Introduction
Drug-induced liver injury (DILI) is a toxic side
effect of numerous drugs (1–4),
including a number of anti-inflammatory and analgesia drugs
(5–8). DILI is the commonest reason for
withdrawing drugs from the market and/or issuing warnings and
modification of use (9). Data
from prospective DILI registries suggest that antibiotics remain
the most common cause of DILI (10–12). The American DILI Network (DILIN)
reported antibiotics to be implicated in 45.4% of cases (13). Other common drug classes reported
by the American DILIN (14) are
herbal and dietary supplements (16.1%), cardiovascular agents
(9.8%), central nervous system agents (9.1%), anti-neoplastic
agents (5.5%) and analgesics (3.7%). DILI includes the whole
spectrum from asymptomatic elevation in liver tests to acute liver
failure (ALF). In fact, DILI remains the most common cause of ALF
in the UK (14) and USA (15).
Binaprofen is an anti-inflammatory drug that is not
currently in market, and still in clinical study; its chemical
structure is C18H23NO5 and it
relieves fever and analgesia (16–18). It exhibits a notable analgesic
effect. Studies have done about the pharmacological and
toxicological effects of binaprofen (19,20), and it has been clinical licensed
in China.
Our previous study (21) demonstrated that binaprofen induces
liver toxicity and damage similar to acetaminophen (APAP) in
zebrafish. APAP is one of the most widely used antipyretic and
analgesic drugs in the US. It is reported to be regularly consumed
by over 60 million Americans on a weekly basis (22). Though it is safe at therapeutic
doses, an overdose can cause severe liver injury and even ALF in
humans (23). The mechanisms that
underlying APAP-induced liver injury have been extensively studied
(24,25). So APAP was chosen as positive drug
in this paper. To the best of our knowledge, however, the mechanism
underlying liver injury has not yet been revealed. The present
study aimed to determine this mechanism at the genetic level and
provide a basis for potential treatment options.
Materials and methods
Maintenance and breeding of
zebrafish
Male and female AB-line, 1–2 g, adult zebrafish
(Danio rerio, 90–100 days post-fertilization; weight, 1–2 g)
were obtained from Southern Medical University, Guangzhou, China.
Zebrafish were acclimatized for 2 weeks. The animal protocol was
designed according to animal welfare and ethics, which was approved
by animal care and use committee of Guangzhou General
Pharmaceutical Research Institute (Haizhu, China; approval no. was
2012-005). Fish were maintained in aerated water at 23±1°C,
humidity 65%, pH 7.8±1.0, 0.25 g/l hardness, 12/12-h light/dark
cycle and density of 1 fish/l with free access to food and water.
Experiments were performed using a total of 150 animals, including
75 male and 75 female.
Zebrafish exposure to binaprofen
Our previous study (21) demonstrated that binaprofen at 0.8
mM and APAP at 4.0 mM cause notable liver damage in zebrafish.
Therefore, zebrafish were divided into control (untreated),
binaprofen (0.8 mM) and APAP (4.0 mM) groups (n=50/group).
Zebrafish were exposed to drug at 22.8°C for 12, 24 or 48 h, then
euthanized via 2-step hypothermal shock, as described in AVMA
guidelines (26).
Zebrafish were rapidly killed by immersion in 2–4°C
water for 10–20 sec. Exposure was continued for ≥10 min following
loss of operculum movement to ensure death. Followed by rapid
chilling, zebrafish tail was transected as previously described
(27) and 20 µl blood was
collected. Exsanguination was performed to euthanize zebrafish.
Rapid chilling followed by exsanguination was the recommended
2-step euthanasia method in AVMA Guidelines for the Euthanasia of
Animals: 2020 Edition (28).
Serum biomarkers detection
Blood was collected by exsanguination. Serum samples
were collected by tail cutting and capillary collection method
(n=5/group). Briefly, the tail was transected from cranial to the
caudal fin, 100-µl microcapillaries was used to collect zebrafish
blood from the cut surface and blood was placed in 1.5 ml
centrifugal tube. A total of 200 µl blood was collected as one
sample; there were 5 samples/group. Blood was centrifuged for 10
min at 1,800 × g at 4°C. Supernatant was collected to detect
alanine transaminase (ALT), aspartate transaminase (AST) and
lactate dehydrogenase (LDH) (ALT assay kit, cat. no. 130301, AST
assay kit, cat. no. 130201and LDH assay kit, cat. no. 130503, all
kits from Zhejiang Yilikang biotek Company) by biochemical
detection method using a biochemical analyzer (7100; Hitachi,
Ltd.).
Malondialdehyde (MDA) and glutathione
(GSH) detection
The entire zebrafish liver (n=5/group) was collected
and placed in 1.5 ml centrifugal tube. 0.9% NaCl was added and
liver tissue was homogenized and centrifuged for 10 min at 4,000 ×
g at 4°C. Supernatant was collected to detect MDA and GSH (MDA
assay kit, cat. no. 20130515 and GSH assay kit, cat. no. 20130428;
both kits from Nanjing Jiancheng Bioengineering Institute) using a
microplate reader (Elx800; BioTek China) according to the
manufacturer's instructions.
Histological analysis
Liver tissue samples (n=10/group) were fixed in 10%
formalin for at 25°C for 24 h and dehydrated with gradient alcohol.
Tissue was immersed in 60°C paraffin wax for 1 h and moved to −10°C
freezing table for 30 min. Paraffin-embedded samples were sliced (5
µm). Sections were dewaxed with xylene and rehydrated in descending
alcohol. Slices were dyed with 0.5% hematoxylin aqueous solution at
25°C for 3 min and 0.5% eosin staining solution at 25°C for 3 min.
90% neutral balata was used as blocking reagent, slices were
blocked at 25°C for 30 sec. Morphological examination of
hepatocytes was performed using a light microscope (BX51; Olympus
Corporation) at 40X magnification using image analysis system 11.0
(cellSens Standard; both Olympus Corporation). Samples were scored
as previously described (29) by
two independent pathologists who were blinded to the experimental
groups.
DAPI analysis
Liver tissue slices (n=10/group) were prepared as
aforementioned. Slices were stained with 1 µg/ml DAPI at 25°C for
20 min. Apoptosis of hepatocytes was assessed by fluorescence
microscopy (BX51, Olympus Corporation) with fluorescence light, 40×
magnification using image analysis system 11.0 (cellSens Standard;
both Olympus Corporation, Japan). Five visual fields were randomly
selected from each slice to observe the apoptotic cells. Normal
cell: complete nucleus and uniform chromatin; Apoptosis cell:
nuclear enrichment, deep staining, or crescent-shaped aggregation
of nuclear chromatin on one side of the nuclear membrane.
Electron microscopic detection of
mitochondria
Liver tissue samples were fixed in 2% osmium
tetroxide at 4°C for 24 h, dehydrated, embedded in Epon/Araldite
resin at 4°C for 1 h and sectioned (70 nm). Samples were stained
with premixed solutions of 2% uranyl acetate and lead citrate at
4°C for 20 min. Ultrastructure examination of hepatocytes was
performed using a transmission electron microscope (JEM-1400; JEOL,
Ltd.) with 10× magnification. The morphological changes of
hepatocyte mitochondria were examined using image analysis system
11.0 (cellSens Standard; both Olympus Corporation).
Microarray analysis
Livers tissue samples were collected and total RNA
was extracted (RNA extraction kit, Qiagen GmbH) and purified. RNA
quality was assessed before detection. RNA was hybridized into two
gene chip probe arrays (Affymetrix; Thermo Fisher Scientific,
Inc.). RNA was used to synthesize double stranded cDNA (iScipt cDNA
Synthesis kit; Qiagen GmbH, German) and produce biotin-tagged cDNA.
cDNA was fragmented to strands 35–200 bases in length. Fragmented
cRNA was hybridized to gene chip array at 45°C for 16 h using Gene
Chip Hybridization Oven640 (Affymerix; Thermo Fisher Scientific,
Inc., US). Gene chip arrays were washed using Gene Chip IVT
labeling kit (Affymerix; Thermo Fisher Scientific, Inc., US) and
stained in SAPE solution at 25°C for 10 min using Affymetrix
Fluidics Station 450 and scanned using Gene Chip Scanner 3000 (both
Affymetrix; Thermo Fisher Scientific, Inc., US) with SAPE solution
and array holding buffer at 25°C for 10 cycles.
Gene chip data were analyzed using Gene Chip
Operating Software (version 1.4, Thermo Fisher Scientific, Inc.).
The criterion of differentially expressed genes was >2-fold
change compared with control. Gene Ontology (GO analysis) was
performed to determine the function of differentially expressed
genes by using Gene Ontology Resource (geneontology.org).
Kyoto Encyclopedia of Genes and Genomes Pathway
analysis was performed to determine the pathways associated with
differentially expressed genes by KEGG pathway database (https://www.kegg.jp). P<0.05 was considered to
indicate a statistically significant difference. Common different
genes were analyzed using VENN database
(bioinformatics.psb.ugent.be).
Reverse transcription-quantitative
(RT-q)PCR
Microarray data were validated by RT-qPCR using five
samples/group. GAPDH was used as housekeeping for the internal
control. A total of six candidate genes was chosen for RT-qPCR.
Primer 5.0 software (PREMIER Design Lnc.) was used to design
primers (Table I). RNA was
extracted from liver samples and purified (RNA Simple Total RNA
kit; Tiangen Biotech Co., Ltd.). RT-qPCR was performed by cDNA
synthesis (iScipt cDNA Synthesis kit) and amplification using a 2
Real-time Detection system (2X QuantiFast SYBR Green PCR Master
Mix; both Qiagen GmbH). RT kit was used according to the
manufacturer's protocol. Thermocycling conditions were as follows:
Initial denaturation at 94°C for 4 min, followed by 45 cycles of
94°C for 20 sec, 58°C for 25 sec and 72°C for 25 sec. Melting curve
analysis was performed. Following amplification, quantitative
detection was performed using a fluorescence qPCR instrument (cat.
no. DA7600; Daan Gene Co., Ltd.). The relative fold change was
calculated using the 2−ΔΔCq method (30).
 | Table I.Primer design. |
Table I.
Primer design.
| Primer | Sequence,
5′→3′ |
|---|
|
Zebrafish-Zgc136383-F |
GTTCCCATCAATCCAGACGGT |
|
Zebrafish-Zgc136383-R |
TGACAGTTCTGCATCAACACATC |
|
Zebrafish-Zgc123120-F |
CCAGACACCTCCCCTCATT |
|
Zebrafish-Zgc123120-R |
CTCTCCAGCACAACTTCCC |
|
Zebrafish-Eif4ebp3l-F |
AAGAAAGCACATCAGAACATAAA |
|
Zebrafish-Eif4ebp3l-R |
GAAATCCAGGCAAACGAAA |
|
Zebrafish-Cap3-F |
CCGCTGCCCATCACTAGA |
|
Zebrafish-Cap3-R |
ATCCTTTCACGACCATCT |
|
Zebrafish-Loc100330641-F |
TGAGATTGCTAATGGTGTTGGC |
|
Zebrafish-Loc100330641-R |
ACATAGCCGTACCATTGACACTTGC |
|
Zebrafish-Vtg6-F |
TGAGTATGCTAATGGTGTGGTTGGC |
|
Zebrafish-Vtg6-R |
TGTTCTGCGTCTTCTTGAGGTTGAG |
|
Zebrafish-GAPDH-F |
GTGACCCCTTTGCTGTTTCTTT |
|
Zebrafish-GAPDH-R |
GGCACGTGGTGCAAACATT |
Statistical analysis
SPSS software (13.0; SPSS, Inc.) was used for
statistical analysis. The data are presented as mean ± SD (n=5).
Variables with normal distribution were analyzed with Student's
t-test (unpaired) for two groups; for multiple groups, one-way
ANOVA followed Tukey's post hoc test was used. Variables with
abnormal distribution were analyzed with Mann-Whitney test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
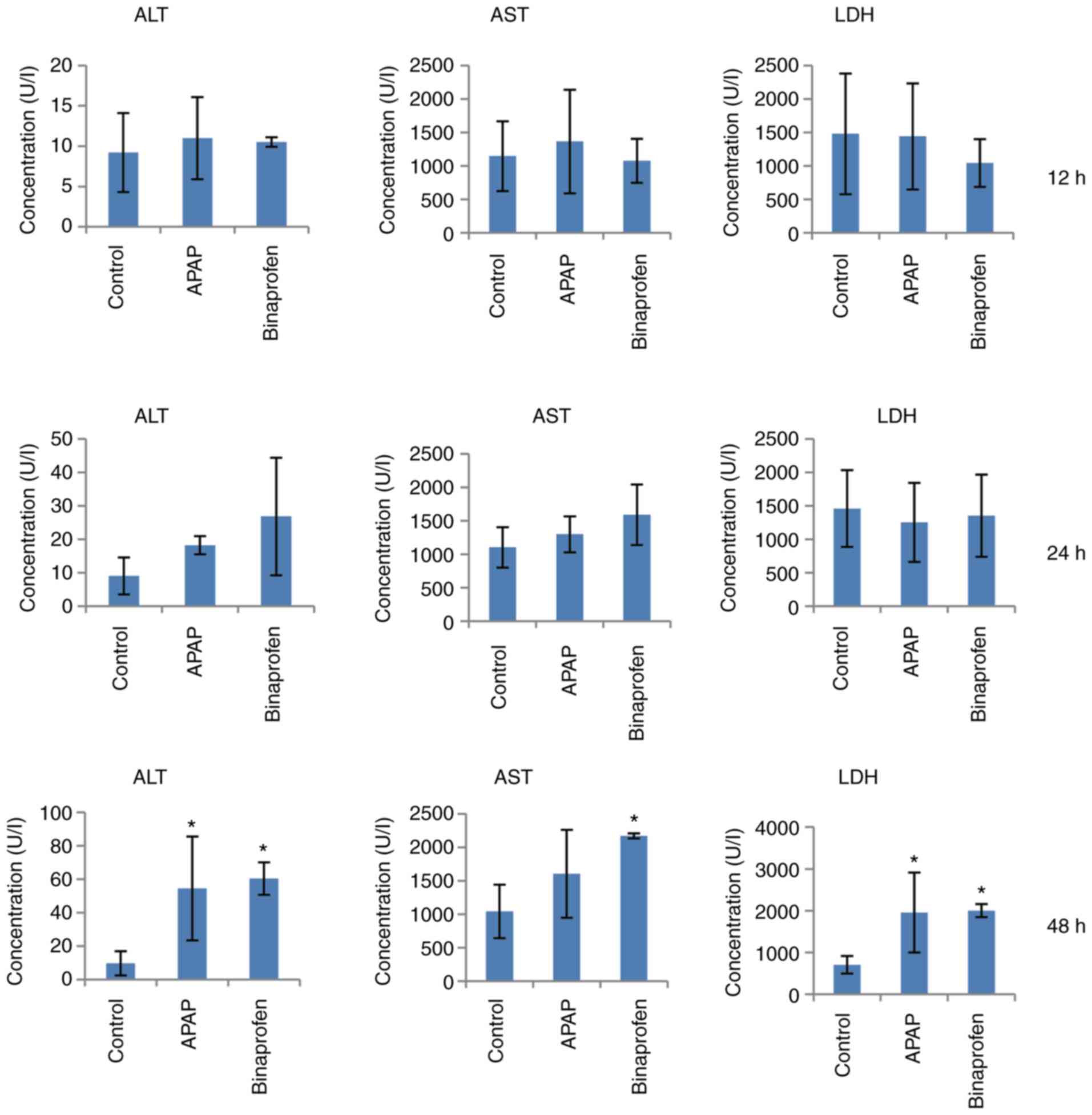
Value of serum biomarkers
Compared with control, APAP increased ALT and LDH
levels at 48 h (Fig. 1).
Binaprofen treatment increased ALT, AST and LDH levels at 48 h.
These results indicated injury or inflammation of liver.
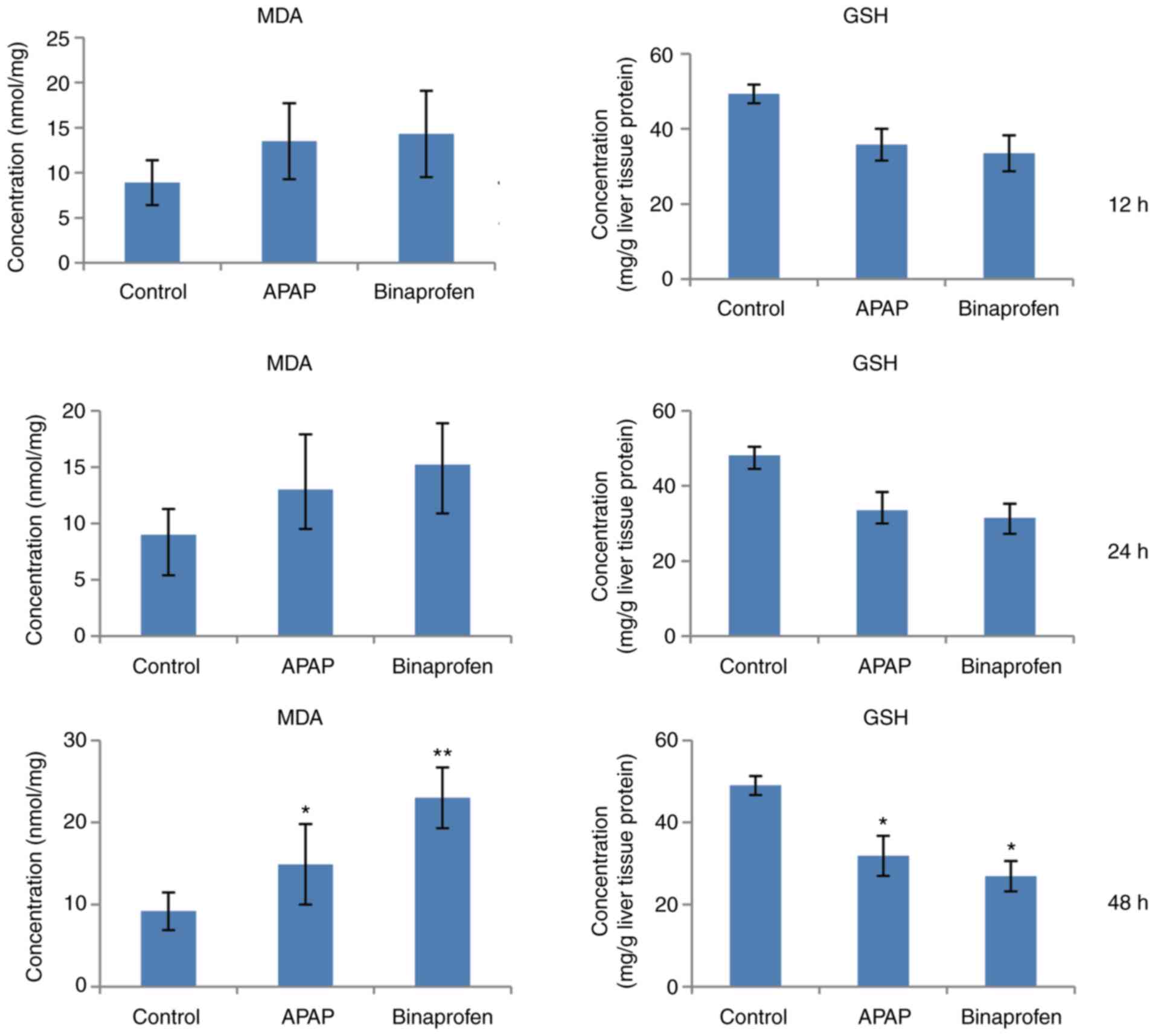
Value of MDA and GSH
Compared with control group, APAP and binaprofen
increased MDA and decreased GSH levels at 48 h (Fig. 2). These results indicated
increased levels of hepatic oxidative products.
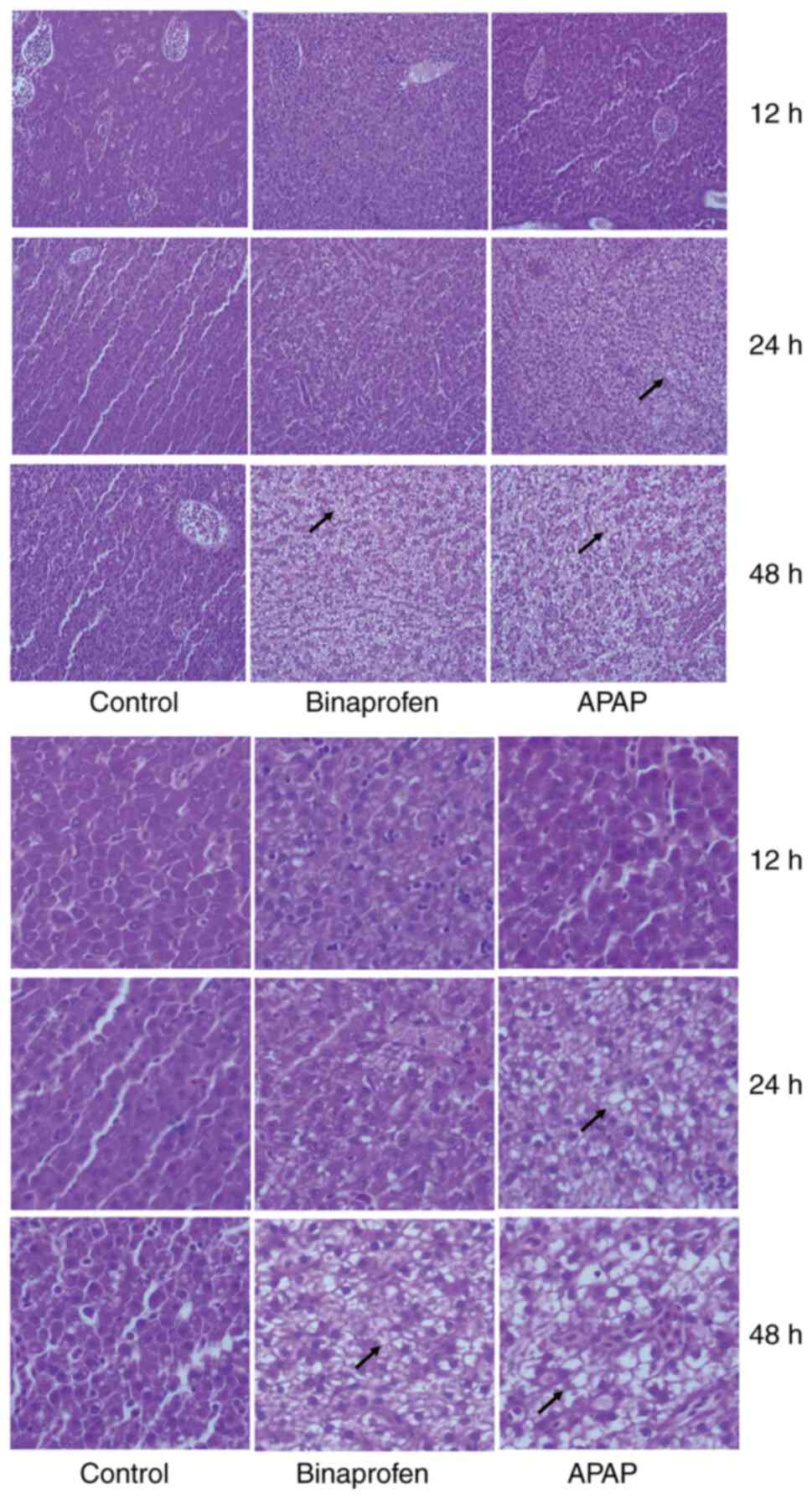
Liver morphological changes from
histological analysis
Compared with control group, APAP caused mild
vacuolization in 2/10 samples at 12 and 24 h and mild to moderate
vacuolization in all samples at 48 h (Fig. 3). Binaprofen caused mild
vacuolization in one sample at 12 h and 2 samples at 24 h and mild
to moderate vacuolization in 5/10 samples at 48 h. These results
indicated morphological changes of liver cell.
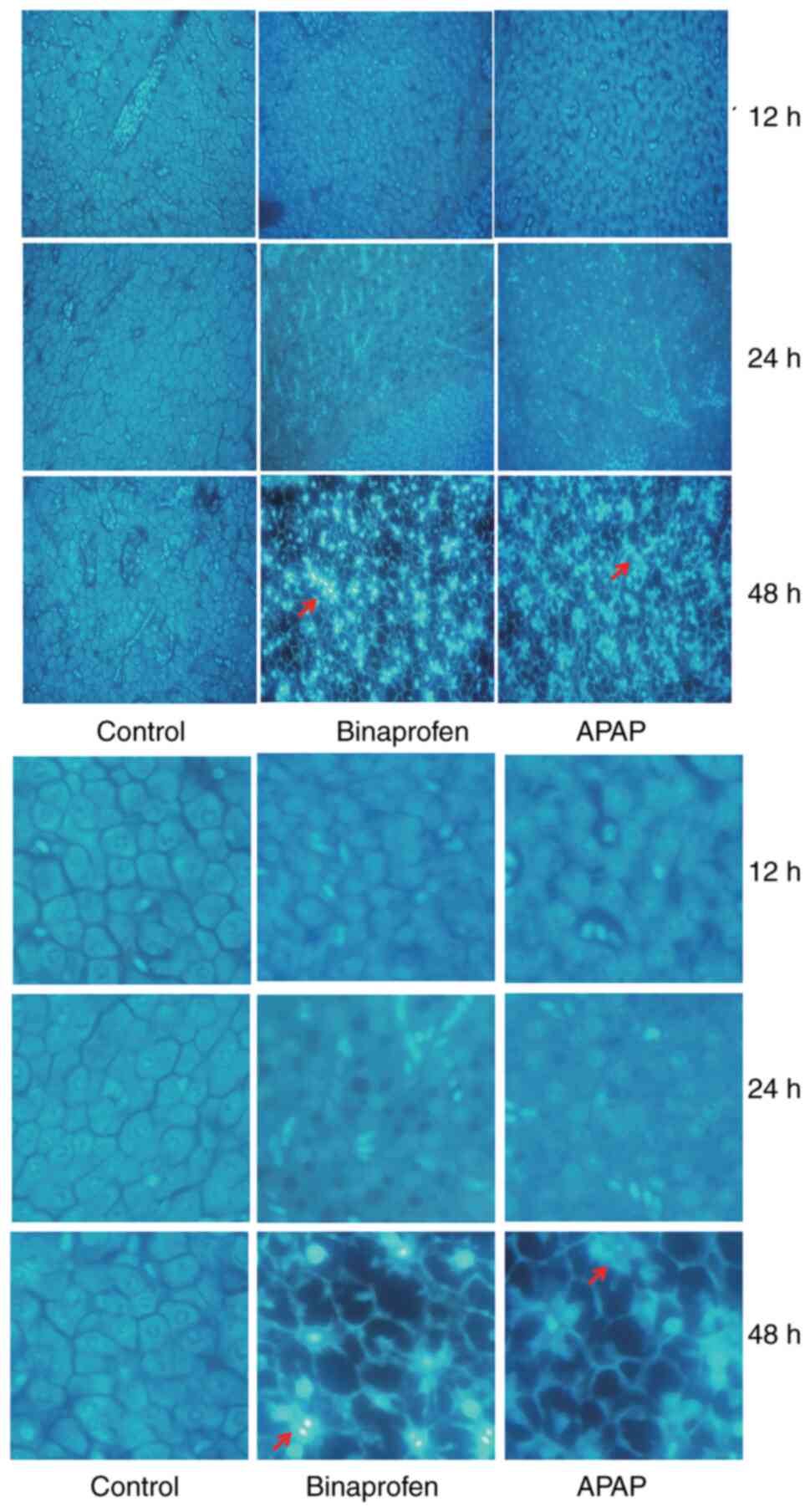
Liver cell apoptosis
Control cells exhibited round nuclei with clear
edges and uniform staining, while the nuclei of apoptotic cells
exhibited irregular edges and concentration of chromosomes
(Fig. 4). Liver cells with strong
staining accompanied by nuclear contraction and nucleosome
fragmentation were considered to indicate apoptosis. In Binaprofen
and APAP groups, DAPI staining is bluish-white fluorescent,
apoptotic cells presented with nuclear enrichment, deep staining,
or crescent-shaped aggregation of nuclear chromatin on one side of
the nuclear membrane. The nucleus broke down to form fragments and
disintegrated. These results indicated that liver cell apoptosis
occurred in binaprofen and APAP groups.
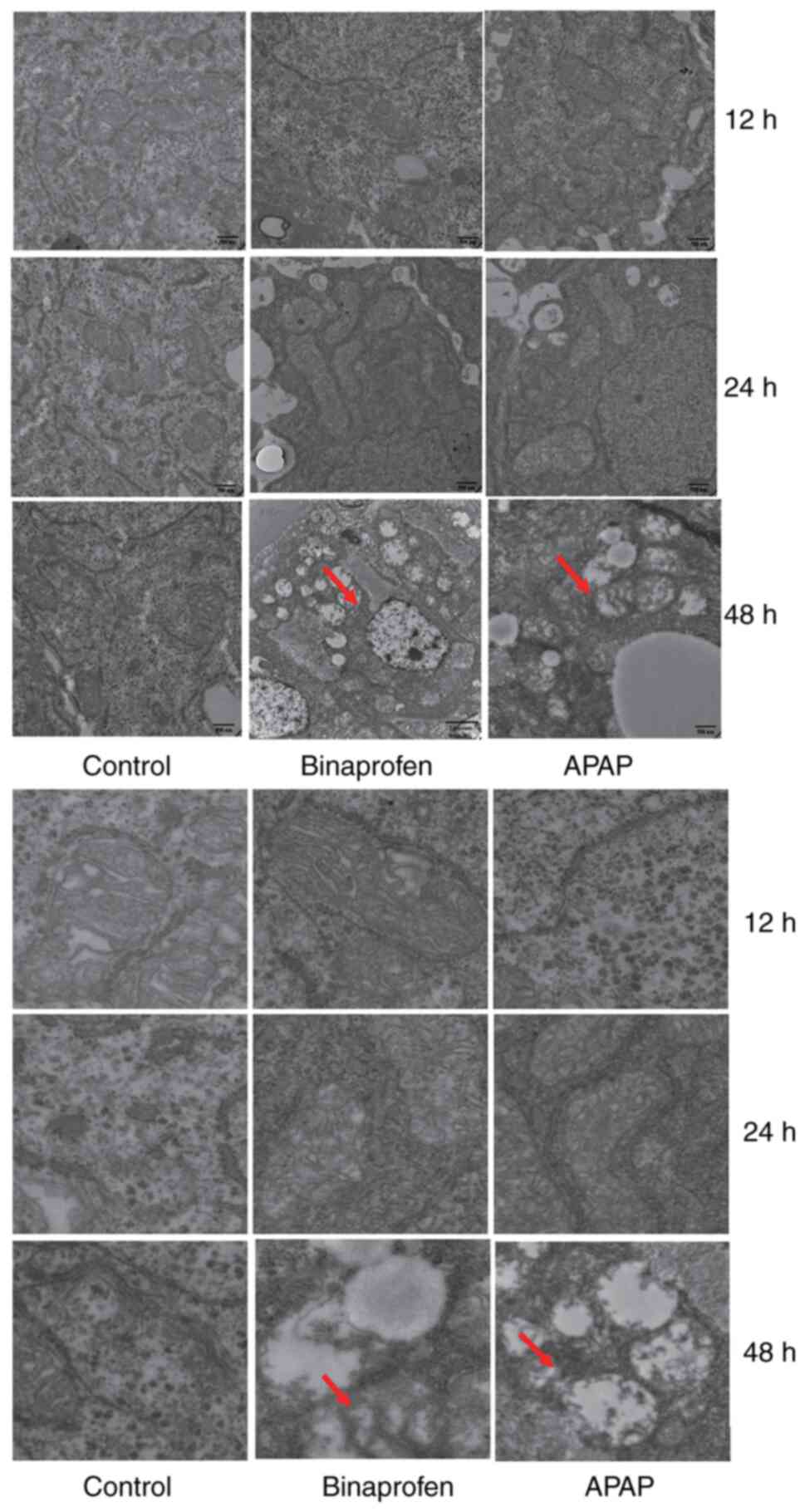
Mitochondrial change
Compared with control, APAP treatment caused
endoplasmic reticulum thickening, mitochondrial swelling and
vacuolation and ruptured cristae at 48 h (Fig. 5). Binaprofen treatment caused
mitochondrial swelling and vacuolation and rupture or disappearance
of cristae at 48 h.
Gene expression profiling
Compared with control group, in binaprofen group,
3,673 genes exhibited fold-change ≥2.0 or ≤0.5 in expression levels
at 12 h. Of these, 2499 genes were up- and 1,174 genes were
downregulated. At 24 h, 3,945 genes exhibited fold-change ≥2.0 or
≤0.5 in expression levels; of these, 2,745 genes were up- and 1,200
genes were downregulated. At 48 h, 5,496 genes exhibited
fold-change ≥2.0 or ≤0.5 in expression levels; of these, 3,851
genes were up- and 1,645 genes were downregulated (Table II, Fig. 6). Venn analysis identified 190
common differentially expressed genes at 12, 24 and 48 h. The
function of downregulated genes was primarily associated with ‘DNA
replication’, ‘DNA metabolic process’, ‘cell cycle’, ‘cell redox
homeostasis’, ‘mitochondrion’ and ‘lipid transport’. The function
of upregulated genes was primarily associated with ‘peroxisome
proliferator’, ‘oxidation activity’, ‘peroxisome’ and ‘apoptosis’.
There was a significant increase in Bcl2 and caspase gene
expression. Expression levels of 8 genes were different at 12, 24
and 48 h; of these, six genes were down- and two were upregulated
(Table III).
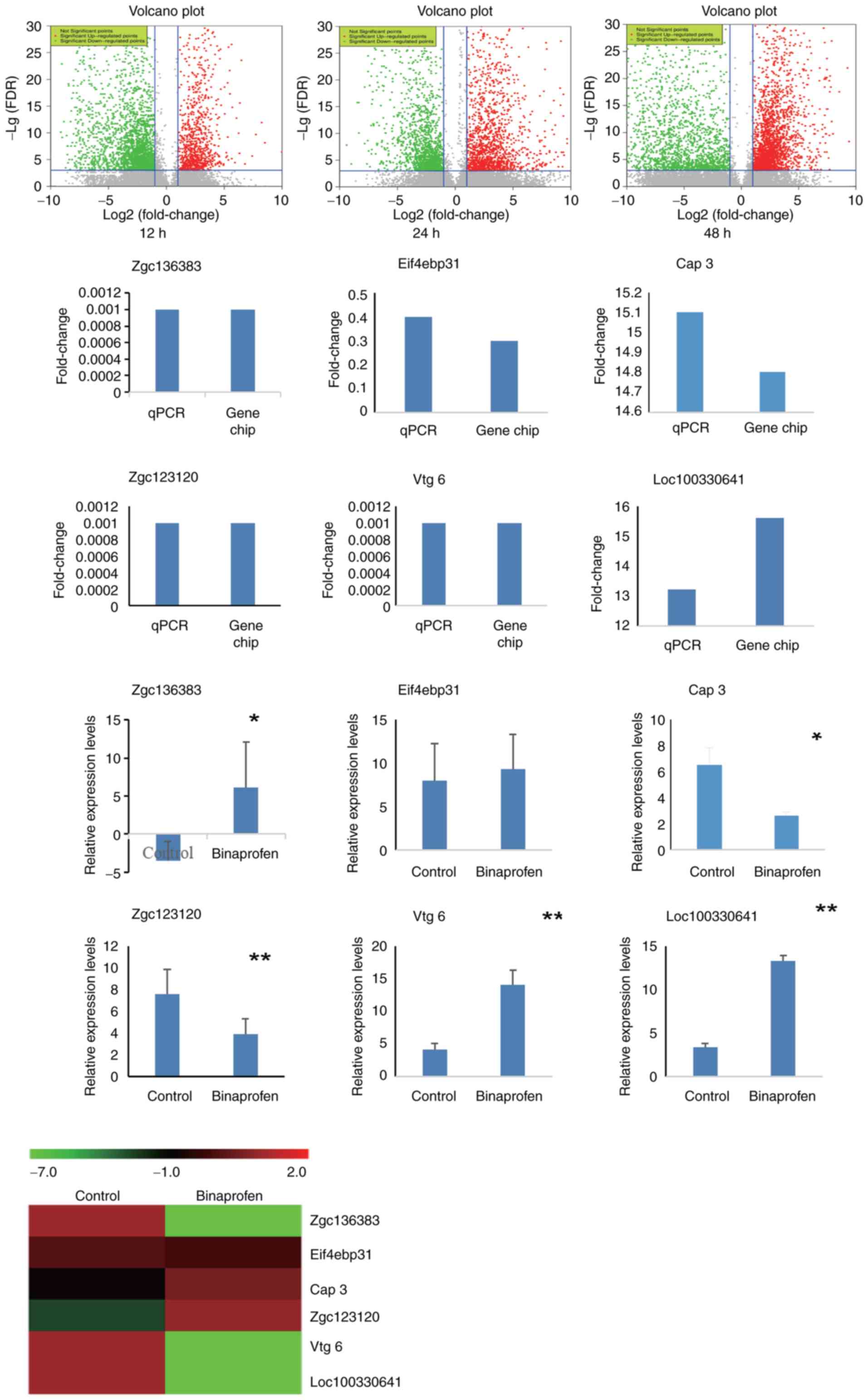 | Figure 6.Microarray and RT-qPCR assay of
zebrafish liver cells following exposure to binaprofen for 48 h.
Volcano graphs showed that 3,673 genes exhibited fold-change ≥2.0
or ≤0.5 in expression levels at 12 h. Of these, 2,499 genes were
up- and 1,174 genes were downregulated. At 24 h, 3,945 genes
exhibited fold-change ≥2.0 or ≤0.5 in expression levels; of these,
2,745 genes were up- and 1,200 genes were downregulated. At 48 h,
5,496 genes exhibited fold-change ≥2.0 or ≤0.5 in expression
levels; of these, 3,851 genes were up- and 1,645 genes were
downregulated. Among these, 6 genes were verified by RT-qPCR assay.
A total of five samples/group was used to detect zgc136383, Vtg 6,
Loc100330641, Eif4ebp31, Zgc123120 and Cap 3. Heatmap of microarray
results was consistent with RT-qPCR results. *P<0.05,
**P<0.01 vs. control. RT-q, reverse transcription-quantitative;
Zgc136383, vitellogenin 4; Zgc123120, BCL2/adenovirus E1B
interacting protein 4; Eif4ebp31, eukaryotic translation initiation
factor 4E binding protein 3; Cap 3, apoptosis-related cysteine
peptidase 3; Loc100330641, vitellogenin-like; Vtg 6, vitellogenin
6. |
 | Table II.Number of differentially expressed
genes following exposure to binaprofen for 12–48 h. |
Table II.
Number of differentially expressed
genes following exposure to binaprofen for 12–48 h.
|
| Gene count |
|---|
|
|
|
|---|
| Gene
regulation | 12 h (n=3,673) | 24 h (n=3,945) | 48 h (n=5,496) |
|---|
| Up | 2,499 | 2,745 | 1,200 |
| Down | 1,174 | 1,200 | 1,645 |
 | Table III.Effect of binaprofen on certain
differentially expressed genes at 12, 24 and 48 h. |
Table III.
Effect of binaprofen on certain
differentially expressed genes at 12, 24 and 48 h.
|
| Fold change |
| Time of greatest
differential expression, h |
|---|
|
|
|
|
|---|
| Gene | 12 h | 24 h | 48 h | Regulation |
|---|
| Zgc136383 | 0.001 | 0.001 | 0.000 |
Down | 48 |
| Vtg6 | 0.0008 | 0.0007 | 0.0005 |
Down | 48 |
| Loc100330641 | 0.001 | 0.001 | 0.000 |
Down | 48 |
| Eif4ebp31 | 0.429 | 0.314 | 0.301 |
Down | 48 |
| Loc1005352888 | 0.0004 | 0.0004 | 0.0003 |
Down | 48 |
| Loc100534731 | 0.442 | 0.414 | 0.276 |
Down | 48 |
| Zgc123120 | 11.459 | 18.673 | 15.656 | Up | 24 |
| Cap 3 | 50.117 | 50.375 | 14.833 | Up | 24 |
GO analysis
GO analysis showed that the function of
downregulated genes was primarily associated with ‘DNA
replication’, ‘DNA metabolic process’, ‘cell cycle’, ‘cell redox
homeostasis’, ‘mitochondrion’ and ‘lipid transport’ (Table IV). The function of upregulated
genes was primarily associated with ‘peroxisome proliferator’,
‘oxidation activity’ and ‘peroxisome’.
 | Table IV.Gene Ontology analysis of
differentially expressed genes following exposure to binaprofen for
12–48 h. |
Table IV.
Gene Ontology analysis of
differentially expressed genes following exposure to binaprofen for
12–48 h.
| A, Downregulated
genes |
|---|
|
|---|
|
| Gene count |
|---|
|
|
|
|---|
| Term | 12 h | 24 h | 48 h |
|---|
| DNA
replication | 32 | 6 | 6 |
| DNA metabolic
process | 71 | 4 | 8 |
| Cell cycle | 47 | 8 | 8 |
| Mitochondrion | 27 | 18 | 18 |
| Lipid
transport | - | 15 | 10 |
|
| B, Upregulated
genes |
|
|
| Gene
count |
|
|
|
| Term | 12 h | 24 h | 48 h |
|
| Peroxisome
proliferator | 28 | - | - |
| Oxidation
activity | - | 4 | 98 |
| Peroxisome | - | 4 | 4 |
| Apoptosis | 12 | 12 | 15 |
Gene pathway analysis
Differentially expressed genes were associated with
biological process according to GO pathway analysis. Pathway
analysis showed that upregulated and downregulated GO pathways were
associated with ‘cell cycle’, ‘DNA replication’, ‘ribosome’,
‘spliceosome’, ‘pyrimidine metabolism’, ‘purine metabolism’, ‘PPAR
signaling pathway’ and ‘p53 signaling pathway’ (Table V).
 | Table V.Pathway analysis of differentially
expressed genes following exposure to binaprofen for 12–48 h. |
Table V.
Pathway analysis of differentially
expressed genes following exposure to binaprofen for 12–48 h.
|
|
| Gene count |
|---|
|
|
|
|
|---|
| KEGG ID | Pathway | 12 h | 24 h | 48 h |
|---|
| dre03030 | DNA
replication | 24 | 24 | 29 |
| dre03010 | Ribosome | - | 28 | 29 |
| dre03040 | Spliceosome | 41 | 6 | - |
| dre04110 | Cell cycle | 41 | 41 | 52 |
| dre00240 | Pyrimidine
metabolism | 31 | - | 34 |
| dre00230 | Purine
metabolism | 34 | - | 37 |
| dre03320 | PPAR signaling
pathway | - | 12 | 18 |
| dre04115 | p53 signaling
pathway | 12 | 12 | 13 |
RT-qPCR
Expression levels of 8 genes changed over time:
Loc100535288, Loc100534731, Zgc136383, vitellogenin (Vtg) 6,
Loc100330641, Vtg-like eukaryotic translation initiation factor 4E
binding protein 31 (Eif4ebp31), Zgc123120 and Cap 3. Because
Loc100535288 and Loc100534731 were uncharacterized geneswithout any
functional information, the other six genes were selected to verify
the results of microarray (Fig.
6). Microarray showed that Zgc136383, Vtg6, Loc100330641,
Eif4ebp31 were downregulated and Zgc123120 and Cap 3 were
upregulated. RT-qPCR showed that Zgc136383, Vtg 6, Loc100330641,
Eif4ebp31 were downregulated and Zgc123120 and Cap 3 were
upregulated. Meanwhile, the fold-change was also similar. The
results of RT-qPCR and microarray were consistent.
Discussion
Binaprofen is not currently commercially available.
Our previous study evaluated the effect of different doses of
binaprofen and APAP on liver injury; both induced liver injury to a
similar extent (21). The
half-maximal lethal concentration (LC50) of binaprofen
was 1.2 mM, which was 2 times higher than its maximum recommend
dose (19); LC50 of
APAP was 5.2 mM, which was 1.6 times higher than its people maximum
recommend dose (31). Therefore,
binaprofen was selected for investigation of the underlying
mechanism of toxicity. APAP was used as a positive control because
of its known ability to induce liver injury (32).
The present study investigated the mechanism of
binaprofen-induced liver injury in zebrafish. Binaprofen increased
levels of liver biomarkers ALT, AST and LDH in a time-dependent
manner, increased MDA and decreased GSH content. Binaprofen induced
hepatocyte vacuolization, as well as mitochondrial swelling,
vacuolization and rupture or disappearance of cristae. Binaprofen
induced hepatocyte apoptosis. Binaprofen induced altered gene
expression at 12, 24 and 48 h. There were 190 common differentially
expressed genes at all three timepoints. The function of
downregulated genes were primarily associated with ‘DNA
replication’, ‘DNA metabolic process’, ‘cell cycle’, ‘cell redox
homeostasis’, ‘mitochondrion’ and ‘lipid transport’. The function
of upregulated genes was primarily associated with ‘peroxisome
proliferator’, ‘oxidation activity’, ‘peroxisome’ and ‘apoptosis’.
GO pathways were associated with ‘cell cycle’, ‘DNA replication’,
‘ribosome’, ‘spliceosome’, ‘pyrimidine metabolism’, ‘purine
metabolism’, ‘PPAR signaling pathway’ and ‘p53 signaling pathway’.
Six genes from microarray were verified, and the results of RT-qPCR
were in accordance with microarray results.
Therefore, the experimental results showed that the
mechanism of hepatotoxicity was associated with lipid peroxidation
and apoptosis.
DILI is associated with inappropriate activation of
apoptotic cell death pathways (33–36). Apoptosis serves a key role in
progression of liver disease, such as cirrhosis (37,38). Apoptosis is mediated by two
central pathways, the intrinsic and extrinsic pathway. The
mitochondrial pathway is the intrinsic pathway and begins with
permeabilization of the mitochondrial outer membrane (39–42). Release of cytochrome c from
mitochondria is key factor to initiate apoptosis (43,44). The released cytochrome c activates
caspase-9; this leads to caspase-3 activation, cellular protein
cleavage and apoptosis (45).
Following binaprofen exposure, there was swelling in mitochondria
and increase in DAPI-positive cells with condensed and fragmented
nuclei. According to reports (46), DAPI stains apoptotic cells with
high labeling efficiency (~100%) and does not change the
ultrastructure of organelles. The present study aimed to determine
the toxicity of binaprofen, therefore, DAPI was used to detect
hepatocyte apoptosis. The results suggested apoptosis occurred;
this was confirmed by altered expression of genes associated with
apoptosis in the microarray. Moreover, electron microscopy showed
liver cell mitochondrial swelling and vacuolization, which
indicated that apoptosis was associated with mitochondria. In
addition, there was a significant increase in Bcl2 family and
caspase gene expression in microarray; this was validated by
RT-qPCR. These data suggested that binaprofen induces zebrafish
liver injury via the mitochondria-mediated apoptosis pathway.
In the present study, DAPI staining of apoptotic
cells was not quantified. TUNEL staining and western blotting were
not performed to confirm levels of apoptosis markers; these
experiments should be performed in future to quantify apoptosis.
Here, it is also found that the signaling pathway of
binaprofen-induced liver injury may be associated with PPAR
signaling pathway and P53 signaling pathway, we will do further
study to clear them.
The mechanism of binaprofen-induced liver injury was
associated with lipid peroxidation and apoptosis. Binaprofen
induced hepatocyte mitochondrial structural damage and activated
apoptosis via the mitochondrial signaling pathway.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Foundation of Ministry of
Science and Technology of People's Republic of China (grant nos.
2018ZX09721004 and 201604046020), High-Level Leading Talent
Introduction Program of Guangdong Academic of Sciences and People's
Republic of China (grant no. 2016GDASRC-0104).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus database
of National Center for Biotechnology Information repository,
ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE199758 (accession no.
GSE199758).
Authors' contributions
QG designed and performed experiments. GC performed
experiments. HO and QN analyzed the data. RJ conceived the study.
RQ and RJ interpreted the data. All authors have read and approved
the final manuscript. QG and GC confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Animal Care and Use Committee of Guangzhou General Pharmaceutical
Research Institute (approval no. 2012-005) and performed in
accordance with international guidelines for the care and use of
laboratory animals. All zebrafish experiments in this study were
performed from 1st of July to 30th of August 2012.
Patient consent for participation
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jee A, Sernoskie SC and Uetrecht J:
Idiosyncratic Drug-induced liver injury: Mechanistic and clinical
challenges. Int J Mol Sci. 22:29542021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Villanueva-Paz M, Morán L, López-Alcántara
N, Freixo C, Andrade RJ, Lucena MI and Cubero FJ: Oxidative stress
in drug-induced liver injury (DILI): From mechanisms to biomarkers
for use in clinical practice. Antioxidants (Basel). 10:3902021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jing J and Teschke R: Traditional chinese
medicine and herb-induced liver injury: Comparison with
drug-induced liver injury. J Clin Transl Hepatol. 6:57–68. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Teschke R: Idiosyncratic DILI: Analysis of
46,266 cases assessed for causality by RUCAM and published from
2014 to early 2019. Front Pharmacol. 10:7302019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Subramanya SB, Venkataraman B, Meeran MFN,
Goyal SN, Patil CR and Ojha S: Therapeutic potential of plants and
plant derived phytochemicals against acetaminophen-induced liver
injury. Int J Mol Sci. 19:37762018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanabar DJ: A clinical and safety review
of paracetamol and ibuprofen in children. Inflammopharmacology.
25:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Donati M, Conforti A, Lenti MC, Capuano A,
Bortolami O, Motola D, Moretti U, Vannacci A, Rafaniello C,
Vaccheri A, et al: Risk of acute and serious liver injury
associated to nimesulide and other NSAIDs: Data from drug-induced
liver injury case-control study in Italy. Br J Clin Pharmacol.
82:238–248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong H, Yuan-Keng H, Yuan-Keng X, Hui-Yu
O and Wei Y: Effect of felbinac trometamol injection on analgesia
and its active site. Central South Pharm. 7:481–484. 2013.(In
Chinese).
|
|
9
|
Kaplowitz N: Idiosyncratic drug
hepatotoxicity. Nat Rev Drug Discov. 4:489–499. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Björnsson ES: Drug-induced liver injury
due to antibiotics. Scand J Gastroenterol. 52:617–623. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katarey D amd Verma S, . Drug-induced
liver injury. Clin Med (Lond). 16 (Suppl 6):s104–s109.
2016.PubMed/NCBI
|
|
12
|
Leise MD, Poterucha JJ and Talwalkar JA:
Drug-induced liver injury. Mayo Clin Proc. 89:95–106. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chalasani N, Bonkovsky HL, Fontana R, Lee
W, Stolz A, Talwalkar J, Reddy KR, Watkins PB, Navarro V, Barnhart
H, et al: Features and outcomes of 899 patients with drug-induced
liver injury: The DILIN prospective study. Gastroenterology.
148:1340–1352.e7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bernal W and Wendon J: Acute liver
failure. N Engl J Med. 369:2525–2534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee WM: Acute liver failure in the United
States. Semin Liver Dis. 23:217–226. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang W, Ou HY, Xiao BQ, Huang YJ, Yang W
and Wang QS: The study of abirritation of a first type of new drug,
felbinac trometamol injection. Chin J Med Guide. 11:1327–1332.
2009.
|
|
17
|
Wang W, Ou H and Liang H: Preliminary
pharmacodynamics and safety studies of a first type of new drug,
felbinac trometamol injection. Chin J Ethnomedicine Ethnopharmacy.
12:3–4. 2009.(In Chinese).
|
|
18
|
Zhang C, Cui X, Yang Y, Gao F, Sun Y, Gu
J, Fawcett JP, Yang W and Wang W: Pharmacokinetics of felbinac
after intravenous administration of felbinac trometamol in rats.
Xenobiotica. 41:340–348. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao BQ, Lei XL, Yang W, Huang YK, Ou HY
and Lian XK: afety pharmacology research of class I new drug:
felbinac trometamol injection. Chin J New Drug. 20:1386–1391.
2011.(In Chinese).
|
|
20
|
Han Z, Ou HY, Sun H, Feng MJ, Xiao BQ and
Yang W: Experiment for security evaluation of class I new drug of
felbinac trometamol injection. Pharm Today. 23:201–204, (In
Chinese).
|
|
21
|
Guo Q, Guo J, Chen G, Han Z, Xiao B, Jin
R, Liang C and Yang W: Biomarkers associated with
binaprofen-induced liver injury. Mol Med Rep. 18:5076–5086.
2018.PubMed/NCBI
|
|
22
|
Herndon CM and Dankenbring DM: Patient
perception and knowledge of acetaminophen in a large family
medicine service. J Pain Palliat Care Pharmacother. 28:109–116.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Altyar A, Kordi L and Skrepnek G: Clinical
and economic characteristics of emergency department visits due to
acetaminophen toxicity in the USA. BMJ Open. 5:e0073682015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mcgill MR and Jaeschke H: Metabolism and
disposition of acetaminophen: Recent advances in relation to
hepatotoxicity and diagnosis. Pharm Res. 30:2174–2187. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hinson JA, Roberts DW and James LP:
Mechanisms of acetaminophen-induced liver necrosis. Handb Exp
Pharmacol. 196:369–405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cima G: AVMA guidelines for the euthanasia
of animal: 2013 Edition. Am Vet Med Assoc. 242:715–716. 2013.
|
|
27
|
Thurman CE, Rasmussen S and Prestia KA:
Effect of 3 euthanasia methods on serum yield and serum cortisol
concentration in zebrafish (Danio rerio). J Am Assoc Lab Anim Sci.
58:823–828. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Köhler A, Collymore C, Finger-Baier K,
Geisler R, Kaufmann L, Pounder KC, Schulte-Merker S, Valentim A,
Varga ZM, Weiss J and Strähle U: Report of workshop on euthanasia
for zebrafish-a matter of welfare and science. Zebrafish.
14:547–551. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schafer KA, Eighmy J, Fikes JD, Halpern
WG, Hukkanen RR, Long GG, Meseck EK, Patrick DJ, Thibodeau MS, Wood
CE and Francke S: Use of severity grades to characterize
histopathologic changes. Toxicol Pathol. 46:256–265. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lv S, Wang Y, Xu W and Dong X: Serum
exosomal miR-17-5p as a promising biomarker diagnostic biomarker
for breast cancer. Clin Lab. 66:2020. View Article : Google Scholar
|
|
31
|
Fisher ES and Curry SC: Evaluation and
treatment of acetaminophen toxicity. Adv Pharmacol. 85:263–272.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shojaie L, Iorga A and Dara L: Cell death
in liver diseases: A review. Int J Mol Sci. 21:96822020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chao X, Wang H, Jaeschke H and Ding WX:
Role and mechanisms of autophagy in acetaminophen-induced liver
injury. Liver Int. 38:1363–1374. 2018.Schwabe RF and Luedde T:
Apoptosis and necroptosis in the liver: a matter of life and death.
Nat Rev Gastroenterol Hepatol. 15:738–752. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iorga A, Dara L and Kaplowitz N:
Drug-induced liver injury: Cascade of events leading to cell death,
apoptosis or necrosis. Int J Mol Sci. 18:10182017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao X, Yang L, Chang N, Hou L, Zhou X,
Yang L and Li L: Neutrophils undergo switch of apoptosis to NETosis
during murine fatty liver injury via S1P receptor 2 signaling. Cell
Death Dis. 11:3792020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ke PY: Diverse Functions of autophagy in
liver physiology and liver diseases. Int J Mol Sci. 20:3002019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ko S, Russell JO, Molina LM and Monga SP:
Liver progenitors and adult cell plasticity in hepatic injury and
repair: Knowns and unknowns. Annu Rev Pathol. 15:23–50. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dong W, Luo B, Qiu C, Jiang X, Shen B,
Zhang L, Liu W and Zhang W: TRIM3 attenuates apoptosis in
Parkinson's disease via activating PI3K/AKT signal pathway. Aging
(Albany NY). 13:735–749. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Q, Liu J, Zhang M, Wei S, Li R, Gao
Y, Peng W and Wu C: Apoptosis induction of fibroblast-like
synoviocytes is an important molecular-mechanism for herbal
medicine along with its active components in treating rheumatoid
arthritis. Biomolecules. 9:7952019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sha L, Ma D and Chen C: Exosome-mediated
Hic-5 regulates proliferation and apoptosis of osteosarcoma via
Wnt/β-catenin signal pathway. Aging (Albany NY). 12:23598–23608.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li RL, Zhang Q, Liu J, Sun JY, He LY, Duan
HX, Peng W and Wu CJ: Hydroxy-α-sanshool possesses protective
potentials on H2O2-stimulated PC12 cells by
suppression of oxidative stress-induced apoptosis through
regulation of PI3K/Akt signal pathway. Oxid Med Cell Longev.
2020:34817582020.PubMed/NCBI
|
|
42
|
Li Y, Ding H, Liu L, Song Y, Du X, Feng S,
Wang X, Li X, Wang Z, Li X, et al: Non-esterified fatty acid induce
dairy cow hepatocytes apoptosis via the mitochondria-mediated
ROS-JNK/ERK signaling pathway. Front Cell Dev Biol. 8:2452020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li W, Li Y, Jiang X, Li X and Yu Z:
Compound ammonium glycyrrhizin protects hepatocytes from injury
induced by lipopolysaccharide/florfenicol through a mitochondrial
pathway. Molecules. 23:23782018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Saito Y, Hikita H, Nozaki Y, Kai Y, Makino
Y, Nakabori T, Tanaka S, Yamada R, Shigekawa M, Kodama T, et al:
DNase II activated by the mitochondrial apoptotic pathway regulates
RIP1-dependent non-apoptotic hepatocyte death via the TLR9/IFN-β
signaling pathway. Cell Death Differ. 26:470–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nguyen SM, Lieven CJ and Levin LA:
Simultaneous labeling of projecting neurons and apoptotic state. J
Neurosci Methods. 161:281–284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wallberg F, Tenev T and Meier P: Analysis
of apoptosis and necroptosis by fluorescence-activated cell
sorting. Cold Spring Harb Protoc. 2016.pdb.prot087387, 2016.
View Article : Google Scholar
|