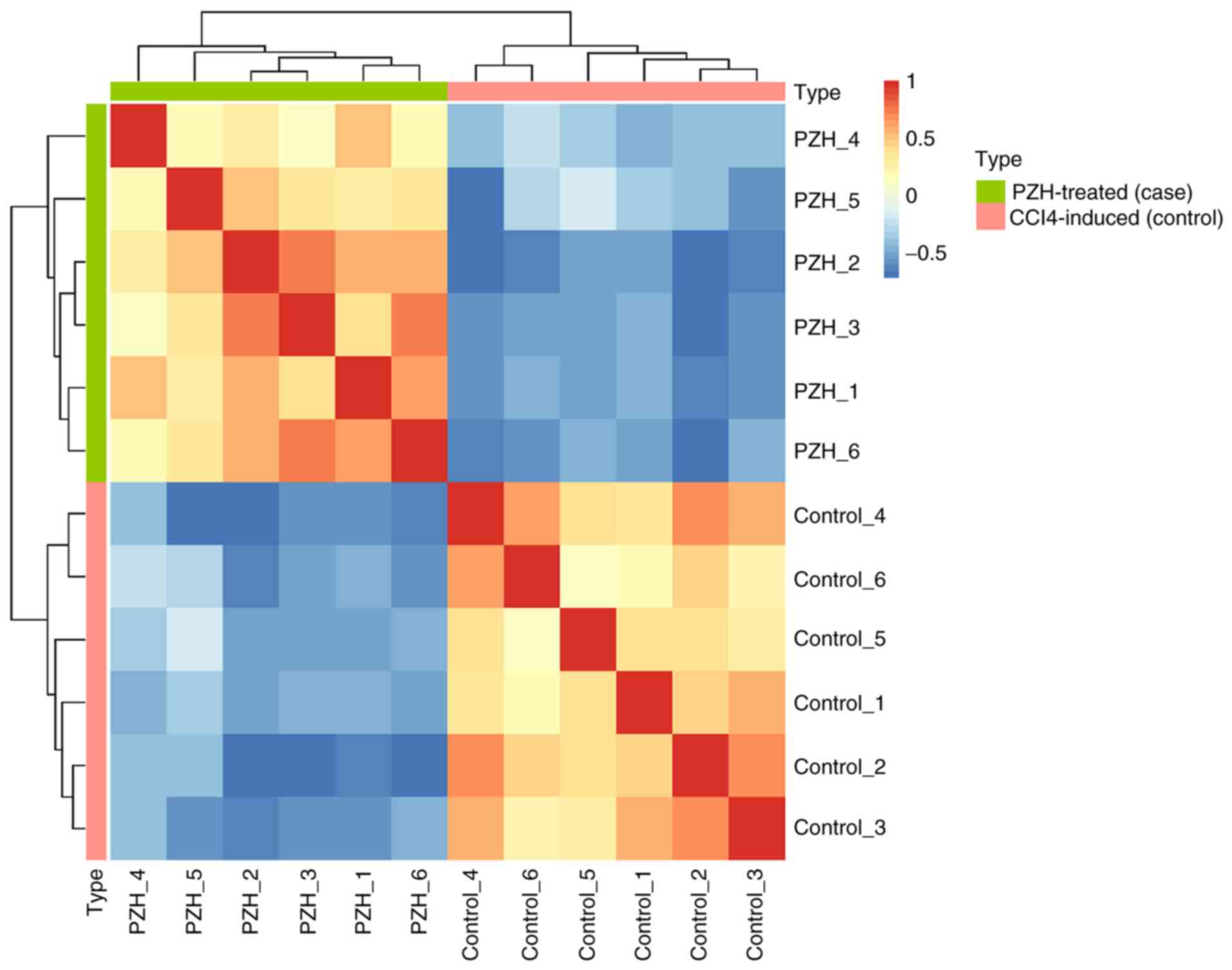
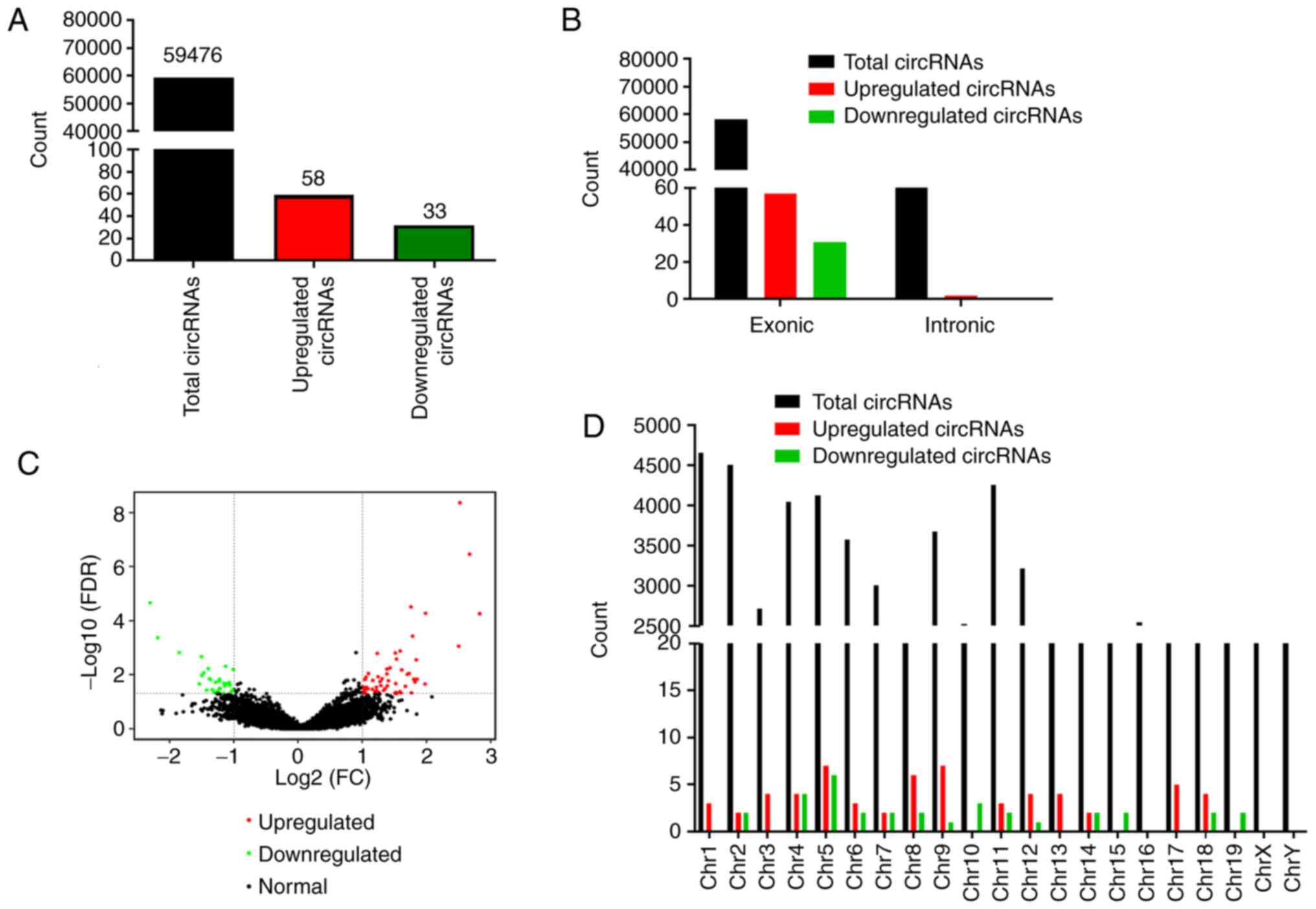
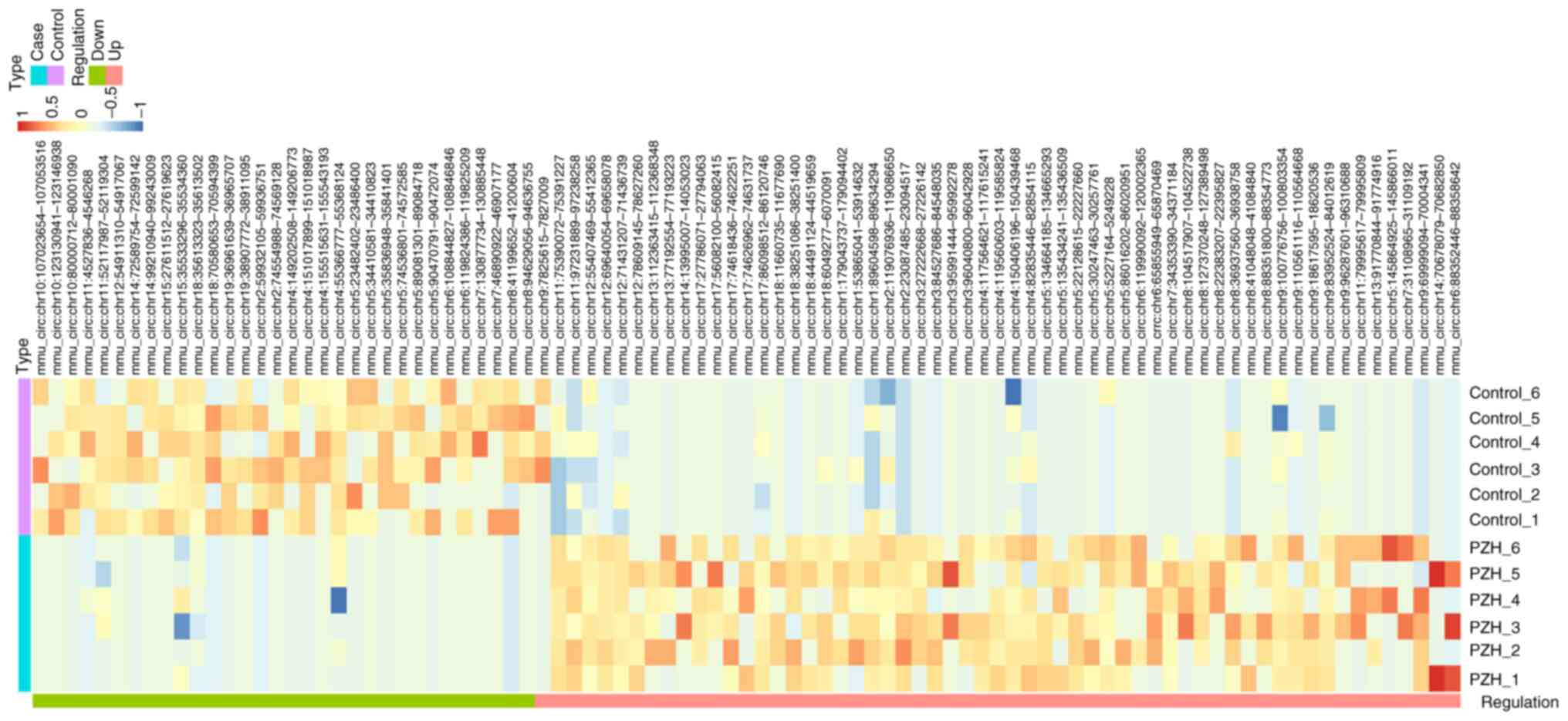
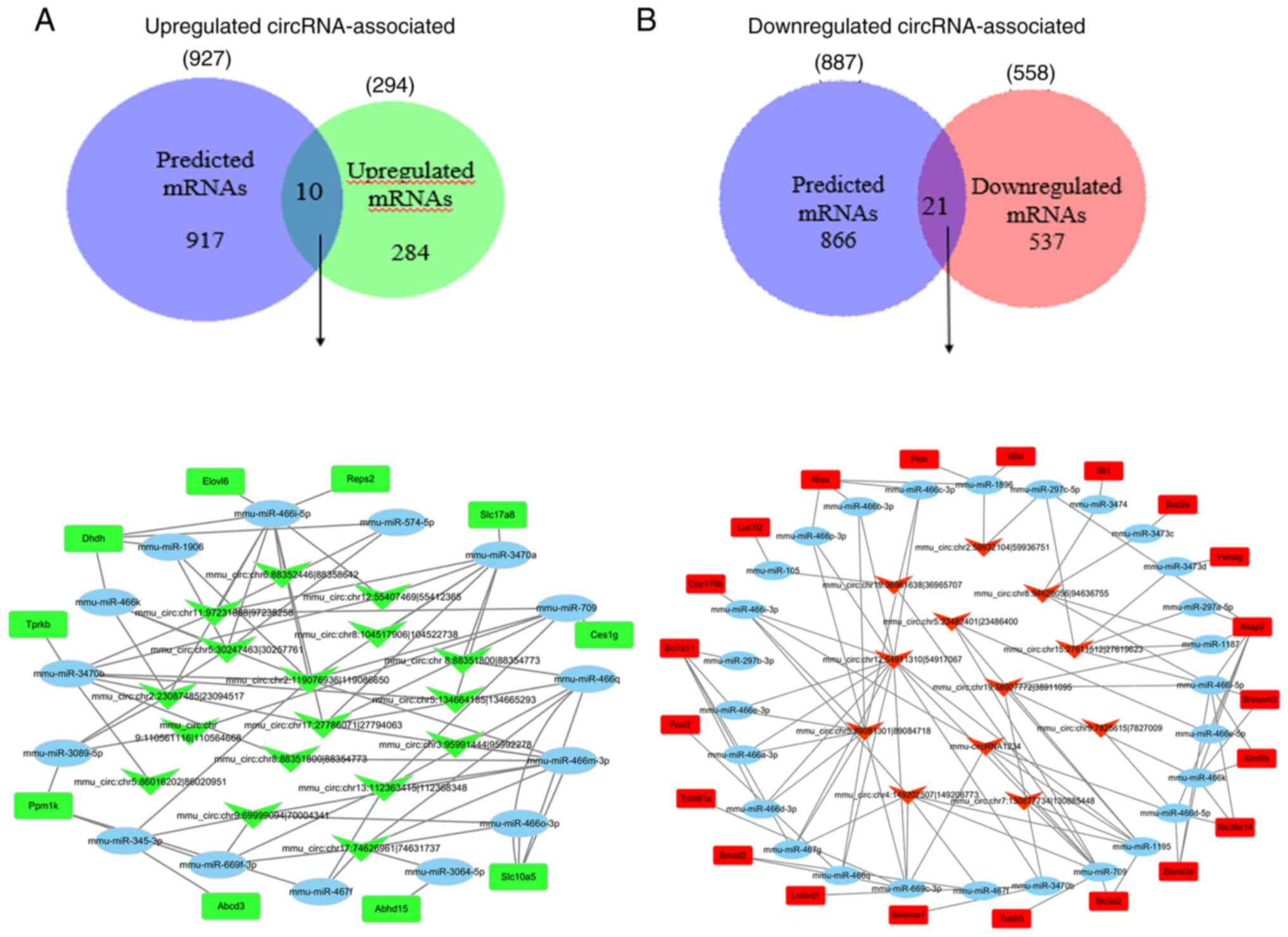
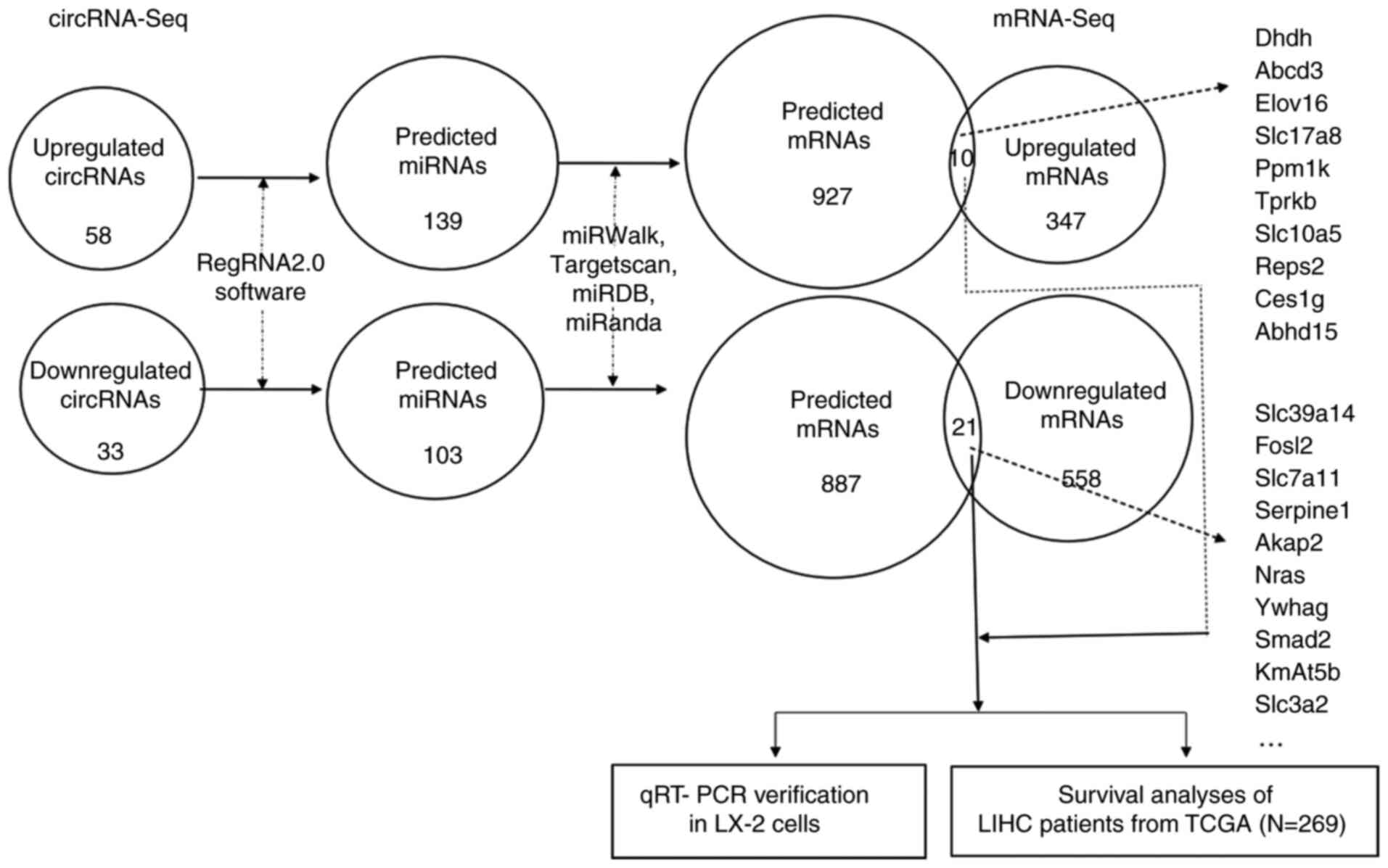
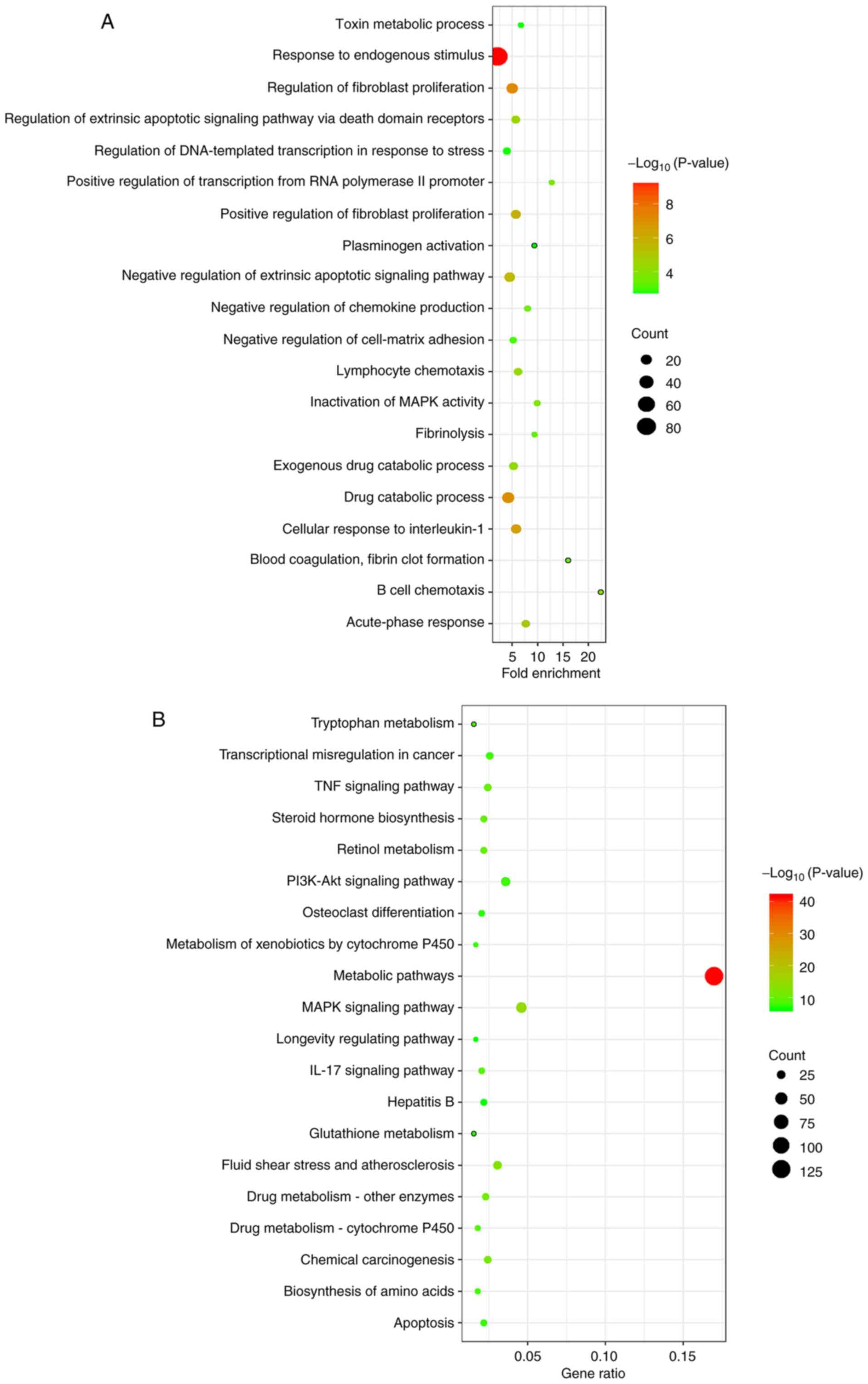
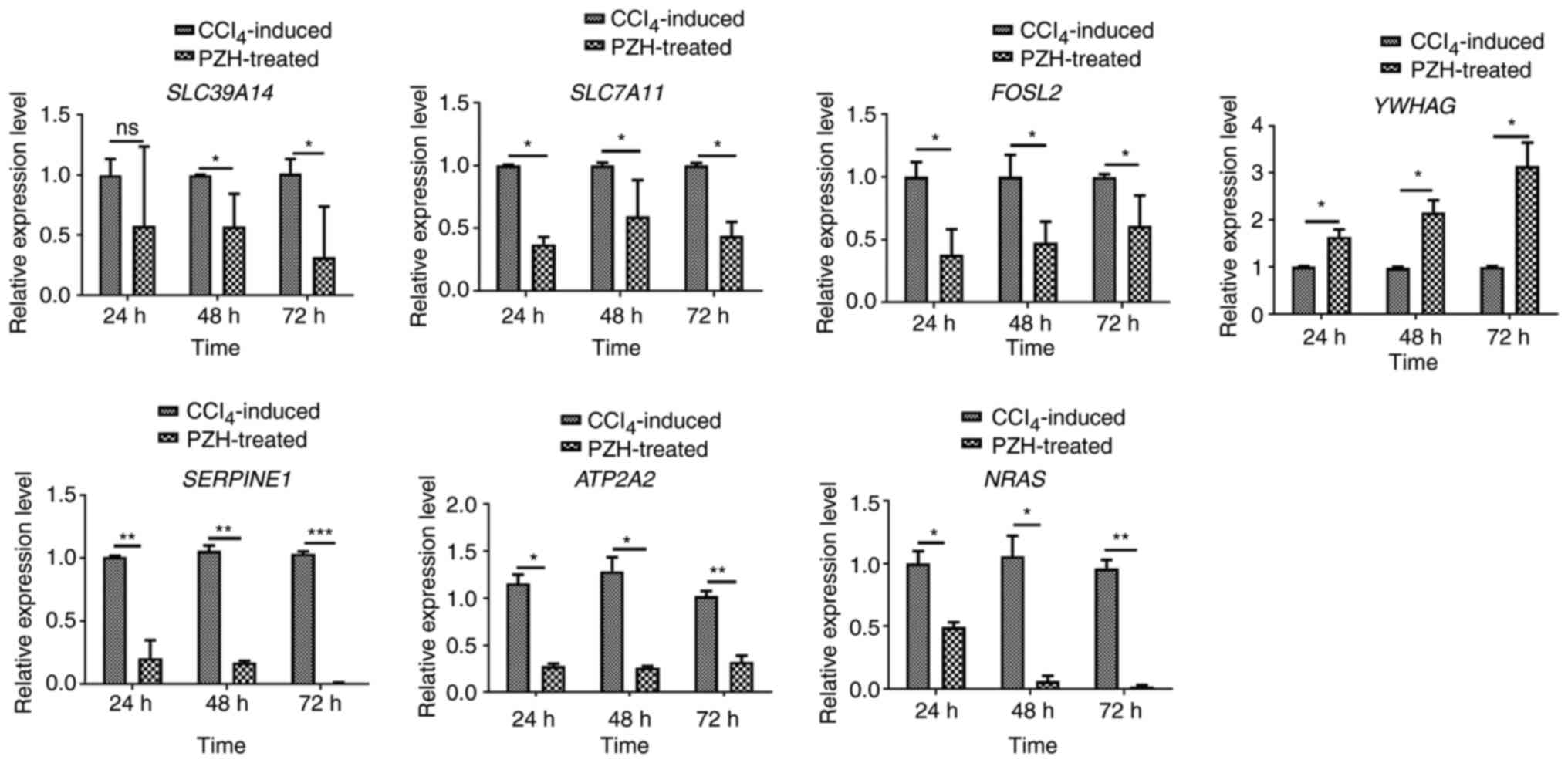
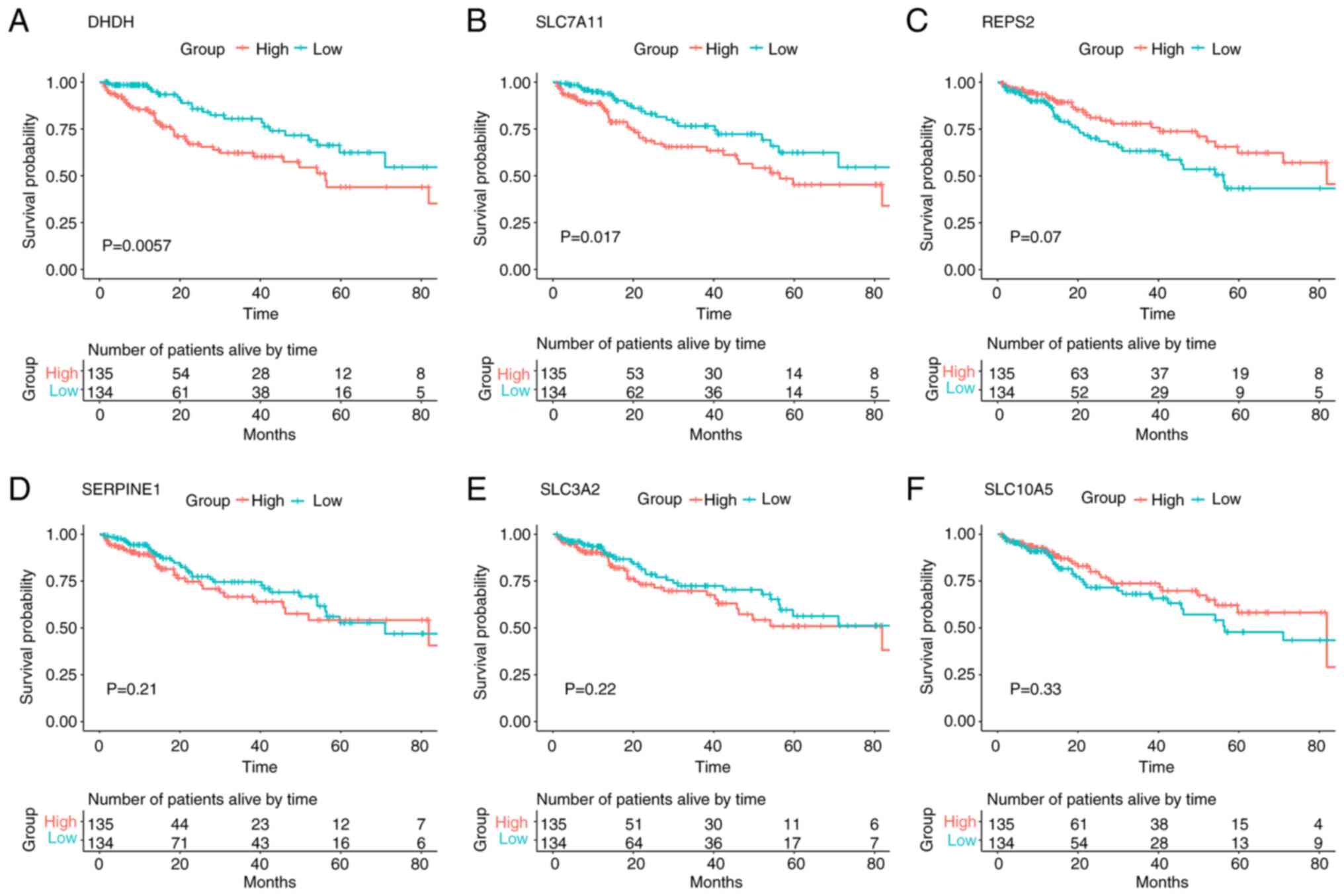
|
1
|
Mederacke I, Hsu CC, Troeger JS, Huebener
P, Mu X, Dapito DH, Pradere JP and Schwabe RF: Fate tracing reveals
hepatic stellate cells as dominant contributors to liver fibrosis
independent of its aetiology. Nat Commun. 4:28232013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schuppan D and Afdhal NH: Liver cirrhosis.
Lancet. 371:838–851. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shrestha N, Chand L, Han MK, Lee SO, Kim
CY and Jeong YJ: Glutamine inhibits CCl4 induced liver fibrosis in
mice and TGF-β1 mediated epithelial-mesenchymal transition in mouse
hepatocytes. Food Chem Toxicol. 93:129–137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeisberg M and Kalluri R: Cellular
mechanisms of tissue fibrosis. 1. Common and organ-specific
mechanisms associated with tissue fibrosis. Am J Physiol Cell
Physiol. 304:C216–C225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wynn TA and Ramalingam TR: Mechanisms of
fibrosis: Therapeutic translation for fibrotic disease. Nat Med.
18:1028–1040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mansour MF, Greish SM, El-Serafi AT,
Abdelall H and El-Wazir YM: Therapeutic potential of human
umbilical cord derived mesenchymal stem cells on rat model of liver
fibrosis. Am J Stem Cells. 8:7–18. 2019.PubMed/NCBI
|
|
8
|
Uehara T, Pogribny IP and Rusyn I: The DEN
and CCl4-induced mouse model of fibrosis and
inflammation-associated hepatocellular carcinoma. Curr Protoc
Pharmacol. 66:14.30.1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Morio LA, Chiu H, Sprowles KA, Zhou P,
Heck DE, Gordon MK and Laskin DL: Distinct roles of tumor necrosis
factor-alpha and nitric oxide in acute liver injury induced by
carbon tetrachloride in mice. Toxicol Appl Pharmacol. 172:44–51.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Steinman L, Martin R, Bernard C, Conlon P
and Oksenberg JR: Multiple sclerosis: Deeper understanding of its
pathogenesis reveals new targets for therapy. Annu Rev Neurosci.
25:491–505. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Watanabe Y, Tsuchiya A, Seino S, Kawata Y,
Kojima Y, Ikarashi S, Starkey Lewis PJ, Lu WY, Kikuta J, Kawai H,
et al: Mesenchymal stem cells and induced bone marrow-derived
macrophages synergistically improve liver fibrosis in mice. Stem
Cells Transl Med. 8:271–284. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Altamirano-Barrera A, Barranco-Fragoso B
and Méndez-Sánchez N: Management strategies for liver fibrosis. Ann
Hepatol. 16:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bozic M and Molleston J: Strategies for
management of pediatric cystic fibrosis liver disease. Clin Liver
Dis (Hoboken). 2:204–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Zhou H, Liu YB, Wang JF, Li H, Ung
CY, Han LY, Cao ZW and Chen YZ: Database of traditional Chinese
medicine and its application to studies of mechanism and to
prescription validation. Br J Pharmacol. 149:1092–1103. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao J, Zhang Y, Wan Y, Hu H and Hong Z:
Pien Tze Huang Gan Bao attenuates carbon tetrachloride-induced
hepatocyte apoptosis in rats, associated with suppression of p53
activation and oxidative stress. Mol Med Rep. 16:2611–2619. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Y, Chen Z, Deng L, Yu J, Wang K,
Zhang X, Ji G and Li F: Pien Tze Huang ameliorates liver injury by
inhibiting the PERK/eIF2α signaling pathway in alcohol and high-fat
diet rats. Acta Histochem. 120:578–585. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin W, Zhuang Q, Zheng L, Cao Z, Shen A,
Li Q, Fu C, Feng J and Peng J: Pien Tze Huang inhibits liver
metastasis by targeting TGF-β signaling in an orthotopic model of
colorectal cancer. Oncol Rep. 33:1922–1928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin YC, Chen YC, Chen TH, Chen HH and Tsai
WJ: Acute kidney injury associated with hepato-protective Chinese
Herb-Pien Tze Huang. J Exp Clin Med. 3:184–186. 2011. View Article : Google Scholar
|
|
19
|
Ragan C, Goodall GJ, Shirokikh NE and
Preiss T: Insights into the biogenesis and potential functions of
exonic circular RNA. Sci Rep. 9:20482019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koh W, Pan W, Gawad C, Fan HC, Kerchner
GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY and Quake SR:
Noninvasive in vivo monitoring of tissue-specific global gene
expression in humans. Proc Natl Acad Sci USA. 111:7361–7366. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44D:D209–D215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang J, Wu X, Sun S, Chen P, Liang X,
Wang J, Ruan J, Zhang S and Zhang X: Circular RNA expression
profile analysis of severe acne by RNA-Seq and bioinformatics. J
Eur Acad Dermatol Venereol. 32:1986–1992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu C, Shi X, Wang AY, Tao Y, Wang Z, Huang
C, Qiao Y, Hu H and Liu L: RNA-Seq profiling of circular RNAs in
human laryngeal squamous cell carcinomas. Mol Cancer. 17:862018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye Z, Kong Q, Han J, Deng J, Wu M and Deng
H: Circular RNAs are differentially expressed in liver
ischemia/reperfusion injury model. J Cell Biochem. 119:7397–7405.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu H, Wang C, Song H, Xu Y and Ji G:
RNA-seq profiling of circular RNAs in human colorectal cancer liver
metastasis and the potential biomarkers. Mol Cancer. 18:82019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bu Q, Long H, Shao X, Gu H, Kong J, Luo L,
Liu B, Guo W, Wang H, Tian J, et al: Cocaine induces differential
circular RNA expression in striatum. Transl Psychiatry. 9:1992019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma X, Zhou Y, Qiao B, Jiang S, Shen Q, Han
Y, Liu A, Chen X, Wei L, Zhou L and Zhao J: Androgen aggravates
liver fibrosis by activation of NLRP3 inflammasome in CCl4-induced
liver injury mouse model. Am J Physiol Endocrinol Metab.
318:E817–E829. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schroeder A, Mueller O, Stocker S,
Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M
and Ragg T: The RIN: An RNA integrity number for assigning
integrity values to RNA measurements. BMC Mol Biol. 7:32006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brown J, Pirrung M and McCue LA: FQC
dashboard: Integrates FastQC results into a web-based, interactive,
and extensible FASTQ quality control tool. Bioinformatics.
33:3137–3139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Utturkar S, Dassanayake A, Nagaraju S and
Brown SD: Bacterial differential expression analysis methods.
Methods Mol Biol. 2096:89–112. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dobin A and Gingeras TR: Optimizing
RNA-seq mapping with STAR. Methods Mol Biol. 1415:245–262. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Glažar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu J, Zhang D, Wang T, Chen Z, Chen L, Wu
H, Huai C, Sun J, Zhang N, Wei M, et al: Target identification of
hepatic fibrosis using Pien Tze Huang based on mRNA and lncRNA. Sci
Rep. 11:169802021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chang TH, Huang HY, Hsu JB, Weng SL, Horng
JT and Huang HD: An enhanced computational platform for
investigating the roles of regulatory RNA and for identifying
functional RNA motifs. BMC Bioinformatics. 14 (Suppl 2):S42013.
View Article : Google Scholar
|
|
41
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Adams JM and Cory S: The Bcl-2 apoptotic
switch in cancer development and therapy. Oncogene. 26:1324–1337.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Austin PC, Lee DS and Fine JP:
Introduction to the analysis of survival data in the presence of
competing risks. Circulation. 133:601–609. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
International Cancer Genome Consortium, .
Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabé RR, Bhan
MK, Calvo F, Eerola I, et al: International network of cancer
genome projects. Nature. 464:993–998. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Piwecka M, Glažar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao J, Hu H, Wan Y, Zhang Y, Zheng L and
Hong Z: Pien Tze Huang Gan Bao ameliorates carbon
tetrachloride-induced hepatic injury, oxidative stress and
inflammation in rats. Exp Ther Med. 13:1820–1826. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Parsons CJ, Takashima M and Rippe RA:
Molecular mechanisms of hepatic fibrogenesis. J Gastroenterol
Hepatol. 22 (Suppl 1):S79–S84. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bai T, Lian LH, Wu YL, Wan Y and Nan JX:
Thymoquinone attenuates liver fibrosis via PI3K and TLR4 signaling
pathways in activated hepatic stellate cells. Int Immunopharmacol.
15:275–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang J, Chu ES, Chen HY, Man K, Go MY,
Huang XR, Lan HY, Sung JJ and Yu J: MicroRNA-29b prevents liver
fibrosis by attenuating hepatic stellate cell activation and
inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget.
6:7325–7338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shu M, Huang DD, Hung ZA, Hu XR and Zhang
S: Inhibition of MAPK and NF-κB signaling pathways alleviate carbon
tetrachloride (CCl4)-induced liver fibrosis in Toll-like receptor 5
(TLR5) deficiency mice. Biochem Biophys Res Commun. 471:233–239.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ghallab A, Myllys M, Holland CH, Zaza A,
Murad W, Hassan R, Ahmed YA, Abbas T, Abdelrahim EA, Schneider KM,
et al: Influence of liver fibrosis on lobular zonation. Cells.
8:15562019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Meng F, Wang K, Aoyama T, Grivennikov SI,
Paik Y, Scholten D, Cong M, Iwaisako K, Liu X, Zhang M, et al:
Interleukin-17 signaling in inflammatory, Kupffer cells, and
hepatic stellate cells exacerbates liver fibrosis in mice.
Gastroenterology. 143:765–776.e3. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang Y, Huang D, Gao W, Yan J, Zhou W,
Hou X, Liu M, Ren C, Wang S and Shen J: Lack of IL-17 signaling
decreases liver fibrosis in murine Schistosomiasis japonica. Int
Immunol. 27:317–325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hyun J, Wang S, Kim J, Rao KM, Park SY,
Chung I, Ha CS, Kim SW, Yun YH and Jung Y: MicroRNA-378 limits
activation of hepatic stellate cells and liver fibrosis by
suppressing Gli3 expression. Nat Commun. 7:109932016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sunaga H, Matsui H, Ueno M, Maeno T, Iso
T, Syamsunarno MR, Anjo S, Matsuzaka T, Shimano H, Yokoyama T and
Kurabayashi M: Deranged fatty acid composition causes pulmonary
fibrosis in Elovl6-deficient mice. Nat Commun. 4:25632013.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shiu TY, Huang SM, Shih YL, Chu HC, Chang
WK and Hsieh TY: Correction: Hepatitis C virus core protein
down-regulates p21Waf1/Cip1 and inhibits curcumin-induced apoptosis
through MicroRNA-345 targeting in human hepatoma cells. PLoS One.
12:e01812992017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shen G, Rong X, Zhao J, Yang X, Li H,
Jiang H, Zhou Q, Ji T, Huang S, Zhang J and Jia H: MicroRNA-105
suppresses cell proliferation and inhibits PI3K/AKT signaling in
human hepatocellular carcinoma. Carcinogenesis. 35:2748–2755. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
An F, Gong B, Wang H, Yu D, Zhao G, Lin L,
Tang W, Yu H, Bao S and Xie Q: miR-15b and miR-16 regulate TNF
mediated hepatocyte apoptosis via BCL2 in acute liver failure.
Apoptosis. 17:702–716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Koo JH, Lee HJ, Kim W and Kim SG:
Endoplasmic reticulum stress in hepatic stellate cells promotes
liver fibrosis via PERK-mediated degradation of HNRNPA1 and
up-regulation of SMAD2. Gastroenterology. 150:181–193.e8. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Du K, Oh SH, Dutta RK, Sun T, Yang WH, Chi
JT and Diehl AM: Inhibiting xCT/SLC7A11 induces ferroptosis of
myofibroblastic hepatic stellate cells but exacerbates chronic
liver injury. Liver Int. 41:2214–2227. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang L, Huang Y, Ling J, Zhuo W, Yu Z,
Luo Y and Zhu Y: Overexpression of SLC7A11: A novel oncogene and an
indicator of unfavorable prognosis for liver carcinoma. Future
Oncol. 14:927–936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lodder J, Denaës T, Chobert MN, Wan J,
El-Benna J, Pawlotsky JM, Lotersztajn S and Teixeira-Clerc F:
Macrophage autophagy protects against liver fibrosis in mice.
Autophagy. 11:1280–1292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Eferl R, Hasselblatt P, Rath M, Popper H,
Zenz R, Komnenovic V, Idarraga MH, Kenner L and Wagner EF:
Development of pulmonary fibrosis through a pathway involving the
transcription factor Fra-2/AP-1. Proc Natl Acad Sci USA.
105:10525–10530. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Rajalingam K, Schreck R, Rapp UR and
Albert S: Ras oncogenes and their downstream targets. Biochim
Biophys Acta. 1773:1177–1195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Carlson CM, Frandsen JL, Kirchhof N,
McIvor RS and Largaespada DA: Somatic integration of an
oncogene-harboring sleeping beauty transposon models liver tumor
development in the mouse. Proc Natl Acad Sci USA. 102:17059–17064.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Rudalska R, Dauch D, Longerich T, McJunkin
K, Wuestefeld T, Kang TW, Hohmeyer A, Pesic M, Leibold J, von Thun
A, et al: In vivo RNAi screening identifies a mechanism of
sorafenib resistance in liver cancer. Nat Med. 20:1138–1146. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yue C, Ren Y, Ge H, Liang C, Xu Y, Li G
and Wu J: Comprehensive analysis of potential prognostic genes for
the construction of a competing endogenous RNA regulatory network
in hepatocellular carcinoma. Onco Targets Ther. 12:561–576. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang L, Huang Y, Zhu Y, Yu Z, Shao M and
Luo Y: Identification and characterization of cadmium-related genes
in liver carcinoma. Biol Trace Elem Res. 182:238–247. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Divella R, Mazzocca A, Gadaleta C, Simone
G, Paradiso A, Quaranta M and Daniele A: Influence of plasminogen
activator inhibitor-1 (SERPINE1) 4G/5G polymorphism on circulating
SERPINE-1 antigen expression in HCC associated with viral
infection. Cancer Genomics Proteomics. 9:193–198. 2012.PubMed/NCBI
|
|
75
|
Czekay RP, Aertgeerts K, Curriden SA and
Loskutoff DJ: Plasminogen activator inhibitor-1 detaches cells from
extracellular matrices by inactivating integrins. J Cell Biol.
160:781–791. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Roca C, Primo L, Valdembri D, Cividalli A,
Declerck P, Carmeliet P, Gabriele P and Bussolino F: Hyperthermia
inhibits angiogenesis by a plasminogen activator inhibitor
1-dependent mechanism. Cancer Res. 63:1500–1507. 2003.PubMed/NCBI
|
|
77
|
Wu B, Wang Y, Yang XM, Xu BQ, Feng F, Wang
B, Liang Q, Li Y, Zhou Y, Jiang JL and Chen ZN: Basigin-mediated
redistribution of CD98 promotes cell spreading and tumorigenicity
in hepatocellular carcinoma. J Exp Clin Cancer Res. 34:1102015.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang L, Zhang Z, Li M, Wang F, Jia Y,
Zhang F, Shao J, Chen A and Zheng S: P53-dependent induction of
ferroptosis is required for artemether to alleviate carbon
tetrachloride-induced liver fibrosis and hepatic stellate cell
activation. IUBMB Life. 71:45–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ghosh AK and Vaughan DE: PAI-1 in tissue
fibrosis. J Cell Physiol. 227:493–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ji J, Yu F, Ji Q, Li Z, Wang K, Zhang J,
Lu J, Chen L, E Q, Zeng Y and Ji Y: Comparative proteomic analysis
of rat hepatic stellate cell activation: A comprehensive view and
suppressed immune response. Hepatology. 56:332–349. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Khambu B, Yan S, Huda N, Liu G and Yin XM:
Autophagy in non-alcoholic fatty liver disease and alcoholic liver
disease. Liver Res. 2:112–119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Gao M and Liu D: CRISPR/Cas9-based Pten
knock-out and sleeping beauty transposon-mediated Nras knock-in
induces hepatocellular carcinoma and hepatic lipid accumulation in
mice. Cancer Biol Ther. 18:505–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Cao W, Li Y, Li M, Zhang X and Liao M:
Txn1, Ctsd and Cdk4 are key proteins of combination therapy with
taurine, epigallocatechin gallate and genistein against liver
fibrosis in rats. Biomed Pharmacother. 85:611–619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li R, Wang Y, Song X, Sun W, Zhang J, Liu
Y, Li H, Meng C, Zhang J, Zheng Q and Lv C: Potential regulatory
role of circular RNA in idiopathic pulmonary fibrosis. Int J Mol
Med. 42:3256–3268. 2018.PubMed/NCBI
|