Introduction
Intervertebral disc (IVD) degeneration (IDD) is
major cause of degenerative spinal disease (1) and can lead to disability, heavy
economic burden and a lower quality of life (2,3).
Previous studies showed cellular and biochemical changes in IVD
tissue in patients with IDD (4,5),
including loss of living cells in the nucleus pulposus (NP) and
destruction of extracellular matrix (ECM) in the IVD (6,7).
Multiple biological approaches have been developed to prevent early
IDD by promoting repair and regeneration of the ECM (8,9).
Inflammation participates in the pathogenesis of ECM metabolism
(7,10); proinflammatory cytokines inhibit
ECM production in NP cells and increase levels of enzymes involved
in ECM degradation, such as matrix metalloproteinases (MMPs) and a
disintegrin and metalloproteinase with thrombospondin motifs
(ADAMTS) (6,7). Additionally, the production of
collagen and proteoglycan increases during disc degeneration
(11,12). Therefore, inhibition of cell
proliferation and repair of ECM synthesis dysfunction is a
promising potential treatment strategy for IDD.
microRNAs (miRNAs or miRs) and long non-coding RNAs
(lncRNAs) serve key roles in IDD production via regulation of
proliferation or apoptosis of NP cells (13,14). Wan et al (15) investigated levels of lncRNAs in
human IDD compared with normal NP tissue using lncRNA-mRNA
microarrays and found that 67 lncRNAs were upregulated and 49
lncRNAs were downregulated in IDD tissue (15). In addition to the lncRNAs
identified by Wan et al (15), additional lncRNAs have been shown
to be involved in progression of IDD, including lncRNA membrane
associated guanylate kinase, WW and PDZ domain-containing 2 (MAG12)
antisense RNA 3 (MAG12-AS3) (16)
and lncRNA prostate androgen regulated transcript 1 (PART1)
(17). LINC00284 has been
reported as a potential target for management of ovarian and
gastric cancer (18,19) due to its ability to promote cell
proliferation and angiogenesis; however, its role in IDD remains
unclear. The prevailing hypothesis holds that lncRNAs function as
competing endogenous RNAs (ceRNAs) to sponge and regulate the
availability of miRNAs (13). In
previous studies, miR-205 has been reported to suppress
proliferation of renal carcinoma cancer (20) and acute lymphoblastic leukemia
cells (21). Chen et al
(22) suggested that miR-205
accelerates ECM accumulation in mesangial cells; thus it is key to
determine whether LINC00284 competitively binds with miR-205-3p to
regulate proliferation of NP cells.
In NP cells, different signaling pathways associated
with ECM metabolism are regulated by inflammation, such as the Wnt,
NF-κB and ERK1/2 signaling pathways (8,23–25). Wnt signaling has been implicated
in regulation of the inflammatory process in the musculoskeletal
system (26,27), where it is involved in formation
of tissue patterns during embryogenesis and carcinogenesis
(28). Moreover, studies have
shown that the Wnt signaling pathway is involved in pathogenesis of
osteoarthritis and rheumatoid arthritis (29,30); however, the role of Wnt signaling
in ECM metabolism in IDD remains unclear.
In the present study, the role and mechanisms of
LINC00284 and miR-205-3p in IDD were investigated. It was
hypothesized that LINC00284 possessed a potential binding site for
miR-205-3p, which was detected using a dual luciferase reporter
assay, and their interaction in cells of the NP was also
investigated. Furthermore, the proliferation, apoptosis of NP cells
as well as ECM synthesis were also investigated. Finally, a
miR-205-3p/Wnt/β-catenin axis was also explored in proliferation of
NP cells and in ECM synthesis mediated by LINC00284. In summary,
the present study will provide a novel experimental and theoretical
basis for targeted therapy of IDD.
Materials and methods
Human tissue samples
Degenerated IVD tissue was collected from 30
patients (14 male, 16 female; median age, 62.3 years; age range,
51–71 years) with IDD and normal NP tissue was obtained from 30
patients (17 male, 13 female; median age, 40.6 years; age range,
32–49 years) with spinal cord injury between June 2018 and December
2020 at the Shanghai Changzheng Hospital (Shanghai, China). The
collected tissues were rapidly frozen in liquid nitrogen and stored
at −80°C. The present study was approved by the Shanghai Changzheng
Hospital (Shanghai, China), and followed the guidelines described
in the Helsinki Declaration (31). Written informed consent was
obtained for tissue sample collection from each participant.
Cell culture and transfection
Human NP cells were purchased from ScienCell
Research Laboratories, Inc. (cat. no. 4800) and cultured in NP Cell
Medium (both ScienCell Research Laboratories, Inc.) at 37°C with 5%
CO2 in a humidified incubator. NP cells had been
initially isolated from NP of the human IVD; each vial contained
>5×105 cells in 1 ml and NP cells were allowed to
expand for 15 passages according to the manufacturer's
instructions. Human embryonic kidney 293T cells (American Type
Culture Collection; cat. no. CRL-1573) were cultured in DMEM
containing 10% FBS (both Thermo Fisher Scientific, Inc.) at 37°C in
a humidified incubator with 5% CO2. LINC00284 specific
small interfering (si)RNA (si-LINC00284) was used to knockdown
LINC00284 expression and scramble siRNA was used as negative
control (si-NC) and human miR-205-3p and scrambled miR-205-3p mimic
(mi-NC) were transfected into cells at the concentration of 20 µM
using Lipofectamine® 3000 (Thermo Fisher Scientific,
Inc.) at 37°C for 24 h according to manufacturer's instructions.
All siRNAs, miRNA mimics and NCs were obtained from Shanghai
GenePharma Co., Ltd. The sequences of constructs were as follows:
si-LINC00284 sense, 5′-GCAUGUUAAUUCACUAUUATT-3′ and antisense,
5′-UAAUAGUGAAUUAACAUGCTT-3′; si-NC sense,
5′-GUUGAAUUAACUUACACUUTT-3′ and antisense,
5′-AAGUGUAAGUUAAUUCAACTT-3′; miR-205-3p mimic sense,
5′-GAUUUCAGUGGAGUGAAGUU-3′ and antisense,
5′-AACUUCACUCCACUGAAAUC-3′ and mi-NC sense,
5′-GGUAGUGCGGAAAGUUUAUU-3′ and antisense,
5′-AAUAAACUUUCCGCACUACC-3′.
When cell confluence reached ~80%, transfected NP
cells were plated into 6- or 96-well plates at 1×105/ml
for serum starvation overnight, then stimulated at 37°C using IL-1β
(Prospec-Tany TechnoGene, Ltd.) at 5, 10 or 20 ng/ml for 24 h.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from tissue or cells using
TRIzol® (Thermo Fisher Scientific, Inc.). LINC00284,
MMP-3, aggrecan and collagen II mRNA levels were detected using
SYBR-Green qPCR kit (Takara Bio, Inc.) according to the protocol.
Briefly the one-step RT-qPCR reaction was: 10 µl of 2X One step TB
Greem RT-PCR Buffer III, 0.4 µl of TaKaRa Ex Taq HS (5 U/µl), 0.4
µl of PrimeScript RT enzyme Mix II, 0.4 µl of forward primer (10
µM), 0.4 µl of reverse primer (10 µM), 2 µl of total RNA, and 6 µl
of RNase Free dH2O, then subjected to reverse
transcription for 5 min at 42°C and initially denatured at 95°C for
10 sec, and then to 40 cycles of amplification with the condition
of 95°C denature for 5 sec, and 60°C annealing for 30 sec. β-actin
was used as the housekeeping gene. The mature miR-205-3p expression
levels in tissue and cells were detected using
Hairpin-it™ miRNA One-step qPCR-PCR SYBR Green kit
(Shanghai GenePharma Co., Ltd.) according to the protocol. Briefly
the reaction system was setup as: 10 µl of One-Step SYBR Mix, 0.5
µl of Enzyme Mix, 0.5 µl of forward primer (2 µM), 0.4 µl of
reverse primer (5 µM), 0.4 µl of ROX Reference Dye, 2 µl of RNA
template, 6.2 µl of RNase Free dH2O, then subjected to
reverse transcription for 40 min at 42°C and initially denatured at
94°C for 3 min, and then to 40 cycles of amplification with the
condition of: 94°C denature for 12 sec, 62°C annealing for 30 sec,
and 72°C extension for 30 sec. Small U6 RNA was used as the
endogenous control. RT-qPCR results were analyzed using the
2−ΔΔCq method (32).
The primers were synthesized by Biomics Biotechnologies Co. Ltd.
and the sequences are listed in Table
I.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Name | Sequence,
5′→3′ |
|---|
| LINC00284 | F:
TGTGGGTGCCAGGTTATGAC |
|
| R:
TGCGCTCATCTTCTCCTCAC |
| MMP-3 | F:
CTCTTCCTTCAGGCGTGGAT |
|
| R:
AGGGAAACCTAGGGTGTGGA |
| Aggrecan | F:
GATGATCTGGCACGAGAAGGG |
|
| R:
CGTTTGTAGGTGGTGGCTGTG |
| Collagen II | F:
CCGTGCTCCTGCCGTTT |
|
| R:
GACATCCTGGCCCTGACAC |
| β-actin | F:
GCCGTTCCGAAAGTTGCCT |
|
| R:
ATCATCCATGGTGAGCTGGCG |
| miR-205-3p | F:
CCTTCATTCCACCGGAGT |
|
| R:
GGTCCAGTTTTTTTTTTTTTTTCAGA |
| U6 RNA | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
Western blot analysis
The protein levels of MMP-3, aggrecan, collagen II
and Wnt1, β-catenin and β-actin were detected using western
blotting. Total proteins from IL-1β-treated cells were lysed in
RIPA buffer with 1% PMSF and InStab™ Protease Cocktail
(Shanghai Yeasen Biotechnology Co., Ltd.) and protein concentration
was quantified using a Pierce BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.). Then, 20 µg/lane protein was resolved using 10%
SDS-PAGE and transferred onto a PVDF membrane (MilliporeSigma),
followed by blocking with EZ-Buffers N 1X BLOCK BSA in TBS (Sangon
Biotech Co., Ltd.) for 2 h at room temperature. The blots were
probed with the following antibodies (all 1:1,000; all from Abcam):
Anti-MMP-3 (cat. no. ab52915), anti-collagen II (cat. no.
ab188570), anti-aggrecan (cat. no. ab3778), anti-Wnt1 (cat. no.
ab15251), anti-β-catenin (cat. no. ab223075) or anti-β-actin (cat.
no. ab8226) at 4°C overnight, followed by incubation with
horseradish peroxidase-conjugated goat anti-rabbit (cat. no.
ab7090) for MMP-3, collagen II, Wnt1 and β-catenin detection or
anti-mouse IgG (cat. no. ab47827) for aggrecan and β-actin for 1.5
h at room temperature. ECL Western Blotting Substrate (Thermo
Fisher Scientific, Inc.) was used to visualize the signals. The
mean gray values of blots were measured using ImageJ version 1.8.0
software (National Institutes of Health). β-actin was used for
normalization.
Cell proliferation assay
Cell proliferation was detected using a Cell
Counting Kit (CCK)-8 assay. After seeding into 96-well plates
(5×103 cells/well) for 24 h, cells were transfected at
37°C with si-LINC00284, miR-205-3p mimics or NC for 24, 48 and 72
h. At 48 h post-transfection, 10 µl CCK-8 reagent (Abcam) was added
per well and cells were incubated for 4 h at 37°C. The absorbance
at 450 nm was measured. Untreated NP cells were used as the
control.
Cell apoptosis assay
Annexin V-FITC/Propidium Iodide (PI) staining and
flow cytometry analysis were used to detect NP cell apoptosis. At
48 h post-transfection, 1×106 cells/well were harvested,
washed in PBS and plated in a 6-well plate. Annexin V-FITC
Apoptosis Detection kit (MilliporeSigma) was used to measure cell
apoptosis according to the manufacturer's instructions. Briefly,
after washing with PBS twice, cells were resuspended in Annexin-V
binding buffer, stained with the Annexin V-FITC/PI for 15 min at
room temperature in the dark and analyzed using FACSCalibur and BD
CellQuest™ Pro software version 6.0 (both BD
Biosciences), and the apoptotic rate was calculated as the numbers
of early + late apoptotic cells/total numbers of cell.
Dual-luciferase reporter assay
LINC00284 sponging of miR-205-3p was predicted using
DIANA-LncBase V2 (carolina.imis.athena-innovation.gr) (33). The binding of LINC00284 with
miR-205-3p was validated using dual-luciferase reporter assay.
Wild-type (wt) or mutated (mut) LINC00284 sequence was constructed
and inserted into a pGL3 vector (Promega Corporation) as luciferase
reporter gene vectors. Briefly, 293T cells were cultured at 37°C
overnight to 70–80% confluence, then ~2×105 cells were
seeded into 24-well plates. After culturing at 37°C for 24 h, cells
were co-transfected with wt-LINC00284 or mut-LINC00284 reporter
vector and miR-205-3p mimics or mi-NC using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
according to manufacturer's instructions. At 48 h
post-transfection, luciferase reporter assay was performed using a
Dual-Luciferase Reporter Assay System (Promega Corporation), the
results were normalized with Renilla luciferase
activity).
IDD animal model construction
To investigate the effect of LINC00284 in IDD in
vivo, si-LINC00284 and si-NC were inserted into short hairpin
(sh)RNA lentiviral vectors and packaged as sh-LINC00284 and sh-NC
lentiviruses by Shanghai GenePharma Co., Ltd. A total of 24 male
Sprague-Dawley rats (weight, 200–250 g; age, 3 months) were
obtained from Nantong University (Nantong, China) for in
vivo experiments and were housed under the condition of 18–26°C
and 40–70% humidity, with a 12 h light/dark cycle and free access
to food and water. The rats were randomly divided into three groups
(n=8/group): sham, treated with sh-NC lentivirus (sham + sh-NC);
IDD model, treated with sh-LINC00284 lentivirus (IDD +
sh-LINC00284) and IDD NC, treated with sh-NC lentivirus treatment
(IDD + sh-NC). All surgical procedures were performed as previously
described (34). Briefly, to
induce IDD, rats were anesthetized by intraperitoneal injection of
pentobarbital sodium (40 mg/kg), then a midline longitudinal
incision was made on the back. The left facet joint between the
fourth and fifth lumbar (L4/5) vertebrae was removed to uncover the
L4/5 IVD. Then, a 21-gauge needle was inserted into the IVD
parallel to the endplate to a depth of 3 mm and held in place for
30 sec. The animal experiments were performed in accordance with
the National Institutes of Health Guide for the Care and Use of
Laboratory Animals (35) and were
approved by the Animal Care and Use Committee of Nantong University
(Nantong, China).
The lentivirus was injected into the anterior
midline area of IVD 7 days after puncture using a microliter
syringe with a 26-gauge needle (10 µl, Shanghai Gaoge Industry and
Trade Co., Ltd.) via the anterior transperitoneal midline, as
previously described (36,37).
X-ray and MRI examination
At 7, 14 and 28 days after initial IVD puncture, six
animals were randomly selected for X-ray and MRI examination. The
animals were kept in a prone position with tails straightened on a
molybdenum target radiographic image unit (GE Healthcare). IVD
height was measured according to the disc height index (DHI) as
previously described (38). DHI
change (%) was calculated relative to sham + sh-NC group.
Transverse relaxation time (T2) midsagittal MRI images were
obtained via Siemens 3.0T Magneto Trio MRI System (Siemens
Healthineers). T2 MRI images were analyzed by two independent
radiologists blinded to the experimental conditions according to
Pfirrmann MRI-grade system as previously described (39).
Histological staining and
morphological analysis
IVD tissue was obtained following model rat
sacrifice by intraperitoneal injection of sodium pentobarbital (100
mg/kg) 28 days after initial disc puncture. Tissue was fixed in 10%
formalin for 48 h at room temperature, decalcified using 10% EDTA
solution for 30 days at room temperature and embedded in paraffin
wax. The paraffin blocks of IVD tissue were sectioned into 5 µm
coronal sections containing NP, annulus fibrosus (AF) and
cartilaginous endplate. The sections were heated at 60°C for 30
min, and then dewaxed with xylene twice for 20 min each, and
rehydration in descending alcohol series stained using a
Hematoxylin-Eosin (H&E) Staining kit (Beijing Solarbio Science
& Technology Co., Ltd.) or Modified Safranine O-Fast Green FCF
Cartilage Stain kit (Beijing Solarbio Science & Technology Co.,
Ltd.) according to manufacturer's protocols. For histological
grading, staining was categorized by two independent pathologists
blinded to the experimental conditions on a scale of 4 (normal) to
12 points (severe degeneration) as described by Masuda et al
(40) under a light microscope
(Olympus CX43; Olympus Corporation; magnification, ×100) and
Olympus cellSens software version V3.2 (Olympus; Olympus
Corporation) was used for analysis.
Statistical analysis
All data are presented as the mean ± standard
deviation of three independent experiments and were analyzed using
SPSS version 19.0 (IBM Corp.). Differences between two groups were
evaluated using unpaired Student's t-test; differences between ≥3
groups were compared using one-way ANOVA followed by post hoc
Dunnett's or Tukey's. Spearman's rank correlation analysis was used
to determine the correlation between LINC00284 and miR-205-3p
expression. P<0.05 was considered to indicate a statistically
significant difference.
Results
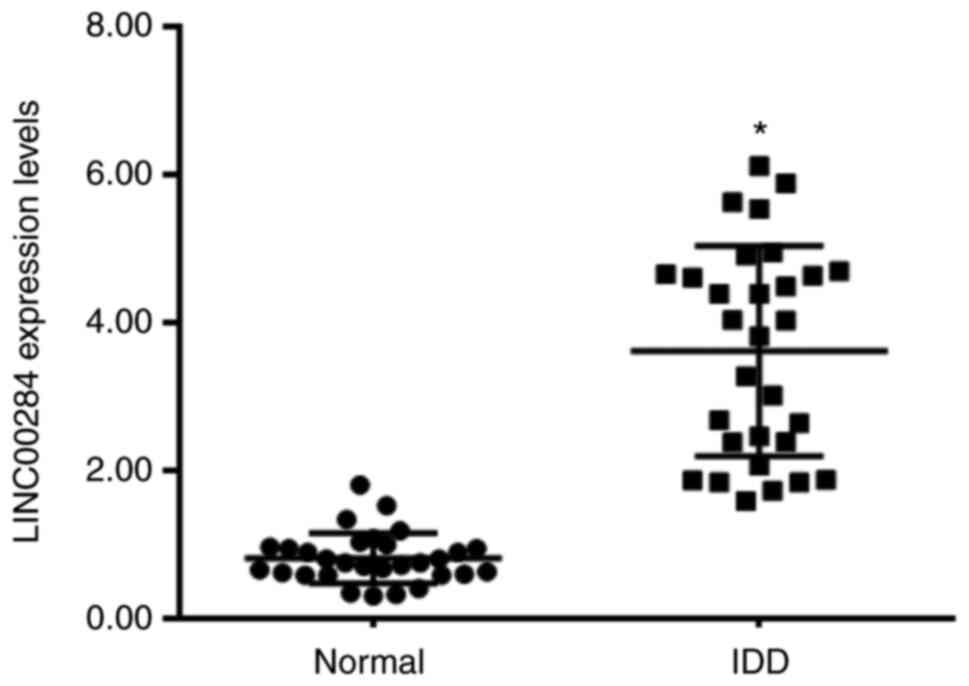
LINC00284 high expression in IDD
tissue
The expression levels of LINC00284 in IDD and normal
tissue was evaluated using RT-qPCR. LINC00284 expression was
significantly upregulated in 30 IDD samples compared with that in
normal NP tissues (30 samples obtained from patients with spinal
cord injury; Fig. 1). The results
indicated that LINC00284 was overexpressed in NP tissues of IDD
patients.
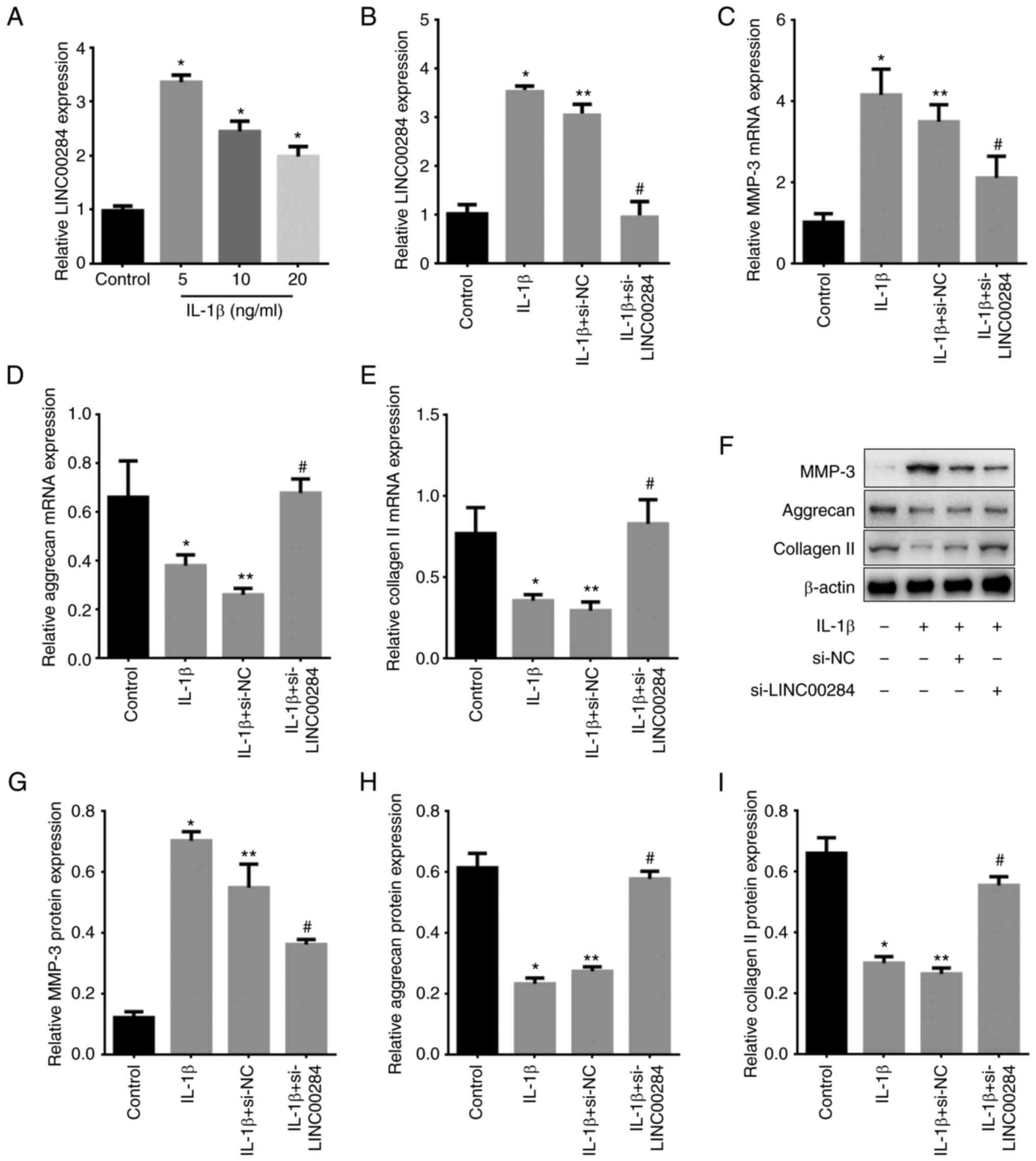
LINC00284 expression increases in
IL-1β-induced NP cells and promotes ECM degradation
Human NP cells were treated with 5, 10 or 20-ng/ml
IL-1β and LINC00284 expression was detected using RT-qPCR.
LINC00284 expression was low in normal NP cells (control) but
significantly increased following IL-1β treatment, with 5 ng/ml
exerting the most significant increase (Fig. 2A). Thus, 5 ng/ml IL-1β was used in
subsequent experiments.
To investigate whether IL-1β promotes IDD by
stimulating NP cells and determine the role of LINC00284 in
development of IDD, LINC00284 expression was knocked down using
siRNA (Fig. 2B) and expression
levels of MMP-3 and other ECM markers were measured using RT-qPCR
and western blotting. IL-1β inhibited expression of aggrecan and
collagen II but increased MMP-3 expression at both the mRNA and
protein level. Additionally, in IL-1β-induced NP cells, LINC00284
knockdown decreased MMP-3 expression but increased aggrecan and
collagen II expression levels compared with si-NC treated cells
(Fig. 2C-I). The results
indicated that LINC00240 may promote the development of IDD.
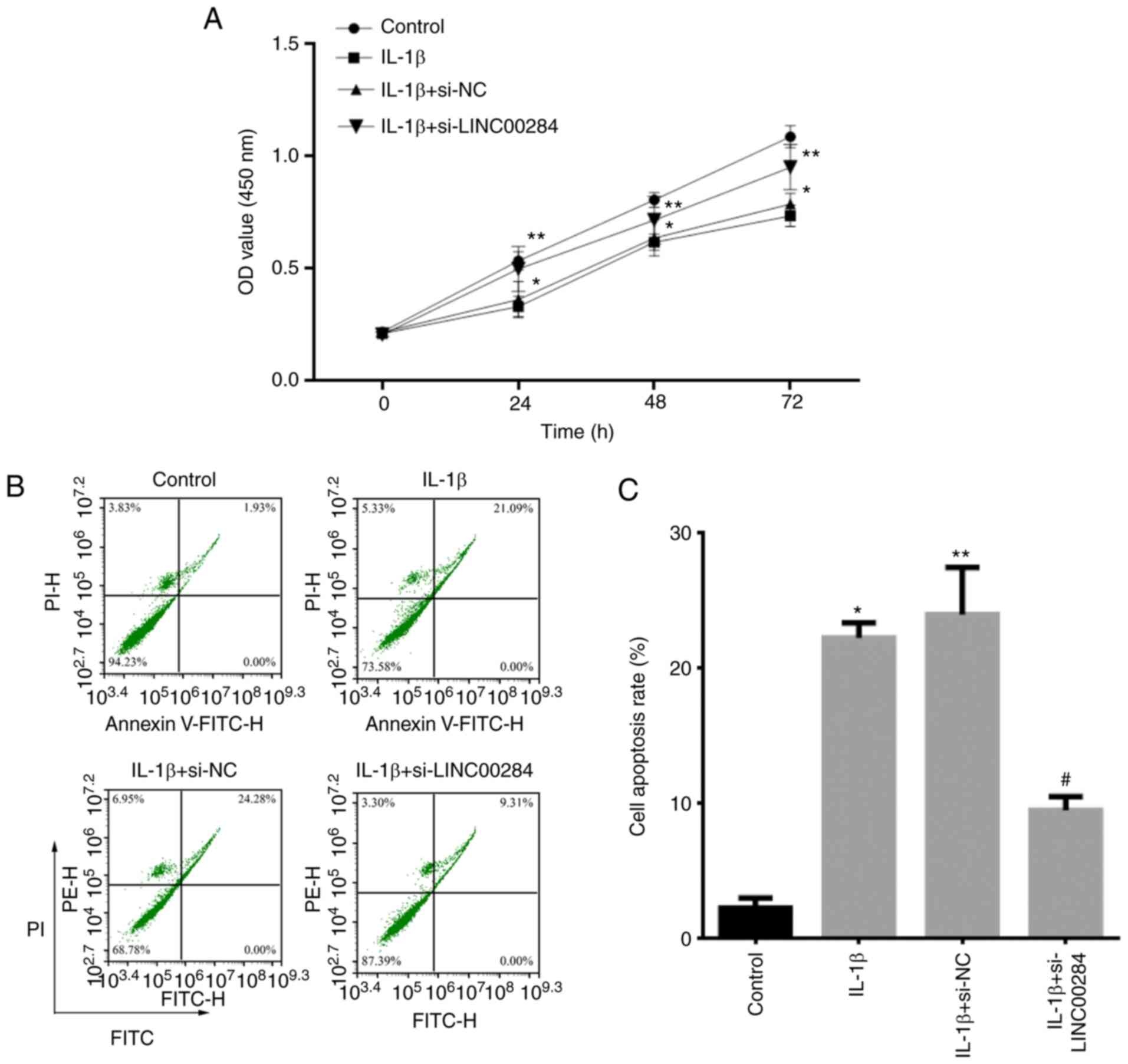
LINC00284 knockdown promotes
proliferation and inhibits apoptosis of IL-1β-induced NP cells
To determine the role of LINC00284 in IDD,
proliferation of IL-1β-induced NP cells following LINC00284
knockdown was detected using CCK-8 assay and Annexin V-FITC/PI
staining with flow cytometry analysis. Compared with the control
group, proliferation was inhibited in cells treated with 5 ng/ml
IL-1β for 24, 48 and 72 h; however, in IL-1β-induced NP cells,
si-LINC00284 promoted cell proliferation (Fig. 3A) but significantly decreased
apoptosis (Fig. 3B and C),
compared with si-NC-treated cells. The results indicated that
LINC00240 may inhibit the proliferation, but promote the apoptosis,
of NP cells in IDD patients.
LINC00284 knockdown improves IDD in an
animal model of IDD
To evaluate the therapeutic effect of LINC00284
knockdown in vivo, intradiscal injection of sh-LINC00284
lentivirus was performed weekly in a needle puncture rat IDD model.
Compared with 7 days after initial puncture, X-ray examination
showed that IDD rats treated with sh-NC exhibited more notable
narrowing of disc height compared with sham rats treated with sh-NC
on days 14 and 28. Following treatment of IDD rats with
sh-LINC00284, the decline in disc height began to slow from days
7–28 (Fig. 4A) and the percentage
DHI in the sh-LINC00284 group was 1.56, 1.28 and 1.32-fold higher
than that in sh-NC IDD rats on days 7, 14 and 28, respectively
(Fig. 4B). According to MRI
examination, 14 and 28 days after initial puncture, intradiscal
injection with sh-NC in IDD rats exhibited weaker MRI signal than 7
days. Compared with sh-NC-treated IDD rats, sh-LINC00284 treatment
notably alleviated weak MRI signal intensity of the disc puncture,
whereas MRI signal decreased significantly (Fig. 4C and D).
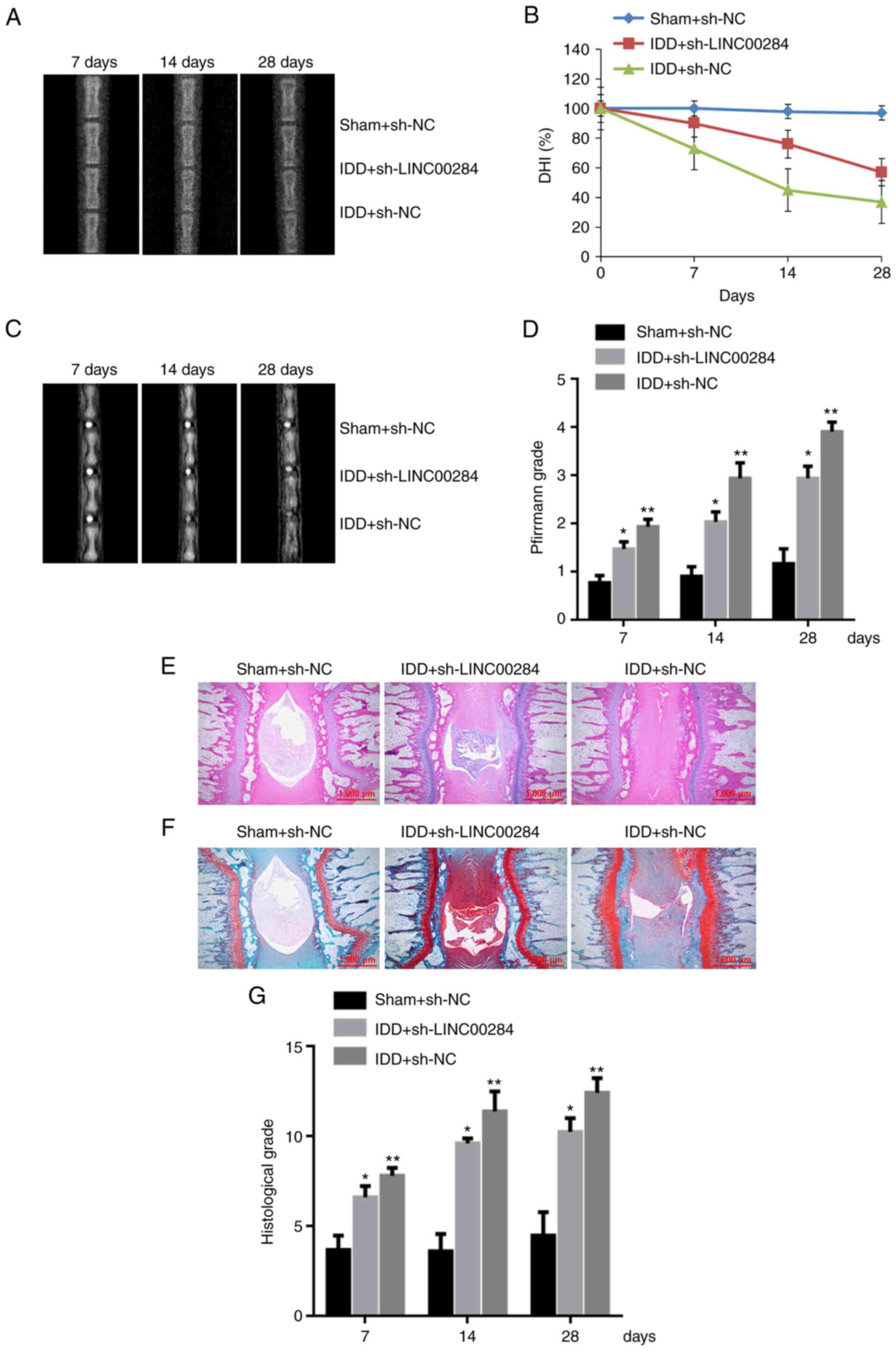 | Figure 4.Downregulation of LINC00284 improves
puncture-induced IDD in vivo. (A) Representative radiographs
of coccygeal vertebrae, (B) DHI change, (C) representative MRI
scans of coccygeal vertebrae and (D) changes in Pfirrmann grade on
days 7, 14 and 28 after initial puncture. (E) Hematoxylin and eosin
staining of whole-tail disc sections from IDD rats on day 28
post-initial puncture. (F) Safranin-O staining of whole-tail disc
sections from IDD rats. (G) Histological grade on days 7, 14 and 28
days post-initial puncture. Scale bar, 1,000 µm. *P<0.05 vs.
sham + sh-NC; **P<0.05 vs. IDD + sh-NC. IDD, intervertebral disc
degeneration; DHI, disc height index; sh, short hairpin; NC,
negative control. |
The histological structure of IVD was observed using
H&E and safranin-O staining. On day 28, the sh-NC group showed
a notable decrease in number of NP cells and destruction of AF
lamella compared with sham (Fig. 4E
and F). Compared with IDD rats treated with sh-NC, sh-LINC00284
treatment preserved the complete structure of the NP and AF;
histological grade of the sh-LINC00284 group on days 7, 14 and 28
increased by 13.1, 15.5 and 17.5% respectively (Fig. 4G). The results indicated that
LINC00240 may be a therapeutic target of IDD.
LINC00284 is the direct target of
miR-205-3p
To investigate whether LINC00284 affected ECM
degradation in IL-1β-induced NP cells, the target ncRNA of
miR-205-3p was predicted using DIANA-LncBase V2 (Fig. 5A) and wt and mut binding sites
between LINC00284 and miR-205-3p were established. The luciferase
activity of wt-LINC00284 and miR-205-3p mimic co-transfected cells
was significantly decreased compared with cells co-transfected with
mi-NC transfected cells and mut-LINC00284 and miR-205-3p mimic
co-transfected cells (Fig. 5B).
These results indicated that LINC00284 was the direct target of
miR-205-3p.
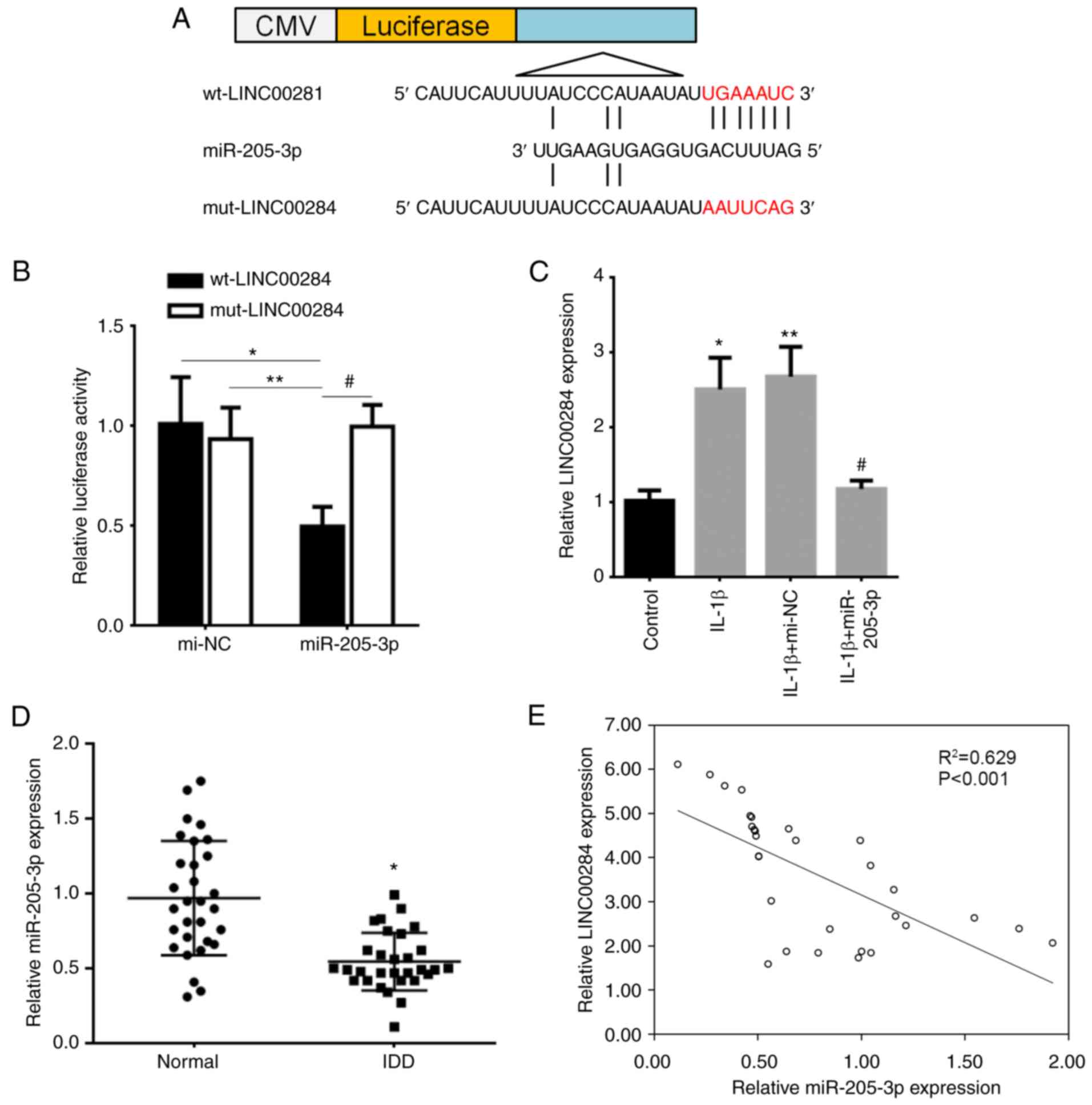 | Figure 5.Correlation between LINC00284 and
miR-205-3p expression in IDD. (A) Binding site (red) between
LINC00284 and miR-205-3p was predicted and assessed using dual
luciferase reporter vectors containing wt-LINC00284 or
mut-LINC00284. (B) Vectors were co-transfected into 293T cells with
miR-205-3p mimics, then luciferase activity was detected using a
dual-luciferase reporter assay. *P<0.05 vs. mi-NC and
wt-LINC00284 co-transfected cells, **P<0.05 vs. mi-NC and
mut-LINC00284 co-transfected cells, #P<0.05 vs. miR-205-3p and
mut-LINC00284 co-transfected cells. (C) Expression levels of
LINC00284 were downregulated by miR-205-3p in IL-1β-induced NP
cells. *P<0.05 vs. control; **P<0.05 vs. IL-1β; #P<0.05
vs. IL-1β + mi-NC. (D) Expression levels of LINC00284 in 30 IDD and
30 normal tissue samples, *P<0.001 vs. normal tissue samples.
(E) Negative correlation between miR-205-3p and LINC00284
expression in IDD tissue. IDD, intervertebral disc degeneration;
mi-NC, miR-negative control; CMV, cytomegalovirus; wt, wild-type;
mut, mutated; miR, microRNA. |
LINC00284 is regulated by miR-205-3p
in NP cells and their expression is correlated in IDD tissue
To validate the interaction between LINC00284 and
miR-205-3p, the effect of transfection of miR-205-3p mimics on
expression of LINC00284 was evaluated in NP cells. IL-1β induced
upregulation of LINC00284 compared with control but miR-205-3p
mimic transfection significantly inhibited LINC00284 expression
levels compared with mi-NC-treated cells (Fig. 5C). RT-qPCR showed that miR-205-3p
levels were significantly downregulated in 30 IDD compared with 30
spinal cord injury tissue samples (Fig. 5D). Moreover, there was a
significant negative correlation between LINC00284 and miR-205-3p
expression in 30 IDD tissue samples (Fig. 5E). The results indicated that
miR-205-3p was low-expressed and was negative correlated with
LINC00240 expression in NP tissues of IDD patients.
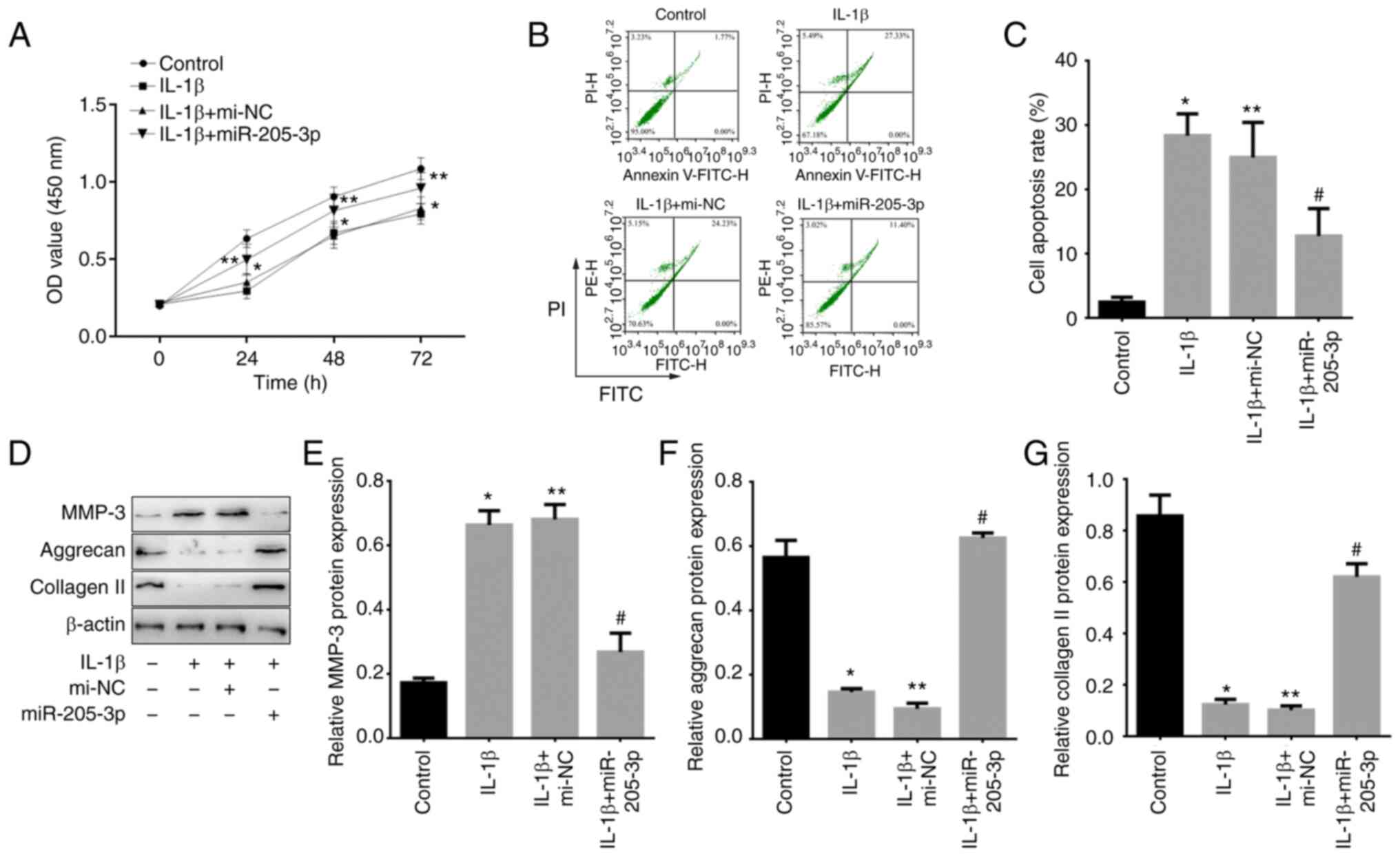
miR-205-3p-mediated LINC00284
downregulation promotes IL-1β-induced proliferation and inhibits
apoptosis of NP cells and promotes ECM synthesis
To confirm the effect of miR-205-3p-mediated
regulation of LINC00284 on proliferation, apoptosis and ECM
synthesis, miR-205-3p mimics were transfected into IL-1β-induced NP
cells. Compared with mi-NC-treated cells, proliferation was
increased by miR-205-3p overexpression (Fig. 6A). Furthermore, the cell apoptosis
were promoted in IL-1β treated NP cells and mi-NC treated
IL-1β-induced NP cells, but a suppressive effect on cell apoptosis
was observed following miR-205-3p upregulation (Fig. 6B and C). Western blot analysis
showed that, compared with control (untreated NP cells), MMP-3
protein levels were upregulated but aggrecan and collagen II
protein levels were downregulated in IL-1β treated NP cells and
mi-NC treated IL-1β-induced NP cells. Compared with mi-NC treated
IL-1β-induced NP cells, MMP-3 protein levels were downregulated by
miR-205-3p, whereas aggrecan and collagen II protein levels were
upregulated in IL-1β-induced NP cells (Fig. 6D-G). The results indicated that
LINC00240 can be inhibited by miR-205-3p in IDD through the
promotion of NP cell proliferation and ECM synthesis.
LINC00284 knockdown promotes
IL-1β-induced ECM synthesis in NP cells via Wnt/β-catenin
signaling
Wnt/β-catenin signaling has been demonstrated to
regulate inflammation in the musculoskeletal system and β-catenin
is controlled by non-genomic mechanisms (26,27). To investigate if this signaling
pathway participated in LINC00284-mediated rescue of ECM
degradation via competitively binding with miR-205-3p, si-LINC00284
and/or miR-205-3p mimics were co-transfected into IL-1β-induced NP
cells. Compared with control, the protein levels of MMP-3, Wnt1 and
β-catenin were increased but aggrecan and collagen II were
decreased in IL-1β treated NP cells and si-NC treated IL-1β-induced
NP cells. Compared with si-NC treated IL-1β-induced cells,
si-LINC00284 resulted in decreased MMP-3, Wnt1 and β-catenin but an
increase in aggrecan and collagen II protein levels.
Co-transfection of si-LINC00284 with miR-205-3p reversed this
effect (Fig. 7). These results
indicated that the effect of LINC00284 on promoting ECM degradation
may partially be mediated via the Wnt/β-catenin signaling
pathway.
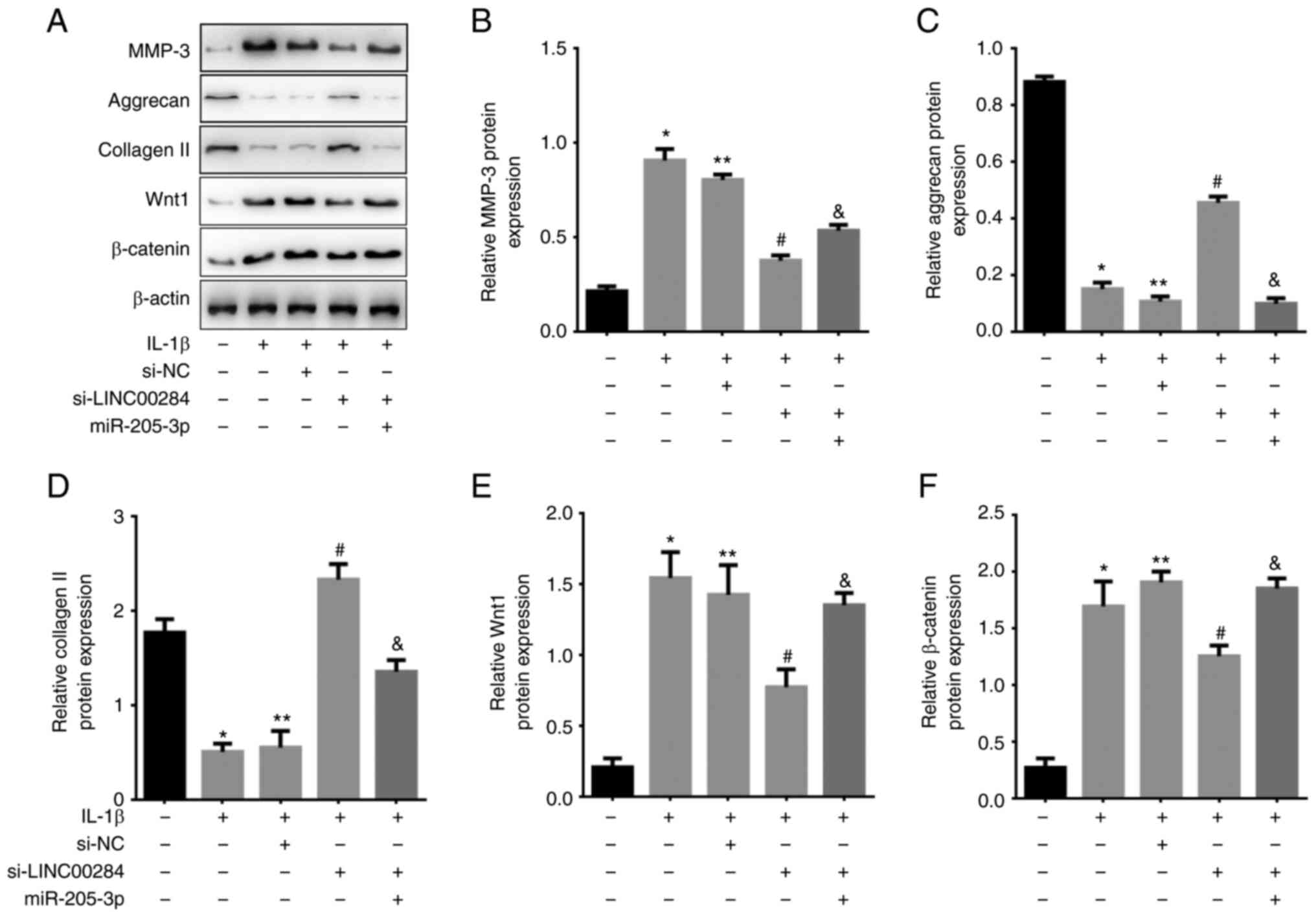 | Figure 7.LINC00284 regulates ECM synthesis in
IL-1β-induced NP cells via the miR-205-3p/Wnt/β-catenin signaling
pathway axis. (A) Expression of MMP-3, ECM markers and
Wnt/β-catenin pathway markers following si-LINC00284 and/or
miR-205-3p transfection in IL-1β-induced NP cells, as determined by
western blotting. (B) MMP-3, (C) aggrecan, (D) collagen II, (E)
Wnt1 and (F) β-catenin protein levels following si-LINC00284 and/or
miR-205-3p transfection. *P<0.05 vs. Control; **P<0.05 vs.
IL-1β; #P<0.05 vs. si-NC; &P<0.05 vs. si-LINC00284. ECM,
extracellular matrix; NP, nucleus pulposus; MMP-3, matrix
metalloproteinase-3; si, small interfering; NC, negative control;
miR, microRNA. |
Discussion
IDD is characterized by disorders in ECM metabolism,
abnormal proliferation and increased expression of numerous
inflammatory factors including IL-1β, IL-8 and TNF-α in NP cells
(41). Although multiple factors
including SIRT7, NTRK2, and CHI3L1 (42) contribute to etiology of IDD, the
molecular mechanism underlying its development remains unclear. An
increasing body of evidence has indicated that ncRNAs, including
lncRNAs and miRNAs, serve key roles in IDD (13). lncRNA00641 regulates autophagy and
IVD degeneration by acting as a ceRNA of miR-153-3p under stress
induced by nutritional deprivation (43). lncRNA taurine upregulated 1
promotes NP cell apoptosis by regulating the miR-26a/high mobility
group box 1 axis (44). In the
present study, LINC00284 expression was significantly upregulated
in IDD tissue and IL-1β-induced NP cells. LINC00284 knockdown
promoted proliferation and decreased apoptosis of IL-1β-induced NP
cells; expression of aggrecan and collagen II, ECM markers of disc
degeneration, was also upregulated.
IVD consists of an outer fibrous layer, AF, an inner
gelatinous core rich in proteoglycans and NP (45). NP cells serve key roles in
maintaining IVD integrity (46);
abnormal proliferation of NP cells results in the appearance of
cell clusters, which are the primary feature of IVD degeneration
(47). Consistent with the
results that LINC00284 promotes cell proliferation in ovarian
cancer (48), in the present
study, LINC00284 inhibited proliferation and promoted NP cell
apoptosis. Furthermore, knockdown of LINC00284 using a targeting
siRNA resulted in promotion of proliferation and inhibition of
apoptosis; in vivo analysis showed that LINC00284 knockdown
attenuated IDD in the rat model. These results suggested that
LINC00284 may be a potent activator of IDD progression.
Additionally, the effect of LINC00284 on needle puncture IDD model
rats was assessed; IDD rats with LINC00284 knockdown showed a
decrease in MRI grade and increase in histological grade. The in
vivo experiments confirmed results of in vitro analysis
and suggested that LINC00284 promoted IDD progression.
lncRNAs serve as sponges as ceRNAs by targeting
miRNAs to participate in regulation of pathophysiological
processes, including in IDD. lncRNA HLA complex group 18 serves as
an endogenous sponge to downregulate miR-146a-5p expression, thus
suppressing proliferation of NP cells (49); lncRNA RNA component of
mitochondrial RNA processing endoribonuclease interacts with
miR-206 to induce upregulation of miR-206, thus promoting
proliferation of NP cells (50).
In papillary thyroid cancer (PTC), Zhao et al (51) demonstrated LINC00284 may serve as
a ceRNA to interact with miR-205 and E2F and increased expression
of LINC00284 was positively correlated with prognosis of PTC.
Consistently, in the present study, it was predicted that
miR-205-3p may possess a binding site for LINC00284 using
DIANA-LncBase V2; it was confirmed that miR-205-3p inhibited
LINC00284 expression via base-pair complementary binding.
miR-205-3p expression was upregulated in IDD tissue and expression
levels of LINC00284 and miR-205-3p were negatively correlated.
Furthermore, the promotive effect of LINC00284 knockdown on
IL-1β-induced cell proliferation and ECM synthesis was partially
reversed by miR-205-3p, suggesting that LINC00284 may act as a
ceRNA to regulate ECM synthesis by sponging miR-205-3p.
The Wnt/β-catenin signaling pathway serves a
regulatory role in IDD (52).
Hiyama et al (53) found
that activation of the Wnt/β-catenin signaling pathway promotes IDD
cell senescence and apoptosis, consistent with the results of the
present study, which showed that LINC00284 knockdown inhibited the
Wnt/β-catenin signaling pathway, as well as the rate of
proliferation and apoptosis. These results suggested that LINC00284
may serve a key role in the progression of IDD via activation of
the Wnt/β-catenin signaling pathway.
In conclusion, LINC00284 served as a ceRNA to
activate the Wnt/β-catenin signaling pathway by sponging
miR-205-3p, resulting in inhibition of cell proliferation in the NP
as well as ECM synthesis in IDD. Thus, LINC00284 knockdown to
rescue miR-205-3p expression may be a promising strategy to treat
IDD.
Acknowledgements
Not applicable.
Funding
The present study was supported by Jiangsu Health and Family
Planning Commission, China (grant no. Y2018099) and Science
Foundation of Nantong City, Jiangsu Province, China (grant no.
JCZ20121).
Availability of data and materials
All the data generated or analyzed during the
present study are included in this published article.
Authors' contributions
MZ and YS conceived and designed the study. XY, HX,
ZW, BW and XL performed the experiments. YZ collected and analyzed
tissue samples. MZ, XY and YS acquired, analyzed and interpreted
the data. HX, ZW, BW and XL reviewed the manuscript. MZ drafted and
revised the manuscript. All authors have read and approved the
final manuscript. MZ and YS confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved and supervised by the
Ethics Committee of Shanghai Changzheng Hospital, Second Military
Medical University. Written informed consent was provided by all
subjects. Animal experiments were approved by the Institutional
Animal Care and Use Committee of Shanghai Changzheng Hospital,
Second Military Medical University. Great effort was made to
minimize the number of animals used and their respective
suffering.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu C, Yang M, Liu L, Zhang Y, Zhu Q,
Huang C, Wang H, Zhang Y, Li H, Li C, et al: Molecular basis of
degenerative spinal disorders from a proteomic perspective
(Review). Mol Med Rep. 21:9–19. 2020.PubMed/NCBI
|
|
2
|
Setton LA and Chen J: Mechanobiology of
the intervertebral disc and relevance to disc degeneration. J Bone
Joint Surg Am. 88 (Suppl 2):S52–S57. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balagué F, Mannion AF, Pellisé F and
Cedraschi C: Non-specific low back pain. Lancet. 379:482–891. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Battié MC, Videman T and Parent E: Lumbar
disc degeneration: Epidemiology and genetic influences. Spine
(Phila Pa 1976). 29:2679–2690. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Battié MC and Videman T: Lumbar disc
degeneration: Epidemiology and genetics. J Bone Joint Surg Am. 88
(Suppl 2):S3–S9. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng H, Danfelter M, Strömqvist B and
Heinegård D: Extracellular matrix in disc degeneration. J Bone
Joint Surg Am. 88 (Suppl 2):S25–S29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roberts S, Caterson B, Menage J, Evans EH,
Jaffray DC and Eisenstein SM: Matrix metalloproteinases and
aggrecanase: Their role in disorders of the human intervertebral
disc. Spine (Phila Pa 1976). 25:3005–3013. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang SZ, Chang Q, Lu J and Wang C: Growth
factors and platelet-rich plasma: Promising biological strategies
for early intervertebral disc degeneration. Int Orthop. 39:927–934.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Richardson SM, Kalamegam G, Pushparaj PN,
Matta C, Memic A, Khademhosseini A, Mobasheri R, Poletti FL,
Hoyland JA and Mobasheri A: Mesenchymal stem cells in regenerative
medicine: Focus on articular cartilage and intervertebral disc
regeneration. Methods. 99:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu H, Pan H, Yang H, Wang J, Zhang K, Li
X, Wang H, Ding W, Li B and Zheng Z: LIM mineralization protein-1
suppresses TNF-α induced intervertebral disc degeneration by
maintaining nucleus pulposus extracellular matrix production and
inhibiting matrix metalloproteinases expression. J Orthop Res.
33:294–303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bach FC, Zhang Y, Miranda-Bedate A,
Verdonschot LC, Bergknut N, Creemers LB, Ito K, Sakai D, Chan D,
Meij BP and Tryfonidou MA: Increased caveolin-1 in intervertebral
disc degeneration facilitates repair. Arthritis Res Ther.
18:592016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fujita K, Ando T, Ohba T, Wako M, Sato N,
Nakamura Y, Ohnuma Y, Hara Y, Kato R, Nakao A, et al: Age-related
expression of MCP-1 and MMP-3 in mouse intervertebral disc in
relation to TWEAK and TNF-α stimulation. J Orthop Res. 30:599–605.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan
YG, Zhang J and Wang WJ: lncRNAs: Novel players in intervertebral
disc degeneration and osteoarthritis. Cell Prolif. 50:e123132017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Ni H, Zhao Y, Chen K, Li M, Li C,
Zhu X and Fu Q: Potential role of lncRNAs in contributing to
pathogenesis of intervertebral disc degeneration based on
microarray data. Med Sci Monit. 21:3449–3458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cui S, Liu Z, Tang B, Wang Z and Li B:
LncRNA MAGI2-AS3 is down-regulated in intervertebral disc
degeneration and participates in the regulation of FasL expression
in nucleus pulposus cells. BMC Musculoskelet Disord. 21:1492020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Z, Huo Y, Zhou Z, Zhang P and Hu J:
Role of lncRNA PART1 in intervertebral disc degeneration and
associated underlying mechanism. Exp Ther Med. 21:1312021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yildiz-Arslan S, Coon JS, Hope TJ and Kim
JJ: Transcriptional profiling of human endocervical tissues reveals
distinct gene expression in the follicular and luteal phases of the
menstrual cycle. Biol Reprod. 94:1382016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xing C, Cai Z, Gong J, Zhou J, Xu J and
Guo F: Identification of potential biomarkers involved in gastric
cancer through integrated analysis of non-coding RNA associated
competing endogenous RNAs network. Clin Lab. 64:1661–1669. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L and Guo Y: Silencing circular
RNA-ZNF652 represses proliferation and EMT process of renal
carcinoma cells via raising miR-205. Artif Cells Nanomed
Biotechnol. 48:648–655. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song Y, Guo NH and Zheng JF: LncRNA-MALAT1
regulates proliferation and apoptosis of acute lymphoblastic
leukemia cells via miR-205-PTK7 pathway. Pathol Int. 70:724–732.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen B, Li Y, Liu Y and Xu Z: circLRP6
regulates high glucose-induced proliferation, oxidative stress, ECM
accumulation, and inflammation in mesangial cells. J Cell Physiol.
234:21249–21259. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frank S, Peters MA, Wehmeyer C, Strietholt
S, Koers-Wunrau C, Bertrand J, Heitzmann M, Hillmann A, Sherwood J,
Seyfert C, et al: Regulation of matrixmetalloproteinase-3 and
matrixmetalloproteinase-13 by SUMO-2/3 through the transcription
factor NF-κB. Ann Rheum Dis. 72:1874–1881. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hiyama A, Gogate SS, Gajghate S, Mochida
J, Shapiro IM and Risbud MV: BMP-2 and TGF-beta stimulate
expression of beta1,3-glucuronosyl transferase 1 (GlcAT-1) in
nucleus pulposus cells through AP1, TonEBP, and Sp1: Role of MAPKs.
J Bone Miner Res. 25:1179–1190. 2010.PubMed/NCBI
|
|
25
|
Mäkitie RE, Niinimäki T, Nieminen MT,
Schalin-Jäntti C, Niinimäki J and Mäkitie O: Impaired WNT signaling
and the spine-Heterozygous WNT1 mutation causes severe age-related
spinal pathology. Bone. 101:3–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ge XP, Gan YH, Zhang CG, Zhou CY, Ma KT,
Meng JH and Ma XC: Requirement of the NF-κB pathway for induction
of Wnt-5A by interleukin-1β in condylar chondrocytes of the
temporomandibular joint: Functional crosstalk between the Wnt-5A
and NF-κB signaling pathways. Osteoarthritis Cartilage. 19:111–117.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reya T and Clevers H: Wnt signalling in
stem cells and cancer. Nature. 434:843–850. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Söderholm S and Cantù C: The WNT/β-catenin
dependent transcription: A tissue-specific business. WIREs Mech
Dis. 13:e15112021.PubMed/NCBI
|
|
29
|
Rauner M, Stein N, Winzer M, Goettsch C,
Zwerina J, Schett G, Distler JH, Albers J, Schulze J, Schinke T, et
al: WNT5A is induced by inflammatory mediators in bone marrow
stromal cells and regulates cytokine and chemokine production. J
Bone Miner Res. 27:575–585. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakamura Y, Nawata M and Wakitani S:
Expression profiles and functional analyses of Wnt-related genes in
human joint disorders. Am J Pathol. 167:97–105. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
World Medical Association, . World medical
association declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Z, Liu H, Yang H, Wang J, Wang H, Zhang
K, Ding W and Zheng Z: Both expression of cytokines and posterior
annulus fibrosus rupture are essential for pain behavior changes
induced by degenerative intervertebral disc: An experimental study
in rats. J Orthop Res. 32:262–272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Council N: Guide for the care and use of
laboratory animals: Eighth edition. Publication. 327:pp963–965.
2010.
|
|
36
|
Zhang J, Li Z, Chen F, Liu H, Wang H, Li
X, Liu X, Wang J and Zheng Z: TGF-β1 suppresses CCL3/4 expression
through the ERK signaling pathway and inhibits intervertebral disc
degeneration and inflammation-related pain in a rat model. Exp Mol
Med. 49:e3792017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Miyagi M, Ishikawa T, Kamoda H, Suzuki M,
Sakuma Y, Orita S, Oikawa Y, Aoki Y, Toyone T, Takahashi K, et al:
Assessment of pain behavior in a rat model of intervertebral disc
injury using the CatWalk gait analysis system. Spine (Phila Pa
1976). 38:1459–1465. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han B, Zhu K, Li FC, Xiao YX, Feng J, Shi
ZL, Lin M, Wang J and Chen QX: A simple disc degeneration model
induced by percutaneous needle puncture in the rat tail. Spine
(Phila Pa 1976). 33:1925–1934. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Masuda K, Aota Y, Muehleman C, Imai Y,
Okuma M, Thonar EJ, Andersson GB and An HS: A novel rabbit model of
mild, reproducible disc degeneration by an anulus needle puncture:
Correlation between the degree of disc injury and radiologi and
histological appearances of disc degeneration. Spine (Phila Pa
1976). 30:5–14. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu ZM, Lu CC, Shen PC, Chou SH, Shih CL,
Chen JC and Tien YC: Suramin attenuates intervertebral disc
degeneration by inhibiting NF-κB signalling pathway. Bone Joint
Res. 10:498–513. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li H, Li W, Zhang L, He J, Tang L, Li Z,
Chen F, Fan Q and Wei J: Comprehensive network analysis identified
SIRT7, NTRK2, and CHI3L1 as new potential markers for
intervertebral disc degeneration. J Oncol. 2022:44075412022.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang XB, Wang H, Long HQ, Li DY and Zheng
X: LINC00641 regulates autophagy and intervertebral disc
degeneration by acting as a competitive endogenous RNA of
miR-153-3p under nutrition deprivation stress. J Cell Physiol.
234:7115–7127. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tang N, Dong Y, Xiao T and Zhao H: LncRNA
TUG1 promotes the intervertebral disc degeneration and nucleus
pulposus cell apoptosis though modulating miR-26a/HMGB1 axis and
regulating NF-κB activation. Am J Transl Res. 12:5449–5464.
2020.PubMed/NCBI
|
|
45
|
Kirnaz S, Singh S, Capadona C, Lintz M,
Goldberg JL, McGrath LB Jr, Medary B, Sommer F, Bonassar LJ and
Härtl R: Innovative biological treatment methods for degenerative
disc disease. World Neurosurg. 157:282–299. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Johnson WE, Eisenstein SM and Roberts S:
Cell cluster formation in degenerate lumbar intervertebral discs is
associated with increased disc cell proliferation. Connect Tissue
Res. 42:197–207. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Daisuke T and Horton P: Inference of
scale-free networks from gene expression time series. J Bioinform
Comput Biol. 4:503–514. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ruan Z and Zhao D: Long intergenic
noncoding RNA LINC00284 knockdown reduces angiogenesis in ovarian
cancer cells via up-regulation of MEST through NF-κB1. FASEB J.
33:12047–12059. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X
and He Y: Long non-coding HCG18 promotes intervertebral disc
degeneration by sponging miR-146a-5p and regulating TRAF6
expression. Sci Rep. 7:132342017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang X, Peng L, Gong X, Zhang X, Sun R and
Du J: LncRNA-RMRP promotes nucleus pulposus cell proliferation
through regulating miR-206 expression. J Cell Mol Med.
22:5468–5476. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao Y, Wang H, Wu C, Yan M, Wu H, Wang J,
Yang X and Shao Q: Construction and investigation of
lncRNA-associated ceRNA regulatory network in papillary thyroid
cancer. Oncol Rep. 39:1197–1206. 2018.PubMed/NCBI
|
|
52
|
Xie H, Jing Y, Xia J, Wang X, You C and
Yan J: Aquaporin 3 protects against lumbar intervertebral disc
degeneration via the Wnt/β-catenin pathway. Int J Mol Med.
37:859–864. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hiyama A, Sakai D, Risbud MV, Tanaka M,
Arai F, Abe K and Mochida J: Enhancement of intervertebral disc
cell senescence by WNT/β-catenin signaling-induced matrix
metalloproteinase expression. Arthritis Rheum. 62:3036–3047. 2010.
View Article : Google Scholar : PubMed/NCBI
|