Introduction
According to the Warburg effect, tumour cells obtain
energy from the glycolysis pathway, which includes active glucose
uptake and excessive lactate production (1). A growing body of studies on cancer
metabolism (2) sparked our
interest in the new relationship between lactate and tumour
biology. In the glycolytic tumour microenvironment, a high level of
lactate accumulation is associated with a poor survival rate and a
higher incidence of cell metastasis (3–6).
Numerous experimental studies have proposed that lactate may be not
just a byproduct of metabolic reprogramming. Indeed, as a
signalling molecule, lactate plays a crucial role in almost every
step of tumourigenesis and development, such as angiogenesis,
migration, metastasis, immune evasion and cancer stem cell
formation (7–9). However, the mechanism regulating
this process remains unclear.
Hydroxycarboxylic acid receptor 1 (HCAR1; also known
as GPR81) is a member of the GPCR family, and lactate has been
identified as its endogenous ligand (10). HCAR1 is mainly expressed in
adipocytes and was originally reported to be associated with
lipolysis inhibition in adipose tissue (11,12). Studies have shown that HCAR1
expression is strikingly high in several solid tumours, such as
breast, cervical and pancreatic cancers (13,14). HCAR1 is closely associated with
tumour growth and metastasis. In breast cancer, a high level of
HCAR1 expression promoted cell proliferation and angiogenesis
through a PI3K/Akt-dependent pathway (14). HCAR1 and HCAR3 are essential for
breast cancer cells to control their lipid/fatty acid metabolism
(15,16). Roland et al (13) revealed that HCAR1 expression was
positively associated with pancreatic cancer progression. Moreover,
the interference of HCAR1 expression dramatically prevented tumour
proliferation and metastasis in vitro and in vivo.
Surprisingly, lactate-induced PD-L1 expression in tumour cells is
mediated by its receptor HCAR1, thus providing an effective means
for tumour cells to evade the immune system through an autocrine
mechanism (8). Lactate also
controls immune evasion through activation of HCAR1 on stromal
dendritic cells in a paracrine manner (17,18). Collectively, these findings
suggested that HCAR1 engagement stimulates intracellular signalling
pathways that facilitate tumour growth, immune evasion and
metastasis.
In the present study, it was found that breast
cancer cell lines displayed high expression of HCAR1, which is
involved in cell survival and migration. Furthermore, new insight
was provided into the role of the lactate-HCAR1 pathway in
maintaining energy metabolism balance. These findings suggested
that autocrine activation of HCAR1 by lactate plays a key role in
reprogramming cancer cell metabolism to meet the high requirements
for rapid cell growth and migration. The discovery of HCAR1
provides novel ideas for the research and development of new
antitumour drugs.
Materials and methods
Cell culture
The lung cancer cell lines (A549 and NCI-H1299),
hepatoma cell lines (Hep3B, Huh7, HepG2 and HCCLM3), bladder cancer
cell lines (UMUC-3 and T24), colorectal cancer cell lines (T84,
LoVo and SW480) and pancreatic cancer cell lines (CFPAC-1 and
PANC-1) were purchased from Procell Life Science & Technology
Co., Ltd. The tongue squamous cell carcinoma cell lines (HN3, HN4),
hepatoma cell line (SNU-449), melanoma cell line (WM35), cervix
carcinoma cell line (Hela), epithelial carcinoma cell line (A431),
breast cancer cell line (MCF7) and HEK293 cell were maintained by
our laboratory. Cell lines were authenticated using short tandem
repeat (STR) profiling. MCF7 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (FBS) and 1X
penicillin/streptomycin and all cells were incubated at 37°C in a
humidified 5% CO2 incubator.
Reverse transcription-quantitative
(RT-q)PCR and semi–quantitative PCR
The total RNA from all the aforementioned cancer
cell lines was extracted using TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.) and converted into cDNA by the
PrimeScript™ RT reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocol. For RT-qPCR, cDNA templates were amplified
on the ABI7500 System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the SYBR Green PCR Kit (Takara Bio, Inc.).
The qPCR thermocycling conditions were as follows: 95°C for 2 min,
followed by 40 cycles of 94°C for 20 sec, 58°C for 20 sec and 72°C
for 30 sec. Relative levels of mRNA expression were calculated
using the 2−ΔΔCq method (19). Primer (10 µM for each gene)
sequences for each gene were as follows: HCAR1 forward,
5′-GCCCAGCACTGTTTACCTTTTC-3′ and reverse,
5′-CCCCAAAAGCCCAGTGTCTAC-3′; phosphofructokinase muscle type (PFKM)
forward, 5′-GAGTGACTTGTTGAGTGACCTCCAGAAA-3′ and reverse,
5′-CACAATGTTCAGGTAGCTGGACTTCG-3′; PFK liver type (PFKL) forward,
5′-GGCATTTATGTGGGTGCCAAAGTC-3′ and reverse,
5′-CAGTTGGCCTGCTTGATGTTCTCA-3′; hexokinase (HK)2 forward,
5′-GGGCATCTTGAAACAAG-3′ and reverse, 5′-GGTCTCAAGCCCTAAG-3′;
pyruvate kinase M2 (PKM2) forward, 5′-GTGGCTCTGGATACAAAGGG-3′ and
reverse, 5′-ACTTCTCCATGTAAGCGTTGTC-3′; and β-actin forward,
5′-ACAATGTGGCCGAGGACTTT-3′ and reverse, 5′-TGGGGTGGCTTTTAGGATGG-3′.
The β-actin gene was used as an internal control.
Semi-quantitative PCR was performed using Taq PCR
mastermix (Tiangen Biotech, Co., Ltd.), The PCR was conducted using
the following thermal cycles: Initial denaturation at 94°C for 5
min, followed by 28 cycles of 98°C for 10 sec, 55°C for 15 sec and
72°C for 1 min, and then a final 5-min extension at 72°C. Primer
(10 µM for each gene) sequences for semi-quantitative PCR were as
follows: HCAR1 forward, 5′-GGAGCATCGTGTTCCTTAC-3′ and reverse,
5′-TTCTTCATCCGAGCCTGT-3′, which amplify a 349-bp fragment; and
β-actin forward, 5′-TCTACAATGAGCTGCGTGTG-3′ and reverse,
5′-CAACTAAGTCATAGTCCGCC-3′, which amplify a 878-bp fragment. After
amplification, the PCR products were visualized on 1.2% agarose
gels containing GelRed (US Everbright, Inc.). The DNA bands were
quantitated using ImageJ 1.52a software (National Institutes of
Health).
CRISPR/Cas9-mediated HCAR1 knockout
(KO)
To knock out HCAR1 via CRISPR/Cas9, three
sgRNAs were designed using the CRISPR Design Tool (http://crispr.mit.edu/): HCAR1 sgRNA-1,
targeting CAGCACGACCCGTTGTACA; sgRNA-2, targeting
GGTCGTGCTGCCGCATCGA; and sgRNA-3, targeting CACACAGGACCCGCATCCT.
Oligos were annealed at 37°C for 30 min and 95°C for 5 min, and
then the temperature was ramped down to 25°C at 5°C/min. The
annealed fragments were treated with BpiI endonuclease and
incorporated into the pSpCas9(BB)-2A-Puro(pX459) plasmid (Addgene,
Inc.). MCF7 cells were seeded in six-well plates and transfected
with 1 µg of pX459-HCAR1 plasmid by FuGENE® HD
transfection Reagent (Roche Diagnostics) according to the
manufacturer's protocol. Following 3 days after transfection, cells
were selected using 2 µg/ml puromycin for 2 weeks. The KO
efficiency was validated by genomic sequencing and western blot
analysis.
Immunoblot analysis
Breast cancer cells were lysed for 30 min on ice in
RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% Na
deoxycholate, 0.1% SDS, 1 mM PMSF, 1 mM
Na3VO4, 10 mg/ml leupeptin and 10 mg/ml
aprotinin). Protein content was calculated using BCA Protein Assay
Kit (Beyotime Institute of Biotechnology). Whole-cell lysates
containing 50 µg of proteins were separated on 10% gels using
SDS-PAGE and transferred to PVDF transfer membrane (MilliporeSigma)
using a semidry transfer system (Bio-Rad Laboratories, Inc.). After
blocking in a 5% BSA (Biosharp Life Sciences) solution in 1X TBST
for 1 h at room temperature, the membrane was probed with
anti-HCAR1 (1:1,000; cat. no. SAB1300090; Sigma-Aldrich; Merck
KGaA) or β-actin (1:1,000; ca. no. 4967; Cell Signalling
Technology, Inc.) antibody for 1 h at room temperature and
subsequently with HRP-conjugated secondary antibodies (1:2,000;
cat. nos. 7076/7074; Cell Signalling Technology, Inc.) for 1 h at
37°C. Immunoreactive bands were detected using an enhanced
chemiluminescent (ECL) reagent (Thermo Fisher Scientific, Inc.) by
using Azure Biosystem C600 (Azure Biosystems, Inc.). Western blots
were quantitated using ImageJ 1.52a software (National Institutes
of Health).
Cell Counting Kit-8 (CCK-8) assay
Cell viability was assessed using a CCK-8 assay
(Dojindo Laboratories, Inc.). Briefly, cells were seeded at 1,000
cells/well in 96-well plates and were incubated for different time
periods (24, 48, 72 and 96 h) at 37°C. CCK-8 (10 µl) solution was
added to each well and then incubation followed for 2 h. The
absorbance value (OD) of each well was measured at 450 nm with a
microplate reader. Each sample was assayed in four replicates.
Three independent experiments were performed.
EdU assay
The EdU incorporation assay was carried out by a
Click-iT® EdU Alexa Fluor® 555 Imaging Kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Briefly, MCF7 cells were seeded into
24-well plates at 1×104 cells/well and incubated at 37°C
overnight. After treatment with 20 mM lactate or DMEM for 24 h, the
cells were treated with EdU (10 mmol/l) for 1 h. Then, the cells
were fixed at room temperature for 15 min, permeabilized and
incubated with the Click-iT reaction cocktail (Invitrogen; Thermo
Fisher Scientific, Inc.), followed by Hoechst 33342 (Invitrogen;
Thermo Fisher Scientific, Inc.) staining, and observed using a
fluorescence microscope (Nikon Corporation). The number of
EdU-positive cells and Hoechst 33342-positive cells were counted
using ImageJ 1.52a software (National Institutes of Health).
Colony formation assay
The colony formation ability of HCAR1-KO and
control MCF7 cells was estimated by a colony formation assay.
Briefly, cells pretreated with lactate (50 mM, with a stronger
effect) for 24 h were seeded in a six-well plate at a density of
300 cells/well, followed by ten days of culture. Then, the clones
(a colony was defined as >50 cells) were stained with 0.1%
crystal violet (Solai Bao Technology Co., Ltd.) for 30 min at room
temperature and counted using ImageJ 1.52a software. The data shown
represent the average of three independent experiments.
Cell cycle analysis
Cell cycle distribution was assessed with a Cell
Cycle Analysis kit (BD Biosciences) by flow cytometry. Cells were
harvested and washed with PBS containing 1% FBS. Then, the cells
were fixed with 70% ethanol at 4°C overnight, washed twice with PBS
and incubated with propidium iodide (PI)/RNase A staining solution
(5 µg/ml PI, 250 µg/ml RNase A in PBS) for 15 min at 37°C in the
dark. The level of PI incorporation was analyzed by FACScan (BD
FACS Canto II). The percentage of cells in the respective cell
cycle phase was determined using Modfit LT version 3.2 software
(Verity Software House, Inc.).
Transwell migration assay
Transwell chambers (MilliporeSigma) were used to
investigate cell migration ability. A total of 1×105
cells were washed with serum-free medium and treated with 100 ng/ml
pertussis toxin (PTX) for 6 h at 37°C. Cells were plated in the
upper chambers (8.0 µm). Medium containing 10% FBS with 2 or 20 mM
lactate was added to the lower chambers to serve as a
chemoattractant. After 24 h of incubation, migratory cells in the
lower chambers were fixed, stained with 0.1% crystal violet
solution (Solai Bao Technology Co., Ltd.) for 30 min and quantified
using light microscopy. The cell numbers were counted in 5
different random fields (magnification, ×200).
Measurement of enzyme activities
MCF7 cells were seeded at a density of
2.0×105/well in six-well plates. The cells were then
collected and the enzyme activity of PFK, HK and PK were determined
using test kits from Nanjing Jiancheng Bioengineering Institute
(catalog nos. A129-1-1, A077-3-1 and A076-1-1) according to the
manufacturer's instructions.
Oxidative phosphorylation (OXPHOS) and
glycolysis assay
The oxygen consumption rate (OCR) and extracellular
acidification rate (ECAR) of cells were evaluated using a Seahorse
XF96 Extracellular Flux Analyzer (Seahorse Bioscience). Briefly,
MCF7 cells were seeded at a density of 1×104 cells/well
into 96-well Seahorse microplates for 24 h, and the calibration
plate was equilibrated overnight in a non-CO2 incubator.
Cells were washed twice with assay running media and equilibrated
in a non-CO2 incubator before starting the test. Once
the probe calibration was completed, the calibration plate was
replaced with the cell plate to simultaneously measure the OCR and
ECAR of the cells. After injection of oligomycin (1 µM), carbonyl
cyanide-p-trifluoromethoxyphenylhydrazone (1 µM), rotenone (1 µM)
and antimycin A (1 µM), OCR and the corresponding ECAR were
determined. Once completed, the protein concentration was
quantified (BCA Protein Assay Kit; Thermo Fisher Scientific, Inc.)
to normalize the OCR and ECAR values.
Statistical analysis
All data were analyzed using SPSS pack 26.0 software
(IBM Corp,), and the graphs were constructed by GraphPad Prism 5.0
software (GraphPad Software, Inc.). Data are presented as the mean
± SD and represent at least three independent experiments. Unpaired
Student's t-tests, or one-way ANOVA with Bonferroni's post hoc test
was used as indicated. P<0.05 was considered to indicate a
statistically significant difference.
Results
HCAR1 is highly expressed in breast
cancer
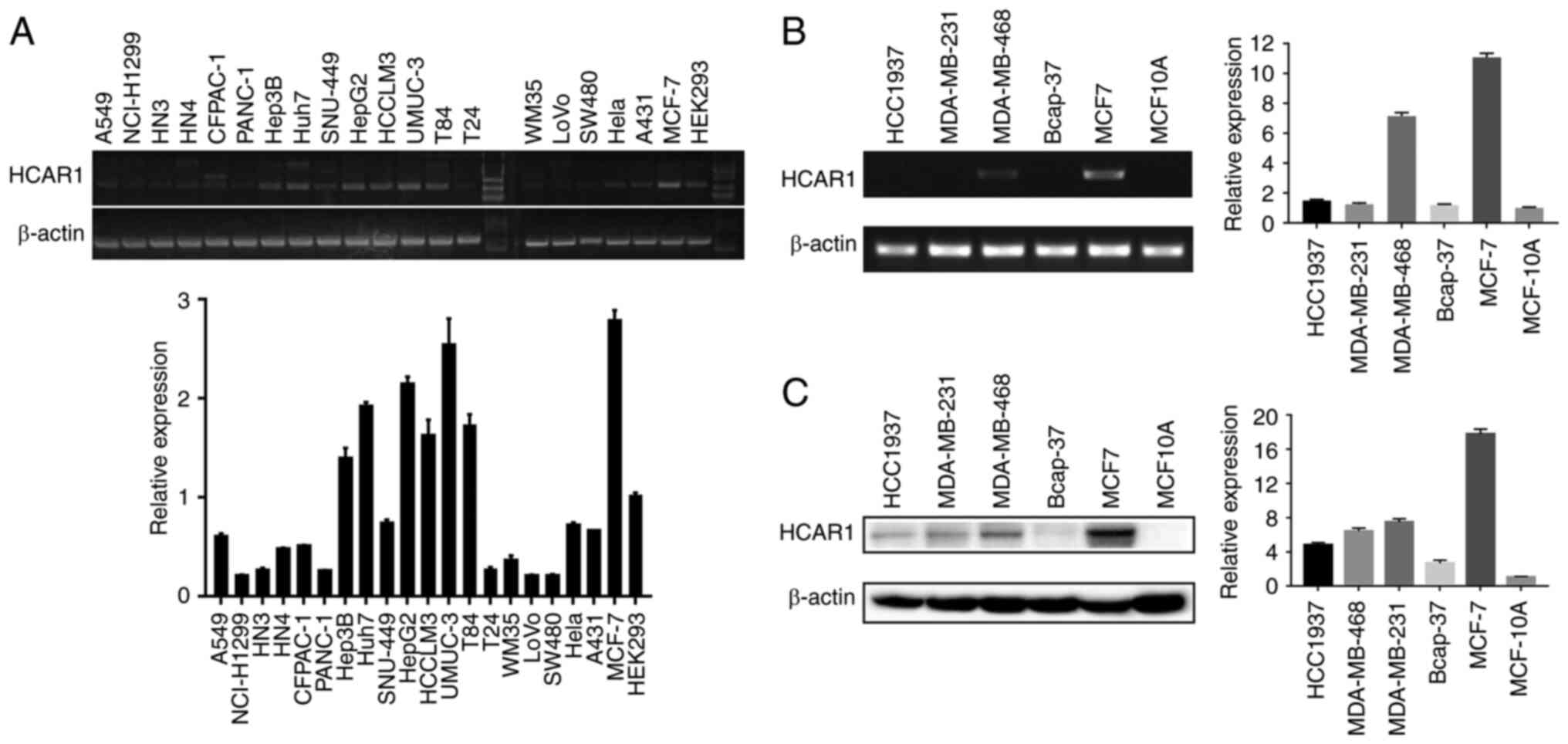
To determine whether HCAR1 was expressed in cancer
cells of solid tumours, whose interior are hypoxic and shows more
lactate accumulation (20,21),
HCAR1 mRNA levels were analyzed in various cancer cell lines. It
was found that HCAR1 was expressed in tongue, lung, breast,
bladder, pancreatic, hepatocellular, colorectal, cervical and
epidermal carcinoma cells (Fig.
1A). Due to high HCAR1 expression, breast cancer cells were
selected for further study. Compared with all kinds of breast
cancer cell lines, a higher level of HCAR1 mRNA and protein
expression was observed in MCF-7 cells (Fig. 1B and C). These results
demonstrated that HCAR1 was expressed in numerous cancer cell types
and in almost all human breast cancer cells.
KO of HCAR1 inhibits breast cancer
cell survival and proliferation
To elucidate the potential effect of HCAR1 on cell
proliferation, HCAR1-KO MCF7 cells were generated by the
CRISPR/Cas9 system. A total of 3 lenti-CRISPR/Cas9-KO constructs
containing nonoverlapping sgRNAs (‘sgRNA1/2/3’) were utilized to
establish stable HCAR1-KO MCF7 cells (Figs. 2C, S1A and B). The survival of HCAR1-KO
MCF7 cells treated with lactate was analyzed by CCK-8 assay
(Fig. 2A). HCAR1 depletion
potently inhibited the viability of MCF7 cells, and lactate
incubation significantly promoted the proliferation of wild-type
(WT) but not HCAR1-KO cells. Consistent with these findings,
colony formation and EdU assay results further demonstrated
that HCAR1 KO inhibited MCF7 cell proliferation, and lactate
treatment did not have a proliferation-promoting effect compared
with WT MCF7 cells (Fig. 2B and
D). As proliferation-suppression phenotypes were observed after
depletion of HCAR1, cell cycle distribution was further
detected by PI staining and flow cytometry. Consistently, a
significant increase in the G1 phase and a decrease in the G2 phase
were observed in MCF7 HCAR1-KO cells (Fig. 2E). These results indicated that
HCAR1 promoted MCF7 cell proliferation.
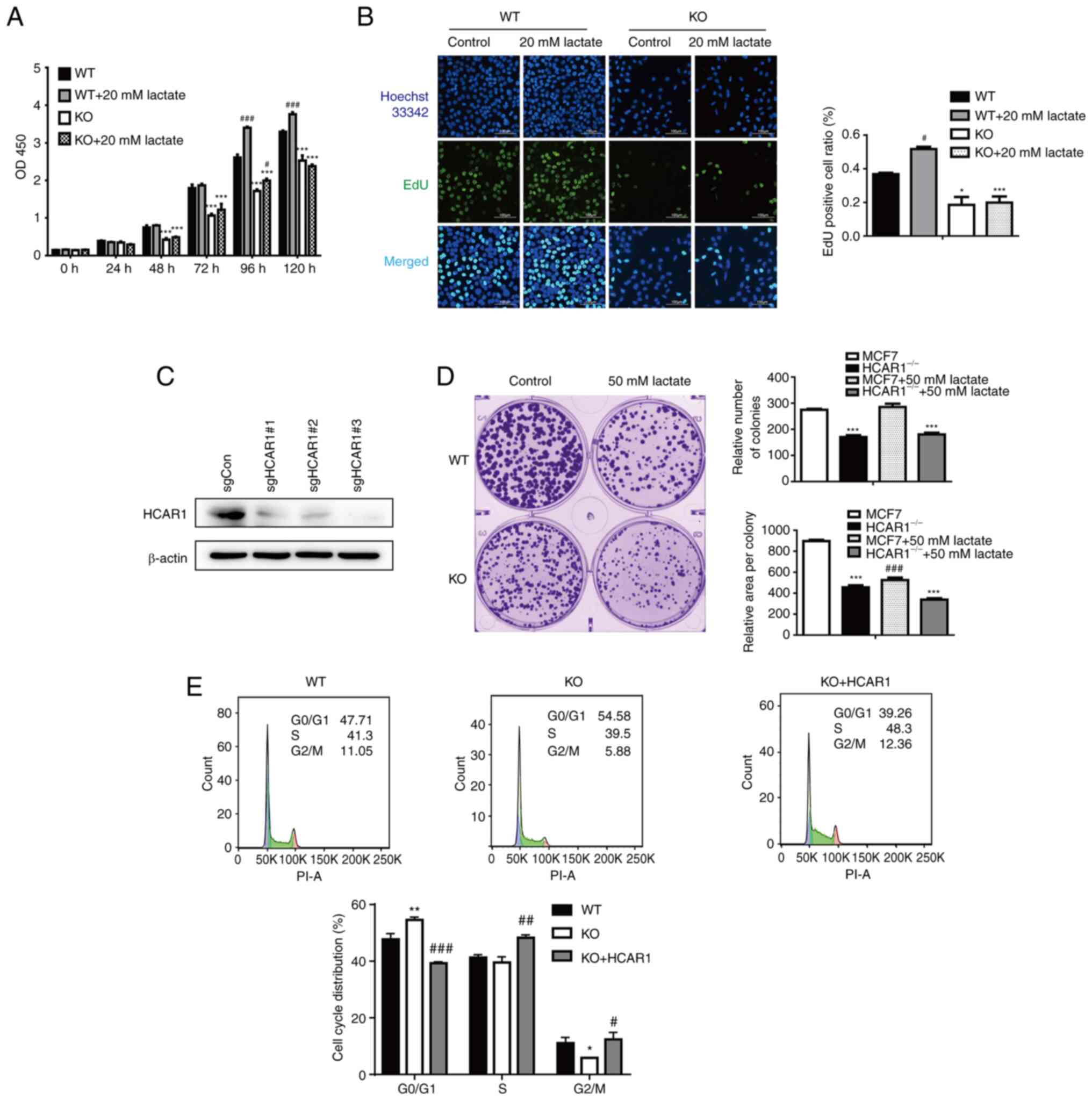 | Figure 2.HCAR1 is an important regulator of
MCF7 cell proliferation and survival. (A) After lactate treatment
of WT and HCAR1-KO cell lines, cell proliferation was
assessed using a Cell Counting Kit-8 (CCK-8). Experiments were
repeated at least three times, and all data are presented as the
mean ± SD. Statistical analyses of the parametric data were carried
out using two-factor analysis of variance (ANOVA). (B) After
Lactate treatment of WT and KO cell lines, cell proliferation was
assessed using an EdU staining proliferation kit. Nuclei were
stained using Hoechst 33342, and EdU was labeled with Alexa
Fluor488 (magnification, ×200). Data are presented as the mean ± SD
(n≥3). (C) HCAR1-KO MCF7 cell clones were generated via
CRISPR/Cas9 technology, and knockout efficiency was detected by
western blot analysis. (D) The colony formation experiment was
performed by staining each cell line with crystal violet and
counting the number and size of effective clones. Data are
presented as the mean ± SD (n≥3). (E) Flow cytometric analysis of
the cell cycle in WT, KO and KO re-overexpressing HCAR1 cells. Data
are representative results from experiments repeated at least three
times. #P<0.05, ##P<0.01 and
###P<0.001 compared with the non-lactate treatment
group; *P<0.05, **P<0.01 and ***P<0.001 compared with the
WT group. HCAR1, hydroxyl carboxylic acid receptor 1; WT,
wild-type; KO, knockout. |
HCAR1 promotes breast cancer cell
metastasis
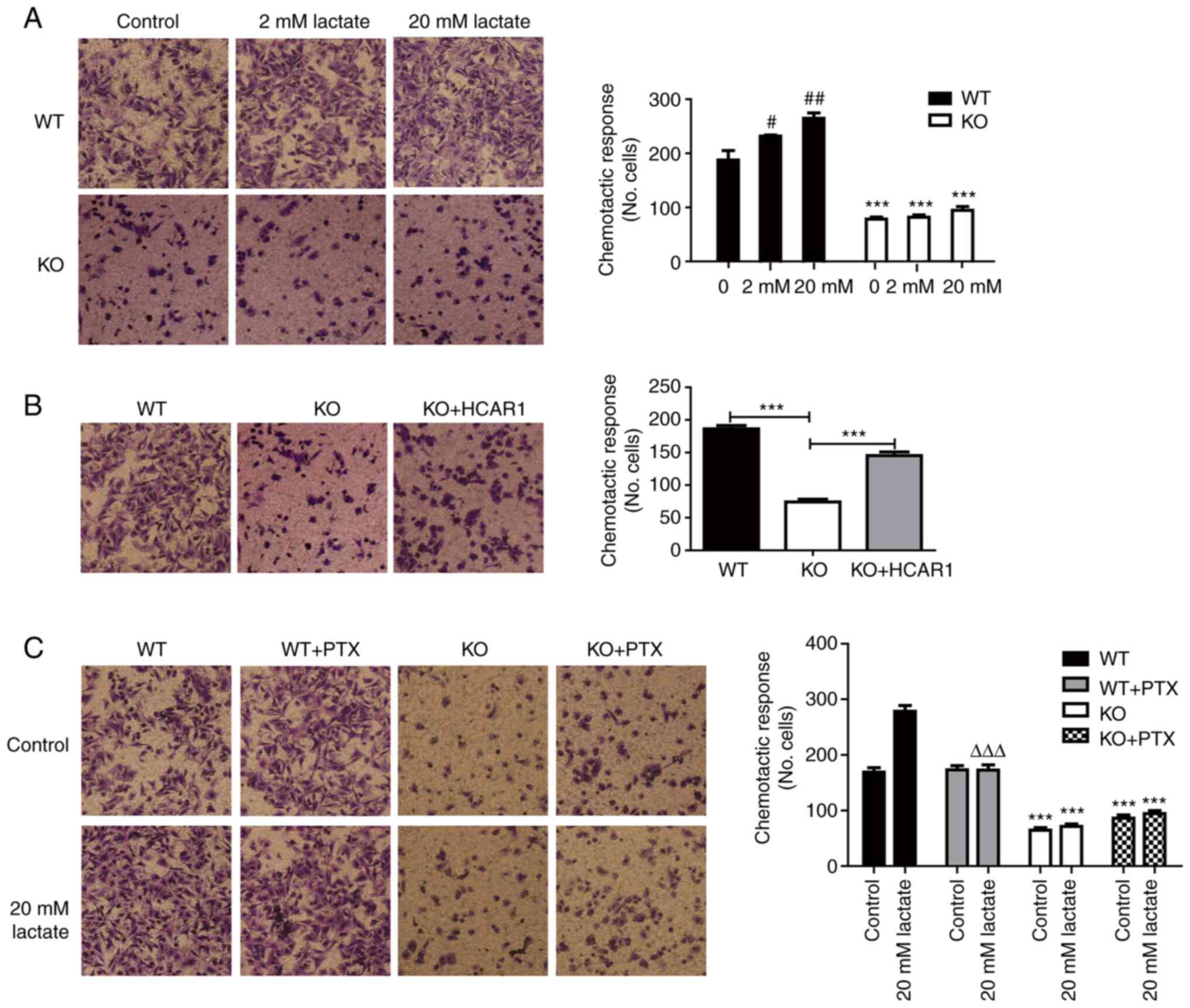
The effects of HCAR1 on the migratory ability of
MCF7 cells were assessed by a Transwell migration assay. The
depletion of HCAR1 in MCF7 cells resulted in a significant
reduction in cell migration (Fig.
3A). However, the inhibition of cell migration was attenuated
after re-overexpression of HCAR1 (Figs. 3B and S2). In addition, lactate promoted WT
MCF7 cell migration in a lactate concentration-dependent manner,
which was inhibited after Gi inhibitor-PTX treatment (Fig. 3C). These data indicated that HCAR1
played a vital role in breast cancer cell migration and that the Gi
protein may participate in the regulation of MCF7 cell
migration.
Effects of HCAR1 on breast cancer cell
glycolysis metabolism
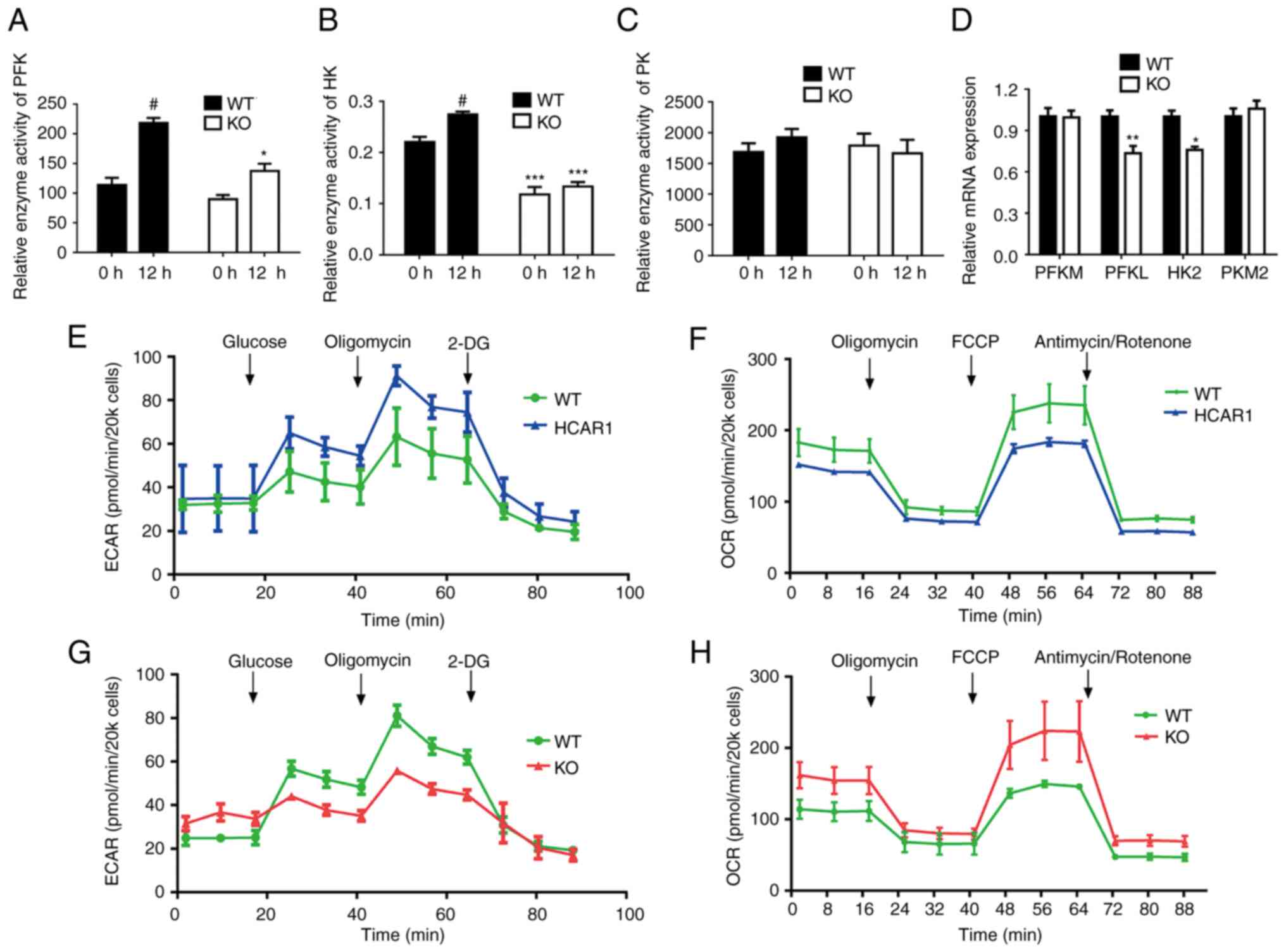
The speed and direction of the metabolic reaction
are associated with rate-limiting enzymes such as PFK, HK and PK
(22,23). Therefore, it was investigated
whether HCAR1 regulated the expression of those key rate-limiting
enzymes in glucose catabolism by RT-qPCR. As revealed in Fig. 4A-D, there was a significant
decrease in the mRNA expression of PFKL and HK2 in MCF7
HCAR1-KO cells, whereas a significant inhibition of the
enzyme activity of PFK and HK was also observed in HCAR1-KO
cells.
KO of HCAR1 reverses the Warburg
effect in MCF7 cells
The glycolysis metabolism results suggested that
HCAR1 could regulate the expression and activity of glycolytic
enzymes. To evaluate whether KO of HCAR1 reversed metabolic
reprogramming consistent with the Warburg effect, cell energy
phenotype assays were conducted using the XFp seahorse bioanalyzer
system. This assay delineated the metabolic phenotypes of MCF7
cells under baseline, KO and overexpression conditions. These
results indicated that HCAR1 KO reduced the ECAR of MCF7
cells and augmented the OCR compared with control cells, which
shifted their energy derivation towards OXPHOS (Fig. 4G and H). Moreover, HCAR1
overexpressing cells had an increased ECAR and a reduced OCR
compared with WT cells (Fig. 4E and
F). These results indicated that HCAR1 could metabolically
control breast cancer by favoring glycolysis over OXPHOS.
Discussion
Since the identification of HCAR1 as a lactate
receptor, studies have revealed that, similar to the other members
of the HCA receptor family, HCAR1 is predominantly expressed in
adipose tissue and suppresses lipolysis by reducing cAMP levels via
a Gi protein-coupled pathway (10,24). However, in contrast to GPR109A
(HCA2), HCAR1 could inhibit lipolysis without provoking skin
flushing (14,25). Therefore, HCAR1 has promising
potential in dyslipidemia. Cancer cells use anaerobic glycolysis
for energy intake, even in normoxic conditions, causing increased
production of lactate (26).
Insufficient tumour blood supply leads to the accumulation of
lactic acid (20–40 mM) (6) in the
tumour microenvironment, which may be sufficient to activate HCAR1.
Altogether, the role of HCAR1 in tumours has markedly attracted
attention.
HCAR1 has been identified to be expressed at high
levels in a variety of tumour cells, where it is able to induce
cancer cells to proliferate and differentiate constantly, thus
enhancing tumour growth. A previous study has shown that HCAR1 is
highly expressed in prostate cancer cells and may inhibit
pancreatic cancer cell progression by regulating lactate
transporter expression (13).
Moreover, daily intraperitoneal injection with isotonic lactate or
sodium lactate in a mouse xenograft model promoted the
proliferation of tumour cells (24). Furthermore, Wanger et al
(27) found that lactate present
in the uterine cervix may participate in the modulation of cellular
DNA damage repair processes and in the resistance of cervical
carcinoma cells to anticancer therapy. In the present study, it was
revealed that after KO of HCAR1, the proliferation, cell
cycle distribution and migration of breast cancer cells were
greatly affected. It is noteworthy that a significant increase in
the G1 phase and a decrease in the G2 phase were observed in
HCAR1-KO cells. K+ channel activity has been
reported to be crucial in cell progression through the G1
checkpoint of the cell cycle (28,29). There are studies regarding
co-localization of GIRK channel and Gi coupled GPCR (muscarinic
receptor) for efficient channel activation (30–32). However, there is no study about
HCAR1 associated K-channel activity. Thus, whether the cell cycle
changes in HCAR1-KO MCF7 cells associated with K-channel
function need to be further explored. Collectively, these results
demonstrated that activation of the HCAR1 receptor was important
for maintaining breast cancer cell survival.
High lactate levels in the tumour microenvironment
play a critical role in promoting cell migration and invasion. On
the one hand, extracellular acidification activated p53-mediated
apoptosis in a caspase-dependent manner in normal cells (33). Cancer cells can survive more
easily in acidified microenvironment due to their high proton
transport activity and low expression level of the P53 gene
(34). On the other hand,
acidification promoted tumour angiogenesis by activating VEGF and
IL-8 release (35,36) and promoting degradation of the
extracellular matrix by proteolytic enzymes to drive tumour
metastasis and invasion (36,37). GPR132 functions as a key
macrophage sensor of the rising lactate in the acidic tumour milieu
to promote the alternatively activated macrophage (M2)-like
phenotype, which facilitates cancer cell migration and invasion
(38). High lactate content
promoted tumour progression by contributing to the phenomenon of
tumour immune escape and by enhancing the migratory potential of
the malignant cell population (39). Less conclusive evidence has been
reported concerning the role of the HCAR1 pathway in breast cancer
cell migration. In the present study, it was found that KO of
HCAR1 resulted in a significant inhibition of MCF7 cell
motility. After re-expressing HCAR1 in the KO cell line, the number
of cells that passed through the Transwell chamber showed a
significant improvement. The results suggested that HCAR1 played an
important role in breast cancer migration. Quite a few G
protein-coupled receptors (GPCRs), such as CXCR4, LPA and PAR1,
participate in the regulation of tumour cell migration (40,41). In the present study, the Gi
protein inhibitor PTX was used to determine whether Gi coupled with
the HCAR1 signalling pathway mediates MCF7 cell migration.
The Warburg effect, a reprogrammed metabolic pathway
that meets the rapidly proliferating tumour cell energy
requirement, has been observed in a variety of malignant tumours
(1,42,43). Enhanced glycolysis leads to
increased glucose uptake and lactate production, which has a
significant influence on the initiation, development and
progression of cancer. In the present study, it was found that the
expression and activity of key glycolytic enzymes were affected by
HCAR1 in breast cancer. After HCAR1 KO in MCF7 cells, the
expression of enzymes, such as PFKL and HK decreased to varying
degrees, and the enzyme activity of PFK and HK showed a significant
reduction, indicating that the levels of glycolysis were to a
moderate extent suppressed by HCAR1 knockdown. Furthermore,
evidence provided by cell energy phenotype assays suggested a
similar result accomplished with the use of MCF7 cells. Initially,
the WT and overexpression HCAR1 cell lines demonstrated a
glycolytic phenotype consistent with the Warburg effect that would
be expected in cancer. However, with CRISPR-Cas9 HCAR1 gene
KO, a more energetic phenotype was demonstrated by reduced ECAR and
increased OCR. The results implied an impairment of cell glycolysis
and showed an oncogenic role for HCAR1.
The limitation to the present study is the
performance of experiments using only one cell line, as the
proliferation, migration and energy metabolism phenotype can vary
in different breast cancer cell types. In the present study,
focused was addressed on MCF7 cells based on higher expression of
HCAR1 in the aforementioned cell line. Further studies are required
to investigate the aforementioned roles of HCAR1 in other breast
cancer cells so as to reveal the relation between HCAR1 and breast
cancer in an improved way.
In conclusion, it was revealed that HCAR1 was
overexpressed in breast cancer cells, particularly in MCF7 cells.
In addition, KO of HCAR1 could substantially inhibit breast
cancer cell proliferation, migration and glycolysis in an in
vitro study. Collectively, the present findings indicated that
HCAR1 may be a tumour promoting factor and that lactate activated
this receptor and hence promoted tumour growth and metastasis by
regulating cellular energy metabolism through glycolysis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Foundation Project for
Science and Technology of Huzhou City (grant no. 2019YZ07).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JQ and LJ conceived and designed the study. LJ, YG,
JC and ZW performed experiments. YJ helped with the collection and
assembly of data. LJ, ZW and JQ analyzed the data and prepared the
figures. LJ and JQ drafted and revised the manuscript. All authors
read and approved the final manuscript. LJ and YJ confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCAR1
|
hydroxyl carboxylic acid receptor
1
|
|
PFK
|
phosphofructokinase
|
|
PK
|
pyruvate kinase
|
|
HK
|
hexokinase
|
|
ECAR
|
extracellular acidification rate
|
|
OCR
|
oxygen consumption rate
|
|
WT
|
wild-type
|
|
KO
|
knockout
|
|
NC
|
negative control
|
|
CCK-8
|
Cell Counting Kit-8
|
|
EdU
|
5-ethynyl-2′-deoxyuridine
|
|
PTX
|
pertussis toxin
|
References
|
1
|
Heiden MGV, Cantley LC and Thompson CB:
Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brizel DM, Schroeder T, Scher RL, Walenta
S, Clough RW, Dewhirst MW and Mueller-Klieser W: Elevated tumor
lactate concentrations predict for an increased risk of metastases
in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 51:349–353.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Walenta S and Mueller-Klieser WF: Lactate:
Mirror and motor of tumor malignancy. Semin Radiat Oncol.
14:267–274. 2004. View Article : Google Scholar
|
|
5
|
Walenta S, Schroeder T and Mueller-Klieser
W: Lactate in solid malignant tumors: Potential basis of a
metabolic classification in clinical oncology. Curr Med Chem.
11:2195–2204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Romero-Garcia S, Moreno-Altamirano MM,
Prado-Garcia H and Sánchez-García FJ: Lactate contribution to the
tumor microenvironment: mechanisms, effects on immune cells and
therapeutic relevance. Front Immunol. 7:522016. View Article : Google Scholar
|
|
7
|
Iñigo SM and Carcinogenesis BGAJ:
Reexamining cancer metabolism: lactate production for
carcinogenesis could be the purpose and explanation of the Warburg
Effect. Carcinogenesis. 119:119–133. 2016.
|
|
8
|
Feng J, Yang H, Zhang Y, Wei H, Zhu Z, Zhu
B, Yang M, Cao W, Wang L and Wu Z: Tumor cell-derived lactate
induces TAZ-dependent upregulation of PD-L1 through GPR81 in human
lung cancer cells. Oncogene. 36:5829–5839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dhup S, Dadhich RK, Porporato PE and
Sonveaux P: Multiple biological activities of lactic acid in
cancer: Influences on tumor growth, angiogenesis and metastasis.
Curr Pharm Des. 18:1319–1330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu C, Wu J, Zhu J, Kuei C, Yu J, Shelton
J, Sutton SW, Li X, Yun SJ, Mirzadegan T, et al: Lactate inhibits
lipolysis in fat cells through activation of an orphan
G-protein-coupled receptor, GPR81. J Biol Chem. 284:2811–2822.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu C, Kuei C, Zhu J, Yu J, Zhang L, Shih
A, Mirzadegan T, Shelton J, Sutton S, Connelly MA, et al:
3,5-Dihydroxybenzoic acid, a specific agonist for hydroxycarboxylic
acid 1, inhibits lipolysis in adipocytes. J Pharmacol Exp Ther.
341:794–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ca TQ, Ren N, Jin L, Cheng K, Kash S, Chen
R, Wright SD, Taggart AKP and Waters MG: Role of GPR81 in
lactate-mediated reduction of adipose lipolysis. Biochem Biophy Res
Commun. 377:987–991. 2008. View Article : Google Scholar
|
|
13
|
Roland CL, Arumugam T, Deng D, Liu SH,
Philip B, Gomez S, Burns WR, Ramachandran V, Wang H,
Cruz-Monserrate Z and Logsdon CD: Cell surface lactate receptor
GPR81 is crucial for cancer cell survival. Cancer Res.
74:5301–5310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee YJ, Shin KJ, Park SA, Park KS, Park S,
Heo K, Seo YK, Noh DY, Ryu SH and Suh PG: G-protein-coupled
receptor 81 promotes a malignant phenotype in breast cancer through
angiogenic factor secretion. Oncotarget. 7:70898–70911. 2016.
View Article : Google Scholar
|
|
15
|
Offermanns S: Hydroxy-carboxylic acid
receptor actions in metabolism. Trends Endocrinol Metab.
28:227–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stäubert C, Broom OJ and Nordström A:
Hydroxycarboxylic acid receptors are essential for breast cancer
cells to control their lipid/fatty acid metabolism. Oncotarget.
6:19706–19720. 2015. View Article : Google Scholar
|
|
17
|
Lundø K, Trauelsen M, Pedersen SF and
Schwartz TW: Why warburg works: Lactate controls immune evasion
through GPR81. Cell Metab. 31:666–668. 2020. View Article : Google Scholar
|
|
18
|
Brown TP, Bhattacharjee P, Ramachandran S,
Sivaprakasam S, Ristic B, Sikder MOF and Ganapathy V: The lactate
receptor GPR81 promotes breast cancer growth via a paracrine
mechanism involving antigen-presenting cells in the tumor
microenvironment. Oncogene. 39:3292–3304. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang J, Huang D, Jiang Y, Hou J, Tian M,
Li J, Sun L, Zhang Y, Zhang T, Li Z, et al: Lactate modulates
cellular metabolism through histone lactylation-mediated gene
expression in non-small cell lung cancer. Front Oncol.
11:6475592021. View Article : Google Scholar
|
|
21
|
Salem A, Asselin MC, Reymen B, Jackson A,
Lambin P, West CML, O'Connor JPB and Faivre-Finn C: Targeting
hypoxia to improve non-small cell lung cancer outcome. J Natl
Cancer Inst. 110:doi: 10.1093/jnci/djx160, 2018.
|
|
22
|
Yu G, Yu W, Jin G, Xu D, Chen Y, Xia T, Yu
A, Fang W, Zhang X, Li Z and Xie K: PKM2 regulates neural invasion
of and predicts poor prognosis for human hilar cholangiocarcinoma.
Mol Cancer. 14:1932015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Wang Y, Liu Z, Guo X, Miao Z and Ma
S: Atractylenolide I induces apoptosis and suppresses glycolysis by
blocking the JAK2/STAT3 signaling pathway in colorectal cancer
cells. Front Pharmacol. 11:2732020. View Article : Google Scholar
|
|
24
|
Goodwin ML, Jin H, Straessler K, Smith-Fry
K, Zhu JF, Monument MJ, Grossmann A, Randall RL, Capecchi MR and
Jones KB: Modeling alveolar soft part sarcomagenesis in the mouse:
A role for lactate in the tumor microenvironment. Cancer Cell.
26:851–862. 2014. View Article : Google Scholar
|
|
25
|
Sakurai T, Davenport R, Stafford S, Grosse
J, Ogawa K, Cameron J, Parton L, Sykes A, Mack S, Bousba S, et al:
Identification of a novel GPR81-selective agonist that suppresses
lipolysis in mice without cutaneous flushing. Eur J Pharmacol.
727:1–7. 2014. View Article : Google Scholar
|
|
26
|
Ward C, Langdon SP, Mullen P, Harris AL,
Harrison DJ, Supuran CT and Kunkler IH: New strategies for
targeting the hypoxic tumour microenvironment in breast cancer.
Cancer Treat Rev. 39:171–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wagner W, Ciszewski WM and Kania KD: L-
and D-lactate enhance DNA repair and modulate the resistance of
cervical carcinoma cells to anticancer drugs via histone
deacetylase inhibition and hydroxycarboxylic acid receptor 1
activation. Cell Commun Signal. 13:362015. View Article : Google Scholar
|
|
28
|
Xu B, Wilson BA and Lu L: Induction of
human myeloblastic ML-1 cell G1 arrest by suppression of K+ channel
activity. Am J Physiol. 271:C2037–C2044. 1996. View Article : Google Scholar
|
|
29
|
Marakhova I, Domnina A, Shatrova A,
Borodkina A, Burova E, Pugovkina N, Zemelko V and Nikolsky N:
Proliferation-related changes in K(+) content in human mesenchymal
stem cells. Sci Rep. 9:3462019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tateyama M and Kubo Y: Gi/o-coupled
muscarinic receptors co-localize with GIRK channel for efficient
channel activation. PLoS One. 13:e02044472018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Days E, Kaufmann K, Romaine I, Niswender
C, Lewis M, Utley T, Du Y, Sliwoski G, Morrison R, Dawson ES, et
al: Discovery and Characterization of a Selective Activator of the
G-Protein Activated Inward-Rectifying Potassium (GIRK) Channel.
Probe Reports from the NIH Molecular Libraries Program [Internet].
National Center for Biotechnology Information (US); Bethesda, MD:
2010
|
|
32
|
Dascal N and Kahanovitch U: The roles of
Gβγ and Gα in gating and regulation of GIRK channels. Int Rev
Neurobiol. 123:27–85. 2015. View Article : Google Scholar
|
|
33
|
Park HJ, Lyons JC, Ohtsubo T and Song CW:
Acidic environment causes apoptosis by increasing caspase activity.
Br J Cancer. 80:1892–1897. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Porporato PE, Dhup S, Dadhich RK, Copetti
T and Sonveaux P: Anticancer targets in the glycolytic metabolism
of tumors: A comprehensive review. Front Pharmacol. 2:492011.
View Article : Google Scholar
|
|
35
|
Shi Q, Abbruzzese JL, Huang SY, Fidler IJ,
Xiong Q and Xie K: Constitutive and inducible interleukin 8
expression by hypoxia and acidosis renders human pancreatic cancer
cells more tumorigenic and metastatic. Clin Cancer Res.
5:3711–3721. 1999.PubMed/NCBI
|
|
36
|
Shi Q, Le X, Wang B, Abbruzzese JL, Xiong
Q, He Y and Xie K: Regulation of vascular endothelial growth factor
expression by acidosis in human cancer cells. Oncogene.
20:3751–3756. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rozhin J, Sameni M, Ziegler GH and Sloane
BF: Pericellular pH affects distribution and secretion of cathepsin
B in malignant cells. Cancer Res. 54:6517–6525. 1994.PubMed/NCBI
|
|
38
|
Xie Q, Zhu Z, He Y, Zhang Z, Zhang Y, Wang
Y, Luo J, Peng T, Cheng F, Gao J, et al: A lactate-induced
Snail/STAT3 pathway drives GPR81 expression in lung cancer cells.
Biochim Biophys Acta Mol Basis Dis. 1866:1655762020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goetze K, Walenta S, Ksiazkiewicz M,
Kunz-Schughart LA and Mueller-Klieser W: Lactate enhances motility
of tumor cells and inhibits monocyte migration and cytokine
release. Int J Oncol. 39:453–463. 2011.
|
|
40
|
Dusaban SS, Purcell NH, Rockenstein E,
Masliah E, Cho MK, Smrcka AV and Brown JH: Phospholipase Cε links G
protein-coupled receptor activation to inflammatory astrocytic
responses. Proc Natl Acad Sci USA. 110:3609–3614. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Liao R, Chen X, Ying X, Chen G, Li
M and Dong C: Twist-mediated PAR1 induction is required for breast
cancer progression and metastasis by inhibiting Hippo pathway. Cell
Death Dis. 11:5202020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar
|
|
43
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|