Introduction
Melanoma is the 19th most common cancer diagnosis
worldwide, representing <5% of skin cancers and accounting for
80% of skin cancer-related deaths (1). Compared with other cancer types, the
global incidence of cutaneous melanoma has been increasing annually
at a more rapid rate (2). Every
year, ~160,000 new cases of invasive melanoma and 48,000 deaths due
to the disease are recorded worldwide (3). The oncogenesis and developmental
mechanisms of melanoma remain unclear and require further
exploration. Furthermore, it is of vital importance to explore
efficient biomarkers for early and accurate diagnosis.
Tumorigenesis and malignancy development are complex
processes affected by various factors. Due to genetic changes and
epigenetic defects, the tumour microenvironment, which consists of
cellular and non-cellular components, also serves an important
regulatory role in cancer progression (4). Tumour hypoxia is a condition wherein
solid tumours have lesser oxygenation than that observed in normal
tissues (5), which results in the
obstruction of therapy and the progression of malignancy (6). Various studies have demonstrated the
crucial role of hypoxia in the malignant behaviour of melanoma
cells, including abnormal proliferation, distant metastasis and
immune evasion (7–9). As a type of RNA that does not encode
proteins and has a length >200 nucleotides, long non-coding RNAs
(lncRNAs) have been explored in the occurrence and development of
numerous tumours (10,11). lncRNAs participate in the
modification of proteins that affect the survival and proliferation
of cancer cells and thereby serve a role in cancer cell survival
and development, even in hypoxic circumstances (12). It has been reported that hypoxia
yield proliferation-associated lncRNA acts as a competing
endogenous RNA (ceRNA) that binds to microRNA (miRNA/miR)-431-5p
under hypoxic conditions to upregulate CDK14 expression, resulting
in gastric cancer cell proliferation (13). In colorectal cancer cells, lncRNA
COL4A2 antisense RNA 1 facilitates growth and glycolysis under
hypoxia via the miR-20b-5p/hypoxia-inducible factor 1α subunit axis
(14). Several studies have also
explored the relationship between malignant melanoma progression
and hypoxia-related lncRNAs, including long intergenic non-protein
coding RNA 518 and non-coding RNA activated by DNA damage (15,16). Therefore, lncRNAs may serve a
crucial role in hypoxia-related malignancy development and poor
melanoma prognosis.
lncRNA-based signatures have recently drawn
attention due to their high predictive accuracy (17,18). However, hypoxia-related lncRNAs as
biomarkers for prognostic prediction in melanoma are yet to be
investigated. The present study aimed to build a
hypoxia-related-lncRNA signature and nomogram that can improve the
overall survival (OS) predictability in patients with melanoma
using bioinformatics. The present study identified a
7-hypoxia-related-lncRNA signature through a comprehensive analysis
of The Cancer Genome Atlas (TCGA) database and two hypoxia-related
gene sets. The present signature was verified and analysed by
combining the clinical features of melanoma, and validated using
clinical samples and in vitro experiments.
Materials and methods
Data collection
The transcriptome expression profiles corrected by
FPKM (fragments per kilobase of exon per million mapped fragments)
and clinical information were downloaded from TCGA database
(https://portal.gdc.cancer.gov). The
details of the clinical characteristics, including OS, survival
time, age, sex, stage, tumour size, distant metastasis and lymph
node metastasis, of the included cohort of 471 patients are listed
in Table SI. Among these 494
patients, 23 patients with incomplete data or vague living status
were excluded from the present study. The hypoxia-related gene
expression profiles KRIEG and WINTER were downloaded from the Gene
Set Enrichment Analysis (GSEA) database (http://www.gsea-msigdb.org) and are listed in Table SII. The data used in the present
study were downloaded on May 21, 2021.
Establishment of a
hypoxia-related-lncRNA signature as a prognostic model
Pearson's correlation analysis was used to identify
hypoxia-related lncRNAs. The correlation between lncRNA expression
and hypoxia-related gene expression was calculated. The selection
criteria included values with |R2|>0.7 and
P<0.001. Univariate Cox regression analysis was used to screen
for prognosis-associated hypoxia-related lncRNAs. A hazard ratio
(HR) <1 represented favourable OS outcomes, and a HR >1
represented poor OS outcomes. lncRNAs with P<0.05 were selected
as hypoxia-related lncRNAs and used to construct the present
prognostic model.
Evaluation of the
hypoxia-related-lncRNA prognostic signature
Multivariate Cox regression analysis was used to
create a signature, and the risk score formula was as follows:
(RiskScore) = ∑i=1n(Expi ⁎Coei).
In the aforementioned formula, ‘n’ indicates the number of
prognostic genes, ‘Expi’ indicates the level of lncRNA I
expression, and ‘Coei’ indicates the regression coefficient of the
corresponding lncRNA obtained using the multivariate Cox regression
model. Patients with melanoma were divided into high- and low-risk
subgroups according to the median risk score. A survival curve was
used to compare the OS of patients between the high- and low-risk
groups. The diagnostic efficacy and clinicopathological
characteristics of the 7-hypoxia-related-lncRNA signature were
evaluated using receiver operating characteristic (ROC) curves. The
risk score efficiency of the present signature to independently
predict the survival rate was assessed using univariate and
multivariate Cox regression analyses.
Estimation of clinical independence
and construction of the nomogram
The ‘rms’ R package was used to integrate the
clinical features with the risk scores obtained. The present
7-hypoxia-related-lncRNA signature was used to construct a nomogram
for further clinical prediction. Variates of clinical features were
categorized into dichotomous variables and defined by Cox
regression analysis. Samples with incomplete clinical information
were excluded to make the results consistent. A horizontal straight
line was drawn to ascertain the points for each variable by
variables feature. Subsequently, the total points of each patient
were calculated by adding the points for all variables together,
followed by normalization to a distribution of 0 to 100. The
performance of the present nomogram was evaluated using calibration
plots and time-dependent ROC curves.
Principal component analysis
(PCA)
PCA was conducted using the ‘scatterplot3D’ R
package to illustrate the level of expression of melanoma samples
in the low- and high-risk groups. Patients were divided into
different subgroups using different gene sets, including the
present 7-hypoxia-related-lncRNA gene set, hypoxia-related-lncRNA
gene set, hypoxia-related gene set and whole gene sets.
Construction of the co-expression
network
The lncRNA-miRNA-mRNA co-expression network was
constructed using Cytoscape (version 3.8.2) to elucidate the
association of the hypoxia-related lncRNAs with their target miRNAs
and downstream mRNAs. The lncRNA-miRNA connection was predicted
using data downloaded from miRcode (http://www.mircode.org). TargetScan (version 8.0)
(https://www.targetscan.org), miRDB
(http://www.mirdb.org) and miRTarBase databases
(http://mirtarbase.mbc.nctu.edu.tw/php/index.php)
were used to screen for miRNAs and their corresponding target
mRNAs. The selected mRNAs were compared with hypoxia-related mRNAs
to obtain reliable hypoxia-related lncRNAs.
GSEA
GSEA was employed to identify enriched gene sets in
the high-risk groups. The Gene Ontology-Biological Process (GO-BP)
ontology gene sets (c5.go.bp.v7.4.symbols.gmt) were downloaded from
the Molecular Signatures Database (http://www.gsea-msigdb.org). Gene set permutations
were performed 1,000 times for each analysis.
Immune cell infiltration analysis
The CIBERSORT algorithm of the R software was used
to determine the profile of tumour-infiltrating immune cells
(TIICs; including 22 immune cells) in all tumour samples. The
relationship between TIICs and risk scores was analysed using the
limma package. Comparisons of the TIIC levels between the high- and
low-risk groups were estimated using the unpaired Student's t
test.
Cell culture
A375 human melanoma cells were purchased from the
Institute of Biochemistry and Cell Biology of the Chinese Academy
of Sciences. Cells were incubated in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). The cells were seeded in cell culture plates and
cultivated in a humidified incubator at 37°C with 5%
CO2.
Human tissue specimens
The human melanoma specimens were provided by the
Department of Dermatology of the Chongqing Medical University
(Chongqing, China) from October 2021 to March 2022. The present
study was performed using 20 melanoma samples and their paired
adjacent tissues, which were collected from 10 males and 10
females, with age range from 49 to 58 years old. Inclusion criteria
were as follows: People who were diagnosed with melanoma by
pathological examination at first time with medical history which
was detailed enough to meet the statistical needs of this trial.
Exclusion criteria were as follows: People who were diagnosed as
melanoma or treated previously; people with other special
conditions (such as severe chronic secondary fungi or bacterial
infection); and people with metastatic melanoma. The expression
levels of three lncRNAs were detected using reverse
transcription-quantitative PCR (RT-qPCR). The present study was
approved (approval no. 2021-487) by the Institute Research Ethics
Committee of the First Affiliated Hospital of Chongqing Medical
University (Chongqing, China). All individuals provided written
informed consent for the use of their samples for clinical
research.
Small interfering RNA (siRNA/si)
interference assay
All siRNAs targeting human MIR205 host gene
(MIR205HG, Gene ID: 642587; si-MIR205HG-1 and si-MIR205HG-2) and
scrambled negative control (si-NC) were designed and synthesized by
Shanghai GenePharma Co., Ltd. and are listed in Table I. The siRNAs were transfected into
A375 cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
In brief, A375 cells were seeded into 6-well plates at a density of
1.2×105 cells per well and routinely cultured until the
confluence reached 70%. Then the medium was changed to 2 ml DMEM
without serum, supplemented with 5 µl Lipofectamine®
2000 and 5 µl (20 µM) siRNA to transfect for 6 h at 37°C with 5%
CO2. Subsequent experimentation was performed 48 h after
transfection. The efficiency of MIR205HG knockdown was verified
using RT-qPCR.
 | Table I.Sequences of small interfering RNAs
and primer sequences for reverse transcription-quantitative
PCR. |
Table I.
Sequences of small interfering RNAs
and primer sequences for reverse transcription-quantitative
PCR.
| Designation | Gene/siRNA | Sequences
(5′-3′) | Organism |
|---|
| Primer | HLA-DQB1-AS1 | F:
AGCAGAGTCCAGGGTGTATTG | Homo
sapiens |
|
|
| R:
GCTGCTAGTGGTCGGGAAG |
|
|
| TRBV11-2 | F:
GTGACCCTGATTGGGCAAAG |
|
|
|
| R:
TATCTGGGAGACTGGGCAAC |
|
|
| MIR205HG | F:
AGGAGTCATTTCTGTTCCGCA |
|
|
|
| R:
CAAATAGTGTCCCAGCCACCT |
|
|
| GAPDH | F:
CAGTGGCAAAGTGGAGATTGTTG |
|
|
|
| R:
TCGCTCCTGGAAGATGGTGAT |
|
| siRNA |
MIR205HG-siRNA-1 | F:
UCUCCUUCAAUUCCACUUUTT | Homo
sapiens |
|
|
| R:
AAAGUGGAAUUGAAGGAGATT |
|
|
|
MIR205HG-siRNA-2 | F:
GAGACAGCCAGAGAGAAAUTT |
|
|
|
| R:
AUUUCUCUCUGGCUGUCUCTT |
|
RT-qPCR
Total RNA was extracted from the two groups using
TRIzol® reagent (Takara Bio, Inc.) according to the
manufacturer's protocol. First-strand cDNA synthesis was performed
using the PrimeScript RT reagent kit (Takara Bio, Inc.). The
following thermocycling conditions were used for RT: 25°C for 5
min, 42°C for 40 min and 85°C for 2 min. Subsequently, qPCR was
performed using an ABI7500 real-time PCR system using SYBR green
(Takara Bio, Inc.). The qPCR thermocycling conditions were as
follows: 94°C for 2 min, followed by 40 cycles of 94°C for 30 sec
and 55°C for 45 sec. All RT-qPCR primers were purchased from
Shanghai GenePharma Co., Ltd. and the sequences are listed in
Table I. Each sample was detected
at least three times independently. The 2−ΔΔCq method
was utilized to quantify the expression levels (19).
Cell proliferation assay
The growth rate of A375 cells was assessed using a
Cell Counting Kit-8 assay (CCK-8; Boster Biological Technology).
A375 cells transfected with different siRNAs were seeded into
96-well plates (5,000 cells per well). After incubation for 48 h at
37°C with 5% CO2, 10 µl CCK-8 reagent was added to each
well. The absorbance of the cell medium at 450 nm was recorded to
assess the cell viability in each group.
Cell cycle analysis
A375 cells cultured in six-well plates were
transfected with different siRNAs. After transfection for 48 h,
cells were collected and fixed with 70% cooling ethanol at 4°C
overnight. Following staining with propidium iodide solution
containing RNase A, cells were washed with PBS three times. After
an incubation of 30 min at room temperature, the cell cycle
distribution was analysed using a FACSCalibur flow cytometer (BD
Accuri C6 Plus). CytExpert software (version 2.4.0.28; Beckman
Coulter, Inc.) was employed for analysis.
Western blotting
Total protein was extracted using RIPA lysis buffer
(Beyotime Institute of Biotechnology) containing 1% PMSF and 1
mmol/l β-glycerophosphate sodium salt hydrate, which were both
purchased from Selleck Chemicals. The protein concentration was
calculated using a BCA protein quantitative kit (Beyotime Institute
of Biotechnology). A 10% SDS-polyacrylamide gel (Beyotime Institute
of Biotechnology) was used to separate the same amount of protein
sample (20 µg) and protein was transferred to nitrocellulose
membranes (MilliporeSigma). Membranes were blocked with 5% skim
milk in Tris-buffered saline containing 1% Tween 20 for 2 h at room
temperature. Membranes were incubated overnight at 4°C with the
following primary antibodies: Anti-phospho-GSK3-β (1:1,000; cat.
no. 5558; Cell Signaling Technology, Inc.), anti-GAPDH (1:10,000;
cat. no. 10494-1-AP; ProteinTech Group, Inc.), anti-c-Myc (1:1,000;
cat. no. A5011; Bimake), anti-β-catenin (1:1,000; cat. no. 8480;
Cell Signaling Technology, Inc.), anti-phospho-β-catenin (1:1,000;
cat. no. 4176; Cell Signaling Technology, Inc.), anti-GSK3-β
(1:1,000; cat. no. 121456; Cell Signaling Technology, Inc.).
Following primary incubation, membranes were incubated with
HRP-labeled Goat Anti-Rabbit IgG (H+L) (1:1,000; cat. no. A0208;
Beyotime Institute of Biotechnology) at room temperature for 2 h.
Subsequently, enhanced chemiluminescence reagents (MilliporeSigma)
were employed to assess protein expression and protein expression
was normalized to that of GAPDH. The blots were quantified using
Evolution (version 18.0.8.0, Viber Lourmat).
Statistical analysis
The R software (version 3.5.1) with corresponding
packages and GraphPad Prism 7 (GraphPad Software, Inc.) were used
for statistical analyses. Unless specified otherwise above,
P<0.05 was considered to indicate a statistically significant
difference. The PERL programming language (version 5.30.2;
http://www.perl.org) was employed to process
data. The Kaplan-Meier method and log rank test were performed to
compare the OS between the high- and low-risk groups. The results
of analyses were presented as the mean ± standard deviation. The
Student's t-test was utilized to analyze the differences between
two groups according to the specific. Dunnett's test was the post
hoc test used following one-way ANOVA to compare three or more
groups.
Results
Construction of the
hypoxia-related-lncRNA signature
RNA expression data from 471 patients with melanoma
and 1 normal adjacent tissue were extracted, and two
hypoxia-related gene sets (KRIEG and WINTER) were selected to
identify hypoxia-related oncogenes. A total of 882 hypoxia-related
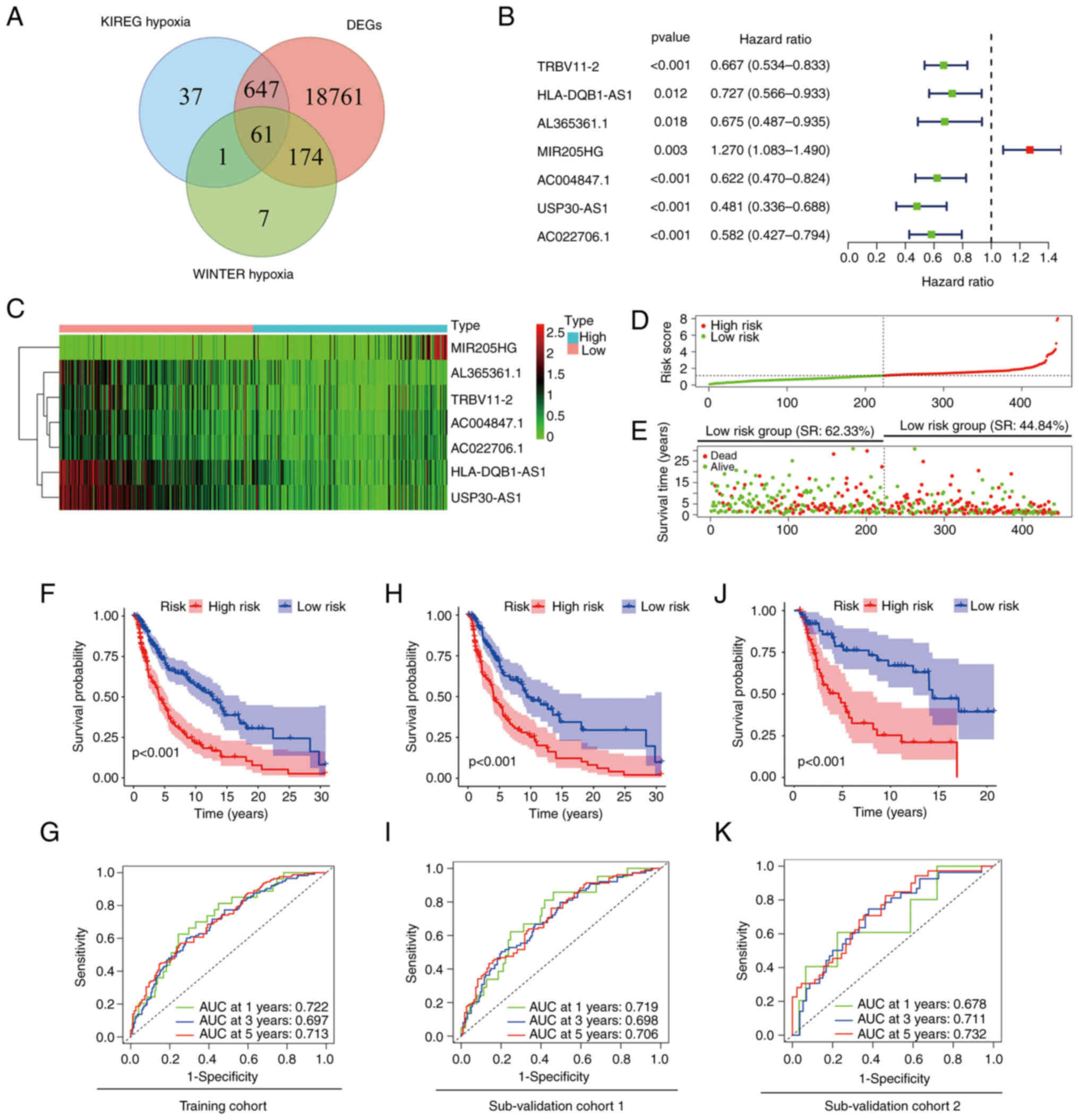
mRNAs were acquired (Fig. 1A).
Using Pearson's correlation analysis between the lncRNA samples
from 471 patients with melanoma and hypoxia-related genes, 72
hypoxia-related lncRNAs were identified. The selection criteria
were |R2|>0.7 and P<0.001. Univariate Cox
regression analysis was then performed to analyse the 72 lncRNAs.
The survival data revealed that 23 hypoxia-related-lncRNAs were
independent prognostic factors in patients with melanoma. To
eliminate the mutual influence among lncRNAs, multivariate Cox
regression analysis was used (Fig.
1B). Finally, a hypoxia-related-lncRNA signature that included
7 lncRNAs was constructed, with one unfavourable (MIR205HG) and six
favourable [T cell receptor β variable 11-2 (TRBV11-2), HLA-DQB1
antisense RNA 1 (HLA-DQB1-AS1), AL365361.1, AC004847.1, ubiquitin
specific peptidase 30 (USP30) antisense RNA 1 (USP30-AS1) and
AC022706.1] lncRNAs as prognostic factors for melanoma (Fig. 1C; Table II).
 | Table II.Hypoxia-related long non-coding RNAs
associated with the overall survival of patients with melanoma. |
Table II.
Hypoxia-related long non-coding RNAs
associated with the overall survival of patients with melanoma.
| Gene symbol | Aliases | Ensemble ID | Location | P-value | HR |
|---|
| TRBV11-2 | Lnc-MGAM2-1 |
ENSG00000241657 |
chr7:142,433,895-142,434,394 | 0.000361432 | 1.330058039 |
| HLA-DQB1-AS1 | HSALNG0049432 |
ENSG00000223534 |
chr6:32,659,879-32,660,729 | 0.012388052 | 0.726654702 |
| AL365361.1 | lnc-KCNA3-3 |
ENSG00000259834 |
chr1:110653560-110657040 | 0.018247458 | 0.675041646 |
| MIR205HG | NPC-A-5 |
ENSG00000230937 |
chr1:209,425,755-209,433,438 | 0.003259287 | 1.270371188 |
| AC004847.1 | Lnc-MYO1G-1 |
ENSG00000260997 |
chr7:44,958,998-44,960,909 | 0.032147891 | 1.76373733 |
| USP30-AS1 | HSALNG0093872 |
ENSG00000256262 |
chr12:109,051,790-109,054,033 | 0.0000617 | 0.481152446 |
| AC022706.1 | Lnc-SLFN5-1 |
ENSG00000267364 |
chr17:33,640,521-33,651,919 | 0.000622238 | 1.508349735 |
Using the present 7-hypoxia-related-lncRNA
signature, patients with melanoma were classified into two groups
based on their risk scores (Fig.
1D). The OS time and survival rate were longer and higher,
respectively, in the low-risk group compared with the high-risk
group (survival rate, 62.33% in the low-risk group vs. 44.84% in
the high-risk group; Fig. 1E),
which was also demonstrated in survival analysis (P<0.001;
Fig. 1F). The diagnostic efficacy
of the signature was estimated using time-dependent ROC curves,
wherein the area under the curve (AUC) was >0.7 for both 1 and 5
years (Fig. 1G). To validate the
reliability and stability of the present risk model, the training
cohort was divided into two sub-validation cohorts (n=314 in
sub-validation cohort 1 and n=132 in sub-validation cohort 2) at
random. Kaplan-Meier survival analysis revealed a worse prognosis
in high-risk group patients in both cohorts (P<0.001 and
P<0.001; Fig. 1H and J).
Time-dependent ROC curves showed promising accuracy of the present
risk model, particularly at 5 years in both sub-validation cohort 1
(AUC=0.706) and sub-validation cohort 2 (AUC=0.732) (Fig. 1I and K). These results suggested
that the present signature could predict the survival period of
patients with melanoma.
Association of the
hypoxia-related-lncRNA signature with the clinicopathological
features of melanoma
The association between the risk scores and
clinicopathological features was assessed. The results suggested
that the 7-hypoxia-related-lncRNA signature was closely related to
melanoma progression. Specifically, patients with American Joint
Committee on Cancer (AJCC) stage III–IV, T stage 3–4 and N stage ≥1
had higher risk scores than patients with AJCC stage I–II
(P=0.0246), T stage 0–2 (P<0.001) and N stage <1 (P=0.0104)
(Fig. 2C, E and F). However,
differences in age, sex and M stage were not observed (P>0.05;
Fig. 2A, B and D). The results
indicated that the present 7-hypoxia-related-lncRNA signature was
closely related to the progression of melanoma.
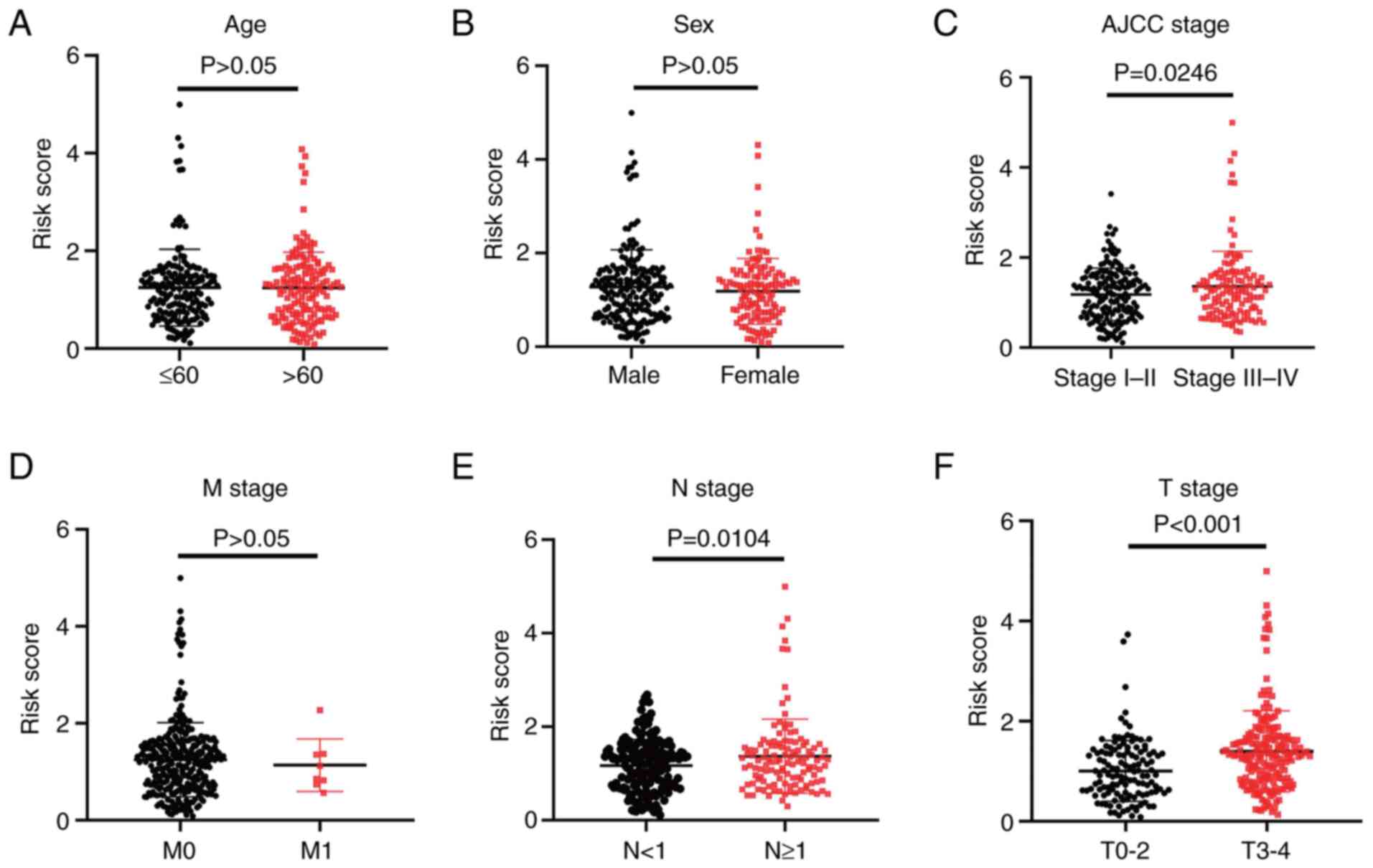 | Figure 2.Association of the
7-hypoxia-related-long non-coding RNA signature with
clinicopathological features in melanoma. Association between the
signature risk scores and the clinicopathological features,
including (A) age (>65 vs. ≤65; unpaired Student's t-test,
P=0.9469), (B) sex (female vs. male; unpaired Student's t-test,
P=0.2866), (C) American Joint Committee on Cancer stage (stage I–II
vs. stage III–IV; unpaired Student's t-test, P=0.0246), (D) M stage
(stage 0 vs. stage 1; unpaired Student's t-test, P=0.6898), (E) N
stage (stage <1 vs. stage ≥1; unpaired Student's t-test,
P=0.0104) and (F) T stage (stage 0–2 vs. stage 3–4; unpaired
Student's t-test, P<0.001). |
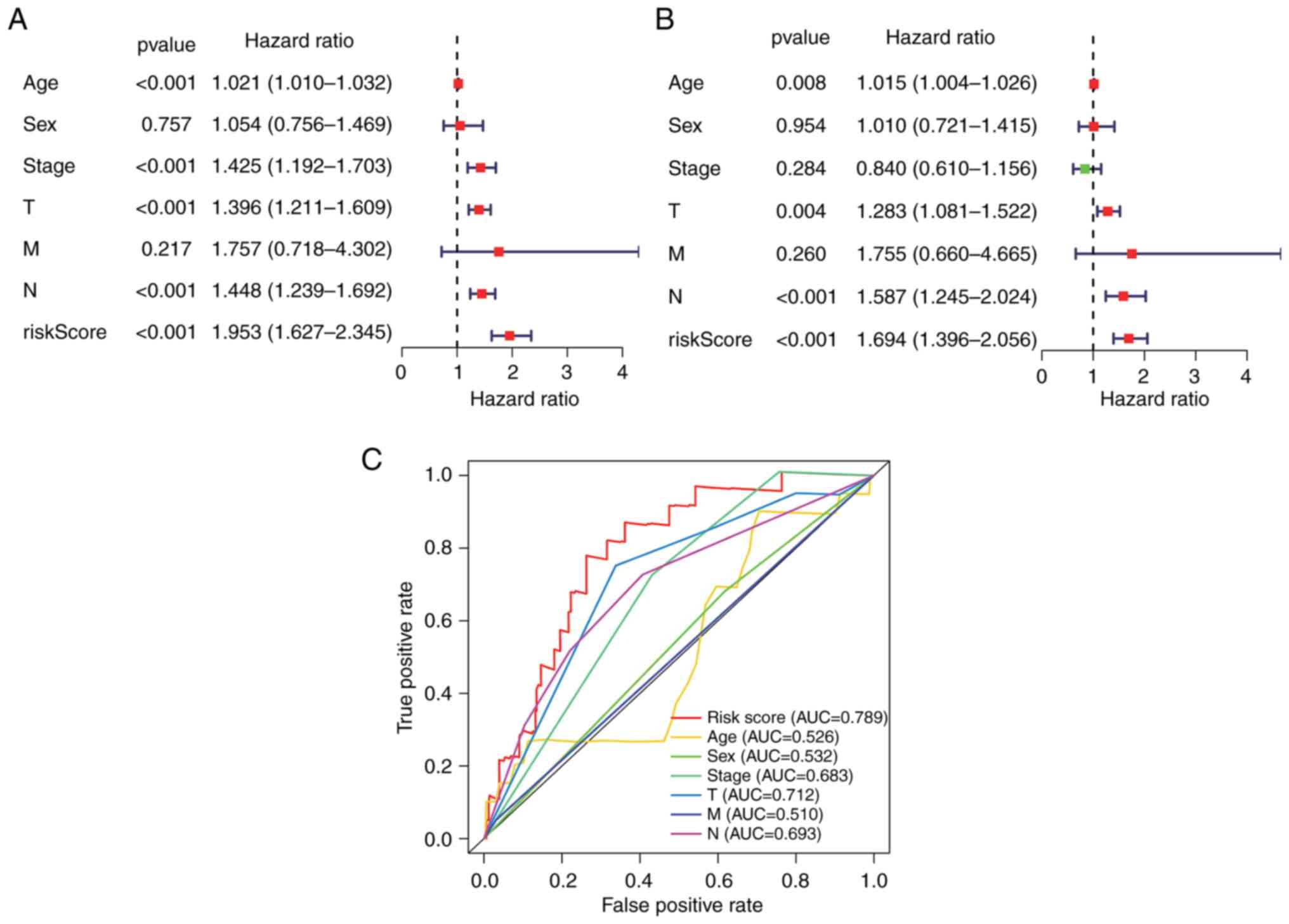
Independent risk characteristics for
the hypoxia-related-lncRNA signature
The prognostic ability of the
7-hypoxia-related-lncRNA signature in patients with melanoma was
assessed via univariate and multivariate Cox regression analyses.
The results demonstrated that the risk score of the present
signature [HR, 1.694; 95% confidence interval (95% CI),
1.396-2.056; P<0.001], age (HR, 1.015; 95% CI, 1.004-1.026;
P=0.008), T stage (HR, 1.283; 95% CI, 1.081-1.522; P=0.004) and N
stage (HR, 1.587; 95% CI, 1.245-2.024; P<0.001) were
significantly related to the OS of patients with melanoma as
independent prognostic indicators (Fig. 3A and B). The time-dependent ROC
curve also revealed a 0.789 AUC value of the risk score, indicating
appropriate sensitivity and specificity of the present
7-hypoxia-related-lncRNA signature in predicting the survival of
patients with melanoma compared with other clinicopathological
parameters (Fig. 3C).
Stratification analysis of clinical
features using the hypoxia-related-lncRNA signature
A stratified analysis of patients with melanoma
based on clinical features, including sex, age, AJCC stage and TNM
stages, was performed. The Kaplan-Meier survival curve analysis
illustrated that patients with melanoma had higher
hypoxia-related-lncRNA signature-predicted risk scores and a
shorter OS period compared with the low-risk groups for most
clinical and clinicopathological features (Fig. S1A-K); however, the patients at
the M1 stage (P=0.959) were an exception, probably due to the small
sample size (Fig. S1L). These
results indicated that the present 7-hypoxia-related-lncRNA
signature could accurately predict the survival period of patients
with melanoma.
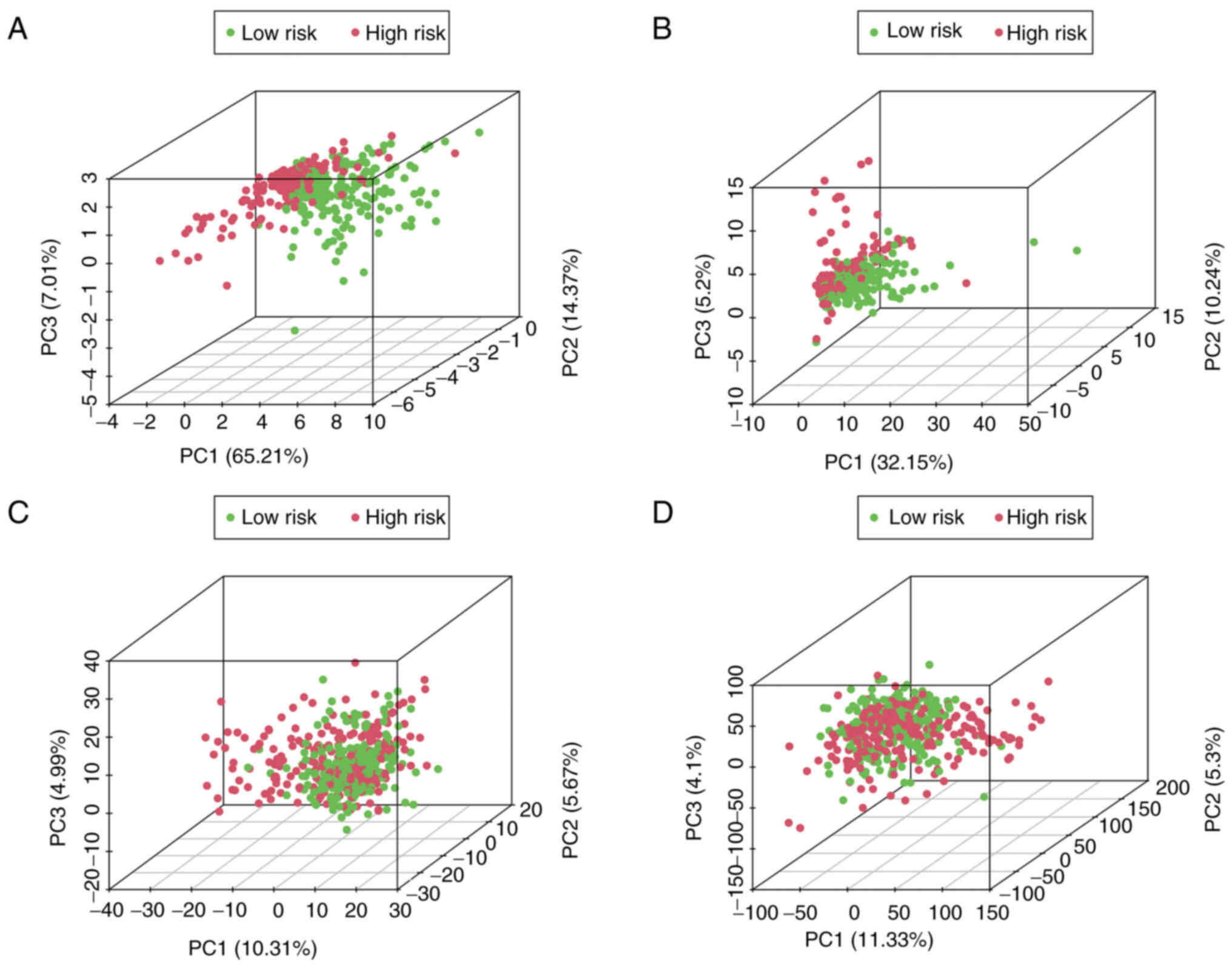
PCA using the hypoxia-related-lncRNA
signatures
PCA was employed to detect the different
distribution patterns between the low- and high-risk groups of
patients with melanoma. The 7-hypoxia-related-lncRNA gene set
(Fig. 4A) was used to create a
low- and high-risk group. A significant separation of the risk
score based on the hypoxia-related lncRNA gene set (Fig. 4B), hypoxia-related gene set
(Fig. 4C) and all gene sets
(Fig. 4D) was not observed,
indicating that, compared with other distribution patterns, the
present 7-hypoxia-related-lncRNA signature could accurately divide
patients with melanoma into risk groups.
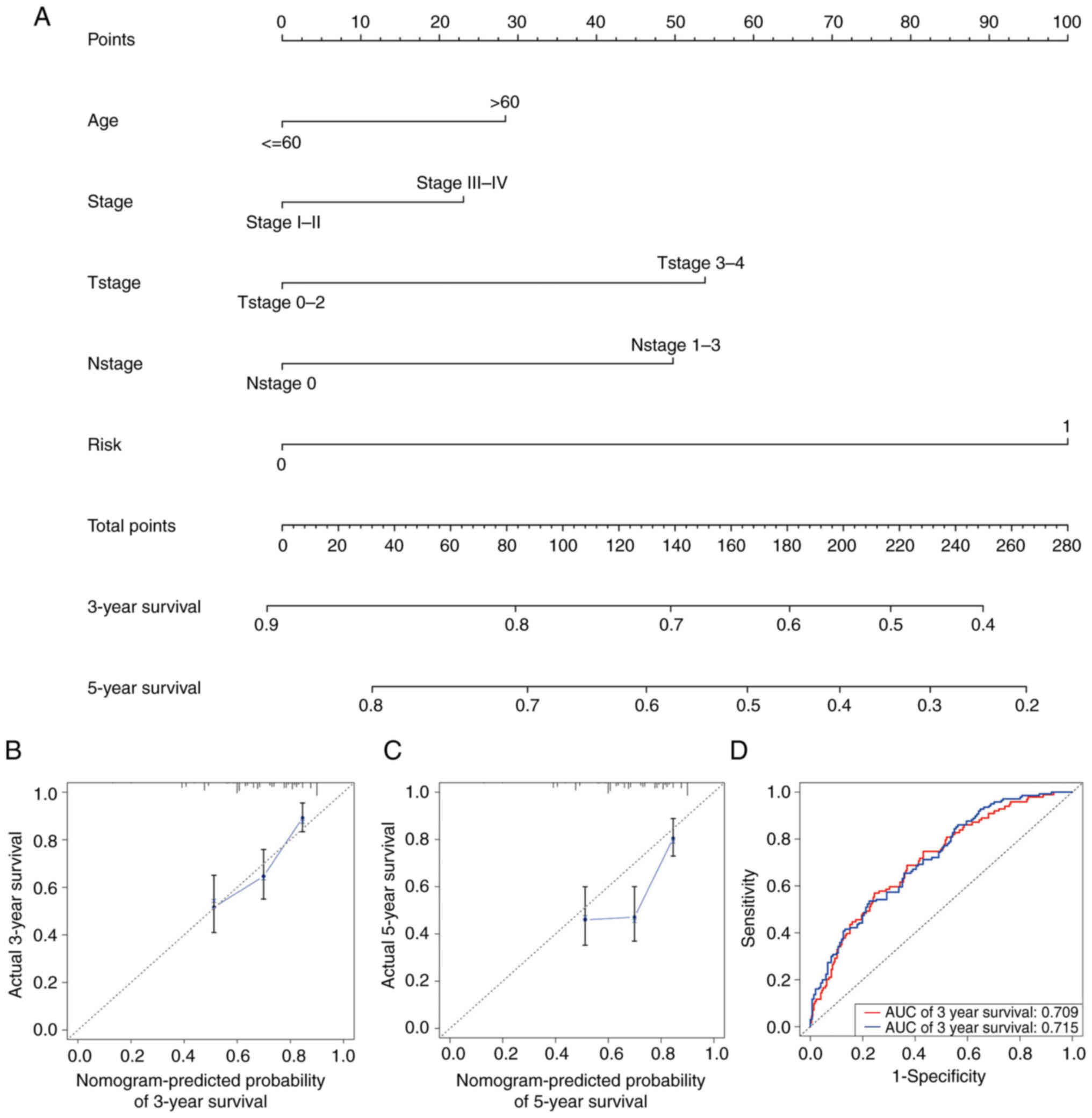
Establishment and validation of the
nomogram
To diagnose or predict the onset or progression of
melanoma, a nomogram was established based on the risk score and
other clinical features, including age, AJCC stage, T stage and N
stage (Fig. 5A). The calibration
plots satisfactorily predicted the 3-year OS rate compared with the
ideal model (Fig. 5B) but
unsatisfactorily predicted the 5-year OS rate (Fig. 5C). Time-dependent ROC curves
revealed that the AUC values of the nomogram at 3 and 5 years were
0.709 and 0.715, respectively (Fig.
5D).
Construction of the ceRNA network and
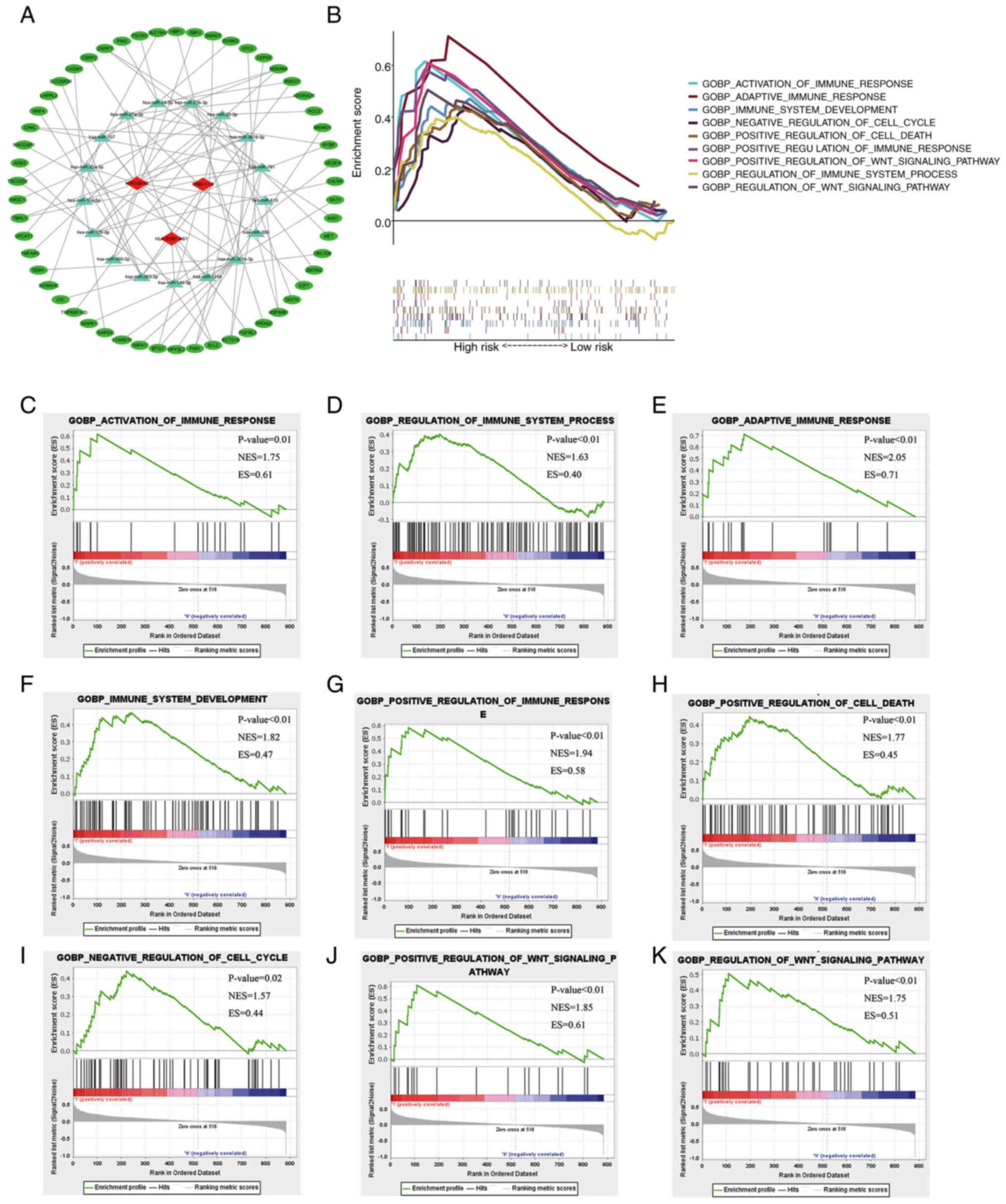
GSEA
To explore the mechanism of the present
7-hypoxia-related-lncRNA signature, the 7 hypoxia-related lncRNAs
and their corresponding miRNAs were first predicted using miRcode.
The results indicated that 92 miRNAs could interact with lncRNA
MIR205HG, lncRNA TRBV11-2 and lncRNA HLA-DQB1-AS1 (Table SIII). Additionally, the
corresponding miRNA target proteins were predicted using three
databases (TargetScan, miRDB and mieTarBase). Via comparison with
the obtained hypoxia-related mRNAs, it was determined that the
present signature targeted 17 miRNAs and 54 mRNAs, and a
co-expression RNA network was constructed (Fig. 6A). To explore the relationship
between the expression of the 7-hypoxia-related-lncRNA signature
and melanoma classification, including biological processes
involved in progression, GSEA was employed to identify key Gene
Ontology terms and pathway enrichment analysis of high-risk score
patients with melanoma in the GO-BP gene sets. Genes associated
with a high-risk score were mainly enriched in the activation of
immune response [normalized enrichment score (NES)=1.75; P=0.012],
regulation of immune system process (NES=1.63; P=0.002), adaptive
immune response (NES=2.05; P<0.001), development of immune
system (NES=1.82; P<0.001), positive regulation of immune
response (NES=1.94; P<0.001), positive regulation of cell death
(NES=1.77; P=0.002), negative regulation of cell cycle (NES=1.57;
P=0.028), positive regulation of Wnt signalling pathway (NES=1.85;
P<0.001) and regulation of Wnt signalling pathway (NES=1.75;
P=0.004) (Fig. 6B-K).
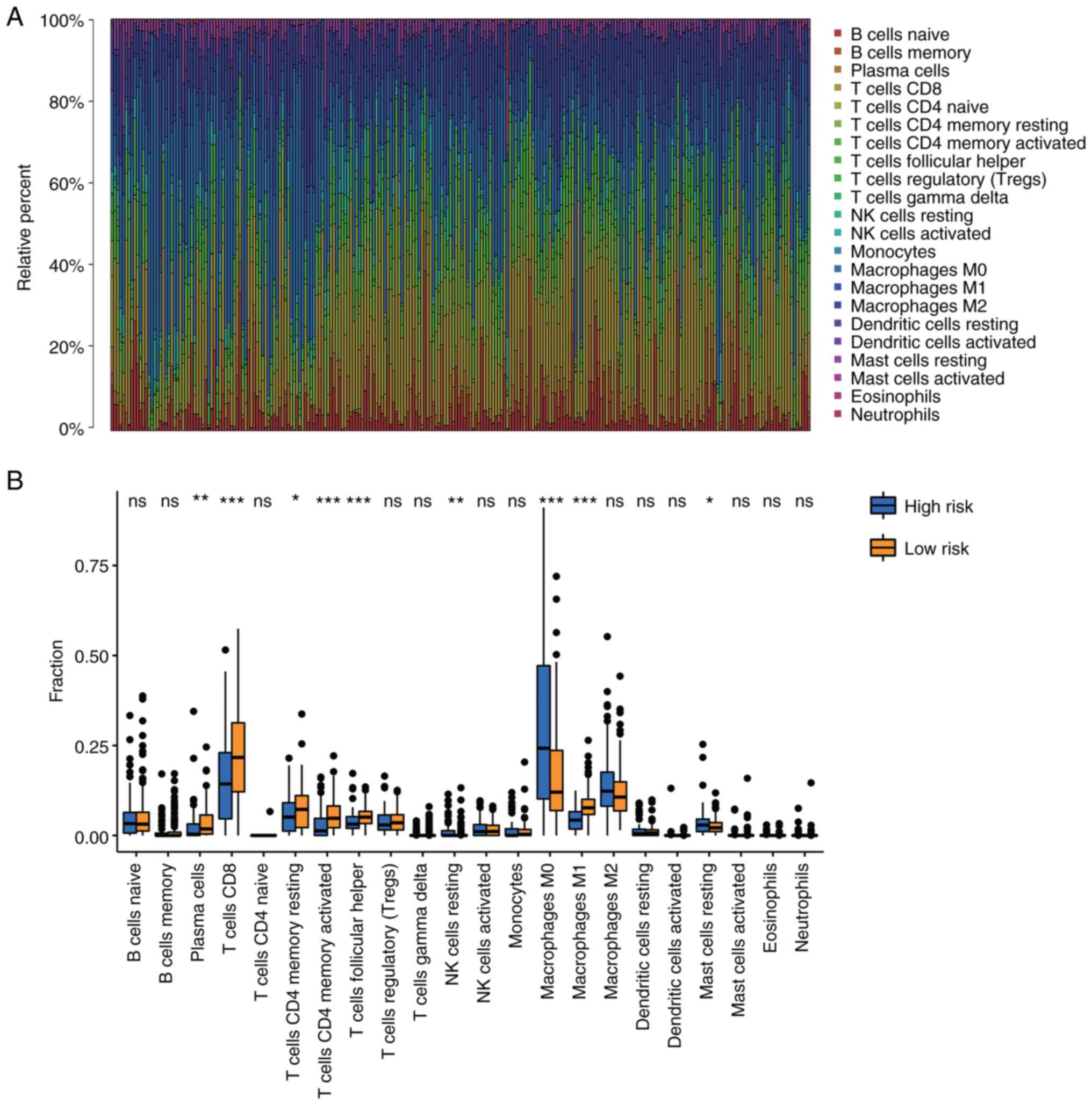
Regulation of TIICs by the
7-hypoxia-related lncRNAs
Based on the GSEA results in Fig. 6C-G, the present study explored the
association between immune cell infiltration and the risk score.
The CIBERSORT algorithm was used to validate the association of the
hypoxia-related lncRNAs with TIICs in patients with melanoma. The
proportion of TIICs in each patient with melanoma was investigated
using the CIBERSORT algorithm (Fig.
7A). A comparison of the TIIC levels between the high- and
low-risk groups showed an increased level of M0 macrophages
(P<0.001) and resting mast cells (P=0.046) in the high-risk
group, and a decreased number of plasma cells (P=0.002),
CD8+ T cells (P<0.001), resting memory
CD4+ T cells (P=0.036), activated memory CD4+
T cells (P<0.001), follicular helper T cells (P<0.001),
resting natural killer (NK) cells (P=0.007) and M1 macrophages
(P<0.001) in the high-risk group (Fig. 7B). These results revealed the
important role of the hypoxia-related lncRNAs in regulating
TIICs.
MIR205HG silencing inhibits melanoma
cell proliferation via the canonical Wnt/β-catenin signalling
pathway
The expression levels of human leukocyte antigen
(HLA), MIR205HG-1 and TRBV11 in melanoma and paired adjacent
tissues of 20 clinical samples were detected using RT-qPCR. The
expression levels of HLA and TRBV11 were elevated, whereas the
expression levels of MIR205HG-1 were decreased in the normal
tissues (Fig. 8A-C). These
results suggested the specificity of siRNA to MIR205HG. To
investigate the role of MIR205HG in melanoma cells, A375 cells were
transfected with two sequences of siRNAs that target MIR205HG and
negative control (NC) siRNA. As shown in Fig. 8D, the knockout efficiency was
detected using RT-qPCR. Then si-MIR205HG-1 was selected for further
experimentation since it significantly inhibited the expression of
MIR205HG in A375 cells compared with the NC group. The results of
cell cycle distribution analysis using flow cytometry indicated
that the percentage of cells in the S-phase was greater in the
siMIR205HG-1 group than in the NC group in A375 cells (Fig. 8E). Additionally, detection of cell
viability using a CCK-8 assay also demonstrated the proliferation
of A375 cells, which was inhibited to a greater extent compared
with the NC group cells after si-MIR205HG-1 transfection (Fig. 8F). Based on the aforementioned
GSEA results, the potential relationship between the MIR205HG and
the Wnt/signaling pathway was investigated. Subsequently, the
expression levels of relative proteins were detected by western
blotting (Fig. 8G). The results
revealed that the expression levels of p-β-catenin and p-GSK3-β in
A375 cells were increased after siMIR205HG-1 transfection, while
the expression of c-Myc and β-catenin was decreased. The ratio of
phosphorylated protein to total protein (p/t, phosphorylated/total)
was calculated. The results demonstrated that phosphorylation
modification on GSK3-β was enhanced (p/t, NC. vs. si-MIR205HG-1,
0.8782 vs. 1.219, P=0.014) and the same trend was also observed in
β-catenin (p/t, NC. vs. si-MIR205HG-1, 0.4397 vs. 1.262, P=0.002).
Therefore, these results indicated that silencing MIR205HG could
partly inhibit the proliferation abilities of melanoma cells
through the Wnt/β-catenin signalling pathway.
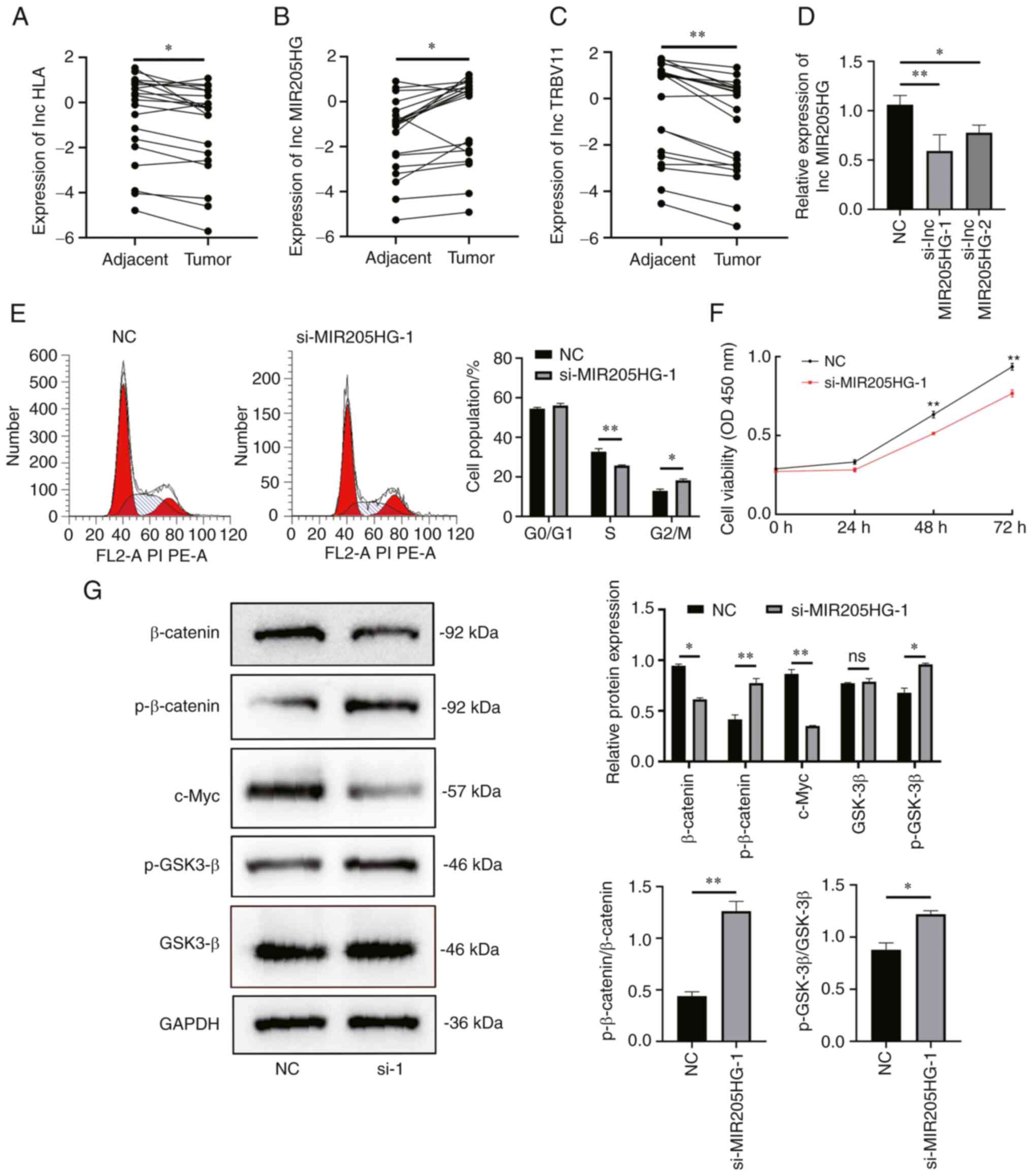 | Figure 8.Silencing of MIR205HG inhibits
proliferation of melanoma via the canonical Wnt/β-catenin signaling
pathway. (A-C) RT-qPCR to detect the expression levels of
HLA-DQB1-AS1, MIR205HG-1 and TRBV11-2 in melanoma or paired tissues
from 20 patients (Paired Student's t-test; *P<0.05 and
**P<0.01). (D) RT-qPCR to detect the relative silencing levels
of lncMIR205HG (Data are presented as the mean ± SD; n=3; one-way
ANOVA and Dunnett's test; *P<0.05 and **P<0.01). (E) Flow
cytometric analysis of the cell cycle distribution in A375 cells
after transfection with NC or MIR205HG siRNA. The percentages of
cells in G0/G1, S and G2/M phase
are shown in the bar graph (n=3; unpaired Student's t-test;
*P<0.05 and **P<0.01). (F) Proliferation of A375 cells
transfected with NC or si-MIR205HG-1 was assessed using a Cell
Counting Kit-8 assay (n=3; one-way ANOVA and Dunnett's test;
**P<0.01). (G) β-catenin, p-β-catenin, c-Myc, GSK3-β and
p-GSK3-β levels in A375 cells transfected with siRNAs determined by
western blotting, ratio of phosphorylated protein to total protein
(p/t, phosphorylated/total) about GSK3-β and β-catenin were
calculated (n=3; unpaired Student's t-test; *P<0.05 and
**P<0.01). MIR205HG, MIR205 host gene; NC, negative control; p-,
phosphorylated; RT-qPCR, reverse transcription-quantitative PCR;
siRNA/si, small interfering RNA; ns, non-significant. |
Discussion
The incidence of melanoma has increased rapidly over
the last 50 years worldwide (20). Surgical excision and molecular
targeted drugs, which improve the OS and progression-free survival
of patients, are the main treatment options for cutaneous melanoma
(21). Insufficient understanding
of melanoma molecular markers poses a hindrance to melanoma
diagnosis and treatment. Therefore, identifying sensitive and
specific biomarkers and understanding the mechanisms underlying
melanoma development are required to improve the survival rate of
patients with melanoma.
Functioning differently in comparison with protein
coding sequences, lncRNAs serve an important role in regulating
gene expression, which affects chromatin modification, as well as
RNA splicing and protein activity. Emerging evidence suggests that
lncRNAs could have an effect on the occurrence, development,
prognosis and chemotherapy resistance of tumour cells. Recent
studies have reported that lncRNAs, as novel biomarkers, could be
used to predict cancer prognosis (22,23). lncRNAs have been reported to be
involved in melanoma development by regulating the hypoxia pathway
(23,24). Due to consistent overexpression of
HOX transcript antisense RNA (HOTAIR), the incidence of
lymph node metastasis has been increased compared with primary
lesions. However, HOTAIR knockdown suppresses the motility
and invasion of melanoma cells in vitro (25). Aberrant metastasis associated lung
adenocarcinoma transcript 1 (MALAT1) upregulation promotes
melanoma metastasis by stimulating cell migration and
de-suppressing miR-140-mediated snail family transcriptional
repressor 2 and ADAM metallopeptidase domain 10 inhibition,
suggesting that MALAT1 serves as an oncogenic lncRNA in the
development of melanoma (26).
BRAF-activated non-protein coding RNA knockout markedly reduces the
proliferation of melanoma cells via the MAPK signalling pathway
(27), which may act as a
prognostic predictor and possible drug target. FGD5 antisense RNA 1
could promote the prognosis in patients with melanoma (28). Furthermore, several lncRNAs, such
as antisense RNA in the INK4 locus, SPRY-IT1, Llme23, urothelial
cancer associated 1, SRA-like non-coding RNA and survival
associated mitochondrial melanoma specific oncogenic non-coding
RNA, have been demonstrated to be differentially expressed in
patients with melanoma, and to serve a role as potent melanoma
progression and metastasis regulators (29). Although recent studies have
illustrated the mechanism of melanoma development, the involvement
of regulatory lncRNAs in melanoma prognosis under hypoxic
conditions remains unclear.
LncRNAs can modify the regulatory effect of miRNAs
on mRNAs (30). Additionally,
miRNAs can regulate translation and stability of mRNAs by
controlling cellular processes, such as differentiation, apoptosis
and migration. In cancer, miRNAs serve an important role in
controlling metastasis, invasion and proliferation (31). Zhang et al (32) revealed that miR431 can silence
DAB2 interacting protein, which is a Ras GTPase activating protein
tumor suppressor, to activate Ras/Erk and promote metastasis of
pancreatic neuroendocrine tumors. Feng et al (33) revealed that the upregulation of
miR-409-3p and Kruppel like factor 17 could inhibit the invasion
and migration of gastric cancer cells. In the present study, three
lncRNAs from the 7-hypoxia-related-lncRNA signature were further
investigated, and these may serve essential roles in the prognosis
of patients with melanoma by interacting with the 17 miRNAs and 54
mRNAs predicted. But the potential role of these crucial miRNAs
that effect on the prognosis of melanoma remains unknown. Previous
studies proved that miRNAs could regulate the biological process of
cancer cells through Ras GTPase activity, pErk or EMT process
(34,35). In order to render the present
study more complete, close attention shall be addressed on how our
7 hypoxia-related-lncRNA affects Ras GTPase activity, as well as
the relationship between our signature and pErk and EMT
markers.
The present study was based on data downloaded from
TCGA database, which contains complete clinical and survival data
of patients with melanoma. In the present study, a
7-hypoxia-related-lncRNA signature was constructed, and this
included one unfavourable and six favourable lncRNAs as prognostic
factors for melanoma. Certain of the lncRNAs have been previously
reported to influence tumour prognosis (36). However, there remains a lack of
focus on hypoxia-related lncRNAs in melanoma. It has been reported
that abnormal HLA expression may influence cytotoxic T lymphocyte
and NK cell responses in uveal melanoma (37). According to a study by Serana
et al (38), TRBV
contributes to the promotion of the antitumour T-cell responses in
patients with melanoma. Due to its association with multiple
tumour-related pathways, MIR205HG-1 was also investigated,
particularly in epidermis development and the immune response
(39,40). The USP30-mediated stabilisation of
dynamin-related protein 1 has been reported to serve a critical
role in the development of hepatocellular carcinoma (41). As Lv et al (42) reported, a prognostic model based
on the 7 lncRNAs, including AL365361, could effectively predict
early recurrence after surgical resection of hepatocellular
carcinoma. AC022706.1 has also been investigated as a useful
biomarker for promoting antitumour immunity and the immunotherapy
response in gastric cancers (43). In the present study, a signature
of prognosis-related lncRNAs in patients with melanoma was
successfully constructed, which was demonstrated to be associated
with melanoma progression. The internal validation also
demonstrated the accuracy of the present signature. Univariate and
multivariate Cox regression analyses demonstrated that the present
signature could act as an independent predictor for prognosis
prediction of patients with melanoma. Time-dependent ROC curve
analysis suggested that the present risk model had the best
sensitivity and specificity compared with three other
clinicopathological parameters. A novel nomogram based on several
crucial clinical features and the present risk score was
successfully constructed and validated. These results suggested
that the present risk model could be an independent predictor of
the survival period in patients with melanoma.
Based on the prediction of different databases and
hypoxia-related mRNAs, a co-expression RNA network, including 17
miRNAs and 54 mRNAs, was constructed. Using GSEA, it was revealed
that the gene sets enriched in the high-risk group included the
activation of immune response, regulation of immune system process,
adaptive immune response, development of immune system, negative
regulation of cell cycle, positive regulation of cell death,
positive regulation of immune response, positive regulation of the
Wnt signalling pathway and regulation of the Wnt signalling
pathway. Immunotherapy is an important mean of systemic treatment
of melanoma, which could improve the prognosis of melanoma, and
thus, the present study investigated the relationship between the
present risk model and TIICs. The results demonstrated that M0
macrophages, resting mast cells, plasma cells, CD8+ T
cells, resting memory CD4+ T cells, activated memory
CD4+ T cells, follicular helper T cells, NK cells and M1
macrophages exhibited markedly different infiltration patterns in
melanoma. The present results suggested that the
7-hypoxia-related-lncRNA signature could partly reflect the immune
infiltration level of melanoma and provide valuable information for
immunotherapy.
To validate the prediction of the present signature,
the expression levels of HLA, MIR205HG and TRBV11 were detected in
20 clinical samples. The results demonstrated high expression
levels of MIR205HG and low expression levels of HLA and TRBV11 in
melanoma tissues. The increased MIR205HG expression in melanoma
tissues attracted our attention. To investigate the relationship
between poor prognosis and aberrant MIR205HG expression, siRNA was
used to silence its expression, resulting in suppressed
proliferation of melanoma A375 cells. These results suggested that
MIR205HG is a target that can promote melanoma proliferation. The
dysregulation of cell cycle control is an obstacle to the
suppression of melanoma growth (44). Agents targeting the
G1/S and G2/M checkpoints have shown
promising preclinical activity in patients with melanoma (45,46). Because si-MIR205HG-1 suppressed
proliferation in A375 cells, cell cycle arrest was investigated
using flow cytometry in si-MIR205HG-1 cells. Cell aggregation that
decreased at the S phase but increased at the G2/M phase
was observed after treatment with si-MIR205HG-1. These results
demonstrated that silencing MIR205HG may inhibit melanoma cell
proliferation at the G2/M phase, which could partly
explain the relationship between the present risk model based on
the expression of 7 hypoxia-related lncRNAs and poor prognosis of
patients with melanoma.
The aberrant activation of canonical Wnt/β-catenin
signalling is observed in multiple malignant tumours and is
recognised as an attractive therapeutic target for molecular drugs
(47). It has been reported that
Wnt serves a crucial role in tumorigenesis and malignancy, which
contributes to tumour proliferation and differentiation, survival,
stress response and resistance (48). To elucidate the molecular
mechanisms of the effect of si-MIR205HG-1 on cell proliferation,
regulatory proteins of the Wnt/β-catenin signalling pathway were
examined in A375 cells using western blot analysis. Increased
p-GSK3-β levels in A375 cells decreased β-catenin and c-Myc
expression, resulting in low si-MIR205HG-1 expression. These
results demonstrated that the silencing of MIR205HG-1 inhibited the
proliferation of melanoma via the canonical Wnt/β-catenin
signalling pathway. LncRNA can influence tumor cell migration and
invasion. Chen et al (16)
revealed that NORAD may play critical roles in tumorigenesis and
progression of malignant melanoma by regulating of the MIR205-EGLN2
pathway. Xu et al (49)
revealed that lncRNA CRNDE can promote the migration and invasion
of melanoma by sponging MIR205 and releasing CCL18. And in the
present study, it was primarily identified that the overexpression
of MIR205HG could inhibit the proliferation of melanoma cells
through the canonical Wnt/β-catenin signalling pathway. In order to
render the present study more thorough, it will be investigated in
future studies how the dose of our 7 hypoxia-related-lncRNA
influences melanoma by cell migration and invasion.
The present study has several limitations. The
present prognostic model should be further validated using a larger
clinical sample size with adequate follow-up duration. In addition,
the underlying mechanisms of how these 7 lncRNAs or some of them
influence the proliferation of melanoma should be more
comprehensively investigated. The detailed mechanisms of how
MIR205HG inhibits the proliferation of melanoma cells via the
canonical Wnt/β-catenin signalling pathway require more exploration
and validation. Furthermore, the present study lacks animal
experiments, which will be verified in further exploration.
At present, there were kinds of lncRNA signatures
used to predict the prognosis of melanoma. Although hypoxia is an
important circumstance for melanoma cells surviving, the study
focused on hypoxia-related-lncRNA signature has not been reported
yet. Moreover, MIR205HG played an important role in influencing the
pluripotency, proliferation (50), and dissemination of malignant
melanoma (51). Nevertheless, few
of them paid attention to investigate and verify that MIR205HG
could be a prognostic biomarker and an actionable target for
inhibiting proliferation of melanoma.
In conclusion, a novel 7-hypoxia-related-lncRNA
signature was preliminarily constructed and validated to promote
the prognostic prediction of patients with melanoma. Based on this
risk model, a nomogram and ceRNA network were successfully
constructed to provide a new sight of investigating the underlying
molecular mechanism of melanoma. Among these lncRNAs, supressing
the overexpression of MIR205HG inhibited the proliferation of
melanoma cells via the canonical Wnt/β-catenin signalling pathway,
which could partly explain the relationship between the present
risk model and poor prognosis of patients with melanoma.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL and AJC designed and implemented experiments. THL
and HGZ collected data and performed statistical analysis. YL and
THL helped perform the analysis with constructive discussions and
revised the manuscript carefully. All authors read and approved the
final manuscript. YL and AJC confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
This article meets the requirements of TCGA and
other websites for publication and does not contain any studies
involving animals performed by any of the authors. The study was
approved (approval no. 2021-487) by the Ethics Committee of the
First Affiliated Hospital of Chongqing Medical University. All
individuals provided written informed consent for the use of their
samples for clinical research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Leonardi GC, Falzone L, Salemi R, Zanghì
A, Spandidos DA, Mccubrey JA, Candido S and Libra M: Cutaneous
melanoma: From pathogenesis to therapy (Review). Int J Oncol.
52:1071–1080. 2018.PubMed/NCBI
|
|
2
|
Davey MG, Miller N and McInerney NM: A
review of epidemiology and cancer biology of malignant melanoma.
Cureus. 13:e150872021.PubMed/NCBI
|
|
3
|
Carr S, Smith C and Wernberg J:
Epidemiology and risk factors of melanoma. Surg Clin North Am.
100:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu X and Kang Y: Hypoxia and
hypoxia-inducible factors: Master regulators of metastasis. Clin
Cancer Res. 16:5928–5935. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Y, Ciotti GE and Eisinger-Mathason
TSK: Hypoxia and the tumor secretome. Adv Exp Med Biol. 1136:57–69.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Graham K and Unger E: Overcoming tumor
hypoxia as a barrier to radiotherapy, chemotherapy and
immunotherapy in cancer treatment. Int J Nanomedicine.
13:6049–6058. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jing X, Yang F, Shao C, Wei K, Xie M, Shen
H and Shu Y: Role of hypoxia in cancer therapy by regulating the
tumor microenvironment. Mol Cancer. 18:1572019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiu H, Chen F and Chen M: MicroRNA-138
negatively regulates the hypoxia-inducible factor 1α to suppress
melanoma growth and metastasis. Biol Open. 8:bio0429372019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong L, You S, Zhang Q, Osuka S, Devi NS,
Kaluz S, Ferguson JH, Yang H, Chen G, Wang B, et al:
Arylsulfonamide 64B inhibits hypoxia/HIF-induced expression of
c-Met and CXCR4 and reduces primary tumor growth and metastasis of
uveal melanoma. Clin Cancer Res. 25:2206–2218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu M, Zhong J, Zeng Z, Huang K, Ye Z,
Deng S, Chen H, Xu F, Li Q and Zhao G: Hypoxia-induced feedback of
HIF-1α and lncRNA-CF129 contributes to pancreatic cancer
progression through stabilization of p53 protein. Theranostics.
9:4795–4810. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Piao HY, Liu Y, Kang Y, Wang Y, Meng XY,
Yang D and Zhang J: Hypoxia associated lncRNA HYPAL promotes
proliferation of gastric cancer as ceRNA by sponging miR-431-5p to
upregulate CDK14. Gastric Cancer. 25:44–63. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Z, Wang Y, Deng J, Liu D, Zhang L, Shao
H, Wang Z, Zhu W, Zhao C and Ke Q: Long non-coding RNA COL4A2-AS1
facilitates cell proliferation and glycolysis of colorectal cancer
cells via miR-20b-5p/hypoxia inducible factor 1 alpha subunit axis.
Bioengineered. 12:6251–6263. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, He D, Xiao M, Zhu Y, Zhou J and Cao
K: Long noncoding RNA LINC00518 induces radioresistance by
regulating glycolysis through an miR-33a-3p/HIF-1α negative
feedback loop in melanoma. Cell Death Dis. 12:2452021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Cao K, Li J, Wang A, Sun L, Tang
J, Xiong W, Zhou X, Chen X, Zhou J and Liu Y: Overexpression of
long non-coding RNA NORAD promotes invasion and migration in
malignant melanoma via regulating the MIR-205-EGLN2 pathway. Cancer
Med. 8:1744–1754. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu L, Liu G, He YW, Chen R and Wu ZY:
Identification of a pyroptosis-associated long non-coding RNA
signature for predicting the immune status and prognosis in skin
cutaneous melanoma. Eur Rev Med Pharmacol Sci. 25:5597–5609.
2021.PubMed/NCBI
|
|
18
|
Chen H, Pan Y, Jin X and Chen G:
Identification of a four hypoxia-associated long non-coding RNA
signature and establishment of a nomogram predicting prognosis of
clear cell renal cell carcinoma. Front Oncol. 11:7133462021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Raimondi S, Suppa M and Gandini S:
Melanoma epidemiology and sun exposure. Acta Derm Venereol.
100:adv001362020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pavri SN, Clune J, Ariyan S and Narayan D:
Narayan, malignant melanoma: Beyond the basics. Plast Reconstr
Surg. 138:330e–340e. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Qin C, Liu HW, Guo X and Gan H:
An effective hypoxia-related long non-coding RNAs assessment model
for prognosis of clear cell renal carcinoma. Front Oncol.
11:6167222021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong PJ, Shao YC, Huang SR, Zeng YF, Yuan
XN, Xu JJ, Yin WN, Wei L and Zhang JW: Hypoxia-associated
prognostic markers and competing endogenous RNA Co-expression
networks in breast cancer. Front Oncol. 10:5798682020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang J, Liu H, Zhang W, Li Y, Fan Z,
Jiang H and Luo J: Identification of lncRNA-mRNA regulatory module
to explore the pathogenesis and prognosis of melanoma. Front Cell
Dev Biol. 8:6156712020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun L, Sun P, Zhou QY, Gao X and Han Q:
Long noncoding RNA MALAT1 promotes uveal melanoma cell growth and
invasion by silencing of miR-140. Am J Transl Res. 8:3939–3946.
2016.PubMed/NCBI
|
|
27
|
Yu X, Zheng H, Chan MT and Wu WKK: BANCR:
A cancer-related long non-coding RNA. Am J Cancer Res. 7:1779–1787.
2017.PubMed/NCBI
|
|
28
|
Gao Y, Zhu H and Mao Q: Expression of
lncRNA FGD5-AS1 correlates with poor prognosis in melanoma
patients. J Gene Med. 22:e32782020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu X, Zheng H, Tse G, Chan MT and Wu WK:
Long non-coding RNAs in melanoma. Cell Prolif. 51:e124572018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karagkouni D, Karavangeli A,
Paraskevopoulou MD and Hatzigeorgiou AG: Characterizing
miRNA-lncRNA interplay. Methods Mol Biol. 2372:243–262. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lai X, Eberhardt M, Schmitz U and Vera J:
Systems biology-based investigation of cooperating microRNAs as
monotherapy or adjuvant therapy in cancer. Nucleic Acids Res.
47:7753–7766. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang T, Choi S, Zhang T, Chen Z, Chi Y,
Huang S, Xiang JZ and Du YN: miR-431 promotes metastasis of
pancreatic neuroendocrine tumors by targeting DAB2 interacting
protein, a ras GTPase activating protein tumor suppressor. Am J
Pathol. 190:689–701. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Feng J, Li K, Liu G, Feng Y, Shi H and
Zhang X: Precision hyperthermia-induced miRNA-409-3p upregulation
inhibits migration, invasion, and EMT of gastric cancer cells by
targeting KLF17. Biochem Biophys Res Commun. 549:113–119. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salminen A, Kaarniranta K, Kauppinen A,
Ojala J, Haapasalo A, Soininen H and Hiltunen M: Impaired autophagy
and APP processing in Alzheimer's disease: The potential role of
Beclin 1 interactome. Prog Neurobiol. 106-107:33–54. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Su J, Morgani SM, David CJ, Wang Q, Er EE,
Huang YH, Basnet H, Zou Y, Shu W, Soni RK, et al: TGF-β
orchestrates fibrogenic and developmental EMTs via the RAS effector
RREB1. Nature. 577:566–571. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang B, Tang B, Gao J, Li J, Kong L and
Qin L: A hypoxia-related signature for clinically predicting
diagnosis, prognosis and immune microenvironment of hepatocellular
carcinoma patients. J Transl Med. 18:3422020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Souri Z, Wierenga APA, Mulder A, Jochemsen
AG and Jager MJ: HLA expression in uveal melanoma: An indicator of
malignancy and a modifiable immunological target. Cancers (Basel).
11:11322019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Serana F, Sottini A, Caimi L, Palermo B,
Natali PG, Nisticò P and Imberti L: Identification of a public CDR3
motif and a biased utilization of T-cell receptor V beta and J beta
chains in HLA-A2/Melan-A-specific T-cell clonotypes of melanoma
patients. J Transl Med. 7:212009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo J, Gan Q, Gan C, Zhang X, Ma X and
Dong M: LncRNA MIR205HG regulates melanomagenesis via the
miR-299-3p/VEGFA axis. Aging (Albany NY). 13:5297–5311. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu N, Liu Z, Liu X and Chen H:
Comprehensive analysis of a competing endogenous RNA network
identifies seven-lncRNA signature as a prognostic biomarker for
melanoma. Front Oncol. 9:9352019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gu L, Zhu Y, Lin X and Li Y, Cui K,
Prochownik EV and Li Y: Amplification of glyceronephosphate
O-acyltransferase and recruitment of USP30 stabilize DRP1 to
promote hepatocarcinogenesis. Cancer Res. 78:5808–5819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lv Y, Wei W, Huang Z, Chen Z, Fang Y, Pan
L, Han X and Xu Z: Long non-coding RNA expression profile can
predict early recurrence in hepatocellular carcinoma after curative
resection. Hepatol Res. 48:1140–1148. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He Y and Wang X: Identification of
molecular features correlating with tumor immunity in gastric
cancer by multi-omics data analysis. Ann Transl Med. 8:10502020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu W and McArthur G: Cell cycle regulation
and melanoma. Curr Oncol Rep. 18:342016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Barnaba N and LaRocque JR: Targeting cell
cycle regulation via the G2-M checkpoint for synthetic lethality in
melanoma. Cell Cycle. 20:1041–1051. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Afrang N and Honardoost M: Cell cycle
regulatory markers in melanoma: New strategies in diagnosis and
treatment. Med J Islam Repub Iran. 33:962019.PubMed/NCBI
|
|
47
|
Nishiya N: Screening for chemical
suppressors of the Wnt/β-catenin signaling pathway. Yakugaku
Zasshi. 137:133–136. 2017.(In Japanese). View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xue G, Romano E, Massi D and Mandalà M:
Wnt/β-catenin signaling in melanoma: Preclinical rationale and
novel therapeutic insights. Cancer Treat Rev. 49:1–12. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu L, Zhang Y, Zhao Z, Chen Z, Wang Z, Xu
S, Zhang X, Liu T and Yu S: The long non-coding RNA CRNDE competed
endogenously with miR-205 to promote proliferation and metastasis
of melanoma cells by targeting CCL18. Cell Cycle. 17:2296–2308.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sahranavardfard P, Madjd Z, Emami Razavi
AN, Ghanadan AR, Firouzi J, Khosravani P, Ghavami S, Ebrahimie E
and Ebrahimi M: An integrative analysis of the Micro-RNAs
contributing in stemness, metastasis and B-Raf pathways in
malignant melanoma and melanoma stem cell. Cell J. 23:261–272.
2021.PubMed/NCBI
|
|
51
|
Sánchez-Sendra B, Serna E, Navarro L,
González-Muñoz JF, Portero J, Ramos A, Murgui A and Monteagudo C:
Transcriptomic identification of miR-205 target genes potentially
involved in metastasis and survival of cutaneous malignant
melanoma. Sci Rep. 10:47712020. View Article : Google Scholar : PubMed/NCBI
|