Introduction
Liver fibrosis may be caused by acute or chronic
liver injury and has been associated with severe morbidity and
mortality (1). The progression
from hepatitis to liver fibrosis, cirrhosis and even hepatic
carcinoma has been reported to be accelerated by the occurrence of
metabolic liver disease, alcoholism and viral hepatitis (2,3).
The development of liver fibrosis is a result of the interaction
among various types of resident hepatocytes, inflammatory cells,
mediators and extracellular matrix (ECM) components, which are
usually characterized by increased matrix protein production and
decreased matrix remodeling (4,5).
Liver fibrosis accelerates the progression of acute liver disease
to its chronic form by disrupting the normal liver parenchyma
(6). Furthermore, animal models
that simulate liver fibrosis have improved our understanding of
liver injury, and carbon tetrachloride (CCl4)-induced
liver fibrosis is widely used in mouse models to study hepatotoxic
mechanisms (3,7). CCl4-induced acute liver
injury leads to the complex regulation of cellular responses. It
has previously been established that continuous exposure of mice to
a low dose of CCl4 (0.2 ml/kg), beginning at 8 weeks of
age and lasting up to 6 weeks, leads to the development of liver
fibrosis and compensatory cell proliferation (8). A CCl4-induced liver
fibrosis mouse model has been demonstrated to be able to
effectively simulate the formation of liver injury in vivo,
which has been widely used in the study of the mechanism of
hepatotoxicity (3,8–11).
Currently, treatments for liver fibrosis mainly
include eliminating the primary causes and suppressing
inflammation. However, there is still a lack of effective
preventative methods or therapeutic drugs for liver fibrosis
(12,13). The drug discovery process has paid
great attention to the investigation of the efficacy drugs used in
traditional medicine, as they are cheaper and have fewer side
effects. Pien Tze Huang (PZH), a complex combination of Panax
notoginseng (85%); Calculus bovis (5%); snake
gallbladder (7%), obtained from the dry gall bladder of snake; and
musk (3%), obtained from the preputial gland located in the abdomen
of male musk deer, has been used as a traditional medicine with
anti-inflammatory and soothing effects on skin boils and abscesses
(14). A previous study has
demonstrated that PZH exerts an important protective effect on
CCl4-induced liver injury in mice (15). Additional studies have also
suggested that PZH is effective in inhibiting liver damage caused
by excessive drinking (16),
improving histopathological damage in the liver and positively
affecting physiological and biochemical indexes, such as serum
alanine aminotransferase and aspartate aminotransferase, to a
certain extent (15,17). Cell necrosis and swelling,
microvesicular steatosis and lymphocyte infiltration in the injured
liver have been reported to be significantly reduced following PZH
treatment for liver disease (18).
Circular RNAs (circRNAs) are a recently discovered
type of non-coding RNAs (ncRNAs). Most circRNAs are endogenous
ncRNAs, and their formation does not have a covalent ring structure
at the 3′ and 5′ end (19). Owing
to their special structure, circRNAs cannot be easily degraded by
nucleic acid exoenzyme ribonuclease R (RNase R) and are more stable
than linear RNA (20).
Furthermore, circRNAs regulate alternative splicing and
transcription in a variety of diseases by acting as efficient
microRNA (miRNA/miR) sponges, which can efficiently prevent the
suppression of their mRNA targets (21,22). A previous study confirmed the
regulatory role of circRNAs in circRNA/miRNA-gene regulatory
networks by building a database of circRNAs derived from
transcriptome sequencing data (23). Therefore, the interactions among
circRNA, miRNA and mRNA have been considered important in
transcriptional and post-transcriptional regulation.
An increasing number of studies have demonstrated
that circRNAs can act as promising and technically suitable
biomarkers for the occurrence and development of various diseases
(24–28). Recently, Xu et al (29) analyzed the regulatory role of the
circRNA/miRNA/mRNA network and revealed that circRNA_0001178 and
circRNA_0000826 may be potential diagnostic markers for colorectal
cancer metastasis. Furthermore, a recent study revealed that Mus
musculus (mmu)_circRNA_002381 may influence the regulatory
process of the transcription of certain genes affected by
cocaine-induced neuroplasticity via the circRNA/miRNA interaction
network (30). However, it
remains unclear as to how the circRNA/miRNA/mRNA network modulates
improvements in the fibrotic liver tissue of PZH-treated
subjects.
The present study investigated the hepatoprotective
activity of PZH against CCl4-induced liver fibrosis, and
aimed to determine the circRNA-based molecular mechanisms by which
it may confer its protection on the mice liver against
CCl4-induced liver damage. Differentially expressed
circRNAs were successfully identified using high-throughput
RNA-sequencing (RNA-seq) in liver tissues from
CCl4-induced and PZH-treated mice. The sponging action
of differentially expressed circRNAs and related miRNA targets was
further examined using multiple bioinformatics tools. Moreover,
functional circRNA/miRNA/mRNA networks were established to provide
novel insights into the treatment of liver fibrosis with PZH. The
functional characteristics of the representative circRNA candidates
were further analyzed by performing reverse
transcription-quantitative PCR (RT-qPCR) on the mRNAs associated
with the differentially expressed circRNAs in hepatic stellate
cells (HSCs).
Materials and methods
Animal experiment
A total of 45 C57BL/6 mice of each group (age, 6
weeks; weight, 20–30 g) were purchased from Shanghai Model
Organisms Center, Inc. The mice were housed at 24±2°C, relative
humidity of 55±15%, kept in clean cages under a 14/10 h light-dark
cycle and fed with standard rodent diet throughout the entire
experimental period. Mice were randomly divided into two groups
named the ‘PZH-treated’ and ‘CCl4-induced’ (PZH-treated,
hepatic fibrosis model using PZH plus CCl4;
CCl4-induced, hepatic fibrosis model using
CCl4 only; n=15 mice/group). Meanwhile, a no-induction
group (n=15 mice/group) was added to confirm the success of the
liver fibrosis model. Mice in the CCl4-induced liver
fibrosis model group were intraperitoneally injected with a solvent
mixture of 10 µl/g body weight CCl4 (Wuhan Yafa
Biological Technology Co., Ltd.) and 10% corn oil twice a week and
intragastrically administered with double-distilled
(dd)H2O once a day. Mice in the PZH-treatment group were
intraperitoneally injected with a solvent mixture of 10 µl/g body
weight CCl4 and 10% corn oil twice a week (31) and intragastrically administered
with 0.25 mg/g PZH-sonicated reagent (Zhangzhou Pientzehuang
Pharmaceutical Co., Ltd.; Chinese Food and Drug Administration
approval no. Z35020242; the PZH drug powder was dissolved in double
distilled water, and its concentration was diluted to 25 mg/ml)
once a day. During the 8-week experimental period, three mice in
each group were anesthetized intraperitoneally with 40 mg/kg
pentobarbital (Sigma-Aldrich; Merck KGaA) and sacrificed by
cervical dislocation every 2 weeks in the first 6 weeks, and their
liver tissue and blood were harvested for detecting the hepatic
fibrosis level and hepatic biochemical index. The remaining 12 mice
were also anesthetized and sacrificed by cervical dislocation in
the 8th week. Mice were subsequently subjected to a laparotomy and
liver tissue was harvested for the following experiments.
Throughout the animal experiment mental state, hair and the body
weight of the mice were closely monitored for toxic effects caused
by CCl4. No significant differences were observed in the
body weight of the mice between CCl4-induced and
PZH-treated groups (P=0.7772; Fig.
S1). The experimental procedures in the present study were
approved by The Institutional Review Board of Shanghai Jiao Tong
University (Shanghai, China; approval no. IACUC.NO:2017-0033).
RNA isolation for high-throughput
sequencing
Total RNA was extracted from mouse liver tissue
samples using TRIzol® reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Samples were then
processed with RNase-free DNase to remove traces of genomic DNA.
The RNA concentration and quality were determined using a
NanoDrop® ND-2000 spectrophotometer (Thermo Fisher
Scientific, Inc.) and the Bioanalyzer 2100 System (Agilent
Technologies, Inc.). The A260/A280 ratio and RNA integrity number
(RIN; Agilent Technologies Deutschland GmbH) were used to assess
RNA purity and integrity, respectively (32). All samples met the following
criteria: A260/A280 ratio >1.8 and RIN >8, which means that
the RNA integrity was high quality for sequencing.
Transcriptome data analysis
RNA libraries were constructed using ribosomal
RNA-depleted RNAs with TruSeq Stranded Total RNA Library Prep Kit
(cat. no. RS-122-2201; Illumina, Inc.) according to the
manufacturer's protocol. Briefly, 5 µg RNA was treated using the
Ribo-Zero™ kit (cat. no. MRZH11124; Illumina, Inc.) to remove all
ribosomal RNAs and linear RNAs were digested using RNase R (cat.
no. RNR07250; Lucigen Corporation). Subsequently, the enriched
circRNAs were fragmented and a double-stranded cDNA library was
synthesized using random primers and adapters. Finally, the cDNAs
were purified and amplified with a thermocycler. All samples were
mixed and underwent bridge amplification to generate clusters using
HiSeq 4000 Paired-End Cluster Kit (cat. no. PE-410-1001-1;
Illumina, Inc.). Quality control was performed using Q30. The
library was subjected to 150-bp paired-end sequencing using HiSeq
4000 SBS Kit (cat. no. FC-410-1001-1; Illumina, Inc.). FastQC
(version v0.11.5; http://www.bioinformatics.babraham.ac.uk/projects/fastqc)
(33) was used to evaluate the
quality of the sequencing data. Trim-galore (version 0.6.0)
(34) with default parameters was
used to remove double-ended adapters and perform quality control on
raw sequencing data.
Identification of differentially
expressed circRNAs between the two groups
The filtered data were first mapped to the reference
genome/transcriptome (version mm10, downloaded from UCSC,
http://genome.ucsc.edu/) using STAR software
version 2.5.3 (35). Only the
common circRNAs predicted by CIRI (version 2.0.6) (36) and CIRCexplorer (37) software were termed ‘identified
circRNAs’ in the subsequent analysis. CircBase (http://www.circbase.org/) (38) and Circ2Traits (39) (http://gyanxet-beta.com/circdb) databases were used to
annotate the identified circRNAs. circRNAs were normalized to the
number of back-splice junctions spanning spliced reads per billion
mapping (SRPBM). The differentially expressed circRNAs were
screened according to a log2(FC)>1 and P<0.05.
RNA-seq data were deposited in National Center for Biotechnology
Information Gene Expression Omnibus (GEO; accession no.
GSE150883).
Target prediction and bioinformatics
analysis
For the specific competing endogenous RNA (ceRNA)
network of significantly dysregulated circRNAs and mRNAs, the
miRNA/mRNA and miRNA/circRNA interactions were predicted using the
RegRNA 2.0 (40) (http://regrna2.mbc.nctu.edu.tw/), miRWalk 2.0
(41) (http://mirwalk.umm.uni-heidelberg.de) and TargetScan
7.2 (42) (http://www.targetscan.org) databases. miRNA-binding
sites on circRNAs were determined using RegRNA 2.0 with a cutoff
score of ≥170 and free energy of ≤-25. The putative target genes of
these miRNAs were identified using the miRWalk algorithm with a
binding energy of ≤-30 (43). The
intersection of predicted target genes identified by both of these
algorithms was selected as the differentially expressed
circRNA-related predicted target genes in the present study for
further analysis. The shared mRNAs between the differentially
expressed circRNA-related predicted target genes in the present
study and the differentially expressed mRNAs from mRNA-seq data
(GEO accession no. GSE133481) were extracted. The overlapping
mRNA-related miRNAs and miRNA binding sites on these circRNAs were
selected to construct the ceRNA regulatory network. Cytoscape
(version 3.6.1) was used to visualize the circRNA/miRNA/mRNA
network interactions (44). The R
package Cluster Profiler (45)
was used for Gene Ontology (GO) analysis (http://geneontology.org/) and the Kyoto Encyclopedia
of Genes and Genomes (KEGG) database (https://www.kegg.jp/) was used for pathway analysis of
the differentially expressed circRNA-related target genes based on
the adjusted P<0.05.
Validation of differentially expressed
circRNA-related target gene candidates by RT-qPCR in LX-2
cells
The expression levels of candidate differentially
expressed circRNA-related target genes in the ceRNA network were
further validated by RT-qPCR in the human hepatic stellate LX-2
cell line (American Type Culture Collection). LX-2 cells were grown
in an incubator with 5% CO2 at 37°C in DMEM (Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS
(MilliporeSigma), 2 mM l-glutamine, 100 U/ml penicillin and 100
µg/ml streptomycin (complete medium; Gibco; Thermo Fisher
Scientific, Inc.). Cells were seeded into 6-well plates
(1×106/well) and divided into the PZH-induced (0.75
mg/ml) and the untreated control groups. The expression levels of
differentially expressed circRNA-related target gene candidates
were determined at 24, 48 and 72 h. Total RNA was extracted from
LX-2 cells using TRIzol reagent, aforementioned. Total RNA was
reversed transcribed into cDNA using 1 µg isolated RNA with the
PrimeScript First Strand cDNA Synthesis Kit (Takara Bio, Inc.)
according to the manufacturer's protocol. qPCR was subsequently
performed using an ABI7500 Real-Time PCR System (Thermo Fisher
Scientific, Inc.) and the FastStart Universal SYBR Green Master
(Roche Diagnostics). Each qPCR reaction contained 25 µl FastStart
Universal SYBR Green Master, 0.3 µM forward primer, 0.3 µM reverse
primer, 50 ng cDNA template and ddH2O to a final volume
of 50 µl. The qPCR conditions were as follows: Initial denaturation
at 95°C for 10 min; followed by 40 cycles at 95°C for 15 and 60 sec
at 60°C. All primers are shown in Table SI. Relative mRNA expression
levels were quantified using the 2−ΔΔCq method and
normalized to the internal reference gene GAPDH (46). At 2−ΔΔCq >1, the
mRNA expression levels were considered to be upregulated and at
2−ΔΔCq <1 downregulated.
Statistical analysis
Data from three independent experiments were used
for analysis. Data are presented as the mean ± SEM. GraphPad Prism
8.0 (GraphPad Software, Inc.) was used for all statistical
analyses. Unpaired Student's t-test and one-way ANOVA with Duncan's
post hoc test were used to determine the statistical differences
between the control and PZH-treated groups. P<0.05 was
considered to indicate a statistically significant difference.
Survival analysis
Survival analysis was performed using the survival
package in R (version 3.6.3) (47) to explore the relationship between
the differentially expressed circRNA-related targets and the status
data of 269 clinical patients with liver hepatocellular carcinoma
from The Cancer Genome Atlas (TCGA) database (48). Patients with expression levels
higher than the median values were assigned to the high-expression
group, and patients with expression levels lower than the median
values were assigned to the low-expression group. Kaplan-Meier
survival analysis was performed to plot survival curves and the
log-rank test was used to assess the statistical difference in
survival rates between the high- and low-expression groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Prediction and expression analysis of
circRNAs
To identify differentially expressed circRNAs in
PZH-treated mice exhibiting liver damage improvement compared with
the CCL4-induced group, RNA-seq was used to profile
circRNA expression levels in the tissue samples from six
PZH-treated mice and six CCL4-induced mice with liver
fibrosis. The quality control results from the sequencing data
demonstrated that the Q30 of all samples reached 95% (Table I). The filtered data was
subsequently mapped to the reference genome/transcriptome
(GRCm38/mm10; Table II). CIRI
and CIRCexplorer software were used to identify the circRNAs in all
samples. Detailed information regarding the circRNAs identified is
displayed in Table SII. A total
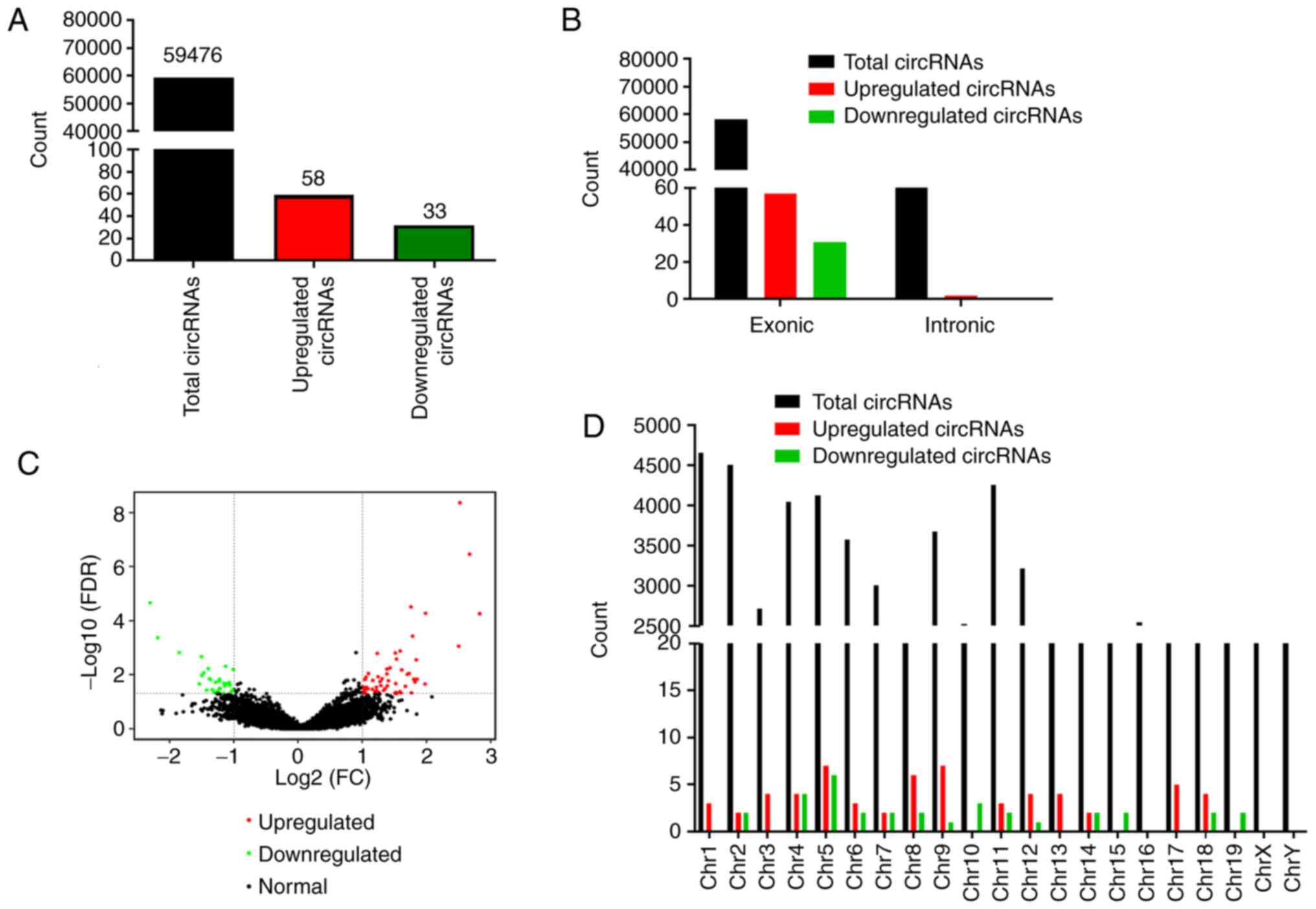
of 59,476 intersection circRNAs were detected by circRNA-seq. Among
all the types of circRNA, the proportion of exon circRNA was as
high as 98.35% (Table SII).
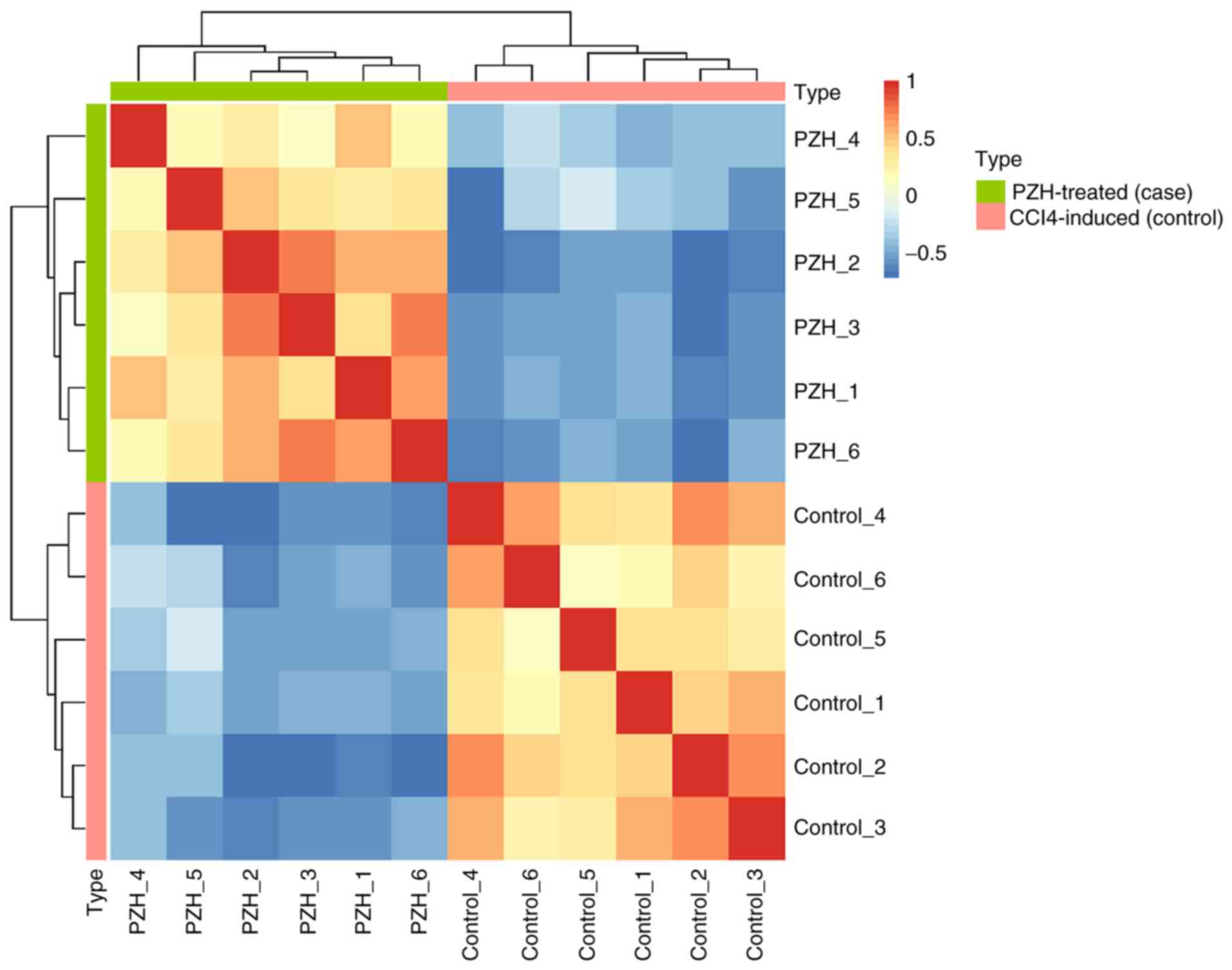
Subsequently, hierarchical clustering was performed using an
expression matrix calculated from each circRNA across 12 samples.
The CCL4-induced and PZH-treated fibrotic liver samples
were classified into different branches (Fig. 1). The SRPBM expression values of
each circRNA for all samples are shown in Table SIII.
 | Table I.Quality control results for all
samples. |
Table I.
Quality control results for all
samples.
| Sample | Reads count | Base count | Error,
%a | Q20, %b | Q30, % | GC, %c |
|---|
| PZH_1 | 81068764 | 10516643897 | 0.0118 | 98.28 | 95.46 | 57.29 |
| PZH_2 | 90141456 | 11798532726 | 0.0119 | 98.25 | 95.41 | 57.20 |
| PZH_3 | 85539612 | 11098006475 | 0.0118 | 98.29 | 95.48 | 57.46 |
| PZH_4 | 89396344 | 11600890156 | 0.0121 | 98.17 | 95.16 | 57.57 |
| PZH_5 | 89704690 | 11682502150 | 0.0121 | 98.14 | 95.14 | 57.70 |
| PZH_6 | 82737206 | 10730442091 | 0.0119 | 98.26 | 95.42 | 57.13 |
| Control_1 | 95957422 | 12581967724 | 0.0120 | 98.16 | 95.26 | 52.03 |
| Control_2 | 79748818 | 10401728633 | 0.0122 | 98.06 | 95.04 | 55.98 |
| Control_3 | 88915072 | 11546488853 | 0.0120 | 98.19 | 95.28 | 56.98 |
| Control_4 | 89463266 | 11634186983 | 0.0120 | 98.12 | 95.26 | 52.62 |
| Control_5 | 99444270 | 12929236537 | 0.0119 | 98.18 | 95.36 | 54.19 |
| Control_6 | 82923014 | 10776422218 | 0.0122 | 97.98 | 94.95 | 53.67 |
 | Table II.Mapping ratio statistics of all clean
data. |
Table II.
Mapping ratio statistics of all clean
data.
| Sample | Reads count | Mapped reads | Mapped ratio,
% | Unique mapped
reads | Unique mapped
ratio, % |
|---|
| PZH_1 | 40534382 | 38952915 | 96.10 | 12552203 | 30.97 |
| PZH_2 | 45070728 | 43433153 | 96.37 | 14150855 | 31.40 |
| PZH_3 | 42769806 | 41221377 | 96.38 | 12478538 | 29.18 |
| PZH_4 | 44698172 | 42680427 | 95.49 | 13259939 | 29.67 |
| PZH_5 | 44852345 | 43051071 | 95.98 | 12857153 | 28.67 |
| PZH_6 | 41368603 | 39789422 | 96.18 | 12193014 | 29.47 |
| Control_1 | 47978711 | 42105288 | 87.76 | 14914179 | 31.08 |
| Control_2 | 39874409 | 36838848 | 92.39 | 11268513 | 28.26 |
| Control_3 | 44457536 | 42229515 | 94.99 | 13088832 | 29.44 |
| Control_4 | 44731633 | 39892267 | 89.18 | 13676170 | 30.57 |
| Control_5 | 49722135 | 45379494 | 91.27 | 15213018 | 30.60 |
| Control_6 | 41461507 | 37464042 | 90.36 | 12430113 | 29.98 |
Identification of differentially
expressed circRNAs
The overall expression profiles of the
differentially expressed circRNAs between the
CCl4-induced and PZH-treated groups are displayed in the
volcano plot in Fig. 2C. In
total, 91 circRNAs, including 58 (63.73%) upregulated and 33
(36.27%) downregulated circRNAs, were demonstrated to be
significantly differentially expressed in liver tissue samples from
the PZH-treated compared with the CCL4-induced group
(Fig. 2A). An analysis of the
source of these circRNAs revealed that, of the 91 differentially
expressed circRNAs, 88 (96.7%) were derived from exons (data not
shown). The results also demonstrated that the most significant
differentially expressed circRNAs were transcribed from the exons
of protein-coding regions (Fig.
2B). The distribution of the identified circRNAs on different
chromosomes was also analyzed. The statistical analysis results
demonstrated that circRNAs were distributed on all chromosomes.
Moreover, all differentially expressed circRNAs were found to be
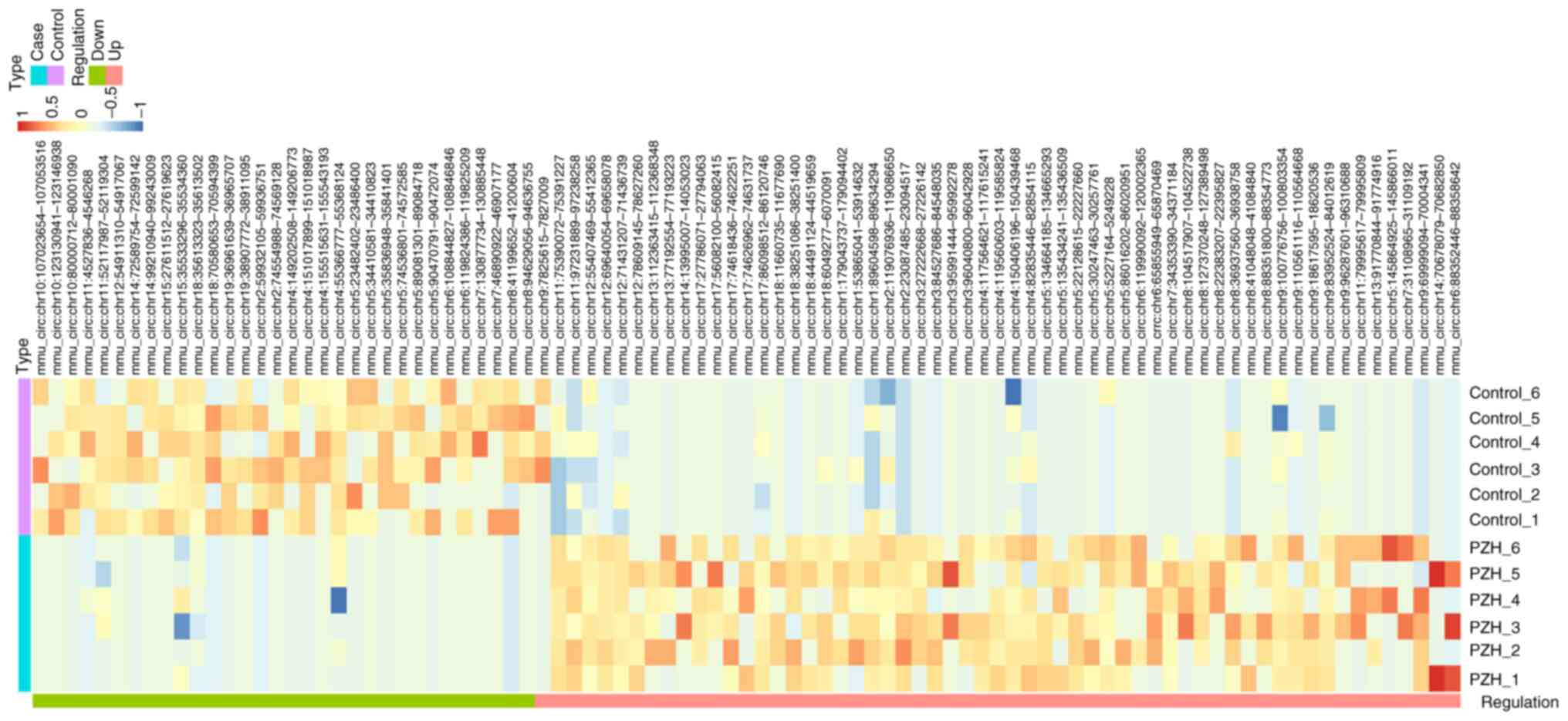
markedly expressed, except for those on chromosome 16 (Fig. 2D). Hierarchical clustering was
also performed to investigate these differentially expressed
circRNAs across 12 samples between PZH-treated and
CCL4-induced groups (Fig.
3). Detailed information on the top 20 differentially expressed
circRNAs is included in Table
SIV. The analysis of the circRNA-seq data suggested that the
expression levels of circRNA differed between PZH treatment and
CCL4-induced fibrotic livers.
Prediction of differentially expressed
circRNA-related target genes and construction of circRNA/miRNA/mRNA
networks
The identification of circRNA target genes is
crucial for characterizing circRNA function. One of the molecular
functions of circRNAs is their role as a ceRNA, which regulate
miRNAs and consequently modulate mRNA expression (24,49). The interactions among circRNA,
miRNA and mRNA were assessed based on the differentially expressed
circRNAs between PZH-treated and the CCl4-inducted
groups; the results showed that the upregulated circRNAs had
interactions with 139 miRNA binding sites and 927 target genes. A
total of 103 potential miRNAs binding sites and 887 target genes of
these miRNAs were also identified on downregulated circRNAs
(Table SV). Multiple
circRNA-related biomarkers associated with liver fibrosis were also
identified using circRNA-seq. However, the number of putative
biomarkers was too large to accurately identify the biomarkers that
may be involved in PZH treatment of liver fibrosis. Therefore, only
the predicted target genes of the differentially expressed circRNAs
were selected for further analysis.
The predicted set of functional genes for up- or
downregulated circRNAs was compared with a set of differentially
expressed mRNA genes enriched in the mRNA-seq data (these mRNAs
were enriched in the same treatment conditions as those in the
present study) (GEO accession no. GSE133481). The shared genes
between these two datasets were recorded. In the upregulated
circRNAs, only 10 overlapping genes from the 927 predicted mRNAs
and 294 upregulated mRNAs from the mRNA-seq data were identified.
Subsequently, the circRNA/miRNA/mRNA regulatory network of these
related miRNAs and the corresponding significantly upregulated
circRNA-binding sites were constructed (Fig. 4A). The results demonstrated that
10 upregulated mRNAs were found to interact with 15 miRNAs, which
were involved in the regulation of 17 circRNAs. Among the 15 miRNAs
in the circRNA/miRNA/mRNA regulatory network, mmu-miR-466i-5p was
targeted by seven identified upregulated circRNAs and exhibited the
largest interaction network. miR-345-3p was predicted to be
negatively modulated by mmu_circ:chromosome
(chr)9:69999094|70004341 and mmu_circ:chr2:119076936|119086650. The
following top five most upregulated circRNAs appeared to be
associated with the largest binding miRNA network:
mmu_circ:chr17:74626961|74631737,
mmu_circ:chr2:119076936|119086650,
mmu_circ:chr13:112363415|112368348,
mmu_circ:chr17:27786071|27794063 and
mmu_circ:chr9:69999094|70004341. In the downregulated circRNAs, 21
overlapping genes from the 887 predicted mRNAs and 558
downregulated mRNAs from the mRNA-seq data were identified.
Similarly, the circRNA/miRNA/mRNA regulatory network of these
related miRNAs and the corresponding significantly downregulated
circRNA-binding sites were constructed (Fig. 4B). The results demonstrated that
21 downregulated mRNAs were found to interact with 27 miRNAs, which
were involved in the regulation of 12 downregulated circRNAs. The
following top five most downregulated circRNAs were found to be
associated with the largest binding miRNA network:
mmu_circ:chr12:54911310|54917067, mmu_circ:chr15:27611512|27619623,
mmu_circ:chr4:149202507|149206773, mmu_circ:chr5:23482401|23486400
and mmu_circ:chr5:89081301|89084718. The expression information for
the dysregulated circRNAs and their related target genes involved
in circRNA/miRNA/mRNA interaction networks are listed in Tables III and IV, respectively. A flowchart
summarizing the circRNA/miRNA/mRNA interaction network associated
with the dysregulated circRNAs, as well as the results of the
subsequent target gene analysis, is displayed in Fig. 5.
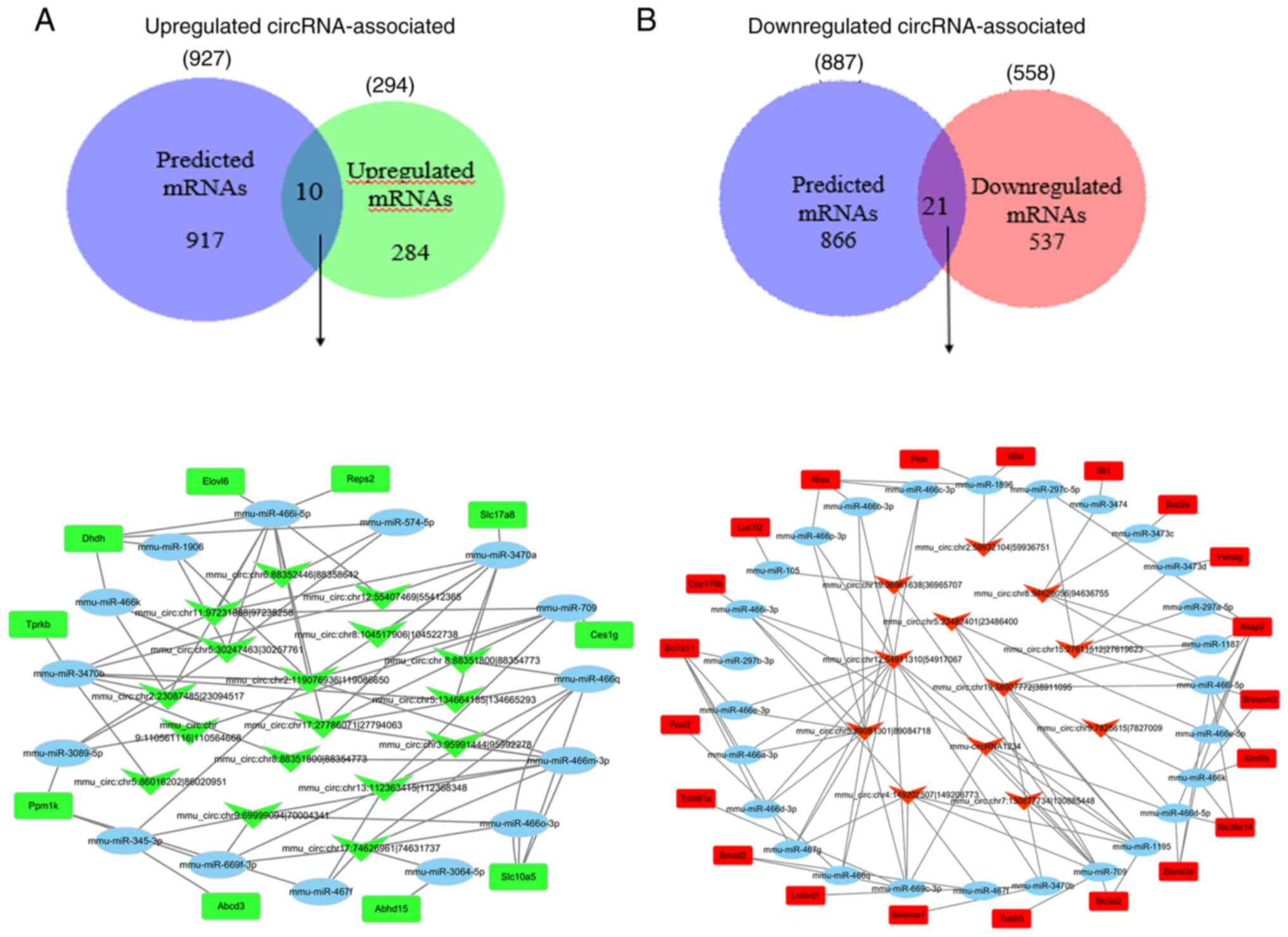 | Figure 4.circRNA-seq data analysis. (A)
Between the upregulated circRNA-related target genes and the
upregulated genes obtained from the mRNA-seq data, 10 overlapping
genes were identified. The circRNA/miRNA/mRNA regulatory network of
their related miRNAs and their corresponding significantly
upregulated circRNA-binding sites was constructed. The network
consisted of 17 circRNAs (arrowheads), 15 miRNAs (ellipses), 10
mRNAs (rectangles) and 75 connections. (B) Between the
downregulated circRNA-related target genes and the downregulated
genes obtained from the mRNA-seq data 21 overlapping genes were
identified. The circRNA/miRNA/mRNA regulatory network of their
related miRNAs and the corresponding significantly downregulated
circRNA-binding sites was constructed. The network consisted of 12
circRNAs (arrowheads), 27 miRNAs (ellipses), 21 mRNAs (rectangles)
and 101 connections. Abcd3, ATP-binding cassette subfamily D,
member 3; Abhd15, abhydrolase domain-containing 15; Akap2,
A-kinase-anchoring protein 2; Baz2a, bromodomain adjacent to
zinc-finger domain 2A; Cep170b, centrosomal protein 170B;
Ces1gES1G, carboxylic ester hydrolase; chr, chromosome; circRNA,
circular RNA; Dhdh, dihydrodiol dehydrogenase; Fktn, fukutin;
Fosl2, FOS-like 2, AP-1 transcription factor subunit; Ifit1,
interferon induced protein with tetratricopeptide repeats 1; Il6st,
interleukin 6 cytokine family signal transducer; Kmt5b, lysine
methyltransferase 5B; Lmbrd1, LMBR1 domain-containing 1; Luc7l2,
putative RNA-binding protein Luc7-like 2; miR, microRNA; seq,
sequencing; mmu, Mus musculus; Nras, NRAS proto-oncogene
GTPase; Ppm1k, protein phosphatase Mg2+/Mn2+
dependent 1K; Serpine1, serpin family E, member 1; Shroom3, shroom
family member 3; Slc3a2, solute carrier family 3, member 2;
Slc7a11, solute carrier family 7, member 11; Slc10a5, solute
carrier family 10, member 5; Slc17a8, solute carrier family 17,
member 8; Slc39a14, solute carrier family 39, member 14; Tprkb,
TP53RK binding protein; TRMT61A, tRNA methyltransferase 61A; Tubb5,
tubulin b class I; Ywhag, tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein g; Zfand2a, zinc-finger
AN1-type-containing 2A Smad2, SMAD family member 2; Trmt61a, tRNA
methyltransferase 61A; Reps2, RALBP1-associated Eps
domain-containing 2; Elov16, ELOVL fatty acid elongase 6. |
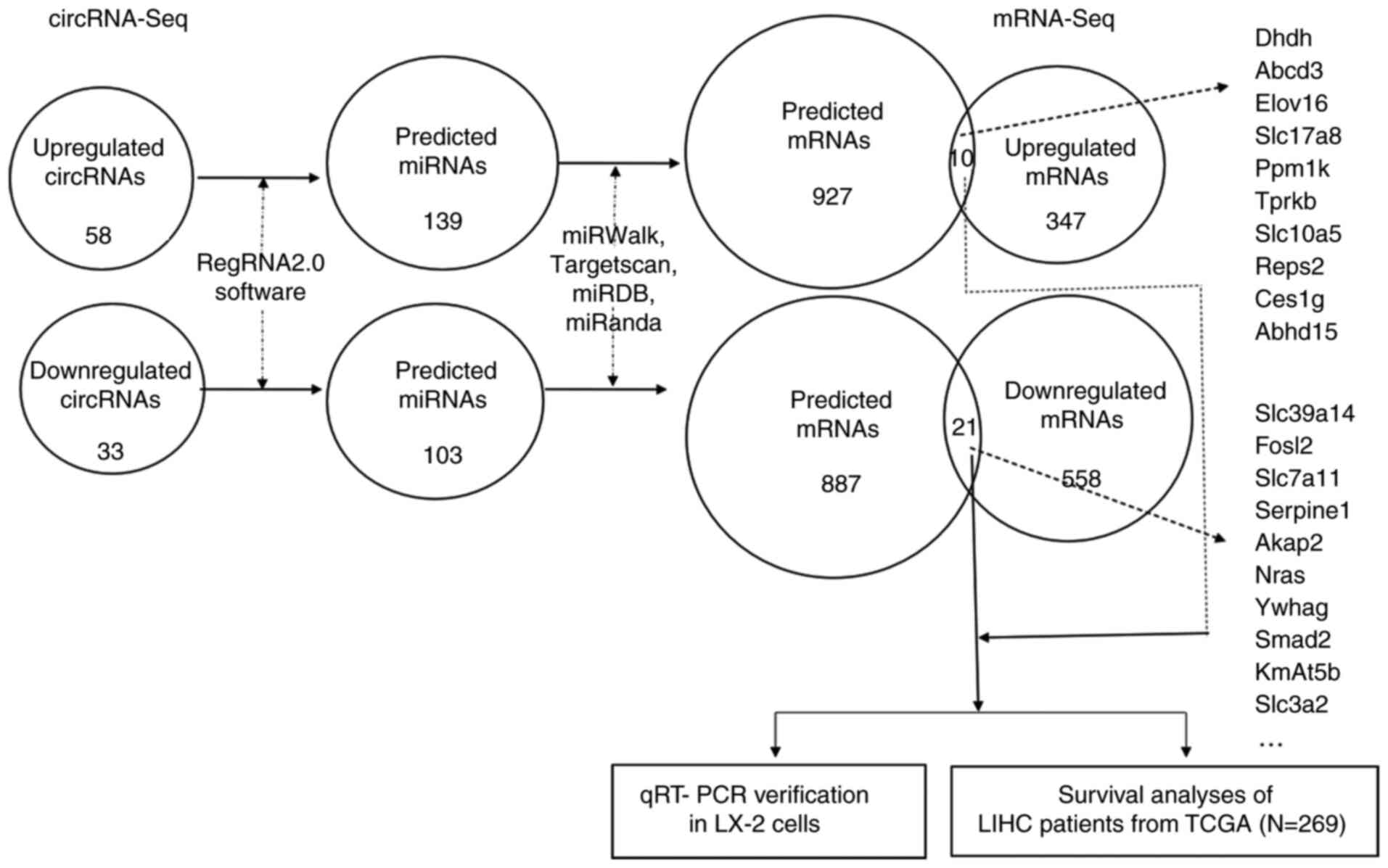 | Figure 5.Bioinformatics analysis and
experimental verification of up- and downregulated circRNAs between
the PZH-treated and untreated control group. The shared mRNAs
between the differentially expressed circRNA-related predicted
target genes in the present study and the differentially expressed
mRNAs from mRNA-seq data (GEO accession no. GSE133481) were
extracted for further analysis. Abcd3, ATP-binding cassette
subfamily D, member 3; Abhd15, abhydrolase domain-containing 15;
Akap2, A-kinase-anchoring protein 2; Ces1g, carboxylic ester
hydrolase; circRNA, circular RNA; Dhdh, dihydrodiol dehydrogenase;
Elov16, ELOVL fatty acid elongase 6; Fosl2, FOS-like 2, AP-1
transcription factor subunit; Kmat5b, lysine methyltransferase 5B;
LIHC, liver hepatocellular carcinoma; miR, microRNA; Nras, NRAS
proto-oncogene GTPase; Ppm1k, protein phosphatase
Mg2+/Mn2+ dependent 1K; PZH, Pien Tze Huang;
Reps2, RALBP1-associated Eps domain-containing 2; RT-qPCR, reverse
transcription-quantitative PCR; Serpine1, serpin family E, member
1; seq, sequencing; Slc3a2, solute carrier family 3, member 2;
Slc7a11, solute carrier family 7, member 11; Slc10a5, solute
carrier family, 10 member 5; Slc17a8, solute carrier family 17,
member 8; Slc39a14, solute carrier family 39, member 14; TCGA, The
Cancer Genome Atlas; Tprkb, TP53RK-binding protein; Ywhag, tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
γ. |
 | Table III.Information on identified
dysregulated circRNAs. |
Table III.
Information on identified
dysregulated circRNAs.
| circRNA ID | Gene | Log2
fold-change | Regulation | P-value |
|---|
|
mmu_circ:chr9:69999094|70004341 | Bnip2 | 2.669154139 | Up |
3.41×10−7 |
|
mmu_circ:chr3:95991444|95992278 | Plekho1 | 1.817786326 | Up |
1.82×10−2 |
|
mmu_circ:chr2:119076936|119086650 | Knl1 | 1.020291023 | Up |
4.89×10−2 |
|
mmu_circ:chr2:23087485|23094517 | Acbd5 | 2.518912306 | Up |
4.36×10−9 |
|
mmu_circ:chr6:88352446|88358642 | Eefsec | 2.828790508 | Up |
5.39×10−5 |
|
mmu_circ:chr11:97231888|97238258 | Npepps | 1.851337918 | Up |
1.47×10−2 |
|
mmu_circ:chr5:30247463|30257761 | Selenoi | 1.219232659 | Up |
2.83×10−2 |
|
mmu_circ:chr17:27786071|27794063 | D17Wsu92e | 1.065565101 | Up |
3.40×10−2 |
|
mmu_circ:chr12:55407469|55412365 | Psma6 | 1.056995708 | Up |
4.91×10−2 |
|
mmu_circ:chr5:134664185|134665293 | Limk1 | 1.514958004 | Up |
1.56×10−3 |
|
mmu_circ:chr8:104517906|104522738 | Nae1 | 1.699428017 | Up |
9.48×10−3 |
|
mmu_circ:chr8:88351800|88354773 | Brd7 | 1.258290693 | Up |
4.82×10−2 |
|
mmu_circ:chr17:74626961|74631737 | Birc6 | 1.408138487 | Up |
1.64×10−2 |
|
mmu_circ:chr13:112363415|112368348 | Ankrd55 | 1.086885064 | Up |
3.34×10−2 |
|
mmu_circ:chr9:110561116|110564668 | Setd2 | 1.583016549 | Up |
4.16×10−2 |
|
mmu_circ:chr5:86016202|86020951 | Cenpc1 | 1.286666981 | Up |
2.53×10−2 |
|
mmu_circ:chr8:88351800|88354773 | Brd7 | 1.258290693 | Up |
4.82×10−2 |
|
mmu_circ:chr5:89081301|89084718 | Slc4a4 | −1.033058375 | Down |
4.03×10−2 |
|
mmu_circ:chr12:54911310|54917067 | Baz1a | −1.044583375 | Down |
4.22×10−2 |
|
mmu_circ:chr2:59932104|59936751 | Baz2b | −1.850754348 | Down |
1.53×10−3 |
|
mmu_circ:chr8:94629056|94636755 | Rspry1 | −1.304377354 | Down |
4.43×10−2 |
|
mmu_circ:chr19:36961638|36965707 | Btaf1 | −1.363049915 | Down |
1.51×10−2 |
|
mmu_circ:chr15:27611512|27619623 | Otulin | −1.229226523 | Down |
1.72×10−2 |
|
mmu_circ:chr19:38907772|38911095 | Tbc1d12 | −1.144664409 | Down |
2.61×10−2 |
|
mmu-circRNA1234 | Fndc3a | −1.384170351 | Down |
1.41×10−2 |
|
mmu_circ:chr9:7825615|7827009 | Birc2 | −1.539082966 | Down |
2.21×10−2 |
|
mmu_circ:chr4:149202507|149206773 | Kif1b | −1.503481469 | Down |
2.16×10−3 |
|
mmu_circ:chr5:23482401|23486400 | Kmt2e | −1.397731511 | Down |
5.86×10−3 |
|
mmu_circ:chr7:130877734|130885448 | Plekha1 | −1.335857428 | Down |
3.53×10−2 |
 | Table IV.Overlapping genes between
dysregulated circular RNA-associated target genes and
differentially expressed mRNAs. |
Table IV.
Overlapping genes between
dysregulated circular RNA-associated target genes and
differentially expressed mRNAs.
| Gene | Log2
fold-change | Regulation | P-value | Full name |
|---|
| Abcd3 | 1.0280 | Up |
7.92×10−6 | ATP-binding
cassette subfamily D, member 3 |
| Elovl6 | 1.1377 | Up |
2.83×10−4 | ELOVL fatty acid
elongase 6 |
| Slc17a8 | 1.2805 | Up |
2.05×10−6 | Solute carrier
family 17, member 8 |
| Ppm1k | 1.0442 | Up |
4.30×10−4 | Protein
phosphatase, Mg2+/Mn2+ dependent 1K |
| Tprkb | 1.3508 | Up |
2.78×10−2 | TP53RK-binding
protein |
| Slc10a5 | 1.6327 | Up |
1.58×10−6 | Solute carrier
family 10, member 5 |
| Reps2 | 1.1568 | Up |
2.75×10−2 | RALBP1-associated
Eps domain-containing 2 |
| Ces1g | 1.2722 | Up |
6.21×10−5 | Carboxylesterase
1G |
| Dhdh | 1.4262 | Up |
1.26×10−6 | Dihydrodiol
dehydrogenase |
| Abhd15 | 1.1437 | Up |
6.69×10−5 | Abhydrolase
domain-containing 15 |
| Nras | −1.0682 | Down |
3.79×10−2 | NRAS
proto-oncogene, GTPase |
| Baz2a | −1.1493 | Down |
2.07×10−2 | Bromodomain
adjacent to zinc-finger domain 2A |
| Luc7l2 | −1.2942 | Down |
2.23×10−2 | LUC7 like 2,
pre-mRNA splicing factor |
| Ywhag | −1.1007 | Down |
5.95×10−6 | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
γ |
| Serpine1 | −3.3643 | Down |
5.52×10−5 | Serpin family E,
member 1 |
| Zfand2a | −1.2588 | Down |
1.21×10−2 | Zinc-finger
AN1-type-containing 2A |
| Smad2 | −1.1590 | Down |
3.52×10−3 | SMAD family member
2 |
| Kmt5b | −1.4307 | Down |
2.56×10−2 | Lysine
methyltransferase 5B |
| Slc3a2 | −1.5998 | Down |
9.27×10−7 | Solute carrier
family 3, member 2 |
| Trmt61a | −1.0673 | Down |
1.30×10−4 | tRNA
methyltransferase 61A |
| Cep170b | −1.5367 | Down |
7.12×10−3 | Centrosomal protein
170B |
| Slc7a11 | −2.2590 | Down |
1.40×10−5 | Solute carrier
family 7, member 11 |
| Il6st | −1.0635 | Down |
4.03×10−5 | Interleukin 6
signal transducer |
| Ifit1 | −1.0547 | Down |
4.36×10−3 | Interferon-induced
protein with tetratricopeptide repeats 1 |
| Tubb5 | −1.2318 | Down |
9.96×10−7 | Tubulin, β5 class
I |
| Lmbrd1 | −1.4803 | Down |
2.68×10−2 | LMBR1
domain-containing 1 |
| Fktn | −1.0167 | Down |
4.39×10−3 | Fukutin |
| Akap2 | −1.2242 | Down |
1.25×10−3 | A-kinase anchoring
protein 2 |
| Fosl2 | −1.2528 | Down |
5.73×10−5 | FOS-like 2, AP-1
transcription factor subunit |
| Slc39a14 | −1.0692 | Down |
5.52×10−4 | Solute carrier
family 39, member 14 |
| Shroom3 | −1.2148 | Down |
1.19×10−2 | Shroom family
member 3 |
Host gene enrichment analysis of
differentially expressed circRNA-related target genes
The function of all differentially expressed
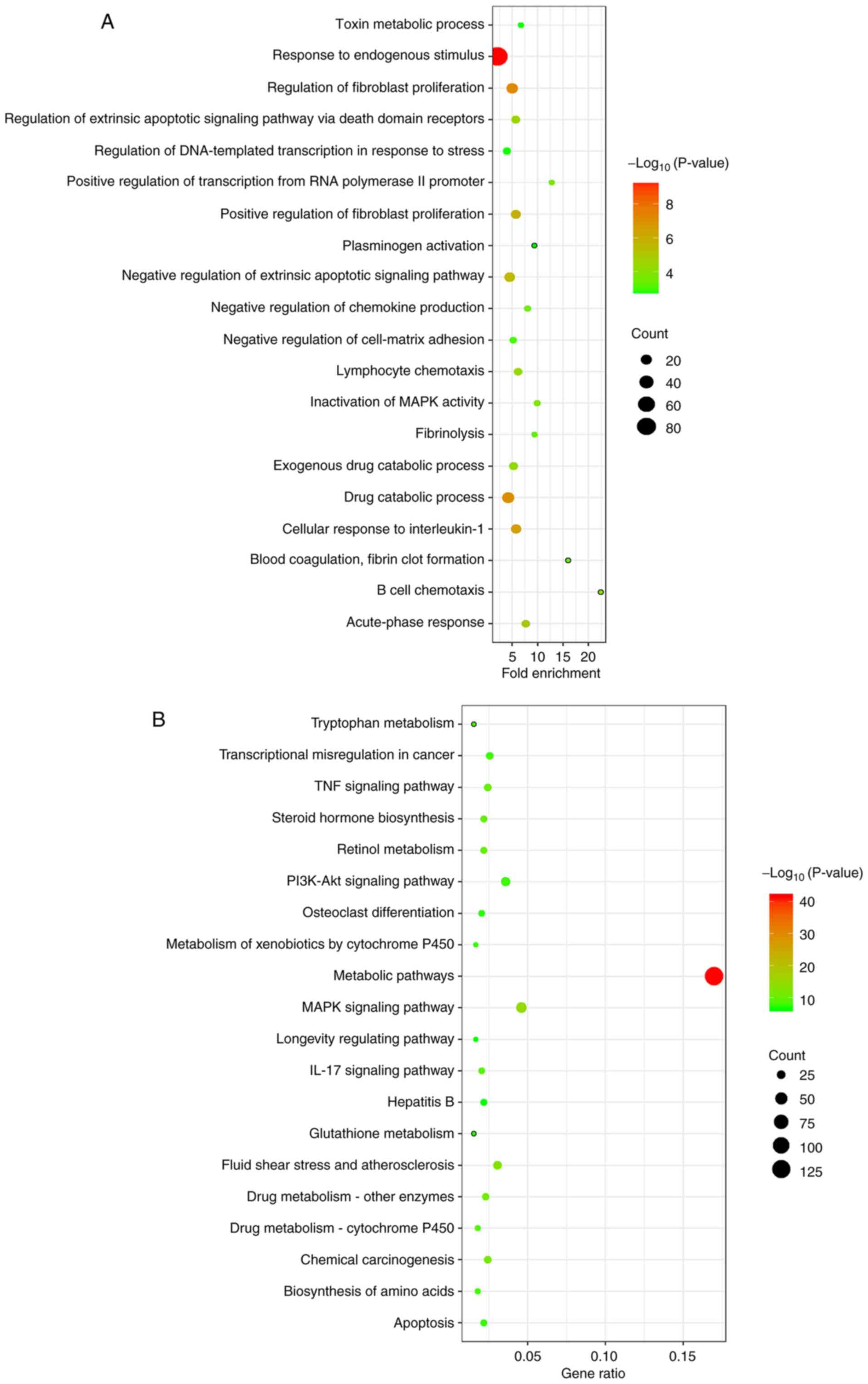
circRNA-related target genes was further explored. The GO analysis
results demonstrated that the target genes mainly participated in
the biological processes of ‘positive regulation of fibroblast
proliferation’, ‘response to endogenous stimulus’, ‘drug catabolic
process’ and ‘regulation of DNA-templated transcription in response
to stress’ (Fig. 6A). KEGG
pathways were also identified for differentially expressed
circRNA-related target genes. KEGG analysis identified the
following enriched pathways: ‘Metabolic pathways’, ‘TNF signaling
pathway’, ‘PI3K-Akt signaling pathway’, ‘IL-17 signaling pathway’,
‘MAPK signaling pathway’ and ‘apoptosis’ (P<0.05; Fig. 6B). Meanwhile, the results of GO
pathway analyses showed that the differentially expressed
circRNA-related target genes in this network were associated with
cellular components, such as ‘fibrinogen complex’, ‘transcription
factor AP-1 complex’ and ‘platelet alpha granule’; and molecular
functions, such as ‘ketosteroid monooxygenase activity’ and ‘MAP
kinase tyrosine/serine/threonine and phosphatase activity’
(Fig. S2).
Validation of candidate
circRNA-related target genes by RT-qPCR
To verify the results of the co-expression network
analysis, seven overlapping circRNA-related target gene candidates
were selected for RT-qPCR in LX-2 cells in the
CCL4-induced and PZH-treated group (Table V). mRNA expression levels of the
seven gene candidates were subsequently detected 24, 48 and 72 h
following PZH treatment (0.75 mg/ml). The seven selected
transcripts included: Tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein γ (YWHAG); solute carrier family
39, member 14 (SLC39A14); FOS-like 2, AP-1 transcription factor
subunit (FOSL2); solute carrier family 7, member 11 (SLC7A11);
serpin family E, member 1 (SERPINE1); ATPase
sarcoplasmic/endoplasmic reticulum Ca2+ (ATP2A2); and
NRAS proto-oncogene GTPase (NRAS). Among these seven, seven
(SLC39A14, FOSL2, SLC7A11, SERPINE1, ATP2A2 and NRAS) were
significantly differentially expressed compared with the control
(Fig. 7). SLC39A14, FOSL2,
SLC7A11, SERPINE1, ATP2A2 and NRAS mRNA expression levels were
significantly downregulated in the PZH-treated group compared with
the control group, while YWHAG was significantly upregulated.
Overall, the results of the RT-qPCR validation of overlapping
candidate circRNA-related target genes were consistent with the
mRNA-seq results.
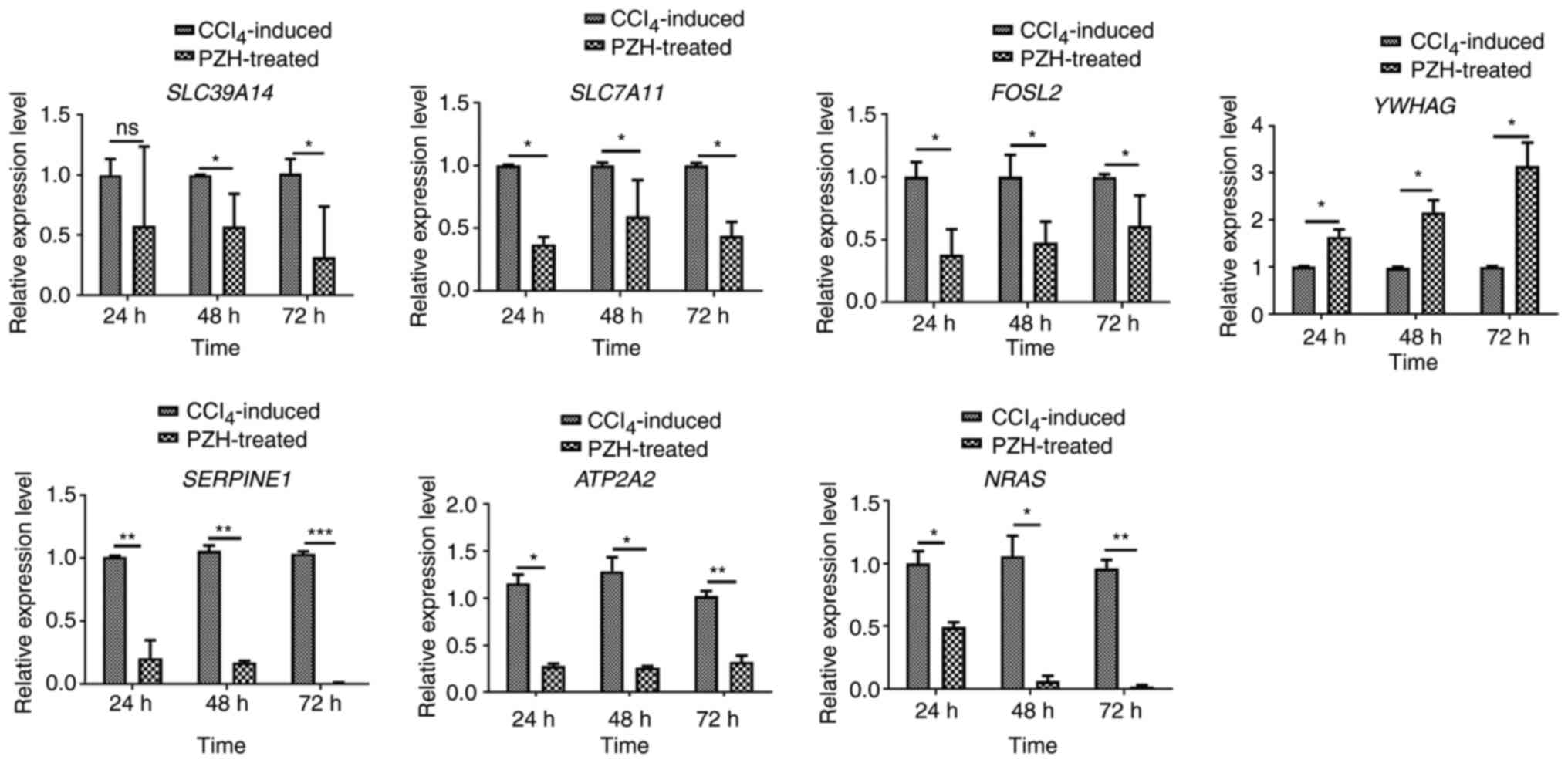 | Figure 7.Validation of target genes displayed
in competing endogenous RNA networks in LX-2 cells by reverse
transcription-quantitative PCR. Data are presented as the median ±
SEM. *P<0.05, **P<0.01 and ***P<0.001. ATP2A2, ATPase
sarcoplasmic/endoplasmic reticulum Ca2+; FOSL2, FOS-like
2, AP-1 transcription factor subunit; NRAS, NRAS proto-oncogene
GTPase; PZH, Pien Tze Huang; SERPINE1, serpin family E, member 1;
SLC7A11, solute carrier family 7, member 11; SLC39A14, solute
carrier family 39, member 14. |
 | Table V.Selected candidate circular
RNA-associated target genes for reverse transcription-quantitative
PCR. |
Table V.
Selected candidate circular
RNA-associated target genes for reverse transcription-quantitative
PCR.
| Genea | Full name | Role in liver
injury | (Refs.) |
|---|
| Slc7a11 | Solute carrier
family 7, member 11 | Induced in
activated hepatic stellate cells | (78) |
| Serpine1 | Serpin family E,
member 1 | Progression of
fibrosis | (79,80) |
| Fosl2 | FOS-like 2, AP-1
transcription factor subunit | Progression of
fibrosis | (67) |
| Atp2a2 | A-kinase anchoring
protein 2 | Progression of
fibrosis | (81) |
| Nras | NRAS
proto-oncogene, GTPase | Progression of
hepatocellular carcinoma | (82) |
| Slc39a14 | Solute carrier
family 39, member 14 | Progression of
fibrosis | (83) |
| Ywhag | Tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein
γ | Progression of
fibrosis | (84) |
Survival analysis between patient
mortality and differentially expressed circRNA-related target gene
expression levels in liver hepatocellular carcinoma (LIHC)
To explore the relationship between the expression
levels of overlapping functional genes associated with
differentially expressed circRNAs and patient survival, 269
patients with LIHC from TCGA database were divided into low- and
high-expression groups based on the median values. The mRNA
expression of 2 out of the 31 overlapping functional genes was
significantly associated with patient survival in LIHC (Fig. 8). These were dihydrodiol
dehydrogenase (DHDH) and SLC7A11. Kaplan-Meier survival analysis
demonstrated that patients with low DHDH expression levels
exhibited a greater survival rate compared with those with high
DHDH expression levels (log-rank test, P=0.0057; Fig. 8A). The results for DHDH were also
consistent with the survival rate of patients with LIHC based on
SLC7A11 mRNA expression analysis (log-rank test, P=0.017; and
Fig. 8B). However, Kaplan-Meier
survival analysis of the remaining genes, including
RALBP1-associated Eps domain-containing 2 (REPS2), SERPINE1, solute
carrier family 3, member 2 (SLC3A2) and solute carrier family 10,
member 5 (SLC10A5), indicated that although the difference in LIPC
survival rates between high-expression and low-expression groups
was not statistically significant (log-rank test, P=0.07, P=0.21,
P=0.22 and P=0.33, respectively), the mRNA expression levels of
these genes were still closely associated with the survival of the
patients (Fig. 8C-F).
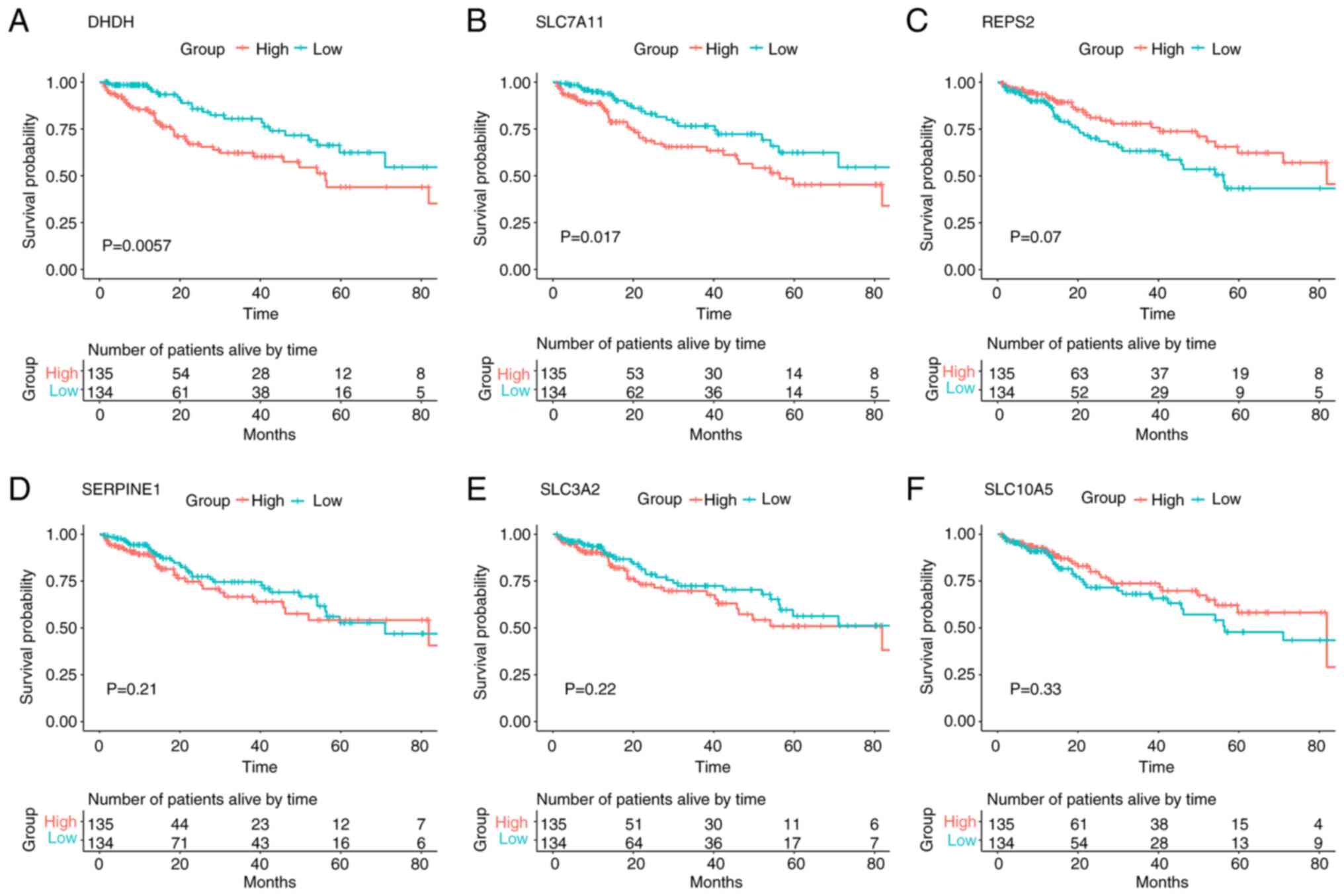 | Figure 8.Kaplan-Meier survival analysis
between the patient clinical outcomes and circular RNA-related
target genes. (A) DHDH, (B) SLC7A11, (C) REPS2, (D) SERPINE1, (E)
SLC3A2 and (F) SLC10A5 gene expression levels in 269 patients with
liver hepatocellular carcinoma from The Cancer Genome Atlas cohort
were used to plot survival curves produced using Kaplan-Meier
survival analysis. Patients exhibiting gene expression levels
higher than the median values were assigned to the high-expression
group, whereas those exhibiting mRNA expression levels lower than
the median values were assigned to the low-expression group. The
log-rank test was used to compare the difference in survival rates
between the high- and low-mRNA expression groups. P<0.05 was
considered to indicate a statistically significant difference.
DHDH, dihydrodiol dehydrogenase; REPS2, RALBP1-associated Eps
domain-containing 2; SLC3A2, solute carrier family 3, member 2;
SLC7A11, solute carrier family 7, member 11; SLC10A5, solute
carrier family 10, member 5; SERPINE1, serpin family E, member
1. |
Discussion
Traditional Chinese Medicine PZH has attracted
considerable attention due to its marked therapeutic effect on
liver injury (15,16). PZH exposure has been reported to
significantly reduce cell necrosis and swelling, microvesicular
steatosis and lymphocyte infiltration in the injured liver
(50). Previous studies have
shown that PZH may affect the immune system by interfering with the
expression of functional genes in immune-related pathways in
CCl4-induced mice (15,50). CCl4 is the mostly used
reagent for the induction of liver injury animal models. Our
previous study demonstrated that the ratio of positive Sirius red
staining against the total area was significantly decreased in the
PZH treatment group compared with the non-treatment group using
Sirius red staining (39). These
results prompted us to explore potential molecular mechanisms
underlying these observed effects in a PZH-treated
CCl4-induced liver injury model. circRNAs have certain
advantages in the development and application of new clinical
diagnostic markers, for example, Ye et al found that
circRNAs participate in the pathogenesis of hepatic injury and
providing efficient targets in the therapy against liver injury
(28), and increasing number of
studies have reported that circRNAs may act as miRNA sponges to
regulate disease occurrence and development (21,24,49). However, to the best of our
knowledge, there are currently no studies on the regulatory
mechanisms of differentially expressed circRNAs in PZH-treated
liver damage using RNA-seq technology. Therefore, mining circRNA
transcript expression profiles using RNA-seq is important for
exploring the pathological mechanism of PZH in the treatment of
liver diseases, especially liver fibrosis. In the present study,
circRNA-seq and bioinformatic analyses were used to explore the
function of circRNAs in PZH-treated mice with liver fibrosis. The
circRNA expression profile identified 91 differentially expressed
circRNAs between the PZH-treated and CCl4-induced
groups, of which 58 were upregulated and 33 were downregulated. To
the best of our knowledge this was the first study to provide
evidence that circRNAs are differentially expressed in PZH-treated
fibrotic liver tissue compared with the controls, which used
CCl4 only.
CircRNAs act as miRNA sponges that regulate target
gene expression by binding to miRNAs (24). The present study predicted that
there were 242 miRNA-binding sites on circRNAs and 994
differentially expressed circRNA-related target genes using related
software, and these results were similar to previous reports in
human studies (26,27,29). To gain insights into the function
of differentially expressed circRNAs, GO term and KEGG pathway
enrichment analyses were performed. From the KEGG pathway
enrichment analysis, it was determined that all differentially
expressed circRNA-related target genes were found to be involved in
several crucial pathways, including ‘metabolic pathways’, ‘TNF
signaling pathway’, ‘PI3K-Akt signaling pathway’, ‘IL-17 signaling
pathway’, ‘MAPK signaling pathway’ and ‘apoptosis’. Previous
studies have reported that the PI3K/Akt signaling pathway is
important for HSC activation and apoptosis, as well as the
regulation of proliferation in liver fibrosis (51–53). Shu et al (54) demonstrated that the inhibition of
the MAPK signaling pathway alleviates CCl4-induced liver
fibrosis in Toll-like receptor 5-deficient mice. Furthermore,
Ghallab et al (55)
identified that the TGF-β and TNF-α inflammatory pathways were
activated by influencing lobular zonation of liver fibrosis.
Moreover, the activation of the IL-17 signaling pathway may inhibit
the development of liver fibrosis (56,57). It can therefore be hypothesized
that the differential expression of circRNAs in these signaling
pathways serves an important role in PZH-induced improvement in the
fibrotic liver.
In the present study, a co-expression network was
constructed based on the circRNA-seq and mRNA-seq data to
investigate their interactions. The predicted set of functional
genes for up- or downregulated circRNAs overlapped with a set of
differentially expressed mRNA genes. In the upregulated circRNAs,
10 overlapping genes were identified and the circRNA/miRNA/mRNA
regulatory network of 15 related miRNAs and a corresponding 17
significantly upregulated circRNA-binding sites on these miRNAs was
constructed. Hyun et al (58) demonstrated that miR-466i-5p was
significantly upregulated in the CCl4-induced liver
fibrosis model group, which further aggravated the activation of
HSCs and induced liver fibrosis. Another report suggested that
ELOVL fatty acid elongase 6 (ELOV16) expression was significantly
downregulated in human lungs with idiopathic pulmonary fibrosis
(59). The results of the present
study indicated that seven of the identified upregulated circRNAs
may serve an important role in the therapeutic effect of PZH in the
fibrotic liver by decreasing the expression levels of miR-466i-5p
and further increasing those of ELOV16. Previous studies have also
reported that miR-345-3p upregulation is involved in the
pathogenesis of liver fibrosis and hepatocellular carcinoma (HCC)
by negatively regulating cyclin-dependent kinase inhibitor 1 in
cancer cells (57,60). This result indicated that the
overexpression of these two circRNAs may induce pulmonary fibrosis
by decreasing miR-345-3p expression levels and consequently
increasing that of ATP-binding cassette subfamily D, member 3. In
the downregulated circRNAs, 21 overlapping genes were identified
and the circRNA/miRNA/mRNA regulatory network of 27 related miRNAs
and the corresponding 12 significantly downregulated
circRNA-binding sites on these miRNAs was constructed. A previous
study reported that miRNA-105 expression is markedly downregulated
in both HCC cell lines and clinical HCC tissues, compared with
normal human hepatocyte and adjacent non-cancerous tissues
(61). The results of the present
study found 12 significantly downregulated circRNA-binding sites in
PZH-treated fibrotic livers in model mice. An et al
(62) demonstrated that miR-467f
is downregulated in acute liver failure compared with mock-treated
livers. Furthermore, the increased expression of SMAD2 has been
demonstrated to serve a vital role in the development of liver
fibrosis (63). The results of
the present study indicated that downregulated
mmu_circ:chr4:149202507|149206773 may participate in the
therapeutic effects of PZH on the fibrotic liver by decreasing the
expression of SMAD2. Overall, these results demonstrated that
differentially expressed circRNAs may alter the expression of
certain functional genes via the circRNA/miRNA/mRNA regulatory
network, therefore mediating PZH-treated liver fibrosis. It was
therefore hypothesized that differentially expressed circRNAs may
act as miRNA sponges and serve an important role in PZH-treated
liver fibrosis by preventing miRNAs from regulating their target
mRNAs.
The RT-qPCR data for the six differentially
expressed circRNA-related target genes in the LX-2 cell model were
consistent with the trends observed in the mRNA-seq data. Among the
verified genes, SLC7A11 was the one most widely investigated in
liver damage. It has previously been reported that inhibiting
SLC7A11 induces ferroptosis in myofibroblastic HSCs and protects
against liver fibrosis (64).
Zhang et al (65) reported
that the upregulation of SLC7A11 is an indicator of unfavorable
prognosis in liver carcinoma. Furthermore, SERPINE1 has been
demonstrated to serve an important role in the development of liver
fibrosis. Lodder et al (66) found that increased expression of
gene SERPINE1 aggravating the degree of liver fibrosis, liver cell
damage and inflammation in myeloid cells. A previous study also
demonstrated that homolog Fos-related antigen 2, encoded by FOSL2,
is a contributing pathogenic factor of pulmonary fibrosis in humans
(67). Furthermore, NRAS is a
proto-oncogene, whose activating mutation has been linked to
several types of human cancer, including HCC (68,69). Previous studies have demonstrated
that sustained NRAS activation resulting from the overexpression of
constitutively active NRAS, induces HCC in genetically compromised
mice (70,71). In the present study, the results
of the RT-qPCR validation of the differentially expressed
circRNA-related target genes (SLC7A11, SERPINE1, FOSL2, NRAS,
SLC39A14 and ATP2A2) corresponded to the mRNA-seq results. These
results further suggested that the differentially expressed
circRNAs acted as miRNA sponges in the regulation of target gene
expression to influence the effects of PZH-treatment on liver
fibrosis.
To explore the association between the
differentially expressed circRNA-related targets and the survival
time of patients with LIHC, Kaplan-Meier survival analysis was
performed using data from the TCGA (n=269). The upregulated genes
in PZH-treated liver fibrosis (SLC7A11 and DHDH) were found to be
significantly positively associated with patient survival. Yue
et al (72) reported that
SLC7A11 served an important role in HCC as a potential prognostic
indicator and its overexpression promoted HCC development.
Furthermore, SLC7A11 may be a prognostic factor for liver
carcinoma, as indicated by the survival analysis (73). Elevated levels of plasminogen
activator inhibitor-1 (the protein product of SERPINE1) have been
reported in patients with viral infection-related HCC (74). A number of studies have also
indicated that SERPINE1 may contribute to cancer dissemination
mechanisms, including the prevention of excessive ECM degradation,
modulation of cell adhesion and stimulation of angiogenesis and
cell proliferation (75,76). Wu et al (77) reported that SLC3A2 was highly
expressed in the human HCC cell membrane and may serve an important
role in promoting tumor metastasis and HCC progression. Overall,
these results indicated that a specific set of differentially
expressed circRNA-related functional genes may act as therapeutic
targets for liver injury.
In conclusion, the present study identified 91
differentially expressed circRNAs in a PZH-treated
CCl4-induced liver fibrosis model and the potential
functions of 6 circRNA-related target genes were validated in LX-2
cells. Kaplan-Meier survival analysis further confirmed that two
target mRNAs had potential clinical prognostic value for LIHC.
Meanwhile, a functional circRNA/miRNA/mRNA network was
systematically established to further investigate the underlying
mechanisms of action of differentially expressed circRNAs. Overall,
the present study provided new insights into the mechanisms
underlying the pathogenesis of liver fibrosis and may provide novel
and potentially efficient therapeutic targets against liver injury.
However, a limitation of the present study was that it did not
include any further molecular biology experiments to validate each
of the differentially expressed circRNAs, and thus this will be
investigated future studies.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by grants from The 863 Program (grant
nos. 2012AA02A515 and 2012AA021802), The National Nature Science
Foundation of China (grant nos. 81773818, 81273596, 30900799 and
81671326), The National Key Research and Development Program (grant
nos. 2017YFC0909303, 2016YFC0905000, 2016YFC0905002, 2016YFC1200200
and 2016YFC0906400), The 4th Three-year Action Plan for Public
Health of Shanghai (grant no. 15GWZK0101), The Shanghai Pujiang
Program (grant no. 17PJD020), The Shanghai Key Laboratory of
Psychotic Disorders (grant no. 13dz2260500), The Natural Science
Foundation of Fujian Province (grant no. 2016J05210), The Anhui
Medical University for Scientific Research (grant no. XJ201607) and
The Anhui Medical University for Scientific Research (grant no.
2017×kj006).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI Gene Expression Omnibus
repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150883
(accession no. GSE150883).
Authors' contributions
SQ, FH, LH and JZ designed the study. TW, JZ, DZ,
HW, LC, QX, NZ, YW and LG performed the experiments. TW, LG, JM, MW
and LL performed the data analysis. TW and JZ drafted the
manuscript. All authors have read and approved the final
manuscript. TW and JZ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The mice used in the study were obtained from the
Shanghai Southern Model Animal Center. All experimental procedures
were performed in accordance with the guidelines for the Care and
Use of Laboratory Animals at the Chinese Academy of Animal
Sciences. All animal experiments were approved by the Institutional
Review Board of Shanghai Jiao Tong University (Shanghai, China;
approval no. IACUC.NO:2017-0033).
Patient consent for publication
Not applicable.
Competing interests
The TCM Pien Tze Huang (PZH) was manufactured by
Zhangzhhou Pien Tze Huang Pharmaceutical Co., Ltd., to which LG and
FH are affiliated as employees. All other authors declare that they
have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
CCl4
|
carbon tetrachloride
|
|
ceRNA
|
competing endogenous RNA
|
|
circRNA
|
circular RNA
|
|
ECM
|
extracellular matrix
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
miRNA/miR
|
microRNA
|
|
PZH
|
Pien Tze Huang
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
seq
|
sequencing
|
References
|
1
|
Mederacke I, Hsu CC, Troeger JS, Huebener
P, Mu X, Dapito DH, Pradere JP and Schwabe RF: Fate tracing reveals
hepatic stellate cells as dominant contributors to liver fibrosis
independent of its aetiology. Nat Commun. 4:28232013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schuppan D and Afdhal NH: Liver cirrhosis.
Lancet. 371:838–851. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shrestha N, Chand L, Han MK, Lee SO, Kim
CY and Jeong YJ: Glutamine inhibits CCl4 induced liver fibrosis in
mice and TGF-β1 mediated epithelial-mesenchymal transition in mouse
hepatocytes. Food Chem Toxicol. 93:129–137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeisberg M and Kalluri R: Cellular
mechanisms of tissue fibrosis. 1. Common and organ-specific
mechanisms associated with tissue fibrosis. Am J Physiol Cell
Physiol. 304:C216–C225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wynn TA and Ramalingam TR: Mechanisms of
fibrosis: Therapeutic translation for fibrotic disease. Nat Med.
18:1028–1040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mansour MF, Greish SM, El-Serafi AT,
Abdelall H and El-Wazir YM: Therapeutic potential of human
umbilical cord derived mesenchymal stem cells on rat model of liver
fibrosis. Am J Stem Cells. 8:7–18. 2019.PubMed/NCBI
|
|
8
|
Uehara T, Pogribny IP and Rusyn I: The DEN
and CCl4-induced mouse model of fibrosis and
inflammation-associated hepatocellular carcinoma. Curr Protoc
Pharmacol. 66:14.30.1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Morio LA, Chiu H, Sprowles KA, Zhou P,
Heck DE, Gordon MK and Laskin DL: Distinct roles of tumor necrosis
factor-alpha and nitric oxide in acute liver injury induced by
carbon tetrachloride in mice. Toxicol Appl Pharmacol. 172:44–51.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Steinman L, Martin R, Bernard C, Conlon P
and Oksenberg JR: Multiple sclerosis: Deeper understanding of its
pathogenesis reveals new targets for therapy. Annu Rev Neurosci.
25:491–505. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Watanabe Y, Tsuchiya A, Seino S, Kawata Y,
Kojima Y, Ikarashi S, Starkey Lewis PJ, Lu WY, Kikuta J, Kawai H,
et al: Mesenchymal stem cells and induced bone marrow-derived
macrophages synergistically improve liver fibrosis in mice. Stem
Cells Transl Med. 8:271–284. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Altamirano-Barrera A, Barranco-Fragoso B
and Méndez-Sánchez N: Management strategies for liver fibrosis. Ann
Hepatol. 16:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bozic M and Molleston J: Strategies for
management of pediatric cystic fibrosis liver disease. Clin Liver
Dis (Hoboken). 2:204–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Zhou H, Liu YB, Wang JF, Li H, Ung
CY, Han LY, Cao ZW and Chen YZ: Database of traditional Chinese
medicine and its application to studies of mechanism and to
prescription validation. Br J Pharmacol. 149:1092–1103. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao J, Zhang Y, Wan Y, Hu H and Hong Z:
Pien Tze Huang Gan Bao attenuates carbon tetrachloride-induced
hepatocyte apoptosis in rats, associated with suppression of p53
activation and oxidative stress. Mol Med Rep. 16:2611–2619. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Y, Chen Z, Deng L, Yu J, Wang K,
Zhang X, Ji G and Li F: Pien Tze Huang ameliorates liver injury by
inhibiting the PERK/eIF2α signaling pathway in alcohol and high-fat
diet rats. Acta Histochem. 120:578–585. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin W, Zhuang Q, Zheng L, Cao Z, Shen A,
Li Q, Fu C, Feng J and Peng J: Pien Tze Huang inhibits liver
metastasis by targeting TGF-β signaling in an orthotopic model of
colorectal cancer. Oncol Rep. 33:1922–1928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin YC, Chen YC, Chen TH, Chen HH and Tsai
WJ: Acute kidney injury associated with hepato-protective Chinese
Herb-Pien Tze Huang. J Exp Clin Med. 3:184–186. 2011. View Article : Google Scholar
|
|
19
|
Ragan C, Goodall GJ, Shirokikh NE and
Preiss T: Insights into the biogenesis and potential functions of
exonic circular RNA. Sci Rep. 9:20482019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koh W, Pan W, Gawad C, Fan HC, Kerchner
GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY and Quake SR:
Noninvasive in vivo monitoring of tissue-specific global gene
expression in humans. Proc Natl Acad Sci USA. 111:7361–7366. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44D:D209–D215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang J, Wu X, Sun S, Chen P, Liang X,
Wang J, Ruan J, Zhang S and Zhang X: Circular RNA expression
profile analysis of severe acne by RNA-Seq and bioinformatics. J
Eur Acad Dermatol Venereol. 32:1986–1992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu C, Shi X, Wang AY, Tao Y, Wang Z, Huang
C, Qiao Y, Hu H and Liu L: RNA-Seq profiling of circular RNAs in
human laryngeal squamous cell carcinomas. Mol Cancer. 17:862018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye Z, Kong Q, Han J, Deng J, Wu M and Deng
H: Circular RNAs are differentially expressed in liver
ischemia/reperfusion injury model. J Cell Biochem. 119:7397–7405.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu H, Wang C, Song H, Xu Y and Ji G:
RNA-seq profiling of circular RNAs in human colorectal cancer liver
metastasis and the potential biomarkers. Mol Cancer. 18:82019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bu Q, Long H, Shao X, Gu H, Kong J, Luo L,
Liu B, Guo W, Wang H, Tian J, et al: Cocaine induces differential
circular RNA expression in striatum. Transl Psychiatry. 9:1992019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma X, Zhou Y, Qiao B, Jiang S, Shen Q, Han
Y, Liu A, Chen X, Wei L, Zhou L and Zhao J: Androgen aggravates
liver fibrosis by activation of NLRP3 inflammasome in CCl4-induced
liver injury mouse model. Am J Physiol Endocrinol Metab.
318:E817–E829. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schroeder A, Mueller O, Stocker S,
Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M
and Ragg T: The RIN: An RNA integrity number for assigning
integrity values to RNA measurements. BMC Mol Biol. 7:32006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brown J, Pirrung M and McCue LA: FQC
dashboard: Integrates FastQC results into a web-based, interactive,
and extensible FASTQ quality control tool. Bioinformatics.
33:3137–3139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Utturkar S, Dassanayake A, Nagaraju S and
Brown SD: Bacterial differential expression analysis methods.
Methods Mol Biol. 2096:89–112. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dobin A and Gingeras TR: Optimizing
RNA-seq mapping with STAR. Methods Mol Biol. 1415:245–262. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Glažar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu J, Zhang D, Wang T, Chen Z, Chen L, Wu
H, Huai C, Sun J, Zhang N, Wei M, et al: Target identification of
hepatic fibrosis using Pien Tze Huang based on mRNA and lncRNA. Sci
Rep. 11:169802021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chang TH, Huang HY, Hsu JB, Weng SL, Horng
JT and Huang HD: An enhanced computational platform for
investigating the roles of regulatory RNA and for identifying
functional RNA motifs. BMC Bioinformatics. 14 (Suppl 2):S42013.
View Article : Google Scholar
|
|
41
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Adams JM and Cory S: The Bcl-2 apoptotic
switch in cancer development and therapy. Oncogene. 26:1324–1337.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Austin PC, Lee DS and Fine JP:
Introduction to the analysis of survival data in the presence of
competing risks. Circulation. 133:601–609. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
International Cancer Genome Consortium, .
Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabé RR, Bhan
MK, Calvo F, Eerola I, et al: International network of cancer
genome projects. Nature. 464:993–998. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Piwecka M, Glažar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao J, Hu H, Wan Y, Zhang Y, Zheng L and
Hong Z: Pien Tze Huang Gan Bao ameliorates carbon
tetrachloride-induced hepatic injury, oxidative stress and
inflammation in rats. Exp Ther Med. 13:1820–1826. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Parsons CJ, Takashima M and Rippe RA:
Molecular mechanisms of hepatic fibrogenesis. J Gastroenterol
Hepatol. 22 (Suppl 1):S79–S84. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bai T, Lian LH, Wu YL, Wan Y and Nan JX:
Thymoquinone attenuates liver fibrosis via PI3K and TLR4 signaling
pathways in activated hepatic stellate cells. Int Immunopharmacol.
15:275–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang J, Chu ES, Chen HY, Man K, Go MY,
Huang XR, Lan HY, Sung JJ and Yu J: MicroRNA-29b prevents liver
fibrosis by attenuating hepatic stellate cell activation and
inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget.
6:7325–7338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shu M, Huang DD, Hung ZA, Hu XR and Zhang
S: Inhibition of MAPK and NF-κB signaling pathways alleviate carbon
tetrachloride (CCl4)-induced liver fibrosis in Toll-like receptor 5
(TLR5) deficiency mice. Biochem Biophys Res Commun. 471:233–239.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ghallab A, Myllys M, Holland CH, Zaza A,
Murad W, Hassan R, Ahmed YA, Abbas T, Abdelrahim EA, Schneider KM,
et al: Influence of liver fibrosis on lobular zonation. Cells.
8:15562019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Meng F, Wang K, Aoyama T, Grivennikov SI,
Paik Y, Scholten D, Cong M, Iwaisako K, Liu X, Zhang M, et al:
Interleukin-17 signaling in inflammatory, Kupffer cells, and
hepatic stellate cells exacerbates liver fibrosis in mice.
Gastroenterology. 143:765–776.e3. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang Y, Huang D, Gao W, Yan J, Zhou W,
Hou X, Liu M, Ren C, Wang S and Shen J: Lack of IL-17 signaling
decreases liver fibrosis in murine Schistosomiasis japonica. Int
Immunol. 27:317–325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hyun J, Wang S, Kim J, Rao KM, Park SY,
Chung I, Ha CS, Kim SW, Yun YH and Jung Y: MicroRNA-378 limits
activation of hepatic stellate cells and liver fibrosis by
suppressing Gli3 expression. Nat Commun. 7:109932016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sunaga H, Matsui H, Ueno M, Maeno T, Iso
T, Syamsunarno MR, Anjo S, Matsuzaka T, Shimano H, Yokoyama T and
Kurabayashi M: Deranged fatty acid composition causes pulmonary
fibrosis in Elovl6-deficient mice. Nat Commun. 4:25632013.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shiu TY, Huang SM, Shih YL, Chu HC, Chang
WK and Hsieh TY: Correction: Hepatitis C virus core protein
down-regulates p21Waf1/Cip1 and inhibits curcumin-induced apoptosis
through MicroRNA-345 targeting in human hepatoma cells. PLoS One.
12:e01812992017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shen G, Rong X, Zhao J, Yang X, Li H,
Jiang H, Zhou Q, Ji T, Huang S, Zhang J and Jia H: MicroRNA-105
suppresses cell proliferation and inhibits PI3K/AKT signaling in
human hepatocellular carcinoma. Carcinogenesis. 35:2748–2755. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
An F, Gong B, Wang H, Yu D, Zhao G, Lin L,
Tang W, Yu H, Bao S and Xie Q: miR-15b and miR-16 regulate TNF
mediated hepatocyte apoptosis via BCL2 in acute liver failure.
Apoptosis. 17:702–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Koo JH, Lee HJ, Kim W and Kim SG:
Endoplasmic reticulum stress in hepatic stellate cells promotes
liver fibrosis via PERK-mediated degradation of HNRNPA1 and
up-regulation of SMAD2. Gastroenterology. 150:181–193.e8. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Du K, Oh SH, Dutta RK, Sun T, Yang WH, Chi
JT and Diehl AM: Inhibiting xCT/SLC7A11 induces ferroptosis of
myofibroblastic hepatic stellate cells but exacerbates chronic
liver injury. Liver Int. 41:2214–2227. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang L, Huang Y, Ling J, Zhuo W, Yu Z,
Luo Y and Zhu Y: Overexpression of SLC7A11: A novel oncogene and an
indicator of unfavorable prognosis for liver carcinoma. Future
Oncol. 14:927–936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lodder J, Denaës T, Chobert MN, Wan J,
El-Benna J, Pawlotsky JM, Lotersztajn S and Teixeira-Clerc F:
Macrophage autophagy protects against liver fibrosis in mice.
Autophagy. 11:1280–1292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Eferl R, Hasselblatt P, Rath M, Popper H,
Zenz R, Komnenovic V, Idarraga MH, Kenner L and Wagner EF:
Development of pulmonary fibrosis through a pathway involving the
transcription factor Fra-2/AP-1. Proc Natl Acad Sci USA.
105:10525–10530. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Rajalingam K, Schreck R, Rapp UR and
Albert S: Ras oncogenes and their downstream targets. Biochim
Biophys Acta. 1773:1177–1195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Carlson CM, Frandsen JL, Kirchhof N,
McIvor RS and Largaespada DA: Somatic integration of an
oncogene-harboring sleeping beauty transposon models liver tumor
development in the mouse. Proc Natl Acad Sci USA. 102:17059–17064.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Rudalska R, Dauch D, Longerich T, McJunkin
K, Wuestefeld T, Kang TW, Hohmeyer A, Pesic M, Leibold J, von Thun
A, et al: In vivo RNAi screening identifies a mechanism of
sorafenib resistance in liver cancer. Nat Med. 20:1138–1146. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yue C, Ren Y, Ge H, Liang C, Xu Y, Li G
and Wu J: Comprehensive analysis of potential prognostic genes for
the construction of a competing endogenous RNA regulatory network
in hepatocellular carcinoma. Onco Targets Ther. 12:561–576. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang L, Huang Y, Zhu Y, Yu Z, Shao M and
Luo Y: Identification and characterization of cadmium-related genes
in liver carcinoma. Biol Trace Elem Res. 182:238–247. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Divella R, Mazzocca A, Gadaleta C, Simone
G, Paradiso A, Quaranta M and Daniele A: Influence of plasminogen
activator inhibitor-1 (SERPINE1) 4G/5G polymorphism on circulating
SERPINE-1 antigen expression in HCC associated with viral
infection. Cancer Genomics Proteomics. 9:193–198. 2012.PubMed/NCBI
|
|
75
|
Czekay RP, Aertgeerts K, Curriden SA and
Loskutoff DJ: Plasminogen activator inhibitor-1 detaches cells from
extracellular matrices by inactivating integrins. J Cell Biol.
160:781–791. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Roca C, Primo L, Valdembri D, Cividalli A,
Declerck P, Carmeliet P, Gabriele P and Bussolino F: Hyperthermia
inhibits angiogenesis by a plasminogen activator inhibitor
1-dependent mechanism. Cancer Res. 63:1500–1507. 2003.PubMed/NCBI
|
|
77
|
Wu B, Wang Y, Yang XM, Xu BQ, Feng F, Wang
B, Liang Q, Li Y, Zhou Y, Jiang JL and Chen ZN: Basigin-mediated
redistribution of CD98 promotes cell spreading and tumorigenicity
in hepatocellular carcinoma. J Exp Clin Cancer Res. 34:1102015.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang L, Zhang Z, Li M, Wang F, Jia Y,
Zhang F, Shao J, Chen A and Zheng S: P53-dependent induction of
ferroptosis is required for artemether to alleviate carbon
tetrachloride-induced liver fibrosis and hepatic stellate cell
activation. IUBMB Life. 71:45–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ghosh AK and Vaughan DE: PAI-1 in tissue
fibrosis. J Cell Physiol. 227:493–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ji J, Yu F, Ji Q, Li Z, Wang K, Zhang J,
Lu J, Chen L, E Q, Zeng Y and Ji Y: Comparative proteomic analysis
of rat hepatic stellate cell activation: A comprehensive view and
suppressed immune response. Hepatology. 56:332–349. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Khambu B, Yan S, Huda N, Liu G and Yin XM:
Autophagy in non-alcoholic fatty liver disease and alcoholic liver
disease. Liver Res. 2:112–119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Gao M and Liu D: CRISPR/Cas9-based Pten
knock-out and sleeping beauty transposon-mediated Nras knock-in
induces hepatocellular carcinoma and hepatic lipid accumulation in
mice. Cancer Biol Ther. 18:505–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Cao W, Li Y, Li M, Zhang X and Liao M:
Txn1, Ctsd and Cdk4 are key proteins of combination therapy with
taurine, epigallocatechin gallate and genistein against liver
fibrosis in rats. Biomed Pharmacother. 85:611–619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li R, Wang Y, Song X, Sun W, Zhang J, Liu
Y, Li H, Meng C, Zhang J, Zheng Q and Lv C: Potential regulatory
role of circular RNA in idiopathic pulmonary fibrosis. Int J Mol
Med. 42:3256–3268. 2018.PubMed/NCBI
|