Introduction
Severe sepsis is one of the most prevalent diseases
in patients admitted to the intensive care unit of hospitals
(1). It is a main cause of death in
critically ill patients and affects millions of individuals around
the world each year, with 11 million sepsis deaths and 48.9 million
cases of sepsis reported in 2017 (2). Despite an overwhelming increase in
knowledge regarding the pathogenesis of sepsis and subsequent
advances in clinical care, the incidence of sepsis is still
increasing in both adults and children, accounting for an
unacceptably high mortality rate ranging between 25–30% depending
on age and disease severity (3). It
is often characterized by a systemic inflammatory response to
infection that is typically bacterial in origin (4). Moreover, sepsis is defined as a
documented or suspected infection in a subset of four findings
(body temperature >38°C or <36°C; heart rate >90
beats/min; hyperventilation evidenced by breathing rate of >20
breaths/min or PaCO2 <32 mmHg; white blood cell count >12,000
cells/µl or <4,000/µl) that describe systemic inflammatory
response syndrome (4,5). Sepsis can progress rapidly, resulting
in organ failure (severe sepsis) or impaired tissue perfusion
(septic shock) (6). Although a
number of biomarkers, such as procalcitonin and C-reactive protein,
have been proposed as candidate markers for the diagnosis,
prognosis and therapeutic guidance of sepsis, each have certain
limitations; for example, low specificity for early diagnosis, and
the lack of definitive evaluation parameters for the severity of
sepsis and the prognosis of patients, which make it difficult to
diagnose sepsis with high sensitivity and specificity (7). Therefore, new biomarkers with high
sensitivity and specificity are urgently required.
MicroRNAs (miRNAs/miRs) are a class of small
noncoding RNAs that pair to sites in mRNAs to regulate gene
expression in eukaryotes (8). To
date, ~1,000 miRNAs have been identified in humans, which may
directly regulate at least 30% of the genes in a cell, therefore
serving important roles in a variety of cellular functions as well
as in several diseases (9,10). Thus, miRNAs are involved in the
regulation of almost all major cellular functions, including cell
development, differentiation, proliferation and apoptosis (11). The abnormal expression of miRNAs may
implicate changes in a wide array of cellular and developmental
processes of disease initiation and progression that can lead to
malignant phenotypes (12–14). This means that in various
pathological conditions, such as inflammation, infection and
sepsis, miRNA levels may change, which is a parameter that can be
quickly detected (15). Thus,
miRNAs isolated from the peripheral blood of patients with sepsis
may be measured by performing genome-wide profiling microarrays in
leukocytes in order to elucidate potential biomarkers of sepsis.
Interestingly, serum miR-16 and miR-483-5p have been identified as
prognostic predictors of patients with sepsis, as they are
associated with sepsis-induced death (16).
To identify putative miRNA biomarkers involved in
the process of sepsis, the current study analyzed the
differentially expressed microRNAs (DEMs) via microarray analysis
followed by verification via reverse transcription-quantitative PCR
(RT-qPCR) and bioinformatics. Additionally, the correlations
between the identified miRNAs and proinflammatory cytokines and
chemokines were evaluated. The results of the present study may
further clarify the roles of miRNAs in the diagnosis and treatment
of sepsis, so as to elucidate a novel miRNA biomarker.
Materials and methods
Patients
A total of 40 patients (21 male patients and 19
female patients; age range, 20–80 years; mean age, 66.5±5.8 years)
admitted to the Department of Respiratory and Critical Care
Medicine of the Third Affiliated Hospital of Inner Mongolia Medical
University (Baotou, China) between January 1st 2015 and October
31st 2015 were enrolled in the current study. The inclusion
criteria were as follows: Patients with sepsis were diagnosed based
on the Sepsis 3.0 guidelines, which can be simplified as follows:
Sepsis = infection + sequential organ failure assessment (SOFA) ≥2
(17). The exclusion criteria were
as follows: i) Patients <18 or >80 years of age; ii) patients
with liver and kidney failure; iii) patients that were pregnant or
lactating; iv) patients with tumors and hematological diseases; v)
an agranulocytosis score of <0.5×109/l; vi) patients with human
immunodeficiency virus-related complications; and vii) patients
administered glucocorticoids up to 4 weeks before enrollment or
that were receiving immunotherapy after organ transplantation. A
total of 40 healthy individuals that donated samples following
routine physical examinations were used as a control group (20
males, 20 females; aged 20–80 years). A further 80 samples
(including 40 samples from patients with sepsis and 40 samples from
healthy controls, whose details are listed in Table I) were obtained from individuals
admitted to the same hospital that were enrolled to the present
study between April 10th 2020 and January 31st 2021. The inclusion
criteria were the same as aforementioned. There was no significant
difference in the mean values of sex and age between the test group
and the control group. All protocols were approved by the Local
Ethics Committee of the Third Affiliated Hospital of Inner Mongolia
Medical University (Baotou, China), and written informed consent
was obtained from all subjects who were permitted to withdraw from
clinical observation at any time for any reason.
 | Table I.Clinical characteristics of patients
with sepsis and healthy controls (n=80). |
Table I.
Clinical characteristics of patients
with sepsis and healthy controls (n=80).
| A, Original
samples |
|---|
|
|---|
| Characteristic | Sepsis | Normal control | P-value |
|---|
| Age (mean ± SD) | 66.5±5.8 | 67.2±4.2 | 0.12 |
| Male (n) | 21 | 20 | 0.80 |
| Female (n) | 19 | 20 | 0.74 |
|
| B, Expanded
samples |
|
|
Characteristic | Sepsis | Normal
control | P-value |
|
| Age (mean ±
SD) | 63.1±1.8 | 61.5±9.4 | 0.17 |
| Male (n) | 18 | 21 | 0.56 |
| Female (n) | 22 | 19 | 0.43 |
Total RNA extraction from blood and
quality control
To compare the DEMs in patients with sepsis and
healthy controls, 2 ml whole blood samples were collected into
sterilized Eppendorf tubes. Total RNA was extracted using the
mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific,
Inc.) in accordance with the manufacturer's protocol. Subsequently,
the concentration and integrity of total RNA was determined using a
spectrophotometer (NanoDrop™; Thermo Fisher Scientific, Inc.) and
agarose gel electrophoresis. The conditions required for microarray
analysis included: An average A260/280 ratio
of total RNA ≥1.8, RNA content ≥1 µg/ml and clear 28S and 18S
electrophoresis bands of total RNA (18).
Microarray assay and data
analysis
Microarray analysis was performed by Sangon Biotech
Co., Ltd. Briefly, total RNA (~200 ng) extracted from the
aforementioned blood samples was subjected to dephosphorylation and
labeling using the miRNA Complete Labeling and Hyb kit (Agilent
Technologies, Inc.), where Cyanine3-pCp was connected to the 3′ end
of RNA using T4 RNA ligase. The labeled reaction products were
concentrated using a vacuum concentrator for 3 h at 45–55°C and
subsequently underwent overnight hybridization using Agilent
SureSelect Capture Technology (Agilent Technologies, Inc.). After
washing with saline-sodium citrate at room temperature, slides were
dried by air and scanned using the Agilent Scanner G2505C (Agilent
Technologies, Inc.), after which the Agilent Feature Extraction
(v10.7; Agilent technologies, Inc.) was utilized to extract data
and analyze the hybridization results. Agilent GeneSpring (version
12.5; Agilent Technologies, Inc.) was used for quartile data
normalization and to determine differences between groups. A
Q-value of ≥5% and a fold change (FC) value of >2.0 or <0.5
were selected as cut-off points for DEMs. Statistical significance
was determined using an unpaired t-test and was represented using a
P-value. To reduce the risk of false positives, values were
adjusted for multiple testing using the Benjamini-Hochberg False
Discovery Rate (FDR) method. The corrected value was represented by
FDR (19). FDR <0.05 was
selected as the cut-off value for DEM screening. The possible
miRNAs that target sepsis were predicted using TargetScan software
(http://www.targetscan.org/vert_80/).
Identification of DEMs in the
peripheral blood of patients with sepsis via RT-qPCR
According to microarray assay, 305 miRNAs with
>2-fold expression changes (patients with sepsis vs. healthy
people) were detected. Subsequently, the top 18 up- and
downregulated miRNAs were selected for microarray analysis
verification via RT-qPCR. The primers used for RT-qPCR were
synthesized by Sangon Biotech Co., Ltd. and are listed in Table II. Total RNA was extracted using
RNAiso Plus (cat. no. 9108; Takara Bio, Inc.), and reverse
transcribed into cDNA using miRNA First Strand cDNA Synthesis
(Stem-loop Method) (cat. no. B532453; Sangon Biotech Co., Ltd.)
according to the manufacturers' instructions. RT-qPCR was performed
using SYBR Premix Ex Taq™ II (Takara Bio, Inc.) with U6 as an
internal control for miRNA detection. The thermocycling conditions
were as follows: Pre-denaturation at 95°C for 10 min, followed by
40 cycles of denaturing at 95°C for 5 sec, annealing at 60°C for 20
sec and elongation at 70°C for 10 sec. The melt curve conditions
were as follows: 60°C to 95°C in increments of 0.3°C; this was used
to determine the melting temperature of the detected miRNAs and
primer dimers. The relative expression of DEMs was calculated using
the comparative 2−ΔΔcq method (20) and the data were analyzed using SPSS
v19.0 software (IBM Corp.).
 | Table II.All primer sequences used in the
current study. |
Table II.
All primer sequences used in the
current study.
| Primer | Sequence (5′ to
3′) |
|---|
|
hsa-miR-3663-3p |
TGAGCACCACACAGGCCG |
| hsa-miR-625-5p |
AGGGGGAAAGTTCTATAGTCC |
| hsa-miR-892b |
CACTGGCTCCTTTCTGGGT |
| hsa-miR-491-5p |
AGTGGGGAACCCTTCCATGA |
| hsa-miR-592 |
GATTGTGTCAATATGCGATGATGT |
|
hsa-miR-3591-3p |
AAACACCATTGTCACACTCCAC |
|
hsa-miR-6514-3p |
CTGCCTGTTCTTCCACTCC |
|
hsa-miR-4694-5p |
ATCCCGTAUCTCTTCGTCTTCGA |
| hsa-miR-362-5p |
AATCCTTGGAACCTAGGTGTGA |
| hsa-miR-1260b |
ATCCCACCACTGCCACCAT |
| hsa-miR-491-3p |
CTTATGCAAGATTCCCTTCTA |
| hsa-miR-501-3p |
GCGGCGGAATGCACCCGGGCAAG |
| hsa-miR-17-3p |
GCCGCAAAGTGCTTACAGTG |
|
hsa-miR-6881-3p |
TCTGGCTTGTATCTAGCGTATGA |
| hsa-miR-501-5p |
AATCCTTTGTCCCTGGGTGAGA |
|
hsa-miR-6756-5p |
TTTTTCCGATTATTGCTCCTGACC |
|
hsa-miR-6752-5p |
TACTGCCCTGACCTGTCCTGTCC |
|
hsa-miR-6786-5p |
TAACCGCACTGTCTGGTAAAGAT |
| Universal
reverse |
GTGCAGGGTCCGAGGT |
| miRNA primer |
|
| Human-TDAG8 |
|
|
Forward |
TTCCTGGGCTACGCAATACC |
|
Reverse |
CCGTAGCTTGGTTGTGCTTC |
| Human TLR4 |
|
|
Forward |
TTCCTGGGCTACGCAATACC |
|
Reverse |
CCGTAGCTTGGTTGTGCTTC |
| GAPDH |
|
|
Forward |
CAGGAGGCATTGCTGATGAT |
|
Reverse |
GAAGGCTGGGGCTCATTT |
| U6 |
|
|
Forward |
GCTTCGGCAGCACATATACTAAAAT |
|
Reverse |
CGCTTCACGAATTTGCGTGTCAT |
RT-qPCR determination of T-cell
death-associated gene 8 (TDAG8) and toll-like receptor 4 (TLR4)
mRNA expression
It has been reported that TDAG8 is involved in the
maintenance of lysosomal function, particularly during pathogen
defense (21). In addition, TLR4
can be activated by lipopolysaccharide (LPS) to induce the
production of proinflammatory mediators to eradicate bacteria or
other pathogens (22).
Dysregulation of the host response to LPS can lead to sepsis, which
is a systemic inflammatory condition (23). The current study therefore detected
the expression levels of TDAG8 and TLR4 mRNA in the peripheral
blood of patients with sepsis. Total RNA was extracted using RNAiso
Plus, and reverse transcribed into cDNA using PrimeScript™ RT
reagent Kit with gDNA Eraser (Perfect Real Time) (cat. no. R047A;
Takara Bio, Inc.) according to the manufacturer instructions. The
primers used for this reaction are listed in Table II. Amplification was performed
using the SYBR Premix Ex Taq™ II under the following thermocycling
conditions: Initial denaturation at 95°C for 30 sec; followed by 50
cycles of denaturation at 95°C for 5 sec, annealing at 60°C for 30
sec and extension at 72°C for 30 sec. The expression of TDAG8 and
TLR4 mRNA was calculated using the 2−ΔΔCq method and
GAPDH was used as an internal control.
TargetScan software assay
The current results revealed that the expression
levels of miR-3663-3p and miR-6881-3p in patients with sepsis were
significantly increased compared with healthy controls. In
addition, our pre-experiment determined that TDAG8 was closely
related to the regulation of the inflammatory response in patients
with sepsis. That is, in the sepsis group, the correlation
coefficients (r) of TDAG8 mRNA expression and IL-6 or CXCL8
concentration were 0.8455 or 0.7117, respectively, indicating that
TDAG8 mRNA expression was positively correlated with IL-6 and CXCL8
levels. In the present study, TargetScan software (www.targetscan.org/vert_72/; TargetScanHuman 7.2)
was applied to predict whether TDAG8 was the biological target of
miR-3663 and miR-6881. After selecting ‘human’ as the species, the
human gene TDAG8 (GPR65) was entered. The database was subsequently
searched for miR-3663-3p and miR-6881-3p.
ELISA
Sepsis often leads to multiple organ failure as a
result of an uncontrolled inflammatory response (24). In the present study, ELISA was
performed to detect the concentrations of various proinflammatory
cytokines, including IL-6 (cat. no. CSB-E04638h; Cusabio Technology
LLC), IL-21 (cat. no. CSB-E11707h; Cusabio Technology LLC), C-X-C
motif chemokine ligand-8 (CXCL8) (cat. no. CSB-E04641h; Cusabio
Technology LLC) and monocyte chemoattractant protein-1 (MCP-1)
(cat. no. CSB-E04655h; Cusabio Technology LLC) in the peripheral
blood of patients with sepsis. All procedures were performed in
accordance with the respective protocols of commercially available
ELISA kits (R&D Systems, Inc.). Each sample was measured three
times and an average value was calculated.
Statistical analysis
Data were obtained from three independent
experiments and were expressed as the mean ± SD. All statistical
analyses were performed using SPSS v19.0 software. The statistical
differences between two groups were detected using unpaired
Student's t-test. The correlation between inflammatory response and
miRNA-3663-3p or TDAG8/TLR4 mRNA expression was determined using
Pearson's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Comparison of clinical characteristics
between patients with sepsis and healthy controls
There were 21 males and 19 females in the sepsis
group, aged between 20–80 years, with a mean age of 66.5±5.8 years.
A total of 40 healthy individuals that donated samples following
routine physical examinations were selected as the control group
(20 males; 20 females; age range, 20–80 years; mean age, 67.2±4.2
years). There were no significant differences between the mean
values of sex and age between the sepsis and healthy control groups
(Table I).
Total RNA extracted from peripheral
blood demonstrates good integrity
The integrity of total RNA was assessed using
agarose gel electrophoresis and subsequent 18S and 28S RNA band
staining for visualization. As presented in Fig. 1, the electrophoretic bands of 28S
and 18S RNA were clear. In addition, the ratio of A260 to A280 in
the extracted RNA was 1.8-2.0. Therefore, total RNA samples with
high purity and good integrity were used for microarray
analysis.
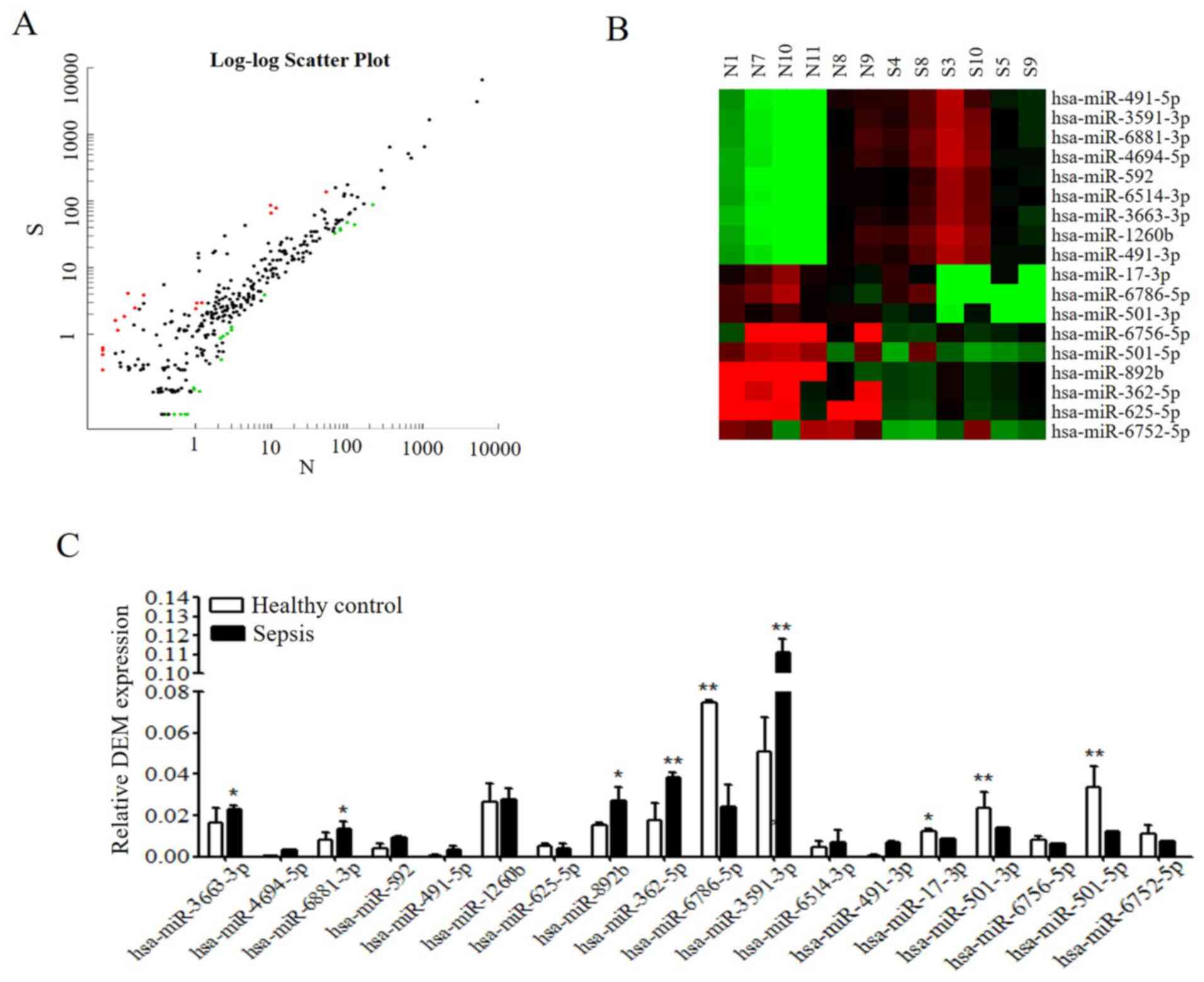
Scatter plot maps and DEM hierarchical
clustering
To clarify the potential function of DEMs,
microarray analysis was performed by Sangon Biotech Co., Ltd.
Agilent GeneSpring was used for the analysis of DEM expression
between patients with sepsis and healthy controls in order to
identify characteristic DEMs. The significance of microarray
analysis was denoted by FDR (FDR <0.05). As presented in
Fig. 2A, the results revealed 305
miRNAs that demonstrated a >2-fold change in expression in
patients with sepsis compared with in healthy individuals,
including 212 upregulated and 93 downregulated DEMs. Among these,
the top 18 up- or downregulated miRNAs, which exhibited significant
differential expression (P<0.05; FC >2.0), were selected for
further analysis. A total of 9 miRNAs demonstrated an increased
expression, while 9 miRNAs exhibited a decreased expression
(Table III). Hierarchical
clustering of the top 18 DEMs was subsequently performed, the
results of which are presented in Fig.
2B.
 | Table III.Differentially expressed microRNAs
obtained from microarray analysis. |
Table III.
Differentially expressed microRNAs
obtained from microarray analysis.
| Systematic
name | P-value (Corr) | P-value | FC (abs) | Expression |
|---|
|
hsa-miR-3663-3p | 0.000472 | 1.320987 | 16.258206 | Upregulated |
|
hsa-miR-4694-5p | 0.372410 | 0.020556 | 9.608086 | Upregulated |
|
hsa-miR-6881-3p | 0.086800 | 0.001669 | 22.589320 | Upregulated |
| hsa-miR-592 | 0.452431 | 0.050030 | 9.550354 | Upregulated |
| hsa-miR-491-5p | 0.353573 | 0.008057 | 9.395253 | Upregulated |
| hsa-miR-1260b | 0.372410 | 0.022981 | 5.556636 | Upregulated |
| hsa-miR-625-5p | 0.238966 | 0.000756 | 10.871430 | Downregulated |
| hsa-miR-892b | 0.452431 | 0.055429 | 5.355109 | Downregulated |
| hsa-miR-362-5p | 0.490820 | 0.099653 | 3.538647 | Downregulated |
|
hsa-miR-6786-5p | 0.519619 | 0.115105 | 2.160317 | Downregulated |
|
hsa-miR-3591-3p | 0.354531 | 0.015007 | 12.917550 | Upregulated |
|
hsa-miR-6514-3p | 0.372414 | 0.025927 | 10.691710 | Upregulated |
| hsa-miR-491-3p | 0.402106 | 0.031632 | 8.345849 | Upregulated |
| hsa-miR-17-3p | 0.321635 | 0.015670 | 4.457891 | Downregulated |
| hsa-miR-501-3p | 0.217634 | 0.024245 | 2.678529 | Downregulated |
|
hsa-miR-6756-5p | 0.032456 | 0.007564 | 3.678321 | Downregulated |
| hsa-miR-501-5p | 0.213489 | 0.012359 | 6.765479 | Downregulated |
|
hsa-miR-6752-5p | 0.316784 | 0.045679 | 7.120635 | Downregulated |
DEM expression from RNA-sequencing
data is validated by RT-qPCR
To validate microarray analysis data, DEM expression
levels were detected by RT-qPCR. As presented in Fig. 2C, the results of RT-qPCR were almost
consistent with those obtained through microarray analysis. Similar
trends in the following miRNAs were detected: miR-625-5p,
miR-6786-5p, miR-17-3p, miR-501-3p, miR-6756-5p, miR-501-5p,
miR-6786-5p, miR-3663-3p, miR-4694-5p, miR-6881-3p, miR-592,
miR-491-5p, miR-1260b, miR-3591-3p, miR-6514-3p and miR-491-3p.
However, the expression levels of miR-892b and miR-362-5p differed
to that of microarray analysis; therefore, a larger sample size for
RT-qPCR analysis is required to validate these results. Integrated
analysis of miRNA and mRNA expression profiles indicated that among
the highly expressed miRNAs, the expression levels of miR-3663-3p,
miR-6881-3p, miR-625-5p, miR-3591-3p and miR-6514-3p were
significantly different compared with the control group [FC (abs)
>10]. Notably, the expression levels of miR-3663-3p were
significantly increased in the peripheral blood of patients with
sepsis according to RT-qPCR. TargetScan software was used to
evaluate the putative target genes of miR-3663-3p. The results
revealed that TDAG8 was predicated to be a potential target gene of
miR-3663-3p (data not shown), as evolutionary conservation was
demonstrated. As the TLR4 signaling pathway has been reported to
serve a role in sepsis (25,26)
and TDAG8 has been reported to be involved in regulation of cell
functions associated with airway inflammation (27), TDAG8 and TLR4 were selected for
further experimentation.
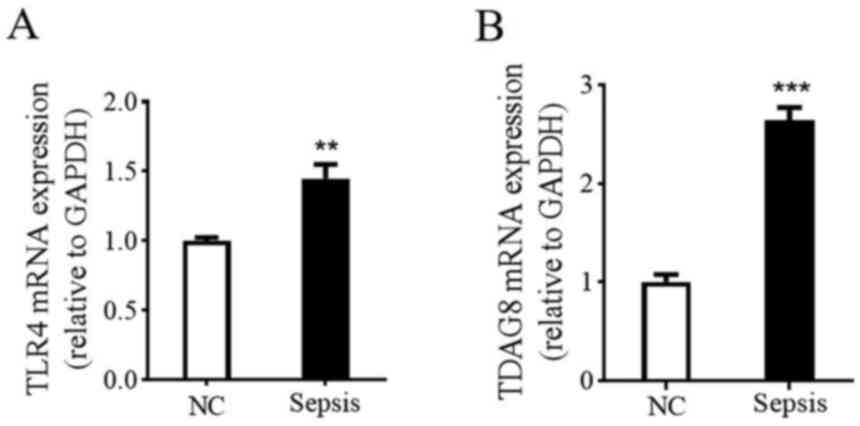
Sepsis upregulates the expression of
TDAG8 and TLR4 mRNA
To further clarify whether TDAG8 and TLR4 are
involved in the occurrence and development of sepsis, the mRNA
expression levels of TDAG8 and TLR4 were determined in human blood
samples via RT-qPCR. As presented in Fig. 3, significant differences were
demonstrated in both TDAG8 and TLR4 mRNA levels between healthy
controls and patients with sepsis. More specifically, the
expression levels of serum TDAG8 (1.0±0.02 vs. 2.7±0.13) and TLR4
(1.0±0.08 vs. 1.5±0.10) mRNA in patients with sepsis were
significantly higher compared with healthy controls. The results
indicated that TDAG8 and TLR4 mRNA expression is upregulated in
patients with sepsis.
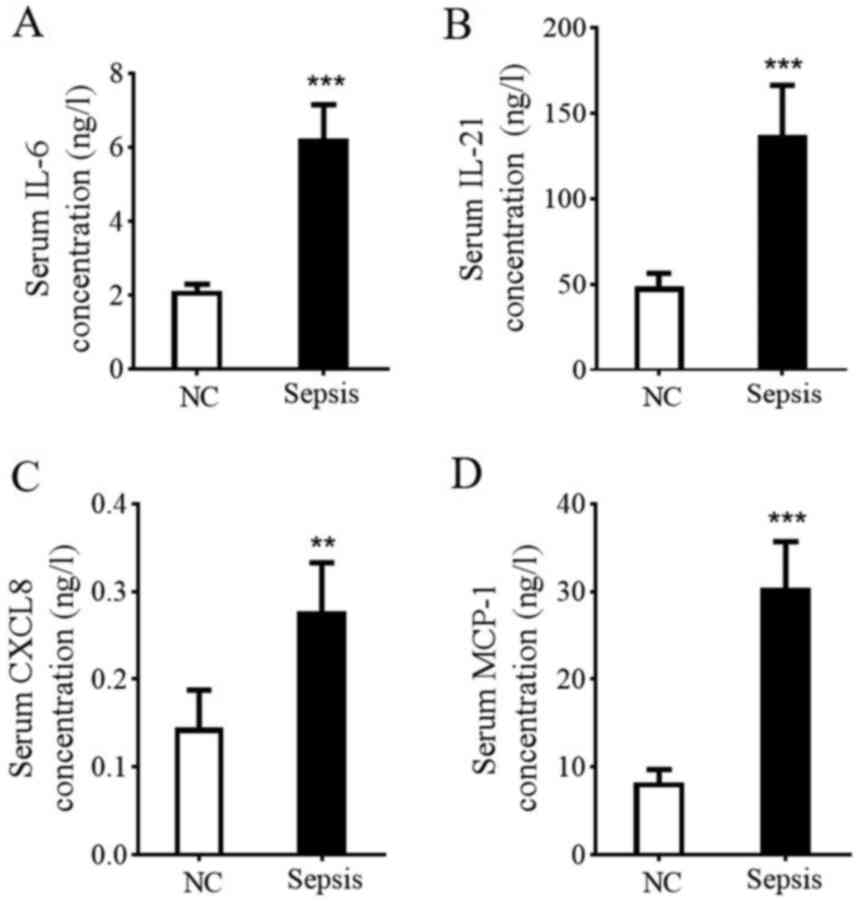
Serum IL-6, IL-21, CXCL8 and MCP-1
content in patients with sepsis
Given the consideration that high expression levels
of TLR4 can result in the production of chemokines and
proinflammatory cytokines (28),
serum IL-6, IL-21, CXCL8 and MCP-1 levels were determined in the
current study by performing ELISA. As presented in Fig. 4, serum IL-6, IL-21, CXCL8 and MCP-1
levels in the healthy control group were 2.12±0.18, 48.50±7.9,
0.15±0.04 and 8.25±1.45 ng/l, while levels in patients with sepsis
were 6.24±0.92, 137.0±29.20, 0.28±0.05 and 30.40±5.30 ng/l,
respectively. The results demonstrated that proinflammatory
cytokines IL-6 and IL-21 and CXCL8 and MCP-1 chemokines were
significantly increased in patients with sepsis (t=6.89, 6.07, 11.8
and 9.03, respectively), suggesting an increased inflammatory
response.
Correlation between the inflammatory
response and miR-3663-3p or TDAG8/TLR4 mRNA expression
To clarify the relationship between miR-3663-3p and
the secretion of proinflammatory cytokines, linear correlation
analysis was performed. The results revealed that, in patients with
sepsis, the correlation coefficients (r) of miR-3663-3p expression
and IL-6, IL-21, CXCL8 and MCP-1 levels were 0.8352, 0.8976, 0.6633
and 0.7661, respectively. Furthermore, the correlation coefficients
(r) of the same miRNA with TDAG8 and TLR4 mRNA expression levels
were 0.7895 and 0.8622, respectively (Fig. 5A). The results indicated a positive
correlation between miRNA-3663-3p and the inflammatory response. As
the secretion of inflammatory factors is closely associated with
the activation of TLR4 (29),
correlation analysis was performed between the expression levels of
TDAG8/TLR4 mRNA and the production of IL-6, IL-21, CXCL8 and MCP-1.
As presented in Fig. 5B and C, the
correlation coefficients (r) of TDAG8 mRNA with the contents of
IL-6, IL-21, CXCL8 and MCP-1 were 0.856, 0.914, 0.515 and 0.902,
respectively. Additionally, the correlation coefficients (r) of
TLR4 mRNA with the contents of IL-6, CXCL8, IL-21 and MCP-1 were
0.940, 0.946, 0.715 and 0.890, respectively. The data indicated a
positive correlation between TDAG8/TLR4 mRNA expression and
proinflammatory cytokine/chemokine secretion. TDAG8 mRNA expression
also revealed a positive correlation with TLR4 mRNA expression,
with a correlation coefficient value (r) of 0.878).
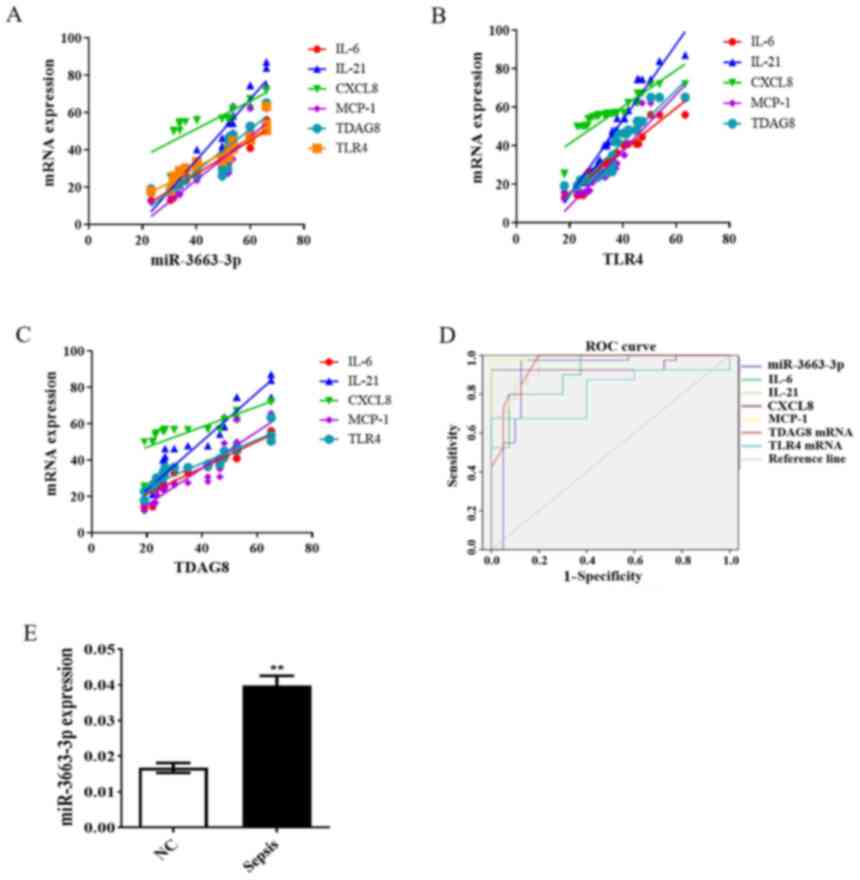 | Figure 5.Relationship between miR-3663-3p
expression and inflammation in sepsis. Correlation analysis of (A)
miR-3663-3p expression with the serum levels of IL-6, IL-21, CXCL8
and MCP-1 and TDAG8 and TLR4 mRNA. Correlation analysis of (B) TLR4
and (C) TDGA8 mRNA expression with the serum contents of IL-6,
IL-21, CXCL8 and MCP-1. (D) ROC curve analysis of miR-3663-3p
expression, total serum IL-6, IL-21, CXCL8 and MCP-1 concentration
and TDAG8 and TLR4 mRNA expression in patients with sepsis. (E)
Expression levels of miR-3663-3p in human peripheral blood samples
detected by reverse transcription-quantitative PCR. **P<0.01 vs.
healthy controls. miR, microRNA; CXCL8, C-X-C motif chemokine
ligand-8; MCP-1, monocyte chemoattractant protein-1; TDAG8, T-cell
death-associated gene 8; TLR4, toll-like receptor 4; ROC, receiver
operating characteristic; NC, normal control. |
Significance of miRNA analysis in the
diagnosis of sepsis
In the current study, miR-3663-3p and the expression
of various cytokines/chemokines (IL-6, IL-21, CXCL8 and MCP-1) were
selected for sensitivity and specificity analysis using a receiver
operating characteristic (ROC) curve. ROC curves revealed that
miR-3663-3p, IL-6, IL-21, CXCL8 and MCP-1 levels, along with TDAG8
and TLR4 mRNA had area under the curve values of 0.908, 0.912,
0.959, 0.944, 0.996, 0.952 and 0.815, respectively, when
distinguishing patients with sepsis from healthy controls (Fig. 5D). When selecting a cut-off value of
0.02 for miR-3663-3p, as determined via ROC curve analysis, the
diagnostic sensitivity and specificity were determined to be 100%.
The cut-off points, diagnostic sensitivities and diagnostic
specificities of IL-6, IL-21, CXCL8 and MCP-1 are listed in
Table IV. The area under the ROC
curve of IL-6, IL-21, CXCL8 and MCP-1 were >0.9, indicating that
they may be used to provide an earlier warning of sepsis when
combined with miR-3663-3p. To validate these findings, an
additional 80 samples (40 samples for healthy controls and 40
samples for patients with sepsis) were obtained for RT-qRCR
analysis. The results revealed that the expression levels of
miR-3663-3p were significantly increased in patients with sepsis
compared with healthy controls (P<0.01; Fig. 5E). The data indicated that
miR-3663-3p could serve as a potentially powerful diagnostic and
predictive biomarker for sepsis, and that miRNA analysis combined
with the measurement of inflammatory cytokine secretion may be a
reliable approach for the fast diagnosis and early identification
of sepsis.
 | Table IV.Diagnostic value of miR-3663-3p,
IL-6, IL-21, CXCL8 and MCP-1 in patients with sepsis. |
Table IV.
Diagnostic value of miR-3663-3p,
IL-6, IL-21, CXCL8 and MCP-1 in patients with sepsis.
| Parameter | AUC | Cut-off value | Sensitivity
(%) | Specificity
(%) |
|---|
| miR-3663-3p | 0.908 | 0.02 | 100.0 | 100.0 |
| IL-6 | 0.912 | 2.84 | 67.7 | 91.7 |
| IL-21 | 0.959 | 57.48 | 79.2 | 100.0 |
| CXCL8 | 0.944 | 14.28 | 100.0 | 95.8 |
| MCP-1 | 0.996 | 14.28 | 97.5 | 100.0 |
| TDAG8 mRNA | 0.952 | 91.29 | 100.0 | 80.0 |
| TLR4 mRNA | 0.815 | 999.00 | 67.5 | 100.0 |
Discussion
The understanding of sepsis and its pathobiology has
improved in recent years, which may improve the definition of
sepsis (17). Severe stage
blood-infection is characterized as sepsis, which may result in
tissue damage, organ failure and death (6). Therefore, the fast diagnosis and early
identification of sepsis (including sepsis, severe sepsis or septic
shock) is crucial for the patient's survival and may be beneficial
when applying the most appropriate treatment protocol. The
regulatory roles of miRNAs make them suitable disease biomarkers,
indicating that they may contribute to the improved prediction of
survival in patients with sepsis (9,10,30).
Previous studies have demonstrated the association
between the expression levels of miRNAs and the mortality of
patients with sepsis (16,31,32).
Furthermore, the altered levels of a miRNA may serve as a
potentially powerful diagnostic and predictive biomarker of sepsis
(33). The present study
investigated the expression of various miRNAs in the context of
sepsis. Considering the rapidity and accessibility of sampling
miRNAs in liquid biopsies, total RNA was extracted from the
peripheral blood of 40 patients with sepsis and 40 healthy controls
to identify novel blood-specific biomarkers of sepsis. A total of
305 DEMs were identified in patients with sepsis, including 212
upregulated and 93 downregulated DEMs. Among these, the top 18 up-
and downregulated miRNAs were selected and validated via
RT-qPCR.
TLR4 plays a key role in the innate immune system
and regulates the secretion of various proinflammatory cytokines,
including TNF-α and IL-6 (29). The
results of the present study confirmed that a significantly
increased expression of TLR4 was detected in patients with sepsis.
Once the TLR4 receptor is activated, its downstream signaling
pathways, including NF-κB, MAPK and STAT, are subsequently
activated. Following TLR4-NF-κB/MAPK/STAT signaling pathway
activation, the abnormal secretion of certain proinflammatory
cytokines, such as IL-6, CXCL8 and MCP-1, is observed (34–36).
In the current study, serum IL-6, IL-21, CXCL8 and MCP-1 levels in
patients with sepsis were significantly increased compared with
healthy controls. TDAG8, which regulates macrophage extracellular
acidification-induced inflammatory cytokine production, is a
receptor with a pronounced immune cell-specific (macrophages, T
cells and microglia) expression profile (37,38).
In the present study, the highly upregulated expression of TLR4 and
TDAG8 mRNA was detected in the peripheral blood of patients with
sepsis, suggesting that TDAG8 and TLR4 expression was closely
associated with the occurrence and development of sepsis. Given
that the current results demonstrated an overproduction of certain
proinflammatory cytokines and chemokines, including IL-6, IL-21,
CXCL8 and MCP-1, accompanied by increased mRNA expressions of TDAG8
and TLR4, the correlation between the inflammatory response and
miR-3663-3p or TDAG8/TLR4 mRNA expression was further analyzed. The
results revealed a positive correlation between miR-3663-3p and the
inflammatory response or TDAG8/TLR4 mRNA. In addition, the
expression of TDAG8/TLR4 mRNA was also positively correlated with
the secretion of proinflammatory cytokines and chemokines.
Furthermore, ROC curve analysis demonstrated that miR-3663-3p had
an area under the curve value of 0.908. With a cut-off point of
0.02, the diagnostic sensitivity and specificity of miR-3663-3p
were 100%, indicating that miR-3663-3p is a potentially powerful
diagnostic and predictive biomarker of sepsis. The ROC curve
analysis of other biomarkers, such as IL-6, CXCL8 and MCP-1, also
demonstrated that miRNA analysis combined with inflammatory
cytokine secretion may be a reliable approach for the fast
diagnosis and early identification of sepsis.
Taken together, the results of the current study
focused on three structurally different types of biomarkers:
Proteins (IL-6, IL-21 and CXCL-1, MCP-1), miRNAs (miR-3663-3p as an
example) and mRNAs (TDAG8 and TLR4), as the combined detection of
several biomarkers in a timely, specific and simultaneous way could
ensure a more accurate diagnosis. The present study has certain
limitations. For instance, further experiments should be conducted
to verify whether miR-3663-3p or other identified DEMs serve a role
in sepsis. Furthermore, Gene Ontology enrichment analysis of
biological processes, cellular components and molecular functions
should be investigated in future studies. However, the validity of
the present results are not affected by these limitations.
The present study elucidated multiple
blood-specific, highly regulated miRNAs that, to the best of our
knowledge, have not yet been associated with sepsis. Most
importantly, the current data demonstrated three structurally
different types of biomarkers (proteins, miRNAs and mRNAs), the
simultaneous and combined detection of which may provide a more
accurate diagnosis for the occurrence and development of sepsis. In
addition, the practicality and applicability of sampling miRNAs in
liquid biopsies will enhance biomarker research and eventually the
clinical management of sepsis.
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science Research
Foundation of Inner Mongolia (grant no. 2016ms0810).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The microarray datasets generated and/or analyzed during
the current study are available in the NCBI GEO repository under
accession no. GSE174507 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174507).
Authors' contributions
XX, BB and HT performed the experiments. XX and BB
analyzed the data and wrote the manuscript. RW and JY designed the
present study and provided experimental materials. JY and XX
confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The protocols of the current study were approved by
the Local Ethics Committee of the Third Affiliated Hospital of
Inner Mongolia Medical University (Baotou, China), and written
informed consent was obtained from all subjects, who were permitted
to withdraw from clinical observation at any time for any
reason.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Papafilippou L, Claxton A, Dark P,
Kostarelos K and Hadjidemetriou M: Nanotools for sepsis diagnosis
and treatment. Adv Healthc Mater. 10:e20013782021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rudd KE, Johnson SC, Agesa KM, Shackelford
KA, Tsoi D, Kievlan DR, Colombara DV, Ikuta KS, Kissoon N, Finfer
S, et al: Global, regional, and national sepsis incidence and
mortality, 1990–2017: Analysis for the global burden of disease
study. Lancet. 395:200–211. 2020. View Article : Google Scholar
|
|
3
|
Cohen J, Vincent JL, Adhikari NK, Machado
FR, Angus DC, Calandra T, Jaton K, Giulieri S, Delaloye J, Opal S,
et al: Sepsis: A roadmap for future research. Lancet Infect Dis.
15:581–614. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maslove DM and Wong HR: Gene expression
profiling in sepsis: Timing, tissue, and translational
considerations. Trends Mol Med. 20:204–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Levy MM, Fink MP, Marshall JC, Abraham E,
Angus D, Cook D, Cohen J, Opal SM, Vincent JL and Ramsay G;
International Sepsis Definitions Conference, : 2001
SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions
conference. Intensive Care Med. 29:530–538. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
American College of Chest
Physicians/Society of Critical Care Medicine Consensus Conference,
. Definitions for sepsis and organ failure and guidelines for the
use of innovative therapies in sepsis. Crit Care Med. 20:864–874.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang R, Wang JM and Gao Y: Advances of
microfluidic technologies applied in diagnosis and treatment of
sepsis. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 31:789–792. 2019.(In
Chinese). PubMed/NCBI
|
|
8
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar
|
|
9
|
Tsitsiou E and Lindsay MA: microRNAs and
the immune response. Curr Opin Pharmacol. 9:514–520. 2009.
View Article : Google Scholar
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nana-Sinkam SP and Croce CM: Non-coding
RNAs in cancer initiation and progression and as novel biomarkers.
Mol Oncol. 5:483–491. 2011. View Article : Google Scholar
|
|
12
|
Liu YY, Jiao WY, Li T and Bao YY:
MiRNA-409-5p dysregulation promotes imatinib resistance and disease
progression in children with chronic myeloid leukemia. Eur Rev Med
Pharmacol Sci. 23:8468–8475. 2019.
|
|
13
|
Slattery ML, Herrick JS, Pellatt DF,
Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS and Wolff
RK: MicroRNA profiles in colorectal carcinomas, adenomas and normal
colonic mucosa: Variations in miRNA expression and disease
progression. Carcinogenesis. 37:245–261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valsecchi V, Boido M, De Amicis E, Piras A
and Vercelli A: Expression of muscle-specific MiRNA 206 in the
progression of disease in a murine SMA model. PLoS One.
10:e01285602015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han Y, Dai QC, Shen HL and Zhang XW:
Diagnostic value of elevated serum miRNA-143 levels in sepsis. J
Int Med Res. 44:875–881. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Zhang P, Chen W, Feng D, Jia Y and
Xie L: Serum microRNA signatures identified by Solexa sequencing
predict sepsis patients' mortality: A prospective observational
study. PLoS One. 7:e388852012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singer M, Deutschman CS, Seymour CW,
Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche
JD, Coopersmith CM, et al: The third international consensus
definitions for sepsis and septic shock (sepsis-3). JAMA.
315:801–810. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Juul SE, Beyer RP, Bammler TK, McPherson
RJ, Wilkerson J and Farin FM: Microarray analysis of high-dose
recombinant erythropoietin treatment of unilateral brain injury in
neonatal mouse hippocampus. Pediatr Res. 65:485–492. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lassen KG, McKenzie CI, Mari M, Murano T,
Begun J, Baxt LA, Goel G, Villablanca EJ, Kuo SY, Huang H, et al:
Genetic coding variant in GPR65 alters lysosomal pH and links
lysosomal dysfunction with colitis risk. Immunity. 44:1392–1405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cochet F and Peri F: The role of
carbohydrates in the lipopolysaccharide (LPS)/toll-like receptor 4
(TLR4) signalling. Int J Mol Sci. 18:23182017. View Article : Google Scholar
|
|
23
|
Plociennikowska A, Hromada-Judycka A,
Borzecka K and Kwiatkowska K: Co-operation of TLR4 and raft
proteins in LPS-induced pro-inflammatory signaling. Cell Mol Life
Sci. 72:557–581. 2015. View Article : Google Scholar
|
|
24
|
Wang DW, Yin YM and Yao YM: Vagal
modulation of the inflammatory response in sepsis. Int Rev Immunol.
35:415–433. 2016. View Article : Google Scholar
|
|
25
|
Farah QY, Ali HA and Neihaya HZ: Using of
TLR2 and TLR4 as biomarker for detection the severity of sepsis.
Int J Psychosoc. 24:4431–4442. 2020.
|
|
26
|
Stan RC, Soriano FG and de Camargo MM: A
mathematical model relates intracellular TLR4 oscillations to
sepsis progression. bioRxiv. 2018.https://doi.org/10.1101/164137
|
|
27
|
Mogi C, Tobo M, Tomura H, Murata N, He XD,
Sato K, Kimura T, Ishizuka T, Sasaki T, Sato T, et al: Involvement
of proton-sensing TDAG8 in extracellular acidification-induced
inhibition of proinflammatory cytokine production in peritoneal
macrophages. J Immunol. 182:3243–3251. 2009. View Article : Google Scholar
|
|
28
|
Miron J, Picard C, Frappier J, Dea D,
Théroux L and Poirier J: TLR4 gene expression and pro-inflammatory
cytokines in Alzheimer's disease and in response to hippocampal
deafferentation in rodents. J Alzheimers Dis. 63:1547–1556. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eidson LN, Inoue K, Young LJ, Tansey MG
and Murphy AZ: Toll-like receptor 4 mediates morphine-induced
neuroinflammation and tolerance via soluble tumor necrosis factor
signaling. Neuropsychopharmacology. 42:661–670. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ahmad S, Ahmed MM, Hasan PMZ, Sharma A,
Bilgrami AL, Manda K, Ishrat R and Syed MA: Identification and
validation of potential miRNAs, as biomarkers for sepsis and
associated lung injury: A network-based approach. Genes (Basel).
11:13272020. View Article : Google Scholar
|
|
31
|
Szilágyi B, Fejes Z, Pócsi M, Kappelmayer
J and Nagy B Jr: Role of sepsis modulated circulating microRNAs.
EJIFCC. 30:128–145. 2019.
|
|
32
|
Huang J, Sun Z, Yan W, Zhu Y, Lin Y, Chen
J, Shen B and Wang J: Identification of microRNA as sepsis
biomarker based on miRNAs regulatory network analysis. Biomed Res
Int. 2014:5943502014.
|
|
33
|
Rahmel T, Schäfer ST, Frey UH, Adamzik M
and Peters J: Increased circulating microRNA-122 is a biomarker for
discrimination and risk stratification in patients defined by
sepsis-3 criteria. PLoS One. 13:e01976372018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang M, Wang C, Wu J, Ha X, Deng Y, Zhang
X, Wang J, Chen K, Feng J, Zhu J, et al: The effect and mechanism
of KLF7 in the TLR4/NF-κB/IL-6 inflammatory signal pathway of
adipocytes. Mediators Inflamm. 2018:17564942018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu GJ, Lin YW, Chuang CY, Tsai HC and Chen
RM: Liver nitrosation and inflammation in septic rats were
suppressed by propofol via downregulating TLR4/NF-κB-mediated iNOS
and IL-6 gene expressions. Life Sci. 195:25–32. 2018. View Article : Google Scholar
|
|
36
|
Wang X, Jiang X, Deng B, Xiao J, Jin J and
Huang Z: Lipopolysaccharide and palmitic acid synergistically
induced MCP-1 production via MAPK-meditated TLR4 signaling pathway
in RAW264.7 cells. Lipids Health Dis. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dai SP, Huang YH, Chang CJ, Huang YF,
Hsieh WS, Tabata Y, Ishii S and Sun WH: TDAG8 involved in
initiating inflammatory hyperalgesia and establishing hyperalgesic
priming in mice. Sci Rep. 7:414152017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tcymbarevich I, Richards SM, Russo G,
Kühn-Georgijevic J, Cosin-Roger J, Baebler K, Lang S, Bengs S,
Atrott K, Bettoni C, et al: Lack of the pH-sensing receptor TDAG8
[GPR65] in macrophages plays a detrimental role in murine models of
inflammatory bowel disease. J Crohns Colitis. 13:245–258. 2019.
View Article : Google Scholar : PubMed/NCBI
|