Introduction
Atrial fibrillation (AF) is a well-known
supraventricular arrhythmia in the general population (1). In animal studies, atrial fibrosis
plays an important role in the induction and perpetuation of AF
(2–4). Atrial fibrosis causes interatrial
inhomogeneity in electrical conduction, creating a substrate for
local re-entry and contributing to the progression of AF (4). The rennin-angiotensin system,
specifically angiotensin II (Ang II), has been suggested to play a
significant role in the development of cardiac remodeling during
AF. However, the precise downstream molecules important in the
genesis of AF-induced atrial fibrosis are currently unclear
(5). The major proteins and
mechanisms involved in the remodeling process of AF include the
altered expression of ion channels (causing a reduction in action
potential duration), Ca2+ handling proteins (impairing
intracellular Ca2+ removal and release), or connexins
(impairing intra-atrial conduction). In addition, spontaneous
sarcoplasmic reticulum Ca2+ release through altered RyR2
can act as a trigger for AF and can induce AF by increasing
fibrosis, which acts as a substrate that promotes re-entry and
maintenance of AF.
miRNAs are small non-coding, single-stranded RNAs
that are18-25 nucleotides in length (6). The expression of various miRNAs has
been linked to cardiovascular disorders, including heart failure
and left ventricular hypertrophy (7,8).
miRNAs represent one of the most actively investigated areas in the
cardiovascular field; they are being investigated as potential
diagnostic and prognostic biomarkers in a number of cardiovascular
pathologies and associated metabolic diseases (9). Urine is a sterile biological fluid
containing the final product produced by the metabolism of proteins
secreted by the kidneys and can be collected in a non-invasive and
simple manner (10). Most urine
miRNAs are derived from renal and urethral cells, and the analysis
of these cells can determine the health status of patients.
Therefore, changes in the miRNAs in AF patients could be confirmed
through urine samples (11,12). In this study, we report a newly
discovered miRNA that is related to fibrosis and Ca2+
handling proteins in the development of AF and attempt to establish
its significance in the diagnosis of AF by examining the changes in
the miRNA through urine samples.
Materials and methods
Urine sample collection
Urine samples were collected from patients diagnosed
with paroxysmal supraventricular tachycardia (PSVT) and AF using a
non-invasive method. Patient samples were recruited from patients
with written informed consent at Ewha Womans University Hospital
from August 2019 to February 2020. The clinical profiles of
patients are shown in Table I.
The study protocol was ethically approved by the local ethics
committee (Institutional Review Board of Ewha Womans University
Hospital (Seoul, Korea); approval no. IRB 2019-10-019). In brief,
after the tube was inserted into the patient's urethra,
approximately 50 ml of initial urine was collected. Then, the old
tube was replaced with a new tube, and experimental urine samples
were collected. Aliquoted urine samples were stored at −70°C until
miRNA isolation.
 | Table I.Clinical profile of patients whose
urine was used for mRNA isolation. |
Table I.
Clinical profile of patients whose
urine was used for mRNA isolation.
| Case | Sex | Age, years | Diagnosis |
|---|
| 1 | Male | 47 | PSVT |
| 2 | Female | 46 | PSVT |
| 3 | Female | 69 | PSVT |
| 4 | Male | 63 | PeAF |
| 5 | Female | 58 | PeAF |
| 6 | Female | 63 | PeAF |
| 7 | Male | 56 | PeAF |
| 8 | Male | 58 | PeAF |
Quantitative gene expression analysis
of urine samples from the AF and PSVT groups
Fibrosis-related gene expression (collagen I/III,
fibronectin 1, and TGF-β) was analyzed using RNA samples isolated
from AF patients' urine and HL-1 cells. To further investigate the
effects of the fibrosis markers, fibrosis-related gene expression
was examined in Ang II-treated cells.
Cell culture
HL-1 is cardiac muscle cell line purchased from
Merck Millipore (Merck Millipore, Germany). Cells were maintained
in Complete Claycomb Medium (Sigma-Aldrich, MO, USA) supplemented
with 10% FBS, 1% penicillin-streptomycin, 100 µM norepinephrine
(Sigma-Aldrich, MO, USA), and 4 mM L-glutamine (Gibco, MA, USA) in
plates coated with fibronectin to 0.02% gelatin solution with a
final concentration of 12.5 µg/ml. (Sigma-Aldrich, MO, USA). Cells
were grown in 5% CO2 incubator at 37°C.
miRNA transfection into HL-1
cells
The negative control miRNA (SMC-2001), miRNA
inhibitor negative control (SMC-2101), miR-423 mimic
(5-AUAAAGGAAGUUAGGCUGAGGGGCAGAGAGCGAGACUUUUCUAUUUUCCAAAAGCUCGGUCUGAGGCCCCUCAGUCUUGCUUCCUAACCCGCGC-3),
and miR-423 inhibitor (5-UGAGGGGCAGAGAGCGAGACUUU-3) were purchased
from Bioneer Corporation (Daejeon, Korea). HL-1 cells were plated
on a 35 mm dish in 1 ml of Claycomb Medium. The transfection of
miRNAs into HL-1 cells was performed with RNAiMax Transfection
Reagent. The cells were transfected with Lipofectamine according to
the manufacturer's protocol (Invitrogen, Grand Island, NY, USA).
Transfection efficacy was evaluated by quantitative PCR. Also, we
showed that transfection of miR-423 was successful using miR-423
mimic, inhibitor and negative controls using RT-qPCR (Fig. S1).
Western blotting
HL-1 cells were homogenized and lysed with RIPA
buffer (Sigma-Aldrich, MO, USA) containing a protease inhibitor
cocktail (Sigma-Aldrich, MO, USA). BCA protein assay kit
(Millipore, Billerica, MA, USA) was used to measure the protein
concentration according to the manufacturer's instructions. Protein
samples (20 µg) of were resolved by SDS-PAGE and then transferred
to polyvinylidene fluoride membrane (Millipore, Billerica, MA,
USA). The membranes were incubated overnight at 4°C with anti-total
CaMKIIδ (1:1,000; Santa Cruz Biotechnology), Thr287 and
Thr306/Thr307 phosphorylated CaMKIIδ (1:1,000; Abcam Reagents),
total phospholamban (PLB) (1:1,000; Santa Cruz Biotechnology),
Thr17 phosphorylated PLB (1:1,000; Santa Cruz Biotechnology),
SERCA2A (1:1,000; Santa Cruz Biotechnology), total RyR2 (1:1,000;
Abcam Reagents), Ser2808 and Ser2814 phosphorylated RyR2 (1:500 and
1:1,000, Badrilla, Leeds, UK) as indicated. The membranes were
incubated with HRP-conjugated mouse or rabbit anti-mouse secondary
antibodies (Santa Cruz), and the protein bands were visualized by
chemiluminescence (Millipore, Billerica, MA, USA).
Quantitative polymerase chain
reaction
Total RNA was extracted from cells using the QIAGEN
miRNeasy Mini kit (QIAGEN, CA, USA) according to the manufacturer's
instructions. Cell-derived total RNA was reverse transcribed using
a miRNA-specific stem-loop real-time reaction with transcription
primers. The concentration of total RNA was quantified using an
UV–Vis spectrophotometer (Nabi, MicroDigital Co., Ltd., Korea) via
the A260/280 ratio; only pure total RNA samples within a ratio were
selected. cDNA was synthesized using the RevertAid™ First Strand
cDNA Synthesis Kit (Thermofisher, NZ, USA). Quantitative PCR was
performed using the iQ5 system (Bio-Rad, ON, USA) with
SYBR® Premix Ex Taq™ II (Agilent Technologies Inc. CA,
USA) for quantification. Triplicates were tested for each sample.
Gene and miRNA expression levels were normalized to GAPDH and U6
snRNA expression levels, respectively. Primer sequences are listed
in Table II.
 | Table II.Primer sequence. |
Table II.
Primer sequence.
| Target gene | Forward primer
(3′-5′) | Reverse primer
(5′-3′) |
|---|
| TGF-β |
TTGCTTCAGCTCCACAGAGA |
TGGTTGTAGAGGGCAAGGAC |
| Collagen I |
GAGCGGAGAGTACTGGATCG |
GCTTCTTTTCCTTGGGGTTC |
| Collagen III |
TGATGGAAAACCAGGACCTC |
CAGTCTCCCCATTCTTTCCA |
| Fibronectin |
GATGCACCGATTGTCAACAG |
TGATCAGCATGGACCACTTC |
| GAPDH |
ACCAGGTATCTGCTGGTTG |
TAACCATGATGTCAGCGTGGT |
|
hsa-miR-3613-5p |
UGUUGUACUUUUUUUUUUGUUC |
|
|
hsa-miR-6763-5p |
CUGGGGAGUGGCUGGGGAG |
|
| hsa-miR-423-5p |
UGAGGGGCAGAGAGCGAGACUUU |
|
|
hsa-miR-1180-5p |
GGACCCACCCGGCCGGGAAUA |
|
|
hsa-miR-6511b-3p |
CCUCACCACCCCUUCUGCCUGCA |
|
| hsa-miR-3197 |
GGAGGCGCAGGCUCGGAAAGGCG |
|
|
hsa-miR-3162-3p |
UCCCUACCCCUCCACUCCCCA |
|
| has-u6 |
CGCAAGGATGACACGCAAATTC |
|
| Universal
reverse |
|
GTGCAGGGTCCGAGGT |
Immunofluorescence staining
For immunofluorescence staining, cells from the
control and experimental groups were grown on 4-chamber slides and
cells were fixed with freshly prepared 4% ice-cold formaldehyde (pH
7.2-7.3) at room temperature. The cells were washed and blocked
with 5% bovine serum albumin (BSA) and 0.3% Triton X-100 in PBS for
30 min. After serial washing and blocking, cells were incubated
with primary antibody against CaMKII and phosphorylated CaMKII
primary antibodies overnight at 4°C to stain cardiomyocyte,
followed by Alexa Fluor 488 and Alexa 555 secondary antibodies
incubation for an additional 30 min at room temperature. Finally,
the cells were counterstained with 4′,6-diamidino-2-phenylindole
(DAPI, 1:1,000) during the final wash. Fluorescence images were
captured using a confocal microscope (LSM800, Zeiss, Germany).
Dynamic Ca2+ imaging
Images were obtained using a confocal microscope
(LSM700; Carl Zeiss, Germany). HL-1 cells were washed three times
with a live cell imaging solution (140 mM NaCl, 2.5 mM KCl, 1.8 mM
CaCl2, 1 mM MgCl2, 20 mM HEPES-Na, 5.6 mM
glucose, pH 7.4) and loaded with 5 µM fluo-4 AM for 20 min at 37°C.
Coverslips were mounted in 1 ml capacity chambers and placed in the
microscope for fluorescence measurements after excitation with a
488 nm wavelength argon laser beam or filter system. Fluo-4 was
excited by a 488 nm line of an Argon laser, and emission signals
over 505 nm were collected. Line-scan images are acquired along the
longitudinal axis of the cells. Each line was composed of 512
pixels spaced at 0.14 µm intervals. After sequential scanning, a
two-dimensional image of 512×10,000 lines or 512×20,000 lines was
generated and stored for offline analysis. Image sequences were
analyzed using the NIH open-access software ImageJ. Intracellular
Ca2+ levels are expressed as the percentage of
fluorescence intensity relative to the basal fluorescence
intensity.
miRNA microarray screening
Microarray analysis was performed with a GeneChip
miRNA 4.0 array containing a set of approximately 2500 human mature
miRNA probes annotated in the miRBase20 database. In the case of
miRNA expression experiment, 100 ng of total RNA sample was used
and biotin-labeling was performed using the poly tail of total RNA.
After loading the biotin-labeled sample onto the GeneChip miRNA 4.0
array, hybridization was performed using the GeneChip hybridization
system. Image processing was performed using an Affymetrix Gene
Array 3000 scanner, and data analysis was performed using
GeneSpring GX 14.9.1 software. Quality control and microarray
analysis of small RNA experiments were performed at Biocore (Seoul,
Korea). A fold change cutoff of 1.5 and a statistical cutoff of
P<0.05 were applied to find differentially expressed miRNAs.
After that, validation experiments were analyzed using RT-qPCR.
Additionally, bioinformatics analysis was performed to predict the
target of miRNA-423. We used Targetscan (http://www.targetscan.org/) and miRDB (http://www.mirdb.org) websites.
Statistical analysis
All quantitative data are expressed as the mean ±
SEM. The normally distributed values are compared using an unpaired
Student's t-test between the two groups. Also, a one-way analysis
of variance (ANOVA) with a post hoc Tukey's test or Bonferroni test
was used to compare the differences among groups when appropriate.
Statistical analyses were performed using SPSS version 23.0 (SPSS,
Inc., IL, USA) and GraphPad Prism 5 program (GraphPad Software,
Inc., CA, USA). A P value of <0·05 was considered statistically
significant.
Results
Quantitative gene expression analysis
of urine samples from the AF and PSVT groups
We analyzed fibrosis-related gene expression using
RNA samples isolated from AF patients' urine and HL-1 cells. In the
urine, the expression of fibrosis-related genes (collagen I/III,
fibronectin 1, and TGF-β) tended to be higher in the AF group than
in the PSVT group (Fig. 1A).
Furthermore, fibrosis-related gene expression was examined in Ang
II (1 µM)-treated cells. the expression of collagen I (Col I),
collagen III (Col III), fibronectin 1 (Fn 1), and Transforming
growth factor-β (TGF-β) was significantly increased in the Ang II
group compared with the control group (Fig. 1B).
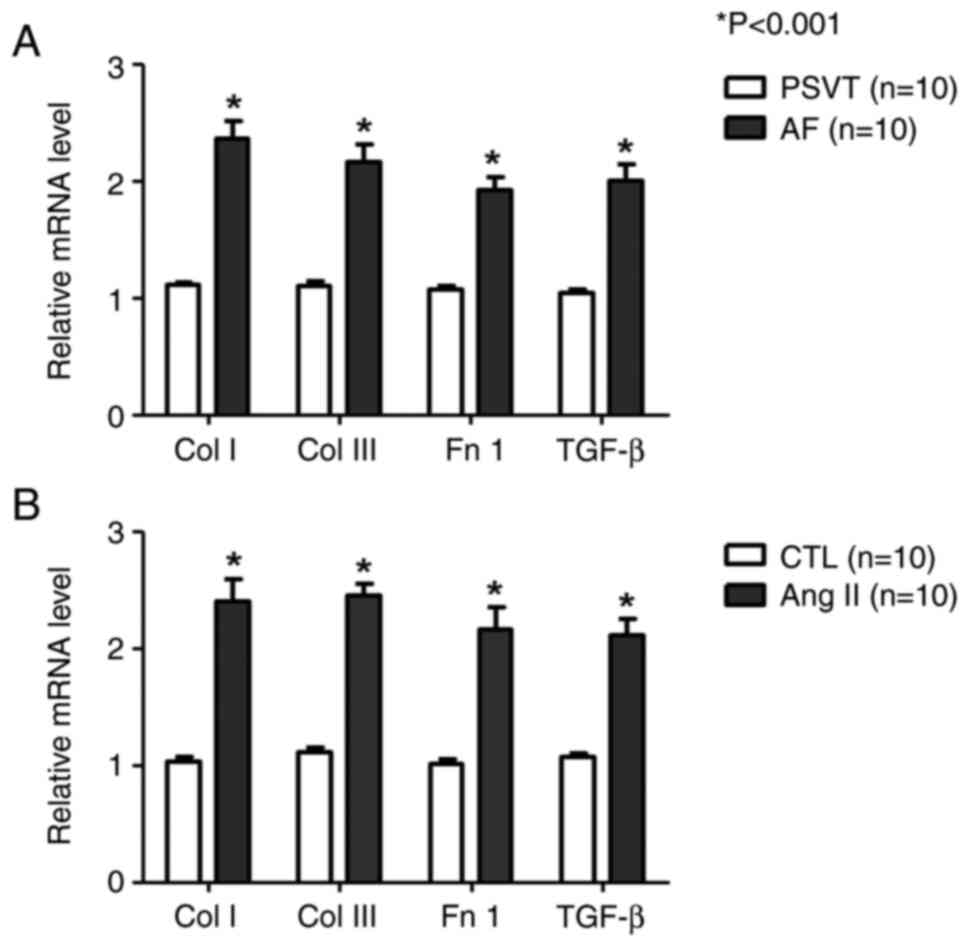 | Figure 1.Examination of the expression levels
of fibrotic markers by quantitative PCR. (A and B) Quantitative PCR
showing the expression levels of Col I, Col III, Fn 1 and TGF-β
mRNA compared with GAPDH mRNA. (A) Human urine. (B) HL-1 cells.
*P<0.001, vs. PSVT and CTL. n=10. AF, atrial fibrillation; Ang
II, angiotensin II; Col I, collagen type 1; Col III, collagen type
III; CTL, Control; Fn 1, fibronectin 1; PSVT, paroxysmal
supraventricular tachycardia; TGF-β, transforming growth
factor-β1. |
miRNA microarray screening
We performed miRNA microarray on two populations of
cells using the Sanger miRBase Version 20 miRNA expression
microarray (LC Sciences, Houston, TX, USA). We analyzed unique
mature miRNAs across biological duplicates of each cell type and
found that 7 miRNAs were significantly downregulated in the AF
group compared with the PSVT group. In particular, miR-423 was
downregulated by approximately 2-fold in live cells (Fig. S2A, B and C). To confirm the
results from miRNA microarray, we selected miR-3613, miR-6763,
miR-423, miR-3162, miR-1180, miR-6511, and miR-3197 for
quantitative PCR analysis. These miRNAs were significantly
downregulated in Ang II-treated HL-1 cells (Fig. 2A and B).
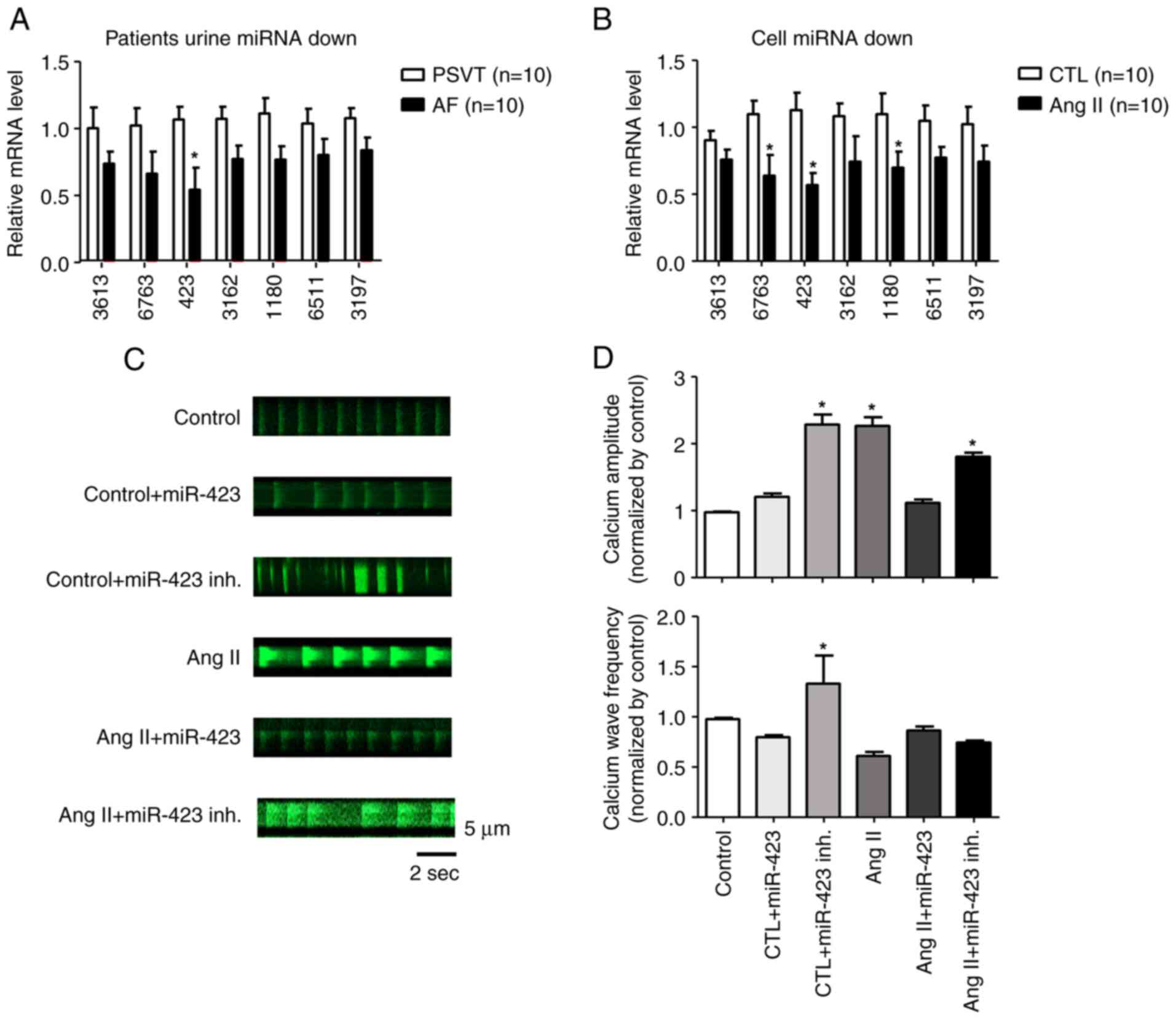 | Figure 2.miRNA microarray screening.
Quantitative PCR of 7 miRNAs in (A) patients and (B) cells. The
expression levels of miR-423 in patients with AF and the AF cell
model were significantly lower than those in the PSVT and control
cell groups, respectively. (C) Representative line-scan of
time-lapse calcium imaging of cells loaded with the intracellular
calcium indicator (Fluo-4 AM). Representative records of
spontaneous Ca2+ transients according to the group. (D)
Summary of the Ca2+ transient amplitude and frequency.
The amplitude was significantly increased in control + miR-423
inhibitor, Ang II and Ang II + miR-423 inhibitor groups.
*P<0.001 vs. control group n=5. AF, atrial fibrillation; Ang II,
angiotensin II; CTL, Control; inh., inhibitor; miRNA/miR, microRNA;
PSVT, paroxysmal supraventricular tachycardia; Fluo-4 AM,
4-(6-Acetoxymethoxy-2,7-difluoro-3-oxo-9-xanthenyl)-4-methyl-2,2-(ethylenedioxy)dianiline-N,N,N,N-tetraacetic
acid tetrakis(acetoxymethyl) ester. |
Fig. 2C shows
representative line-scan images of spontaneous Ca2+
transients obtained from HL-1 cells of the control, control +
miR-423, control + miR 423 inhibitor, Ang II, Ang II + miR423, and
Ang II + miR423 inhibitor groups. Ca2+ amplitude and
wave frequency were increased in the control + miR-423 inhibitor
(P<0.001, 1.0±0.0 F/F0 to 2.2±0.2 F/F0),
Ang II (P<0.001, 1.0±0.0 F/F0 to 2.1±0.1
F/F0), and Ang II + miR-423 inhibitor (P<0.001,
1.0±0.0 F/F0 to 1.8±0.1 F/F0) groups compared
with the control group. In contrast, Ca2+ wave frequency
and amplitude were restored in cells simultaneously treated with
Ang II + miR-423 (1.0±0.0 F/F0 to 1.1±0.1
F/F0) (Fig. 2D). In
this study, we found that miR-423 reduced Ca2+ handling
proteins in AF with the suppression of excessive CaMKII activity
and subsequent restoration of Ca2+ homeostasis-related
proteins.
Inhibition of CaMKII activation and
reduction in dysregulated Ca2+ handling proteins in Ang
II-induced cells following miR-423 administration
To determine whether CaMKII was activated in Ang
II-induced cells, we analyzed the protein levels of
phosphorylated-CaMKII (p-CaMKII, at T287 and T306). As shown in
Fig. 3A, both p-CaMKII and total
CaMKII were significantly increased in Ang II-induced cells but
normalized in the Ang II + miR-423 group, suggesting that CaMKII
was activated under AF conditions. We also evaluated the CaMKII
target phospholamban (PLB). The phosphorylation of PLB by CaMKII at
Thr17 will relieve its inhibition of SERCA2 and consequently
increase SERCA2 Ca2+ uptake activity (Fig. 3B). Ang II-induced cells exhibited
increased protein levels of phosphorylated PLB (p-PLB, at Thr17).
However, the dysregulated Ca2+ handling proteins were
markedly reduced by miR-423 treatment.
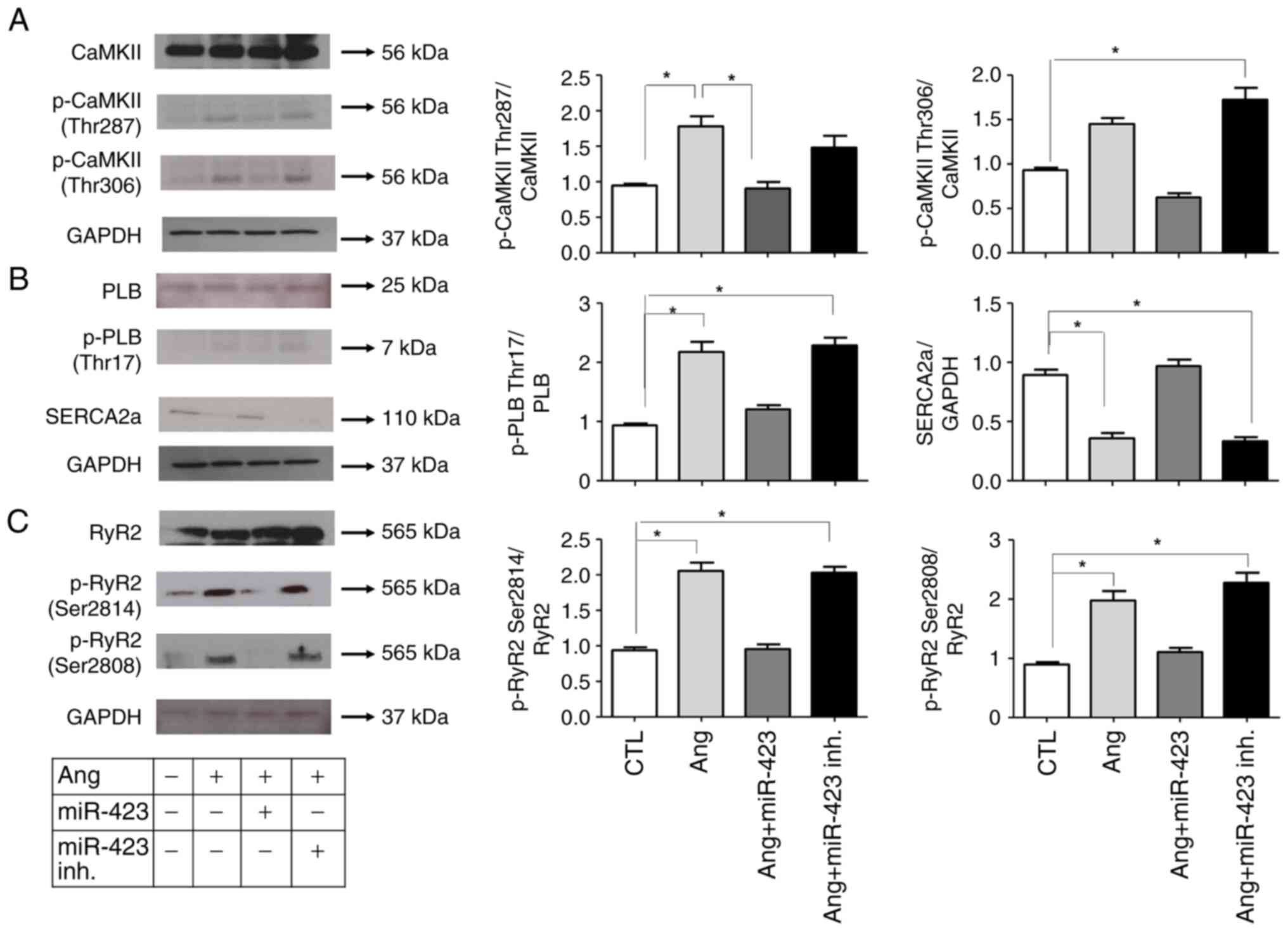 | Figure 3.miR-423 inhibition of CaMKII, PLB and
RyR2 activities in Ang II-induced HL-1 cells. (A-C) (left) Western
blot analysis of (A) CaMKII (Thr 287, Thr 306), (B) PLB (Thr17) and
(C) RyR2 (Ser 2814, Ser 2808). (right) Protein semi-quantification
showing an increase in p-CaMKII, p-PLB and p-RyR2 in Ang II-treated
HL-1 cells and the miR-423 inhibitor-treated group. However, the
phosphorylation of these proteins was decreased in the group
treated with miR-423. n=5-6 per group. *P<0.001. Data are
presented as the mean ± SEM. Ang II, angiotensin II; CaMKII,
calmodulin-dependent protein kinase II; CTL, Control; inh.,
inhibitor; miR, microRNA; p-, phosphorylated; PLB, phospholamban;
RyR2, ryanodine receptor 2; SERCA2a, sarcoplasmic reticulum calcium
ATPase. |
The alteration of CaMKII expression and activation
status could mediate CaMKII-dependent phosphorylation in the
ryanodine receptor 2 gene (RyR2), which encodes a cardiac
sarcoplasmic reticulum Ca2+ release channel, and
phosphorylated RyR2 can increase the probability of opening.
Therefore, we examined the activation state of RyR2. RyR2 contains
several phosphorylation sites, including Ser2814 (phosphorylated by
CaMKII) and Ser2808 (which could be phosphorylated by both CaMKII
and cAMP-dependent protein kinase A). We performed western blotting
with two phospho-specific antibodies (for RyR2-S2814 and
RyR2-S2808). In cells stimulated by Ang II, the phosphorylation of
RyR2 was increased at both Ser2814 and Ser2808, suggesting the
Ca2+ release properties of RyR2. However, these changes
were significantly attenuated by miR-423 treatment (Fig. 3C).
Effects of CaMKII activity in Ang
II-induced HL-1 cells
It is well known that the upregulation of the
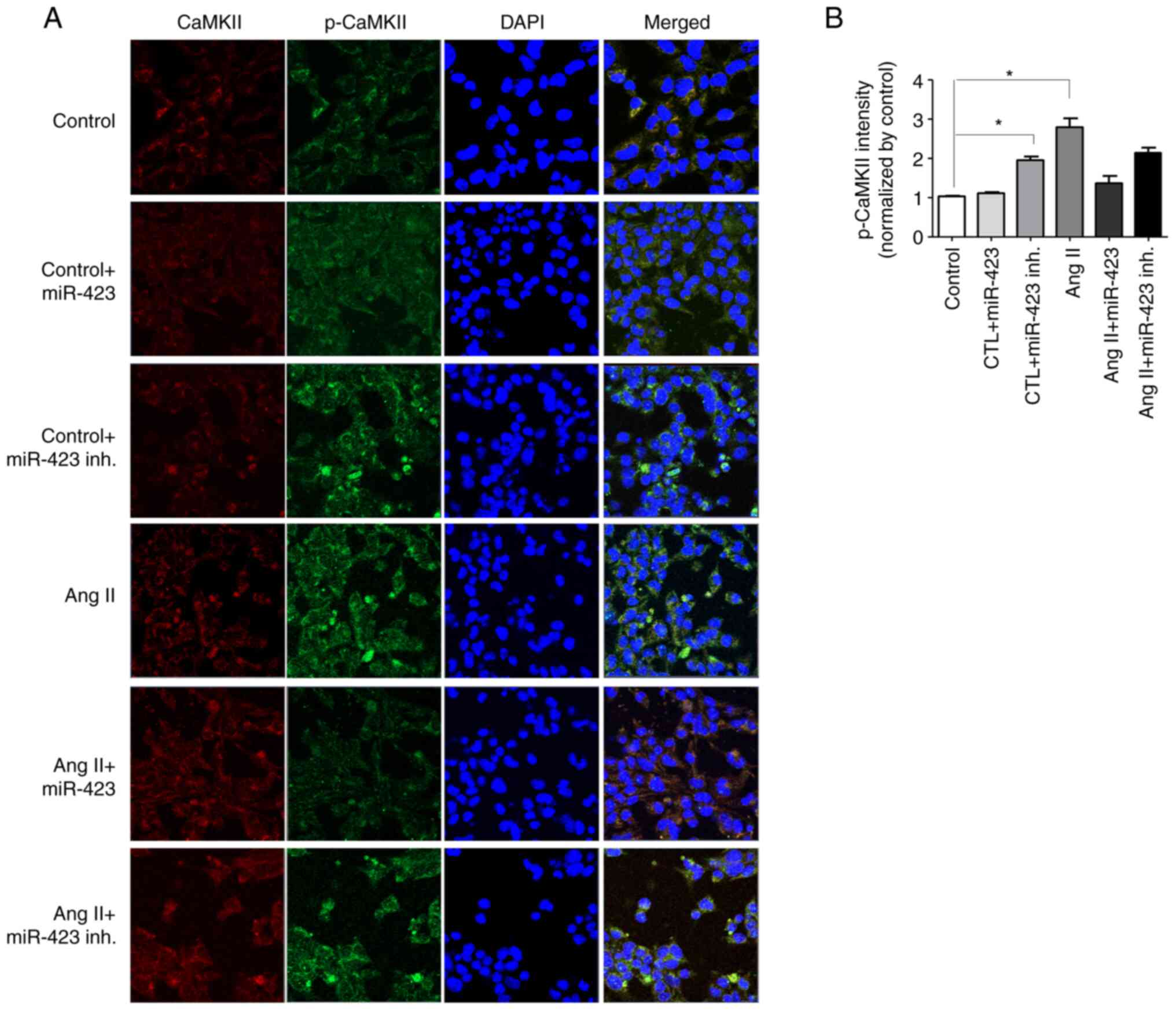
pCaMKII-T286/CaMKII ratio is closely associated with AF. Fig. 4A shows the confocal microscope
images of HL-1 cells using immunofluorescence stain method for
CaMKII and p-CaMKII. Under Ang II treatment, both CaMKII and
p-CaMKII were increased (3.2-fold); however, Ang II + miR-423
effectively prevented the increase in p-CaMKII (1.3-fold,
P<0.001 vs. Ang II group). No effect was observed on CaMKII and
p-CaMKII in the control group (Fig.
4B).
Discussion
Importantly, human studies have shown that soluble
miRNAs can be isolated from a minimal amount of urine supernatant
and still be present at detectable levels. Therefore, the use of
miRNAs as biomarkers for heart disease is a feasible and attractive
option. Unlike proteins or mRNAs, miRNAs are highly stable and
resistant to degradation in clinical urine samples. Studies have
demonstrated the stability of circulating miRNAs. Most commonly,
plasma-derived miRNAs can be used as indicators of cardiovascular
disease (13). However, in the
case of paroxysmal AF, there is a limit to diagnosis using only
electrocardiogram. For this reason, it is useful clinically to
diagnose heart disease using excreted urinary miRNA. Excreted
urinary miRNAs already fulfill some of the prerequisites for a good
biomarker. Nevertheless, further investigation is needed before
preclinical and clinical applications.
Atrial conduction disorder is characterized by a
decrease in action potential duration and impulse propagation rate.
These changes are often induced in the early stages of atrial
remodeling (14). Atrial
dilatation and fibrosis observed in AF are characterized by
structural remodeling, which leads to conduction disturbances.
Until recently, these electrophysiological changes provided the
basis for a method to treat AF. However, AF promotes the remodeling
process of the atria, contributing to the treatment resistance
observed in patients with long-term arrhythmias (15). The molecular basis for the
electrical remodeling of the atrium is changes in ion channels and
exchangers, including NCX1, NKA and L-type Ca2+ channels
and RyR receptors (16). For this
reason, we treated HL-1 cells with Ang II to create an environment
similar to AF. We performed microarray analysis using the urine
from 5 patients with AF and 3 patients with PSVT. The goal of this
study was to identify a miRNA predictor of AF in the patients'
urine. We found miRNAs with different levels of expression in the
AF and PSVT groups. Several miRNAs have been proposed as biomarkers
for AF; however, none of them are currently available for
diagnostic purposes (17). There
are limited data on the association between miRNAs and AF. In
comparison with the control group, AF patients had lower levels of
miR-423. Similarly, in HL-1 cells treated with Ang II, the
expression level of miR-423 was found to be significantly lower
than the level in the control group.
CaMKII mutations are associated with AF in humans.
Previous studies have shown that mice heterozygous for CaMKII
deficiency exhibit atrial electrical dysfunction and increased AF
induction. n this study, the expression of miR-423 was decreased in
the urine of patients with AF compared to patients with PSVT. In
addition, the level of Ca2+ handling protein increased
in HL-1 cells treated with Ang II, resulting in
electrophysiological changes observed in AF. Also, cells with
decreased miR-423 expression exhibited abnormal Ca2+
signaling. Therefore, it was hypothesized that miR-423
downregulation may cause AF. However, the mechanism underlying the
aberrant expression of miR-423 in this experiment were not
clear.
In the present study, various miRNAs (e.g., miR-423)
associated with electrophysiological processes and the expression
of Ca2+ handling proteins were performed. The results
proved that the levels of CaMKII, PLB, and RyR2 were increased in
Ang II-induced HL-1 cells compared with control, and miR-423
treatment reversed this effect. miR-423 was demonstrated in
previous studies was to induce coronary artery disease, it was
associated with cardiac fibrosis (18). This study showed that atrial
fibrillation can be controlled by regulation of miR-423 expression.
In addition, alterations in intracellular Ca2+ signaling
associated with the regulation of miR-423 expression could be
regulated by CaMKII. In Ang II treated cells, overexpression of
miR-423 decreased Ca2+ signaling, whereas inhibition of
miR-423 increased Ca2+ signaling by Ang II. The
impairment of Ca2+ signaling observed in this study
after transfection with miR-423 was similar to the early
electrophysiological changes in AF. Therefore, miR-423 may be
involved in the pathogenesis of AF by modulating CaMKII, which
disrupted Ca2+ signaling.
Limitations of this study is limited to the ability
to determine the clinical value of a diagnostic test capabilities
and AF patient's miRNA signatures because of the small number of
patient's urine samples for analysis. Second, according to the
results of the current study, it is uncertain whether changes in
Ca2+ levels mediate the atrial profibrotic effect of Ang
II or whether this is a related but not causally related event. In
addition, this study was not include the functional analysis on
calcium homeostasis or electrophysiological evaluation. Further
research on an atrial tachypacing-induced AF model will be useful
for defining the causal relationship in atrial structural
remodeling.
In summary, changing miR-423 levels in AF to the
normal range may be a novel strategy for converting AF to sinus
rhythm in the clinical. Considering that this technique is reported
to have good cellular safety and miRNA specificity in in
vitro, the miRNA approach appears to be reliable. Although
miR-423 acted as an important factor in regulating experimental AF
in our study, we do not exclude other mechanisms determining AF and
our findings in experimental methods may not directly apply to
human AF. Nevertheless, our study shows new insights into AF and
other aspects of the mechanism of miR-423 in AF.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by research grants from the
Basic Science Research Program through the National Research
Foundation of Korea (NRF-2017R1E1A1A01078382, 2019R1C1C1003389).
This research was supported by the Korea Medical Device Development
Fund grant funded by the Korean government (Ministry of Science and
ICT, Ministry of Trade, Industry and Energy, Ministry of Health
& Welfare, Ministry of Food and Drug Safety) (Project Number:
9991006899). This research was also supported by the Young Medical
Scientist Research Grant through the Daewoong Foundation
(DY20102P). HP was supported by the RP-Grant 2020 of Ewha Womans
University.
Availability of data and materials
The datasets analyzed during the current study are
available in the GSE190898 repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE190898).
The datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
JP and HyewP confirm the authenticity of all the raw
data. JP and HyewP conceived and designed the study. HyewP and
HyelP participated in the experimental design. JP, HyewP and HyelP
analyzed the data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the IRB of Ewha Womans
University Mokdong Hospital (approval no. EUMC 2019-10-019).
Written informed consent was obtained from all patients before they
were enrolled.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AF
|
atrial fibrillation
|
|
Ang II
|
angiotensin II
|
|
PSVT
|
paroxysmal supraventricular
tachycardia
|
|
Col I
|
collagen type 1
|
|
Col III
|
collagen type III
|
|
Fn1
|
fibronectin 1
|
|
TGF-β
|
transforming growth factor-β1
|
|
CaMKII
|
calmodulin-dependent protein kinase
II
|
|
PLB
|
phospholamban
|
|
RyR2
|
ryanodine receptor 2
|
References
|
1
|
Benjamin EJ, Wolf PA, D'Agostino RB,
Silbershatz H, Kannel WB and Levy D: Impact of atrial fibrillation
on the risk of death: The Framingham Heart Study. Circulation.
98:946–952. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Everett TH IV and Olgin JE: Atrial
fibrosis and the mechanisms of atrial fibrillation. Heart Rhythm. 4
(Suppl 3):S24–S27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Burstein B and Nattel S: Atrial fibrosis:
Mechanisms and clinical relevance in atrial fibrillation. J Am Coll
Cardiol. 51:802–809. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li D, Fareh S, Leung TK and Nattel S:
Promotion of atrial fibrillation by heart failure in dogs: Atrial
remodeling of a different sort. Circulation. 100:87–95. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aldhoon B, Melenovsky V, Peichl P and
Kautzner J: New insights into mechanisms of atrial fibrillation.
Physiol Res. 59:1–12. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang HB, Jiang ZB and Li M: Research on
the typical miRNA and target genes in squamous cell carcinoma and
adenocarcinoma of esophagus cancer with DNA microarray. Pathol
Oncol Res. 20:245–252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Elia L, Kunderfranco P, Carullo P,
Vacchiano M, Farina FM, Hall IF, Mantero S, Panico C, Papait R,
Condorelli G and Quintavalle M: UHRF1 epigenetically orchestrates
smooth muscle cell plasticity in arterial disease. J Clin Invest.
128:2473–2486. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wong LL, Wang J, Liew OW, Richards AM and
Chen YT: MicroRNA and heart failure. Int J Mol Sci. 17:5022016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
D'Alessandra Y, Devanna P, Limana F,
Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC,
Spazzafumo L, De Simone M, et al: Circulating microRNAs are new and
sensitive biomarkers of myocardial infarction. Eur Heart J.
31:2765–2773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang G, Kwan BC, Lai FM, Chow KM, Li PK
and Szeto CC: Urinary miR-21, miR-29, and miR-93: Novel biomarkers
of fibrosis. Am J Nephrol. 36:412–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng L, Quek CY, Sun X, Bellingham SA and
Hill AF: The detection of microRNA associated with Alzheimer's
disease in biological fluids using next-generation sequencing
technologies. Front Genet. 4:1502013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Creemers EE, Tijsen AJ and Pinto YM:
Circulating microRNAs: Novel biomarkers and extracellular
communicators in cardiovascular disease? Circ Res. 110:483–495.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pang H, Ronderos R, Perez-Riera AR,
Femenia F and Baranchuk A: Reverse atrial electrical remodeling: A
systematic review. Cardiol J. 18:625–631. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Verheule S, Tuyls E, Gharaviri A, Hulsmans
S, van Hunnik A, Kuiper M, Serroyen J, Zeemering S, Kuijpers NH and
Schotten U: Loss of continuity in the thin epicardial layer because
of endomysial fibrosis increases the complexity of atrial
fibrillatory conduction. Circ Arrhythm Electrophysiol. 6:202–211.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iwasaki YK, Nishida K, Kato T and Nattel
S: Atrial fibrillation pathophysiology: Implications for
management. Circulation. 124:2264–2274. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Komal S, Yin JJ, Wang SH, Huang CZ, Tao
HL, Dong JZ, Han SN and Zhang LR: MicroRNAs: Emerging biomarkers
for atrial fibrillation. J Cardiol. 74:475–482. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nabialek E, Wanha W, Kula D, Jadczyk T,
Krajewska M, Kowalowka A, Dworowy S, Hrycek E, Włudarczyk W, Parma
Z, et al: Circulating microRNAs (miR-423-5p, miR-208a and miR-1) in
acute myocardial infarction and stable coronary heart disease.
Minerva Cardioangiol. 61:627–637. 2013.PubMed/NCBI
|