesis of diabetic retinopathy.
J Biomed Biotechnol. 2010:1907242010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li H, Li R, Wang L, Liao D, Zhang W and
Wang J: Proanthocyanidins attenuate the high glucose-induced damage
of retinal pigment epithelial cells by attenuating oxidative stress
and inhibiting activation of the NLRP3 inflammasome. J Biochem Mol
Toxicol. 35:e228452021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li W and Xiao H: Dihydromyricetin
alleviates high glucose-induced oxidative stress and apoptosis in
human retinal pigment epithelial cells by downregulating miR-34a
expression. Diabetes Metab Syndr Obes. 14:387–397. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao H and Liu Z: Effects of microRNA-217
on high glucose-induced inflammation and apoptosis of human retinal
pigment epithelial cells (ARPE-19) and its underlying mechanism.
Mol Med Rep. 20:5125–5133. 2019.PubMed/NCBI
|
|
11
|
Kim DI, Park MJ, Lim SK, Choi JH, Kim JC,
Han HJ, Kundu TK, Park JI, Yoon KC, Park SW, et al:
High-glucose-induced CARM1 expression regulates apoptosis of human
retinal pigment epithelial cells via histone 3 arginine 17
dimethylation: Role in diabetic retinopathy. Arch Biochem Biophys.
560:36–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yin L, Sun Z, Ren Q, Su X and Zhang D:
Long non-coding RNA BANCR is overexpressed in patients with
diabetic retinopathy and promotes apoptosis of retinal pigment
epithelial cells. Med Sci Monit. 25:2845–2851. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kutty RK, Samuel W, Duncan T, Postnikova
O, Jaworski C, Nagineni CN and Redmond TM: Proinflammatory cytokine
interferon-γ increases the expression of BANCR, a long non-coding
RNA, in retinal pigment epithelial cells. Cytokine. 104:147–150.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang J, Yang K, Meng X, Liu P, Fu Y and
Wang Y: Silenced SNHG1 inhibited epithelial-mesenchymal transition
and inflammatory response of ARPE-19 cells induced by high glucose.
J Inflamm Res. 14:1563–1573. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Hamblin MH and Yin KJ: The long
noncoding RNA Malat1: Its physiological and pathophysiological
functions. RNA Biol. 14:1705–1714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Biswas S, Thomas AA, Chen S, Aref-Eshghi
E, Feng B, Gonder J, Sadikovic B and Chakrabarti S: MALAT1: An
epigenetic regulator of inflammation in diabetic retinopathy. Sci
Rep. 8:65262018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Radhakrishnan R and Kowluru RA: Long
noncoding RNA MALAT1 and regulation of the antioxidant defense
system in diabetic retinopathy. Diabetes. 70:227–239. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang K, Yu X, Yu Y, Zhang L, Cen Y and
Chu J: Long noncoding RNA MALAT1 promotes high glucose-induced
inflammation and apoptosis of vascular endothelial cells by
regulating miR-361-3p/SOCS3 axis. Int J Clin Exp Pathol.
13:1243–1252. 2020.PubMed/NCBI
|
|
19
|
Gong W, Zhu G, Li J and Yang X: LncRNA
MALAT1 promotes the apoptosis and oxidative stress of human lens
epithelial cells via p38MAPK pathway in diabetic cataract. Diabetes
Res Clin Pract. 144:314–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang Z, Zeng Q, Li D, Ding L, Lu W, Bian
M and Wu J: Long non-coding RNA MALAT1 promotes high
glucose-induced rat cartilage endplate cell apoptosis via the
p38/MAPK signalling pathway. Mol Med Rep. 21:2220–2226.
2020.PubMed/NCBI
|
|
21
|
Zhao X, Wang J, Li P, Tang L and Bai Y:
Casein kinase 2-interacting protein-1 alleviates high
glucose-reduced autophagy, oxidative stress, and apoptosis in
retinal pigment epithelial cells via activating the p62/KEAP1/NRF2
signaling pathway. J Ophthalmol. 2021:66940502021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li T, Niu L, Li M, Liu Y, Xu Z, Gao X and
Liu D: Effects of small interfering RNA-mediated downregulation of
the Krüppel-like factor 4 gene on collagen metabolism in human
hepatic stellate cells. Mol Med Rep. 12:3972–3978. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ho E, Karimi Galougahi K, Liu CC, Bhindi R
and Figtree GA: Biological markers of oxidative stress:
Applications to cardiovascular research and practice. Redox Biol.
1:483–491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fu SH, Lai MC, Zheng YY, Sun YW, Qiu JJ,
Gui F, Zhang Q and Liu F: MiR-195 inhibits the ubiquitination and
degradation of YY1 by Smurf2, and induces EMT and cell permeability
of retinal pigment epithelial cells. Cell Death Dis. 12:7082021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tong P, Peng QH, Gu LM, Xie WW and Li WJ:
LncRNA-MEG3 alleviates high glucose induced inflammation and
apoptosis of retina epithelial cells via regulating miR-34a/SIRT1
axis. Exp Mol Pathol. 107:102–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dong Y, Wan G, Peng G, Yan P, Qian C and
Li F: Long non-coding RNA XIST regulates hyperglycemia-associated
apoptosis and migration in human retinal pigment epithelial cells.
Biomed Pharmacother. 125:1099592020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu X, Luo Y, Chen G, Liu H, Tian N, Zen X
and Liu Q: Long noncoding RNA IGF2AS regulates high-glucose induced
apoptosis in human retinal pigment epithelial cells. IUBMB Life.
71:1611–1618. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lei L, Chen J, Huang J, Lu J, Pei S, Ding
S, Kang L, Xiao R and Zeng Q: Functions and regulatory mechanisms
of metastasis-associated lung adenocarcinoma transcript 1. J Cell
Physiol. 234:134–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abdulle LE, Hao JL, Pant OP, Liu XF, Zhou
DD, Gao Y, Suwal A and Lu CW: MALAT1 as a diagnostic and
therapeutic target in diabetes-related complications: A promising
long-noncoding RNA. Int J Med Sci. 16:548–555. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan B, Tao ZF, Li XM, Zhang H, Yao J and
Jiang Q: Aberrant expression of long noncoding RNAs in early
diabetic retinopathy. Invest Ophthalmol Vis Sci. 55:941–951. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li
YJ, Yan B and Jiang Q: Pathogenic role of lncRNA-MALAT1 in
endothelial cell dysfunction in diabetes mellitus. Cell Death Dis.
5:e15062014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Puthanveetil P, Chen S, Feng B, Gautam A
and Chakrabarti S: Long non-coding RNA MALAT1 regulates
hyperglycaemia induced inflammatory process in the endothelial
cells. J Cell Mol Med. 19:1418–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Postnikova OA, Rogozin IB, Samuel W,
Nudelman G, Babenko VN, Poliakov E and Redmond TM: Volatile
evolution of long non-coding RNA repertoire in retinal pigment
epithelium: Insights from comparison of bovine and human RNA
expression profiles. Genes (Basel). 10:2052019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou RM, Wang XQ, Yao J, Shen Y, Chen SN,
Yang H, Jiang Q and Yan B: Identification and characterization of
proliferative retinopathy-related long noncoding RNAs. Biochem
Biophys Res Commun. 465:324–330. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang S, Yao H, Li M, Li H and Wang F: Long
non-coding RNA MALAT1 mediates transforming growth factor
beta1-induced epithelial-mesenchymal transition of retinal pigment
epithelial cells. PLoS One. 11:e01526872016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu MY, Yiang GT, Lai TT and Li CJ: The
oxidative stress and mitochondrial dysfunction during the
pathogenesis of diabetic retinopathy. Oxid Med Cell Longev.
2018:34201872018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang Q, Li S, Zhou Z, Fu M, Yang X, Hao K
and Liu Y: HDAC6 inhibitor Cay10603 inhibits high glucose-induced
oxidative stress, inflammation and apoptosis in retinal pigment
epithelial cells via regulating NF-κB and NLRP3 inflammasome
pathway. Gen Physiol Biophys. 39:169–177. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li F, Li X, Qiao L, Liu W, Xu C and Wang
X: MALAT1 regulates miR-34a expression in melanoma cells. Cell
Death Dis. 10:3892019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Song Y, Yang L, Guo R, Lu N, Shi Y and
Wang X: Long noncoding RNA MALAT1 promotes high glucose-induced
human endothelial cells pyroptosis by affecting NLRP3 expression
through competitively binding miR-22. Biochem Biophys Res Commun.
509:359–366. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Maugeri G, Bucolo C, Drago F, Rossi S, Di
Rosa M, Imbesi R, D'Agata V and Giunta S: Attenuation of high
glucose-induced damage in RPE cells through p38 MAPK signaling
pathway inhibition. Front Pharmacol. 12:6846802021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Xi X, Mei Y, Zhao X, Zhou L, Ma
M, Liu S, Zha X and Yang Y: High-glucose induces retinal pigment
epithelium mitochondrial pathways of apoptosis and inhibits
mitophagy by regulating ROS/PINK1/Parkin signal pathway. Biomed
Pharmacother. 111:1315–1325. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gordon AD, Biswas S, Feng B and
Chakrabarti S: MALAT1: A regulator of inflammatory cytokines in
diabetic complications. Endocrinol Diabetes Metab. 1:e000102018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gong YP, Zhang YW, Su XQ and Gao HB:
Inhibition of long noncoding RNA MALAT1 suppresses high
glucose-induced apoptosis and inflammation in human umbilical vein
endothelial cells by suppressing the NF-κB signaling pathway.
Biochem Cell Biol. 98:669–675. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen X, Han R, Hao P, Wang L, Liu M, Jin
M, Kong D and Li X: Nepetin inhibits IL-1β induced inflammation via
NF-κB and MAPKs signaling pathways in ARPE-19 cells. Biomed
Pharmacother. 101:87–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang J, Zhou K, Zhang X, Zhou Y, Li Z and
Shang F: Celastrol ameliorates inflammation in human retinal
pigment epithelial cells by suppressing NF-κB signaling. J Ocul
Pharmacol Ther. 35:116–123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sant DW, Camarena V, Mustafi S, Li Y,
Wilkes Z, Van Booven D, Wen R and Wang G: Ascorbate suppresses VEGF
expression in retinal pigment epithelial cells. Invest Ophthalmol
Vis Sci. 59:3608–3618. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qin D and Jiang YR: Tangeretin inhibition
of high-glucose-induced IL-1β, IL-6, TGF-β1, and VEGF expression in
human RPE cells. J Diabetes Res. 2020:94906422020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hwang S, Seong H, Ryu J, Jeong JY, Kang
TS, Nam KY, Seo SW, Kim SJ, Kang SS and Han YS: Phosphorylation of
STAT3 and ERBB2 mediates hypoxia-induced VEGF release in ARPE-19
cells. Mol Med Rep. 22:2733–2740. 2020.PubMed/NCBI
|
|
51
|
Du W, An Y, He X, Zhang D and He W:
Protection of kaempferol on oxidative stress-induced retinal
pigment epithelial cell damage. Oxid Med Cell Longev.
2018:16107512018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yu L, Fu J, Yu N, Wu Y and Han N: Long
noncoding RNA MALAT1 participates in the pathological angiogenesis
of diabetic retinopathy in an oxygen-induced retinopathy mouse
model by sponging miR-203a-3p. Can J Physiol Pharmacol. 98:219–227.
2020. View Article : Google Scholar : PubMed/NCBI
|