Introduction
Small interfering (si)RNA is a promising tool for
inhibiting specific gene expression (1). Cationic liposomes have attracted
attention for siRNA delivery to cells (2,3) as
siRNA/cationic liposome complexes (siRNA lipoplexes) are
efficiently delivered into cells and notably suppress target gene
expression (2). To prepare
cationic liposomes, liposomal formulations are used to combine
cationic and neutral helper lipids, such as phospholipid or
cholesterol (Chol), to increase transfection efficiency and
stability (4). However, the
structures of cationic lipid and phospholipid, such as head group
and alkyl chains, affect the transfection efficiency of siRNA
lipoplexes (5,6). Therefore, discovering an optimal
combination of cationic and neutral helper lipid for liposomal
formulations is key for achieving efficient siRNA transfection.
The most common cationic lipids for siRNA delivery
with cationic liposomes are
1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) (7–10),
1,2-di-O-octadecenyl-3-trimethylammonium-propane (10–12) and dimethyldioctadecylammonium
bromide (DDAB) (9,13) which contain a quaternary ammonium
group in the head group.
1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), which
has a small head group, is unsaturated and has long dialkyl chains;
it is often used as a neutral helper lipid to prepare cationic
liposome (7–12) because DOPE destabilizes siRNA
lipoplexes in endosomes by inducing a change in conformation at
acidic pH when incorporated into the liposomal formulation
(14).
Previously, we selected 6 types of cationic Chol
derivative and 11 types of cationic lipid with dialkyl or trialkyl
chains to prepare 17 types of cationic liposome composed of
cationic lipid with DOPE for siRNA delivery and evaluate gene
knockdown efficacy (5). Among
these cationic liposomes, those composed of dialkyl (DDAB) or
trialkyl cationic lipid
[11-((1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino)-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; TC-1-12] with DOPE notably suppressed targeted mRNA
expression in the mouse lung following systemic injection of siRNA
lipoplexes. TC-1-12 is a novel cationic lipid developed by our
laboratory that exhibits high siRNA transfection efficiency both
in vitro and in vivo (5,6).
However, to the best of our knowledge, few reports have been
published regarding the effect of phospholipids in cationic
liposomal formulations on gene knockdown efficacy using siRNA
lipoplexes (15,16).
To determine the effect of phospholipids in cationic
liposomes on gene knockdown activity of siRNA lipoplexes, three
types of cationic lipid (DOTAP, DDAB and TC-1-12) were selected to
prepare 30 types of cationic liposome composed of cationic lipid
with phosphatidylcholine or phosphatidylethanolamine containing
saturated or unsaturated dialkyl chains (C14, C16 or C18) and in
vitro and in vivo gene knockdown efficacy was evaluated
to find optimal phospholipids in cationic liposome for siRNA
transfection.
Materials and methods
Materials
DDAB (cat. no. DC-1-18) and TC-1-12 were obtained
from Sogo Pharmaceutical Co., Ltd. DOTAP (cat. no. 890895C) was
obtained from Avanti Polar Lipids, Inc.
1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC; cat. no.
MC-8080), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC;
cat. no. MC-6060),
1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC; cat. no.
MC-4040), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC;
cat. no. MC-8181),
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; cat.
no. MC-6081),
1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE; cat.
no. ME-8080),
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE; cat.
no. ME-6060),
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine (DMPE; cat.
no. ME-4040), DOPE (cat. no. ME-8181) and
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE;
cat. no. ME-6081; all COATSOME®) were obtained from NOF
Corporation. All other chemicals were of the highest grade
available.
siRNAs
siRNA sequences of firefly luciferase (Luc siRNA),
non-silencing siRNA [negative control (Cont) siRNA] and cyanine 5
(Cy5)-conjugated Cont siRNA (Cy5-siRNA) were designed as reported
previously (17,18) and synthesized by Sigma-Aldrich
(Merck KGaA). siRNA sequences were as follows: Luc siRNA passenger
strand, 5′-CCGUGGUGUUCGUGUCUAAGA-3′ and guide strand,
5′-UUAGACACGAACACCACGGUA-3′ and Cont siRNA passenger strand,
5′-GUACCGCACGUCAUUCGUAUC-3′ and guide strand,
5′-UACGAAUGACGUGCGGUACGU-3′. For Cy5-siRNA, Cy5 dye was conjugated
at the 5′-end of the passenger strand of Cont siRNA. The siRNA
sequences of mouse Tie2 and luciferase siRNA (Cont 2 siRNA), which
served as a negative control of Tie2 siRNA, were designed as
reported previously (5,19) and synthesized by Japan Bio
Services Co., Ltd. The siRNA sequences were as follows: Tie2
passenger strand, 5′-CcAuCaUuUgCcCaGaUaU-3′ and guide strand,
5′-aUaUcUgGgCaAaUgAuGg-3′ and Cont 2 siRNA passenger strand,
5′-AuCaCgUaCgCgGaAuAcUuCgA-3′ and guide strand,
5′-uCgAaGuAuUcCgCgUaCgUgAu-3′. Lowercase letters represent
2′-O-methyl-modified nucleotides.
Preparation of cationic liposomes and
siRNA lipoplexes
The cationic liposomes were prepared from cationic
lipid and phospholipid at a molar ratio of 1:1. To prepare cationic
liposomes, cationic lipid and phospholipid were dissolved in
chloroform or chloroform/methanol (9:1, v/v). Chloroform and
methanol were evaporated under vacuum on a rotary evaporator at
60°C to obtain a thin film. The thin film was hydrated with water
at 60°C by vortex mixing for 30 sec at 3,000 rpm and sonicated in a
bath-type sonicator (Bransonic® 2510J-MTH, 42 kHz;
Branson UL Trasonics Co.) for 5–10 min at 60°C.
To prepare siRNA lipoplexes, liposome was added to
50 pmol siRNA at a charge ratio (+:-) of 4:1, vortexed for 10 sec
at 3,000 rpm and room temperature and left at room temperature for
15 min, as previously described (9,20).
The charge ratio represents molar ratio of cationic lipid in
cationic liposomes to siRNA phosphate.
The particle size, particle size distribution
[polydispersity index (PDI)] and ζ-potential of cationic liposomes
and siRNA lipoplexes were measured using a light-scattering
photometer (cat. no. ELS-Z2; Otsuka Electronics Co., Ltd.), as
previously reported (21).
Free siRNA levels in siRNA
lipoplexes
siRNA lipoplexes were prepared at charge ratios
(+:-) of 1:1-4:1. The amount of free siRNA in siRNA lipoplexes were
measured using exclusion assay with SYBR® Green I
Nucleic Acid Gel Stain (Takara Bio Inc.) and calculated based on
the standard curves of free siRNA as previously reported (21).
Cell culture
Human breast cancer MCF-7 cells stably expressing
firefly luciferase (MCF-7-Luc), constructed by transfection of
plasmid pcDNA3 containing firefly luciferase (hLuc) gene (GenBank
no. AY535007.1) from plasmid psiCHECK2 (Promega Corporation) were
donated by Dr Kenji Yamato (University of Tsukuba, Tsukuba, Japan).
MCF-7-Luc cells were cultured in RPMI-1640 medium supplemented with
10% heat-inactivated fetal bovine serum (FBS) and 1.2 mg/ml G418
sulfate (geneticin, Wako Pure Chemical Industries, Ltd.) at 37°C in
a humidified atmosphere with 5% CO2.
Gene knockdown in vitro using siRNA
lipoplexes
MCF-7-Luc cells were seeded in 6-well culture plates
at a density of 3×105 cells/well at 37°C. After 24 h,
each siRNA lipoplex with 50 pmol Cont siRNA or Luc siRNA was
diluted in 1 ml RPMI-1640 medium supplemented with 10% FBS and
added to the cells (final concentration, 50 nM). At 48 h
post-transfection, luciferase activity was measured as counts per
second (cps)/µg protein using PicaGene MelioraStar-LT Luminescence
Reagent (Toyo Ink Co. Ltd.) and BCA reagent (Pierce™ BCA
Protein Assay kit; Thermo Fisher Scientific, Inc.) as reported
previously (21). Luciferase
activity (%) was calculated relative to that of untransfected
cells.
Cytotoxicity of siRNA lipoplexes
Each siRNA lipoplex sample with 5 pmol Cont siRNA
was diluted in 100 µl RPMI-1640 medium supplemented with 10% FBS
and added to MCF-7-Luc cells at 50% confluency in 96-well plates
(final concentration, 50 nM). Following 24 h incubation at 37°C,
cell viability was measured via WST-8 assay (Dojindo Laboratories,
Inc.), as previously reported (21). WST-8 substrate was incubated with
cells at 37°C for 60 min.
Biodistribution of siRNA following
intravenous injection of siRNA lipoplexes into mice
Ethical approval for this study was obtained from
the Institutional Animal Care and Use Committee of Hoshi University
(approval no. P21-039). A total of 8 female BALB/c mice (weight,
18–20 g; age, 8 weeks; Sankyo Labo Service Corporation, Inc.) were
housed at 24°C and 55% humidity under 12/12-h light/dark cycle
(lights on at 8:00 a.m.) with food and water ad libitum.
siRNA lipoplexes with 20 µg Cy5-siRNA were administered
intravenously to mice via the lateral tail vein (n=1/siRNA
lipoplex). At 1 h post-injection of siRNA lipoplexes, mice were
sacrificed via cervical dislocation; death was confirmed by
cessation of heartbeat. Tissue (lung, heart, liver, spleen, and
kidney) was analyzed by Cy5 fluorescence imaging using NightOWL
LB981 NC100 system (Berthold Technologies GmbH & Co. KG), as
previously described (21). The
images were analyzed using IndiGo2 software (version 2.0.1.0)
provided with the in vivo imaging system (Berthold
Technologies). Following fluorescence imaging, tissue samples were
frozen on dry ice and sliced into 16 µm sections. The localization
of Cy5-siRNA was examined using a fluorescent microscope (Eclipse
TS100-F; Nikon Corporation) with optical filter Cy5 HQ (excitation,
620/60 nm; dichroic mirror, 660 nm; emission, 700/75 nm; Nikon
Corporation).
Agglutination assay
Erythrocyte suspension was prepared from whole blood
of female BALB/c mice (age, 8 weeks; Sankyo Labo Service
Corporation, Inc.), as previously described (13). siRNA lipoplexes with 2 µg siRNA
were added to 100 µl 2% (v/v) erythrocyte suspension. Following
incubation for 15 min at 37°C, the sample was placed on a glass
plate and agglutination was observed with light microscope at 100×
magnification.
Expression of Tie2 mRNA in the lung
following systemic injection of siRNA lipoplexes
siRNA lipoplexes with 20 µg Cont 2 or Tie2 siRNA
were administered intravenously to 8-week-old female BALB/c mice
via the lateral tail veins (n=3-4/siRNA lipoplex). No siRNA
lipoplexes caused mouse death following systemic injection. The
lung was excised at 48 h post-injection of siRNA lipoplexes and
total RNA was isolated using Isogen II (Nippon Gene Co., Ltd.).
cDNA was synthesized from total RNA using PrimeScript™
RT Master Mix (Takara Bio Inc.) according to the manufacturer's
protocol and quantitative PCR was performed using a Roche Light
Cycler 96 system with FastStart Essential DNA Probes Master (Roche
Diagnostics GmbH) and TaqMan Gene expression assays [Tie2, cat. no.
Mm00443243_m1 and phosphatase and tensin homolog (PTEN), cat. no.
Mm00477208_m1; both Applied Biosystems; Thermo Fisher Scientific,
Inc.; primer sequences not available]. The thermocycling conditions
were as follows: Initial denaturation at 95°C for 600 sec, followed
by 45 cycles of denaturation at 95°C for 10 sec and primer
annealing and extension at 60°C for 30 sec (two-step
amplification). The expression levels of Tie2 mRNA were normalized
to PTEN in each sample as reported previously (19) and analyzed using the comparative
Cq (2−ΔΔCq) method (22).
Statistical analysis
Data are presented as the mean + SD of triplicate
assessments. The statistical significance was determined by
unpaired Student's t test or one-way ANOVA followed by Tukey's post
hoc test using GraphPad Prism version 4.0 (GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Characterization of cationic liposomes
and siRNA lipoplexes
The present study aimed to determine whether alkyl
chain length, saturation and the head group of phospholipids in
cationic liposomes affect gene knockdown efficacy following
treatment with siRNA lipoplexes. DOTAP and DDAB were used as
dialkyl cationic lipids, TC-1-12 was used as a trialkyl cationic
lipid and DSPE, DPPE, DMPE, POPE, DOPE, DSPC, DPPC, DMPC, POPC and
DOPC were used as phospholipids (Fig.
1). Cationic liposomes were prepared from cationic
lipid/phospholipid at a molar ratio of 1:1 (Table I, Table II, Table III).
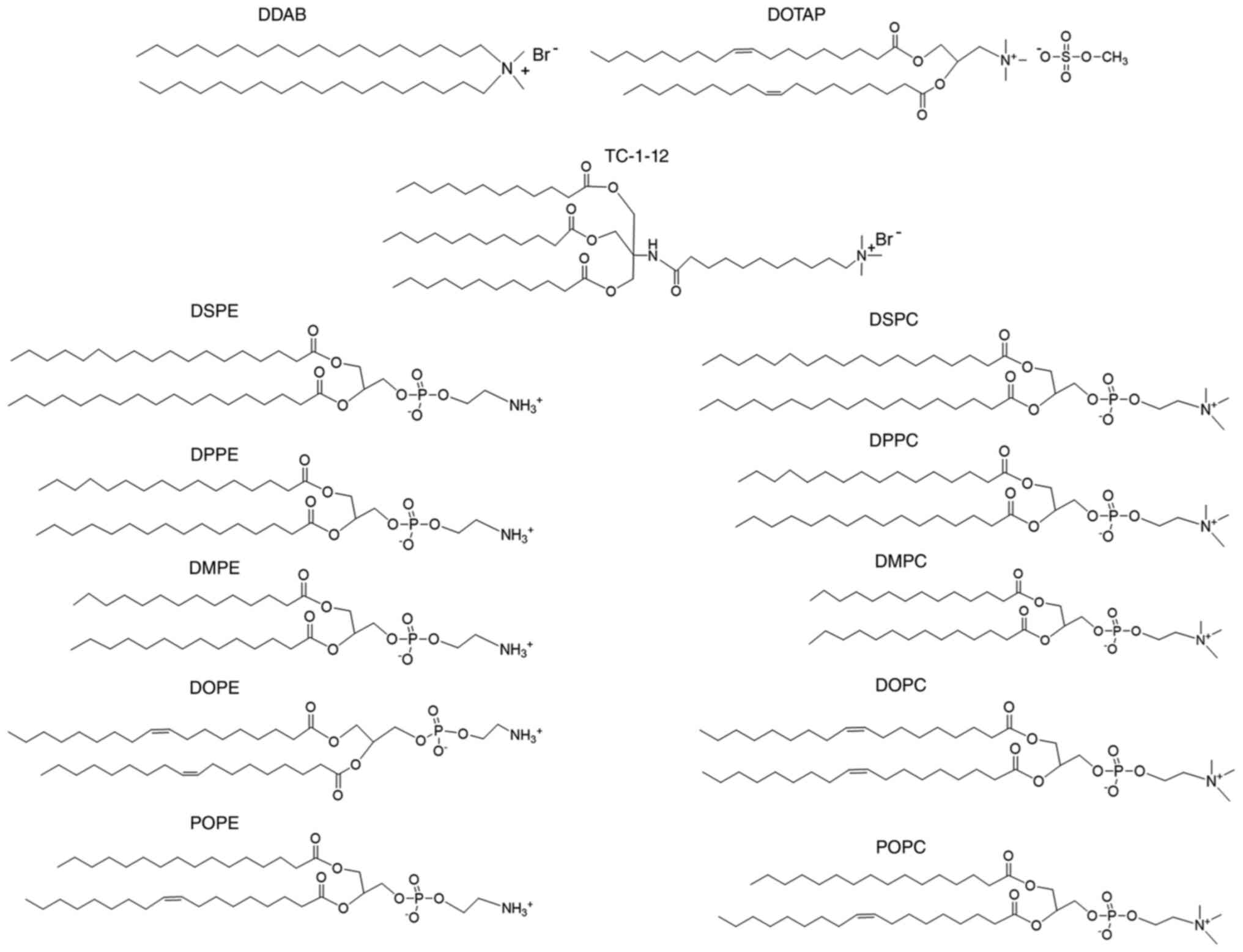 | Figure 1.Structure of cationic lipids and
phospholipids. DDAB, dimethyldioctadecylammonium bromide; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt;
TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DSPC, 1,2-distearoyl-sn-glycero-3-phosphocholine; DPPC,
1,2-dipalmitoyl-sn-glycero-3-phosphocholine; DMPC,
1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DSPE,
1,2-distearoyl-sn-glycero-3-phosphoethanolamine; DPPE,
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine; DMPE,
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
 | Table I.Particle size and ζ-potential of
DDAB-based cationic liposomes and small interfering RNA
lipoplexes. |
Table I.
Particle size and ζ-potential of
DDAB-based cationic liposomes and small interfering RNA
lipoplexes.
|
| Liposome |
Lipoplexb |
|---|
|
|
|
|
|---|
| Liposome | Sizea, nm | PDI |
ζ-potentiala, mV | Sizea, nm | PDI |
ζ-potentiala, mV |
|---|
| LP-DDAB/DSPE | 101.9±0.9 | 0.24±0.01 | 46.9±1.8 | 152.1±3.2 | 0.24±0.01 | 38.7±1.4 |
| LP-DDAB/DPPE | 108.1±1.5 | 0.22±0.01 | 44.7±0.3 | 209.2±5.5 | 0.29±0.01 | 40.2±1.4 |
| LP-DDAB/DMPE | 111.2±2.6 | 0.23±0.01 | 47.3±0.3 | 167.6±1.4 | 0.18±0.01 | 43.8±2.1 |
| LP-DDAB/DOPE | 103.0±1.3 | 0.21±0.01 | 52.2±1.2 | 181.0±4.7 | 0.17±0.02 | 47.1±1.1 |
| LP-DDAB/POPE | 99.6±1.0 | 0.21±0.01 | 46.2±0.7 | 190.2±2.6 | 0.21±0.01 | 42.3±1.7 |
| LP-DDAB/DSPC | 132.4±2.0 | 0.23±0.01 | 45.9±0.7 | Aggregation | ND | ND |
| LP-DDAB/DPPC | 123.1±1.4 | 0.23±0.01 | 47.8±1.7 | 149.7±0.6 | 0.20±0.01 | 41.9±3.0 |
| LP-DDAB/DMPC | 106.7±2.3 | 0.23±0.01 | 45.9±0.2 | 158.1±5.8 | 0.20±0.01 | 37.5±2.0 |
| LP-DDAB/DOPC | 120.0±1.3 | 0.27±0.00 | 45.1±1.8 | 169.3±2.6 | 0.21±0.00 | 40.2±1.8 |
| LP-DDAB/POPC | 110.2±5.4 | 0.20±0.06 | 45.5±4.8 | 178.3±17.7 | 0.17±0.07 | 35.7±0.2 |
 | Table II.Particle size and ζ-potential of
DOTAP-based cationic liposomes and small interfering RNA
lipoplexes. |
Table II.
Particle size and ζ-potential of
DOTAP-based cationic liposomes and small interfering RNA
lipoplexes.
|
| Liposome |
Lipoplexb |
|---|
|
|
|
|
|---|
| Liposome | Sizea, nm | PDI |
ζ-potentiala, mV | Sizea, nm | PDI |
ζ-potentiala, mV |
|---|
| LP-DOTAP/DSPE | 111.8±2.0 | 0.23±0.01 | 50.3±0.5 | 216.3±27.3 | 0.19±0.07 | 48.7±2.7 |
| LP-DOTAP/DPPE | 116.0±0.9 | 0.20±0.01 | 48.3±0.7 | 163.7±6.5 | 0.21±0.01 | 41.1±0.7 |
| LP-DOTAP/DMPE | 113.7±1.8 | 0.28±0.01 | 52.4±2.2 | 184.0±5.6 | 0.20±0.01 | 47.8±2.2 |
| LP-DOTAP/DOPE | 109.2±1.0 | 0.21±0.01 | 53.7±1.6 | 179.1±5.7 | 0.16±0.01 | 42.8±1.5 |
| LP-DOTAP/POPE | 111.7±1.4 | 0.24±0.02 | 49.6±1.0 | 175.8±6.5 | 0.21±0.01 | 40.3±0.4 |
| LP-DOTAP/DSPC | 93.3±0.8 | 0.22±0.01 | 48.9±1.4 | 228.7±1.5 | 0.26±0.00 | 32.2±0.7 |
| LP-DOTAP/DPPC | 112.8±0.8 | 0.26±0.00 | 46.7±0.7 | 200.8±2.8 | 0.23±0.00 | 26.6±2.8 |
| LP-DOTAP/DMPC | 107.2±1.0 | 0.25±0.00 | 46.3±2.7 | 240.5±1.9 | 0.28±0.02 | 31.3±1.1 |
| LP-DOTAP/DOPC | 102.2±0.9 | 0.24±0.01 | 48.6±3.0 | 227.3±4.1 | 0.25±0.00 | 35.2±0.1 |
| LP-DOTAP/POPC | 115.5±2.0 | 0.26±0.01 | 46.0±1.6 | 233.1±3.0 | 0.28±0.01 | 31.3±0.6 |
 | Table III.Particle size and ζ-potential of
TC-1-12-based cationic liposomes and small interfering RNA
lipoplexes. |
Table III.
Particle size and ζ-potential of
TC-1-12-based cationic liposomes and small interfering RNA
lipoplexes.
|
| Liposome |
Lipoplexb |
|---|
|
|
|
|
|---|
| Liposome | Sizea, nm | PDI |
ζ-potentiala, mV | Sizea, nm | PDI |
ζ-potentiala, mV |
|---|
|
LP-TC-1-12/DSPE | 195.1±23.6 | 0.17±0.08 | 39.9±0.8 | 192.9±0.3 | 0.13±0.01 | 38.3±0.9 |
|
LP-TC-1-12/DPPE | 111.7±1.0 | 0.24±0.01 | 40.3±1.2 | Aggregation | N.D. | N.D. |
|
LP-TC-1-12/DMPE | 104.2±0.9 | 0.28±0.02 | 45.1±2.2 | Aggregation | N.D. | N.D. |
|
LP-TC-1-12/DOPE | 122.5±0.9 | 0.24±0.00 | 41.6±0.8 | 179.6±3.7 | 0.21±0.01 | 43.4±1.5 |
|
LP-TC-1-12/POPE | 104.0±1.7 | 0.25±0.01 | 40.3±1.3 | 182.5±3.2 | 0.24±0.01 | 32.6±4.0 |
|
LP-TC-1-12/DSPC | 101.5±8.1 | 0.25±0.04 | 42.0±1.0 | 781.1±187.3 | 0.34±0.07 | 40.1±0.5 |
|
LP-TC-1-12/DPPC | 110.2±2.0 | 0.28±0.01 | 43.4±0.4 | 485.2±28.0 | 0.25±0.01 | 40.0±1.0 |
|
LP-TC-1-12/DMPC | 122.5±1.6 | 0.28±0.01 | 44.8±1.0 | 426.1±5.4 | 0.20±0.00 | 35.5±0.8 |
|
LP-TC-1-12/DOPC | 138.9±2.8 | 0.27±0.02 | 41.5±1.0 | 272.4±13.5 | 0.23±0.05 | 37.0±1.2 |
|
LP-TC-1-12/POPC | 113.8±1.2 | 0.25±0.01 | 42.7±3.1 | 380.9±21.4 | 0.17±0.01 | 31.3±1.3 |
The size of cationic liposomes was 93–195 nm (PDI,
0.17-0.28) and ζ-potential was +40–54 mV (Table I, Table II, Table III). Our previous study reported
that the optimal charge ratio (+:-) to prepare the siRNA lipoplexes
was 4:1 for LP-DOTAP/DOPE, LP-DDAB/DOPE and LP-TC-1-12/DOPE
(9,20). Therefore, subsequent experiments
used charge ratio (+:-) of 4:1 to prepare siRNA lipoplexes.
LP-DDAB/DSPC, LP-TC-1-12/DPPE and LP-TC-1-12/DMPE lipoplexes were
aggregated (size, >1 µm) when liposomes were mixed with siRNA
(Tables I and III). When TC-1-12 was combined with
phosphatidylcholine to prepare cationic liposomes, lipoplex size
increased to 270–780 nm (PDI, 0.17-0.34). However, the lipoplex
sizes except for the combination of TC-1-12/phosphatidylcholine
were 150–240 nm (PDI, 0.13-0.29) and ζ-potential was +31–49 mV.
Binding of siRNA to cationic
liposomes
The binding of siRNA to each cationic liposome was
assessed by exclusion assay with SYBR Green I. The addition of
cationic liposomes to siRNA beyond charge ratios of 2:1-3:1 was
found to markedly decrease the fluorescence of SYBR®
Green I in the DDAB-, DOTAP-, and TC-1-12-based liposomes (Fig. 2). These results suggested that
most DDAB-, DOTAP- and TC-1-12-based cationic liposomes exhibited
greatest binding to siRNA at a charge ratio of 4:1.
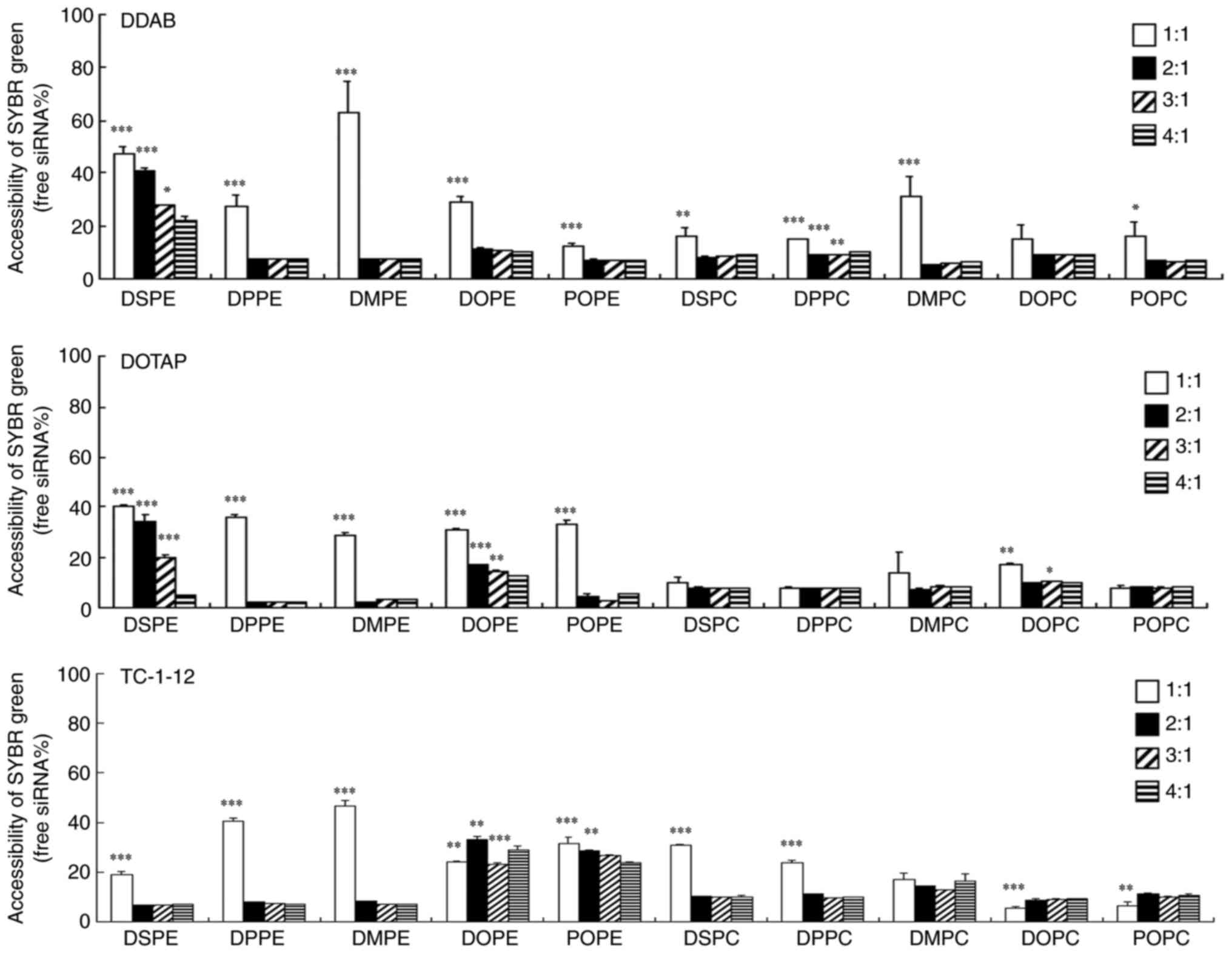 | Figure 2.Effect of phospholipids in cationic
liposomes on binding with siRNA. siRNA lipoplexes were formed at
charge ratios (+:-) of 1:1-4:1 and used in an exclusion assay with
SYBR® Green I Nucleic Acid Gel Stain. The amount of
siRNA available to interact with the SYBR® Green I is
expressed as a percentage of free siRNA without cationic liposome.
Data are presented as the mean + SD (n=3). *P<0.05, **P<0.01,
***P<0.001 vs. 4:1. si, small interfering; DDAB,
dimethyldioctadecylammonium bromide; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt;
TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DSPC, 1,2-distearoyl-sn-glycero-3-phosphocholine; DPPC,
1,2-dipalmitoyl-sn-glycero-3-phosphocholine; DMPC,
1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DSPE,
1,2-distearoyl-sn-glycero-3-phosphoethanolamine; DPPE,
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine; DMPE,
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
Effect of phospholipids in cationic
liposomes on in vitro gene knockdown efficacy
Previously, we reported that inclusion of DOPE in
cationic liposomal formulations induces strong gene knockdown
activity in cells; however, cationic liposomes with higher Chol
content display decreased gene knockdown activity (6). To determine the effect of
phospholipid in cationic liposomes on gene knockdown using siRNA
lipoplexes, each siRNA lipoplex with Luc siRNA was transfected into
MCF-7-Luc cells and gene knockdown efficacy was assessed by
assaying luciferase activity. In DDAB- and DOTAP-based cationic
liposomes, LP-DDAB/DOPE, LP-DOTAP/DOPE and LP-DOTAP/POPE lipoplexes
caused strong gene knockdown efficacy (>80% knockdown compared
with untreated cells; Figs. 3 and
4). LP-DDAB/DOPC and
LP-DOTAP/DPPE lipoplexes exhibited moderate gene knockdown (56 and
49% knockdown, respectively, compared with untreated cells), and
LP-DDAB/DPPE, LP-DDAB/DSPC, and LP-DOTAP/DMPE lipoplexes with Luc
siRNA showed slightly gene knockdown compared with those with Cont
siRNA. In TC-1-12-based liposomes, LP-TC-1-12/DMPE,
LP-TC-1-12/DOPE, LP-TC-1-12/POPE, LP-TC-1-12/DMPC, LP-TC-1-12/DOPC
and LP-TC-1-12/POPC lipoplexes had high gene knockdown efficacy
(>80% knockdown compared with untreated cells; Fig. 5). LP-TC-1-12/DSPE, LP-TC-1-12/DPPE
and LP-TC-1-12/DPPC lipoplexes exhibited moderate gene knockdown
(41–57% knockdown compared with untreated cells), and
LP-TC-1-12/DSPC lipoplexes with Luc siRNA showed slightly gene
knockdown compared with those with Cont siRNA.
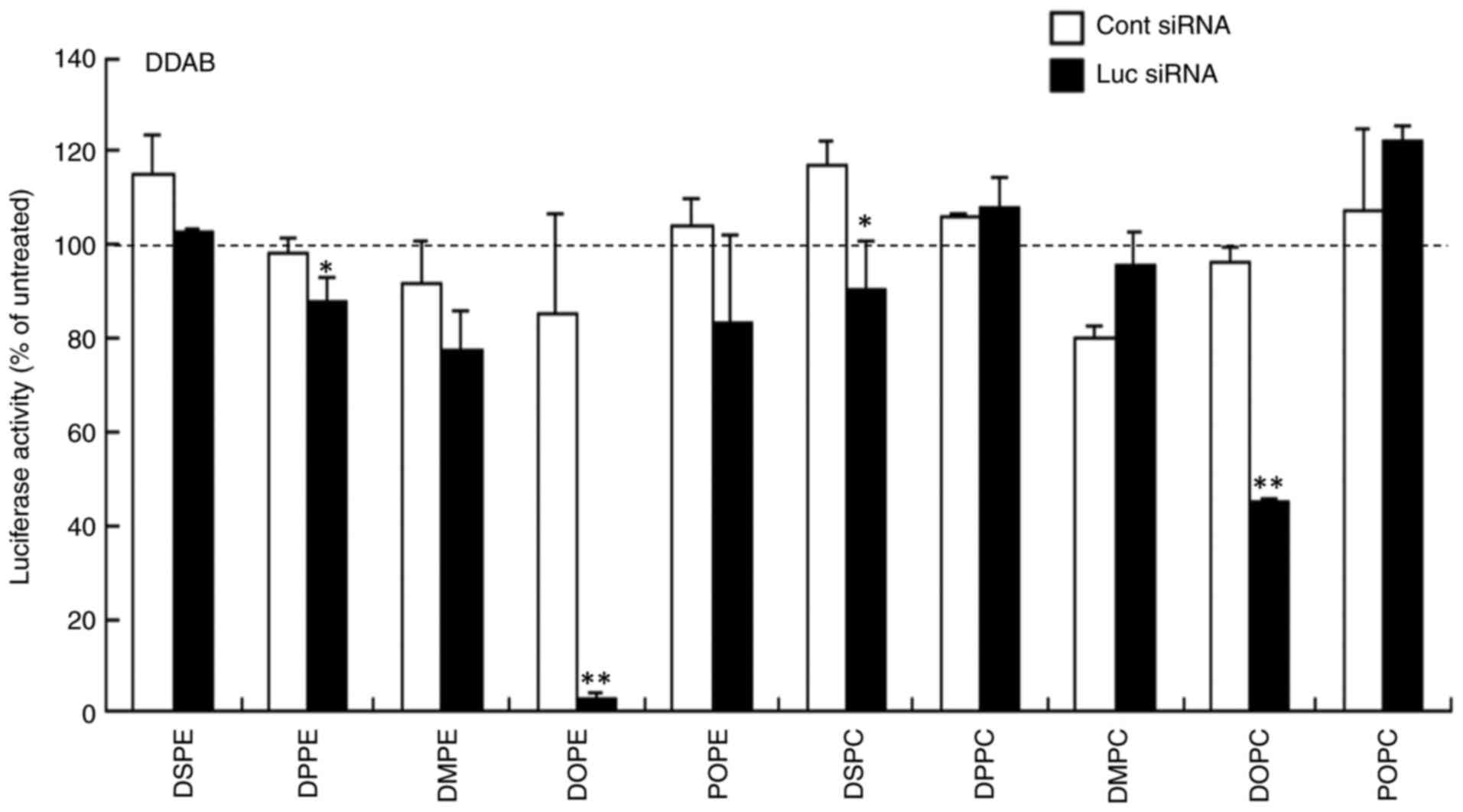 | Figure 3.Effect of phospholipids in DDAB-based
cationic liposomes on gene knockdown using siRNA lipoplexes
following transfection into MCF-7-Luc cells. siRNA lipoplexes with
Cont or Luc siRNA were added to MCF-7-Luc cells at 50 nM and
luciferase assay was performed 48 h post-transfection. Data are
presented as the mean + SD (n=3). *P<0.05, **P<0.01 vs. Cont
siRNA. Cont, control; Luc, luciferase; si, small interfering; DDAB,
dimethyldioctadecylammonium bromide; DSPC,
1,2-distearoyl-sn-glycero-3-phosphocholine; DPPC,
1,2-dipalmitoyl-sn-glycero-3-phosphocholine; DMPC,
1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DSPE,
1,2-distearoyl-sn-glycero-3-phosphoethanolamine; DPPE,
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine; DMPE,
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
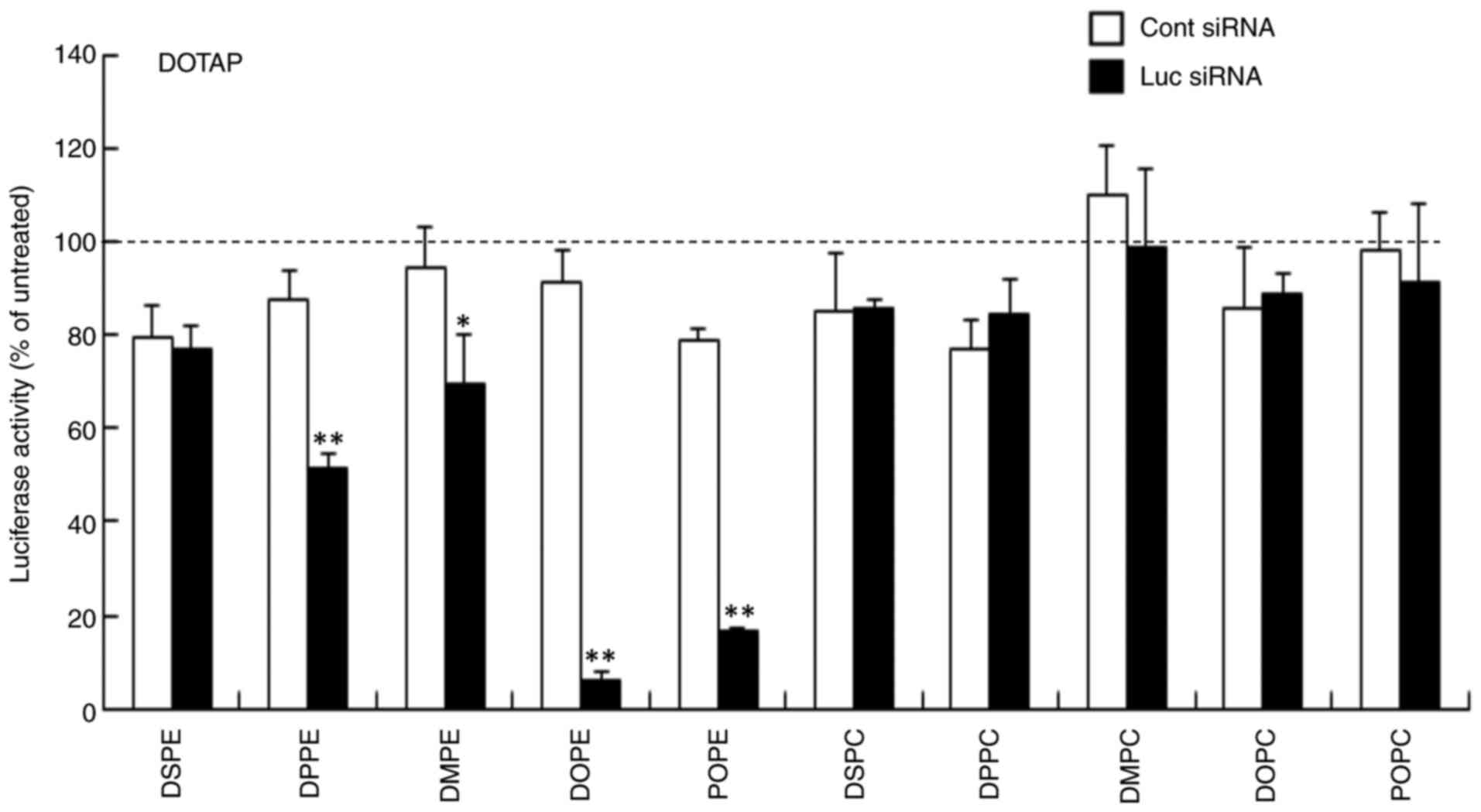 | Figure 4.Effect of phospholipids in
DOTAP-based cationic liposomes on gene knockdown using siRNA
lipoplexes following transfection into MCF-7-Luc cells. siRNA
lipoplexes with Cont or Luc siRNA were added to MCF-7-Luc cells at
50 nM and luciferase assay was performed 48 h post-transfection.
Data are presented as the mean + SD (n=3). *P<0.05, **P<0.01
vs. Cont siRNA. Cont, control; Luc, luciferase; si, small
interfering; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane methyl
sulfate salt; DSPC, 1,2-distearoyl-sn-glycero-3-phosphocholine;
DPPC, 1,2-dipalmitoyl-sn-glycero-3-phosphocholine; DMPC,
1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DSPE,
1,2-distearoyl-sn-glycero-3-phosphoethanolamine; DPPE,
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine; DMPE,
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
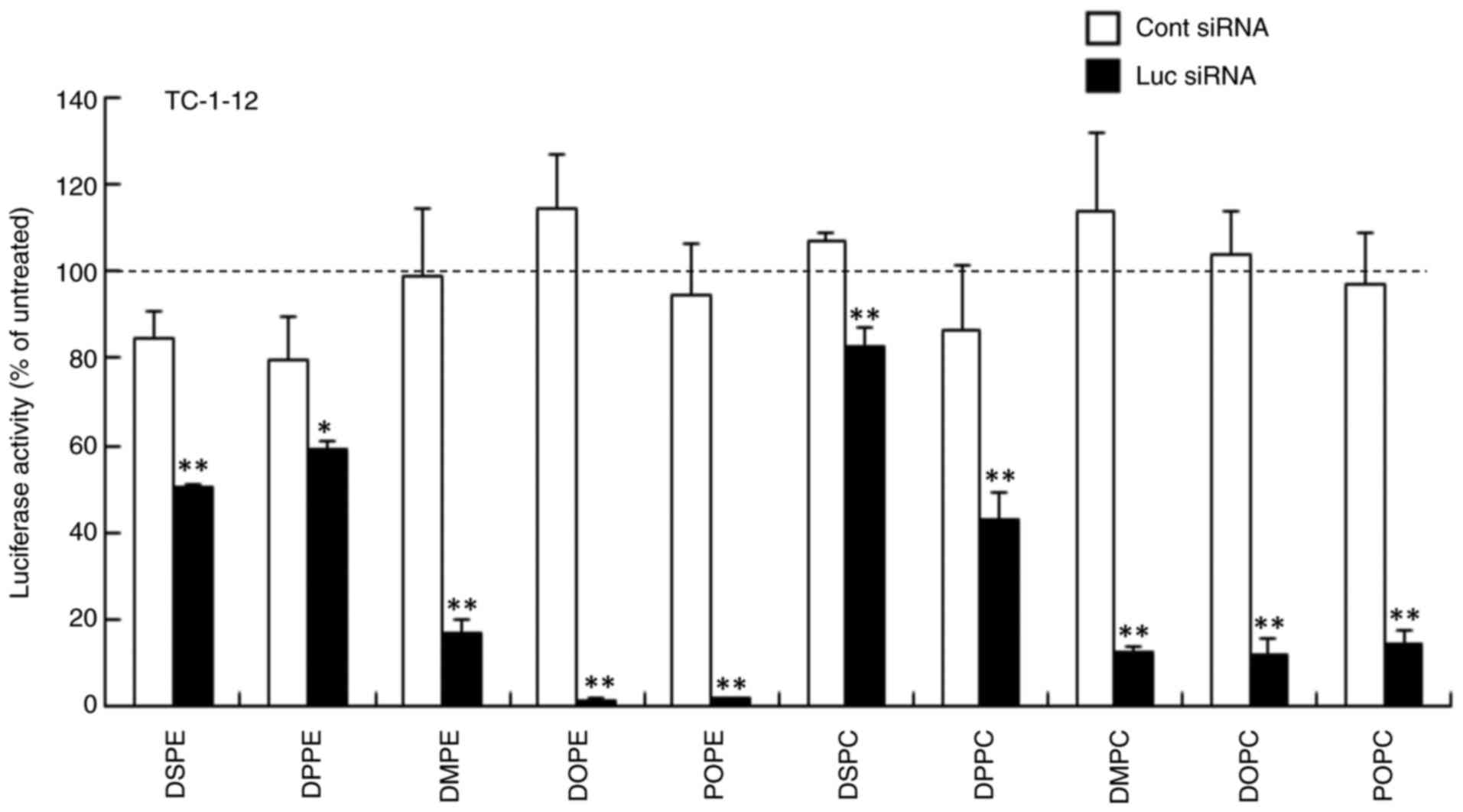 | Figure 5.Effect of phospholipids in
TC-1-12-based cationic liposomes on gene knockdown using siRNA
lipoplexes following transfection into MCF-7-Luc cells. siRNA
lipoplexes with Cont or Luc siRNA were added to MCF-7-Luc cells at
50 nM and luciferase assay was performed 48 h post-transfection.
Data are presented as the mean + SD (n=3). *P<0.05, **P<0.01
vs. Cont siRNA. Cont, control; Luc, luciferase; si, small
interfering; TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DSPC, 1,2-distearoyl-sn-glycero-3-phosphocholine; DPPC,
1,2-dipalmitoyl-sn-glycero-3-phosphocholine; DMPC,
1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DSPE,
1,2-distearoyl-sn-glycero-3-phosphoethanolamine; DPPE,
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine; DMPE,
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
Cytotoxicity induced by siRNA
lipoplexes
The effect of phospholipids in cationic liposomes on
cytotoxicity was evaluated in MCF-7-Luc cells at 24 h
post-transfection of siRNA lipoplexes. LP-TC-1-12/DPPE,
LP-TC-1-12/DMPE and LP-TC-1-12/POPE lipoplexes showed slight
cytotoxicity (75–84% cell viability), while the other lipoplexes
did not exhibit cytotoxicity (>87% cell viability relative to
untreated cells; Fig. 6).
However, the addition of LP-TC-1-12/DPPE, LP-TC-1-12/DMPE and
LP-TC-1-12/POPE without siRNA into the cells did not induce
cytotoxic effects (100–105% cell viability; data not shown). These
findings suggested that cationic liposomes composed of DDAB, DOTAP
or TC-1-12 can be used for efficient siRNA transduction into cells
with minimal toxicity.
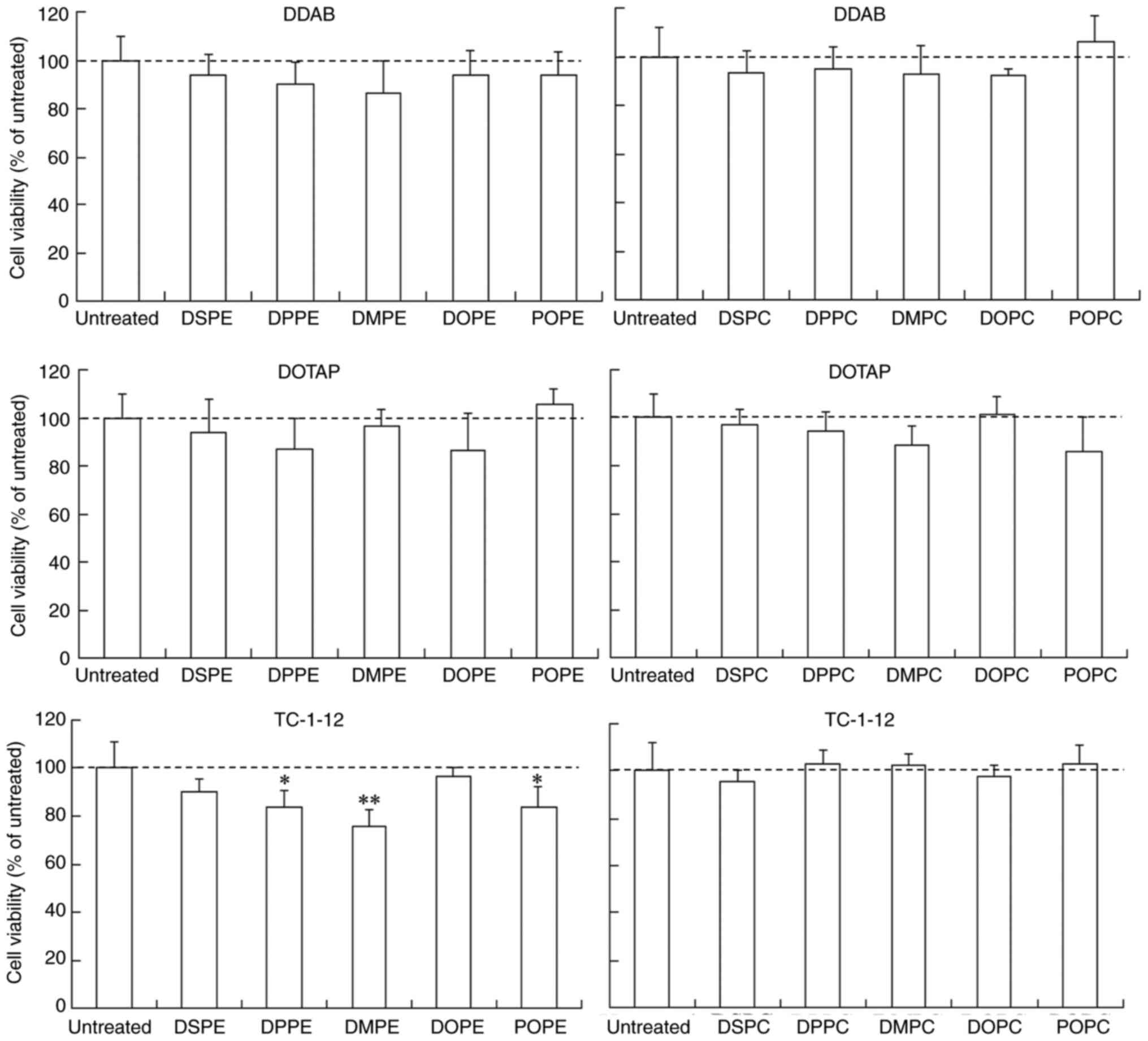 | Figure 6.Effect of phospholipids in cationic
liposomes on cell viability at 24 h post-transfection of siRNA
lipoplexes into MCF-7-Luc cells. siRNA lipoplexes were added to
MCF-7-Luc cells at 50 nM. Data are presented as the mean + SD
(n=4-6). *P<0.05, **P<0.01 vs. untreated. si, small
interfering; DDAB, dimethyldioctadecylammonium bromide; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt;
TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DSPC, 1,2-distearoyl-sn-glycero-3-phosphocholine; DPPC,
1,2-dipalmitoyl-sn-glycero-3-phosphocholine; DMPC,
1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DSPE,
1,2-distearoyl-sn-glycero-3-phosphoethanolamine; DPPE,
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine; DMPE,
1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
Interaction between erythrocytes and
siRNA lipoplexes
Positively charged lipoplexes cause agglutination
via electrostatic interaction with negatively charged erythrocytes
(23); these agglutinates are
effectively trapped by highly extended lung capillaries (24). Therefore, to determine the effect
of phospholipids in cationic liposomes on agglutination of siRNA
lipoplexes with erythrocytes, siRNA lipoplexes were added to
erythrocyte suspensions. LP-DDAB/DOPE, LP-DOTAP/DOPE,
LP-DOTAP/POPE, LP-TC-1-12/DOPE, LP-TC-1-12/POPE, LP-TC-1-12/DMPC,
LP-TC-1-12/DOPC and LP-TC-1-12/POPC were used as their lipoplexes
exhibited high gene knockdown activity in cells (Table IV) and relatively small size
(180–430 nm). All siRNA lipoplexes exhibited agglutination after
mixing with the erythrocyte suspension, regardless of the type of
phospholipid in the liposomal formulation (Fig. 7). In particular, TC-1-12-based
siRNA lipoplexes formed large aggregates following mixing with
erythrocyte suspension.
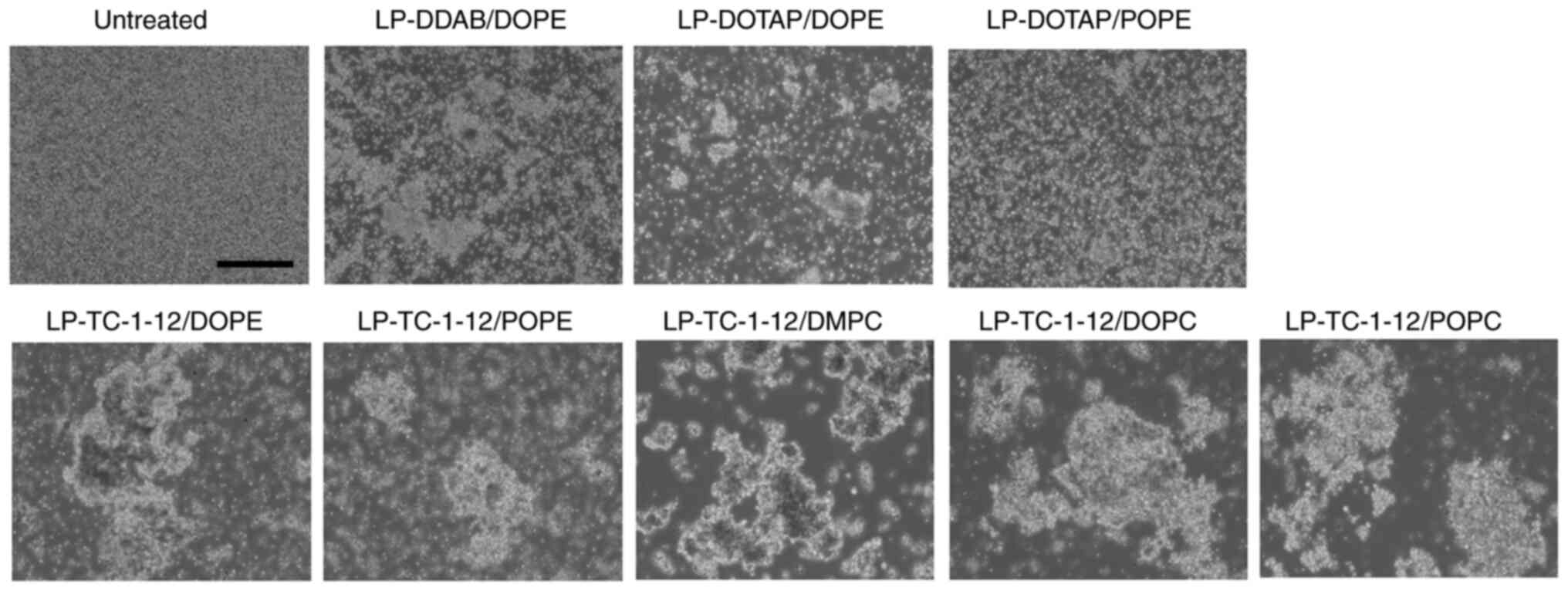 | Figure 7.Effect of phospholipids in cationic
liposomes on agglutination of siRNA lipoplexes with erythrocytes.
Lipoplexes with 2 µg siRNA were incubated with erythrocyte
suspension. Scale bar, 200 µm. si, small interfering; DDAB,
dimethyldioctadecylammonium bromide; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt;
TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
 | Table IV.Summary of in vitro gene
knockdown efficacy following treatment with siRNA lipoplexes. |
Table IV.
Summary of in vitro gene
knockdown efficacy following treatment with siRNA lipoplexes.
|
| Cationic lipid |
|---|
|
|
|
|---|
| Phospholipid | DDAB | DOTAP | TC-1-12 |
|---|
| DSPE | - | - | + |
| DPPE | - | + | - |
| DMPE | - | - | +++ |
| DOPE | +++ | +++ | +++ |
| POPE | - | +++ | +++ |
| DSPC | - | - | - |
| DPPC | - | - | ++ |
| DMPC | - | - | +++ |
| DOPC | ++ | - | +++ |
| POPC | - | - | +++ |
Biodistribution of siRNA following
systemic injection of siRNA lipoplexes
The present study investigated the effect of
phospholipids in liposomal formulation on biodistribution of siRNA
by ex vivo imaging at 1 h after systemic injection of
lipoplexes with Cy5-siRNA. All siRNA lipoplexes caused siRNA
accumulation in the lung (Figs. 8
and 9). In particular,
LP-TC-1-12/DMPC, LP-TC-1-12/DOPC and LP-TC-1-12/POPC lipoplexes
exhibited high siRNA accumulation in the lung, indicating that
relatively large siRNA lipoplexes (270–430 nm) may form large
agglutinations with blood components, resulting in efficient
entrapment in lung capillaries. These results indicated that siRNA
accumulation in the lung following systemic injection of siRNA
lipoplexes may be affected by the size of siRNA lipoplexes, rather
than the type of cationic lipid or phospholipid in cationic
liposomes. In all the lipoplexes, the injected siRNAs were detected
slightly in the liver and spleen, but not in heart. In the kidney,
siRNA lipoplexes containing phosphatidylethanolamine showed high
accumulation compared with the lipoplexes containing
phosphatidylcholine.
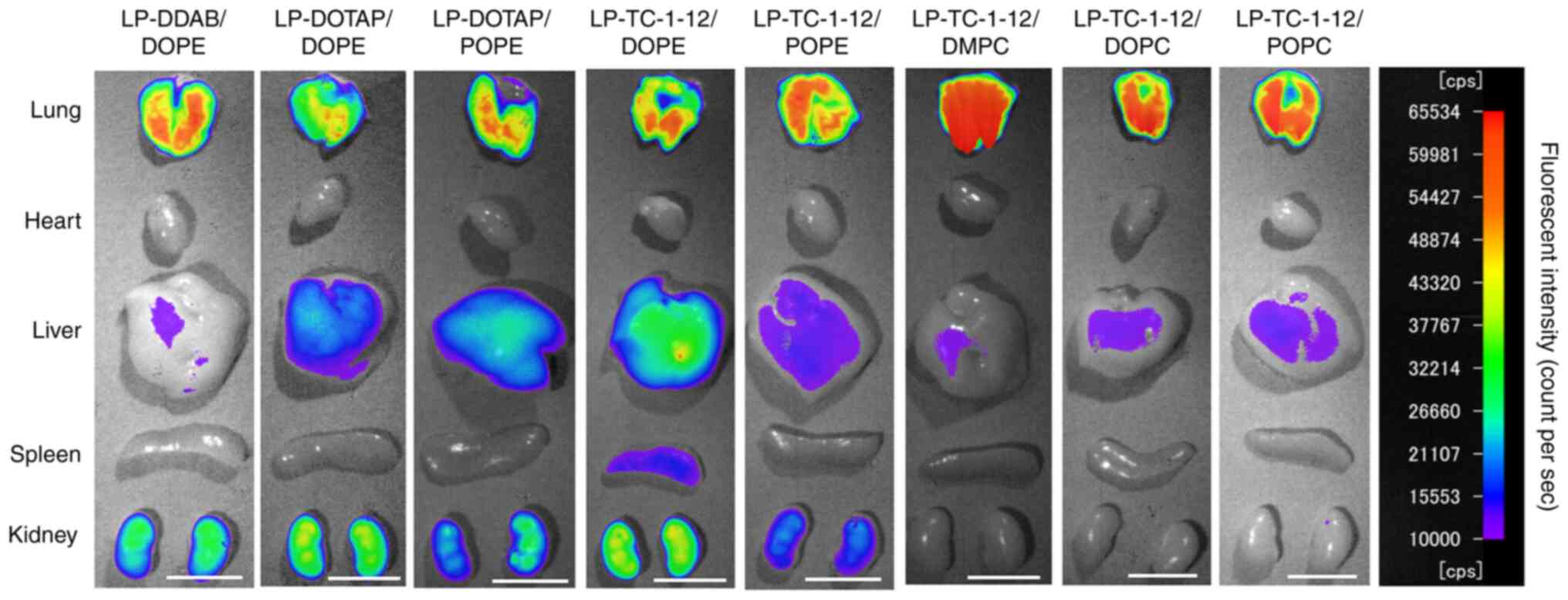 | Figure 8.Effect of phospholipid in cationic
liposomes on biodistribution of siRNA in mice at 1 h after systemic
injection of siRNA lipoplexes. siRNA lipoplexes with 20 µg
Cy5-siRNA were administered intravenously to mice. Cy5 fluorescence
imaging of tissue was performed 1 h post-injection. Fluorescence
intensity is illustrated using a color-coded scale (red, maximum;
purple, minimum). Scale bar, 1 cm. si, small interfering; Cy5,
cyanine 5; DDAB, dimethyldioctadecylammonium bromide; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt;
TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
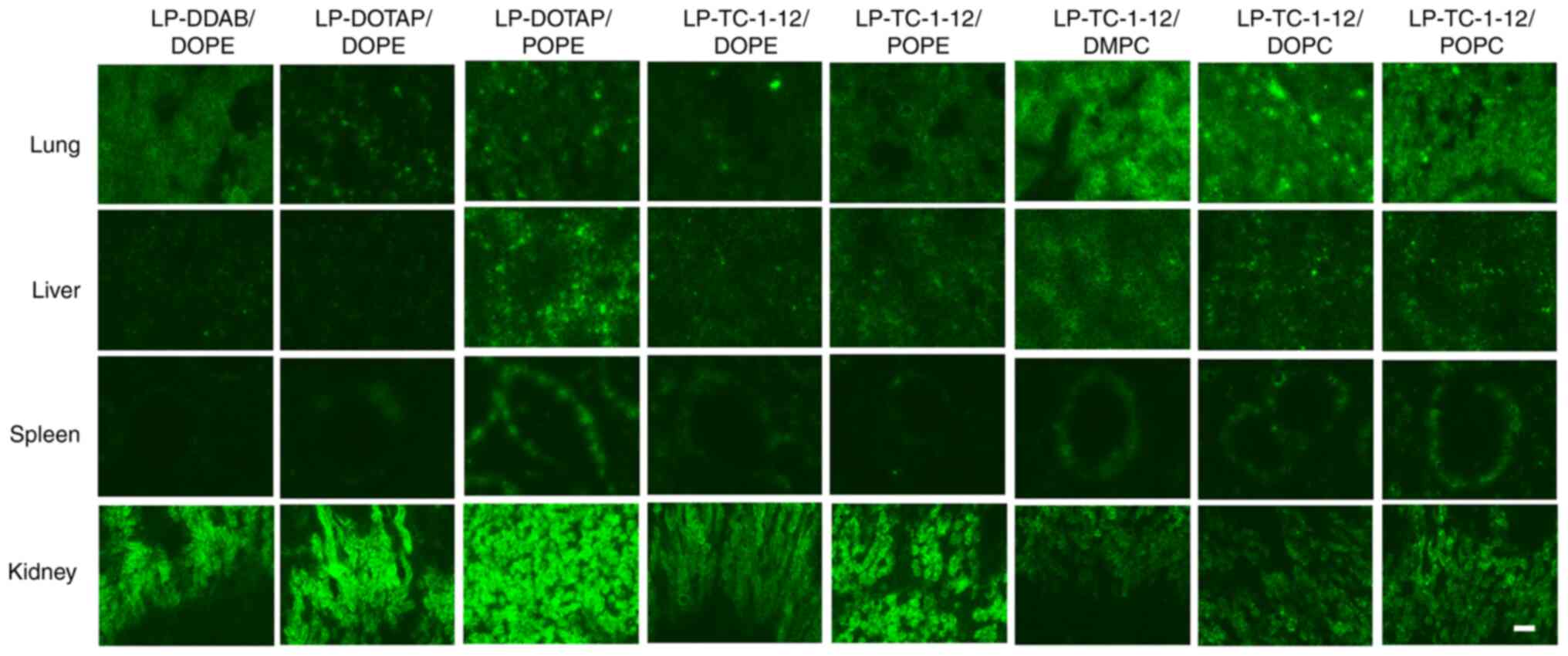 | Figure 9.Effect of phospholipids in cationic
liposomes on biodistribution of siRNA in mice 1 h following
intravenous injection of siRNA lipoplexes. siRNA lipoplex with 20
µg Cy5-siRNA was administered intravenously to mice. At 1 h
post-injection, tissue was frozen and sliced to observe
localization of Cy5-siRNA (green) using a fluorescent microscope.
Scale bar, 100 µm. si, small interfering; Cy5, cyanine 5; DDAB,
dimethyldioctadecylammonium bromide; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt;
TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; POPC,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
Gene knockdown in the lung following
systemic injection of siRNA lipoplexes
Tie2 gene is expressed in vascular endothelium
(25,26) and has previously been used to
evaluate gene knockdown efficacy of siRNA lipoplexes in the lung
(19). The present study
evaluated the effect of phospholipid in cationic liposomes on gene
knockdown of Tie2 mRNA in pulmonary vascular endothelium at 48 h
after single systemic injection of Tie2 siRNA lipoplexes into mice
(Fig. 10). LP-DDAB/DOPE,
LP-DOTAP/POPE, LP-TC-1-12/DOPE, LP-TC-1-12/POPE and LP-TC-1-12/DOPC
were selected for evaluation of knockdown efficacy as their
lipoplexes exhibited high gene knockdown activity (Fig. 3, Fig. 4, Fig. 5; Table IV) and maintained small size
(180–270 nm). LP-DOTAP/DOPE was excluded from the evaluation as
injection of LP-DOTAP/DOPE lipoplexes with 50 µg Tie2 siRNA has
previously been shown not to induce gene knockdown of Tie2 mRNA in
the lung (6). Systemic injection
of LP-DDAB/DOPE, LP-TC-1-12/DOPE and LP-TC-1-12/POPE lipoplexes
with Tie2 siRNA significantly suppressed Tie2 mRNA level (~50, 46
and 33% knockdown, respectively, compared with Cont 2 siRNA).
Combined with the aforementioned results, these data indicated that
accumulation of these siRNA lipoplexes in the lung did not decrease
gene knockdown activity by agglutination with erythrocytes.
However, LP-DOTAP/POPE and LP-TC-1-12/DOPC lipoplexes with Tie2
siRNA did not significantly suppress Tie2 mRNA levels in the
lung.
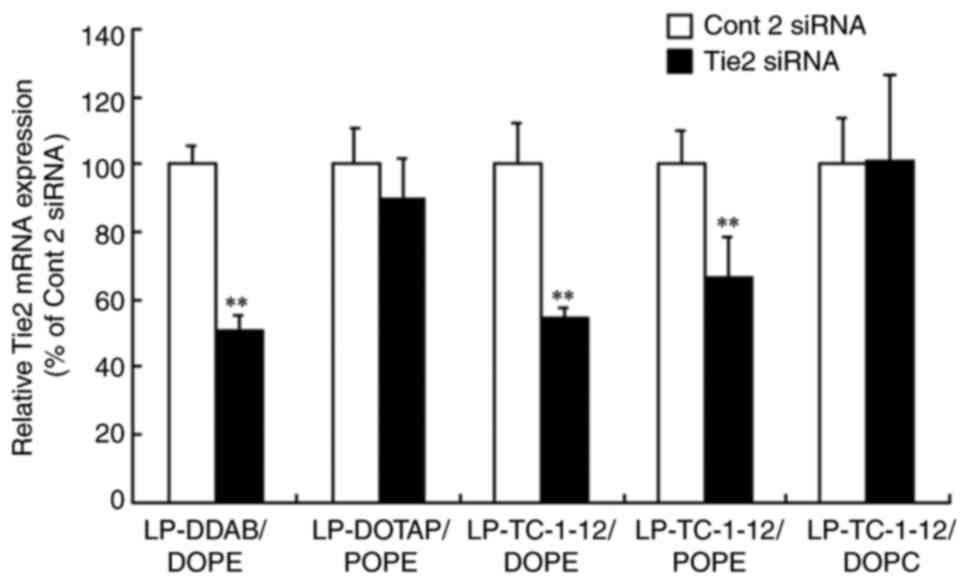 | Figure 10.Effect of phospholipids in cationic
liposomes on knockdown of Tie2 mRNA in the lung following systemic
injection of Tie2 siRNA lipoplexes into mice. Tie2 mRNA levels in
the lung were normalized to those of PTEN at 48 h after systemic
administration of siRNA lipoplex with 20 µg Cont 2 or Tie2 siRNA.
Tie2 expression (%) was calculated relative to that of mice treated
with Cont 2 siRNA. Data are presented as the mean + SD (n=3-4).
**P<0.01 vs. Cont 2 siRNA. si, small interfering; Cont, control;
DDAB, dimethyldioctadecylammonium bromide; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt;
TC-1-12,
11-[(1,3-bis(dodecanoyloxy)-2-((dodecanoyloxy)methyl)propan-2-yl)amino]-N,N,N-trimethyl-11-oxoundecan-1-aminium
bromide; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; POPE,
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine. |
Discussion
Phosphatidylcholine and phosphatidylethanolamine are
phospholipids in the cell membrane of most prokaryotes that are
used as neutral helper lipids to prepare liposomes (15). Cationic liposomes composed of
dialkyl cationic lipids with neutral helper lipids, such as DOPE,
have been evaluated as carriers for siRNA delivery (5,6).
The present study determined the effect of phospholipids in
cationic liposomes on gene knockdown using siRNA lipoplexes.
Following in vitro transfection, inclusion of
phosphatidylcholines in DOTAP or DDAB-based cationic liposomes did
not induce high gene knockdown by siRNA lipoplexes, although
LP-DDAB/DOPC lipoplexes exhibited moderate gene knockdown activity.
This may be because phosphatidylcholine has a larger head group
than phosphatidylethanolamine (15). Du et al (27) reported that DOPC promotes stable
laminar structure, limiting escape of cationic liposomes composed
of DOTAP and DOPC from the endosome. However, DOPE promotes
formation of inverted hexagonal lipid structures and cationic
liposomes composed of DOTAP and DOPE exhibit improved transfection
efficiency by destabilizing the endosomal membrane compared with
those composed of DOTAP and DOPC (27). Such finding suggests that
difference in headgroup structure between phosphatidylethanolamine
and phosphatidylcholine may affect efficiency of siRNA transfection
by cationic liposomes. In the present study, only combination with
DOPE in DDAB-based cationic liposomes induced high gene knockdown
activity. However, combination with DOPE or POPE in DOTAP-based
cationic liposomes induced high gene knockdown in cells, indicating
that saturation of dialkyl chains of cationic lipid and
phospholipids may affect transfection efficiency following
treatment with siRNA lipoplexes. The inclusion of
phosphatidylethanolamine containing unsaturated and long dialkyl
chains in DDAB or DOTAP-based liposomes induced strong gene
knockdown in cells. By contrast, in TC-1-12-based liposomes,
inclusion of phosphatidylcholine or phosphatidylethanolamine
containing saturated and short or unsaturated and long dialkyl
chains caused high gene knockdown by TC-1-12-based cationic
liposomes. DDAB and DOTAP are cationic lipids with long anchors
(C18 dialkyl chains), while TC-1-12 is cationic lipid with short
anchor (C12 trialkyl chains). Thus, the difference in alkyl chain
length of cationic lipids may affect the optimal combination of
cationic lipid with phospholipids for gene knockdown activity. The
phase transition temperatures (Tm) of phosphatidylcholine, DSPC,
DPPC, DMPC, POPC and DOPC are 55, 41, 24, −2 and −17°C,
respectively, and those of phosphatidylethanolamine, DSPE, DPPE,
DMPE, POPE and DOPE are 74, 63, 50, 25 and −16°C, respectively
(28). Phospholipids with
unsaturated and long (POPE, DOPE, POPC and DOPC) or saturated and
short dialkyl chains (DMPC and DMPE) have relatively low Tm and
high membrane fluidity, indicating that high transfection activity
by TC-1-12-based liposomes may be inversely associated with
phospholipid Tm. Koulov et al (29) reported that cationic lipids with
trialkyl chains promote greater vesicle fusion compared with those
with structurally associated dialkyl or monoalkyl chains. Such
findings indicate that TC-1-12-based cationic liposomes may be
effective as vectors for siRNA delivery and exhibit fusogenic
activity. Based on the present results, in vitro gene
silencing activity was markedly affected by the type of
phospholipid in cationic liposomes. The present study used
MCF-7-Luc cells for evaluation of in vitro transfection
efficiency by lipoplexes with Luc siRNA; however, further
experiments are required to evaluate gene knockdown efficacy in
other cancer cell lines using lipoplexes with Luc siRNAs targeting
different sequences in luciferase gene.
In vivo, inclusion of
phosphatidylethanolamine containing unsaturated and long dialkyl
chains into DDAB or TC-1-12-based liposomes induced significant
gene knockdown in the lung. Here, combination of DOTAP with
phospholipid did not induce high in vivo gene knockdown.
However, inclusion of Chol in DOTAP-based cationic liposomes was
previously reported to increase gene knockdown in the lung
(6), indicating that combination
of DOTAP and Chol may be suitable for in vivo transfection.
By contrast, DDAB-based cationic liposomes showed high in
vivo gene knockdown efficacy when DDAB was combined with DOPE.
Our recent study reported that cationic liposomes composed of DDAB
and Chol induce high gene knockdown efficiency of Tie2 mRNA
(>70%) when lipoplexes with 20 µg Tie2 siRNA are systemically
injected into mice (13). DOTAP
has unsaturated dialkyl chains (Tm, −12°C) (30) while DDAB has saturated dialkyl
chains (Tm, 43°C) (31),
indicating that combination of DOTAP and phospholipid may be
unstable in blood circulation (37°C) following systemic injection,
resulting in poor gene knockdown. Although Tm value of TC-1-12 is
unknown, it was hypothesized that this value is low because of the
short alkyl chains. Nonetheless, TC-1-12-based liposomes induced
significant gene knockdown when combined with DOPE or POPE. It is
unclear why TC-1-12 exhibited high in vivo transfection
efficiency when combined with phosphatidylethanolamine containing
unsaturated and long dialkyl chains. TC-1-12 may affect liposome
membrane stability via its trialkyl chains (29). Further study is needed to evaluate
the mechanism underlying in vivo gene knockdown by
TC-1-12-based cationic liposomes. Among the present liposomal
formulations, LP-DDAB/DOPE, LP-TC-1-12/DOPE and LP-TC-1-12/POPE are
potential vectors for delivering siRNA into the lung. The present
study evaluated mRNA levels in the lung following injection of
siRNA lipoplexes; however, future experiments should also evaluate
changes of protein levels in the lung.
In conclusion, the present study determined the
effect of phospholipids in cationic liposomes on gene knockdown in
breast cancer cells and mouse lung using siRNA lipoplexes.
Differences in the structure of head groups and alkyl chains in
phospholipids may have affected gene knockdown efficacy of siRNA
lipoplexes. Overall, the present study provided information
regarding optimal combination of cationic lipid and phospholipid
for efficient siRNA delivery by cationic liposome.
Acknowledgements
The authors would like to thank Ms Ayaka Uchida and
Ms Mayuko Miyauchi (Department of Molecular Pharmaceutics, Hoshi
University, Tokyo, Japan) for assisting with in vitro gene
knockdown using siRNA lipoplexes.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YH conceived and designed the study and wrote the
manuscript. MT, ST, KT, AS, NI, RY and KO performed the
experiments. YH and MT confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by Institutional
Animal Care and Use Committee of Hoshi University (approval no.
P21-039).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wilson RC and Doudna JA: Molecular
mechanisms of RNA interference. Annu Rev Biophys. 42:217–239. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang S, Zhi D and Huang L: Lipid-based
vectors for siRNA delivery. J Drug Target. 20:724–735. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zatsepin TS, Kotelevtsev YV and
Koteliansky V: Lipid nanoparticles for targeted siRNA
delivery-going from bench to bedside. Int J Nanomedicine.
11:3077–3086. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barba AA, Bochicchio S, Dalmoro A and
Lamberti G: Lipid delivery systems for nucleic-acid-based-drugs:
From production to clinical applications. Pharmaceutics.
11:3602019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hattori Y, Nakamura M, Takeuchi N, Tamaki
K, Shimizu S, Yoshiike Y, Taguchi M, Ohno H, Ozaki K and Onishi H:
Effect of cationic lipid in cationic liposomes on siRNA delivery
into the lung by intravenous injection of cationic lipoplex. J Drug
Target. 27:217–227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hattori Y, Tamaki K, Ozaki KI, Kawano K
and Onishi H: Optimized combination of cationic lipids and neutral
helper lipids in cationic liposomes for siRNA delivery into the
lung by intravenous injection of siRNA lipoplexes. J Drug Deliv Sci
Technol. 52:1042–1050. 2019. View Article : Google Scholar
|
|
7
|
Taetz S, Bochot A, Surace C, Arpicco S,
Renoir JM, Schaefer UF, Marsaud V, Kerdine-Roemer S, Lehr CM and
Fattal E: Hyaluronic acid-modified DOTAP/DOPE liposomes for the
targeted delivery of anti-telomerase siRNA to CD44-expressing lung
cancer cells. Oligonucleotides. 19:103–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dakwar GR, Braeckmans K, Ceelen W, De
Smedt SC and Remaut K: Exploring the HYDRAtion method for loading
siRNA on liposomes: The interplay between stability and biological
activity in human undiluted ascites fluid. Drug Deliv Transl Res.
7:241–251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hattori Y, Nakamura A, Arai S, Kawano K,
Maitani Y and Yonemochi E: siRNA delivery to lung-metastasized
tumor by systemic injection with cationic liposomes. J Liposome
Res. 25:279–286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song H, Hart SL and Du Z: Assembly
strategy of liposome and polymer systems for siRNA delivery. Int J
Pharm. 592:1200332021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kudsiova L, Welser K, Campbell F,
Mohammadi A, Dawson N, Cui L, Hailes HC, Lawrence MJ and Tabor AB:
Delivery of siRNA using ternary complexes containing branched
cationic peptides: The role of peptide sequence, branching and
targeting. Mol Biosyst. 12:934–951. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tagalakis AD, He L, Saraiva L, Gustafsson
KT and Hart SL: Receptor-targeted liposome-peptide nanocomplexes
for siRNA delivery. Biomaterials. 32:6302–6315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hattori Y, Saito H, Oku T and Ozaki K:
Effects of sterol derivatives in cationic liposomes on
biodistribution and gene-knockdown in the lungs of mice
systemically injected with siRNA lipoplexes. Mol Med Rep.
24:5982021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rehman Z, Zuhorn IS and Hoekstra D: How
cationic lipids transfer nucleic acids into cells and across
cellular membranes: Recent advances. J Control Release. 166:46–56.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Drescher S and van Hoogevest P: The
phospholipid research center: Current research in phospholipids and
their use in drug delivery. Pharmaceutics. 12:12352020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue HY, Guo P, Wen WC and Wong HL:
Lipid-based nanocarriers for RNA delivery. Curr Pharm Des.
21:3140–3147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hattori Y, Nakamura T, Ohno H, Fujii N and
Maitani Y: siRNA delivery into tumor cells by lipid-based
nanoparticles composed of hydroxyethylated cholesteryl triamine.
Int J Pharm. 443:221–229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hattori Y, Kikuchi T, Nakamura M, Ozaki KI
and Onishi H: Therapeutic effects of protein kinase N3 small
interfering RNA and doxorubicin combination therapy on liver and
lung metastases. Oncol Lett. 14:5157–5166. 2017.PubMed/NCBI
|
|
19
|
Fehring V, Schaeper U, Ahrens K, Santel A,
Keil O, Eisermann M, Giese K and Kaufmann J: Delivery of
therapeutic siRNA to the lung endothelium via novel lipoplex
formulation DACC. Mol Ther. 22:811–820. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hattori Y, Nakamura M, Takeuchi N, Tamaki
K, Ozaki K and Onishi H: Effect of cationic lipid type in PEGylated
liposomes on siRNA delivery following the intravenous injection of
siRNA lipoplexes. Wrld Acd Sci J. 1:74–85. 2019.
|
|
21
|
Hattori Y, Tamaki K, Sakasai S, Ozaki KI
and Onishi H: Effects of PEG anchors in PEGylated siRNA lipoplexes
on in vitro gene-silencing effects and siRNA biodistribution
in mice. Mol Med Rep. 22:4183–4196. 2020.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eliyahu H, Servel N, Domb AJ and Barenholz
Y: Lipoplex-induced hemagglutination: Potential involvement in
intravenous gene delivery. Gene Ther. 9:850–858. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Simberg D, Weisman S, Talmon Y, Faerman A,
Shoshani T and Barenholz Y: The role of organ vascularization and
lipoplex-serum initial contact in intravenous murine lipofection. J
Biol Chem. 278:39858–39865. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Loughna S and Sato TN: Angiopoietin and
Tie signaling pathways in vascular development. Matrix Biol.
20:319–325. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van der Heijden M, van Nieuw Amerongen GP,
Chedamni S, van Hinsbergh VW and Johan Groeneveld AB: The
angiopoietin-Tie2 system as a therapeutic target in sepsis and
acute lung injury. Expert Opin Ther Targets. 13:39–53. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Du Z, Munye MM, Tagalakis AD, Manunta MD
and Hart SL: The role of the helper lipid on the DNA transfection
efficiency of lipopolyplex formulations. Sci Rep. 4:71072014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Phase transition temperatures for
glycerophospholipids. Tech Support at Avanti Polar Lipids, Inc.;
https://avantilipids.com/tech-support/physical-properties/phase-transition-tempsFebruary
1–2022
|
|
29
|
Koulov AV, Vares L, Jain M and Smith BD:
Cationic triple-chain amphiphiles facilitate vesicle fusion
compared to double-chain or single-chain analogues. Biochim Biophys
Acta. 1564:459–465. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hirsch-Lerner D and Barenholz Y: Probing
DNA-cationic lipid interactions with the fluorophore
trimethylammonium diphenyl-hexatriene (TMADPH). Biochim Biophys
Acta. 1370:17–30. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Feitosa E, Alves FR, Niemiec A, Real
Oliveira ME, Castanheira EM and Baptista AL: Cationic liposomes in
mixed didodecyldimethylammonium bromide and
dioctadecyldimethylammonium bromide aqueous dispersions studied by
differential scanning calorimetry, Nile red fluorescence, and
turbidity. Langmuir. 22:3579–3585. 2006. View Article : Google Scholar : PubMed/NCBI
|
























