|
1
|
Slyper M, Porter CBM, Ashenberg O, Waldman
J, Drokhlyansky E, Wakiro I, Smillie C, Smith-Rosario G, Wu J,
Dionne D, et al: A single-cell and single-nucleus RNA-Seq toolbox
for fresh and frozen human tumors. Nat Med. 26:792–802. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu L and Li C: Single-Cell transcriptome
analysis reveals the M2 macrophages and exhausted T cells and
intratumoral heterogeneity in triple-negative breast cancer.
Anticancer Agents Med Chem. 22:294–312. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Q, Wang Z, Zhang Z, Zhang W, Zhang M,
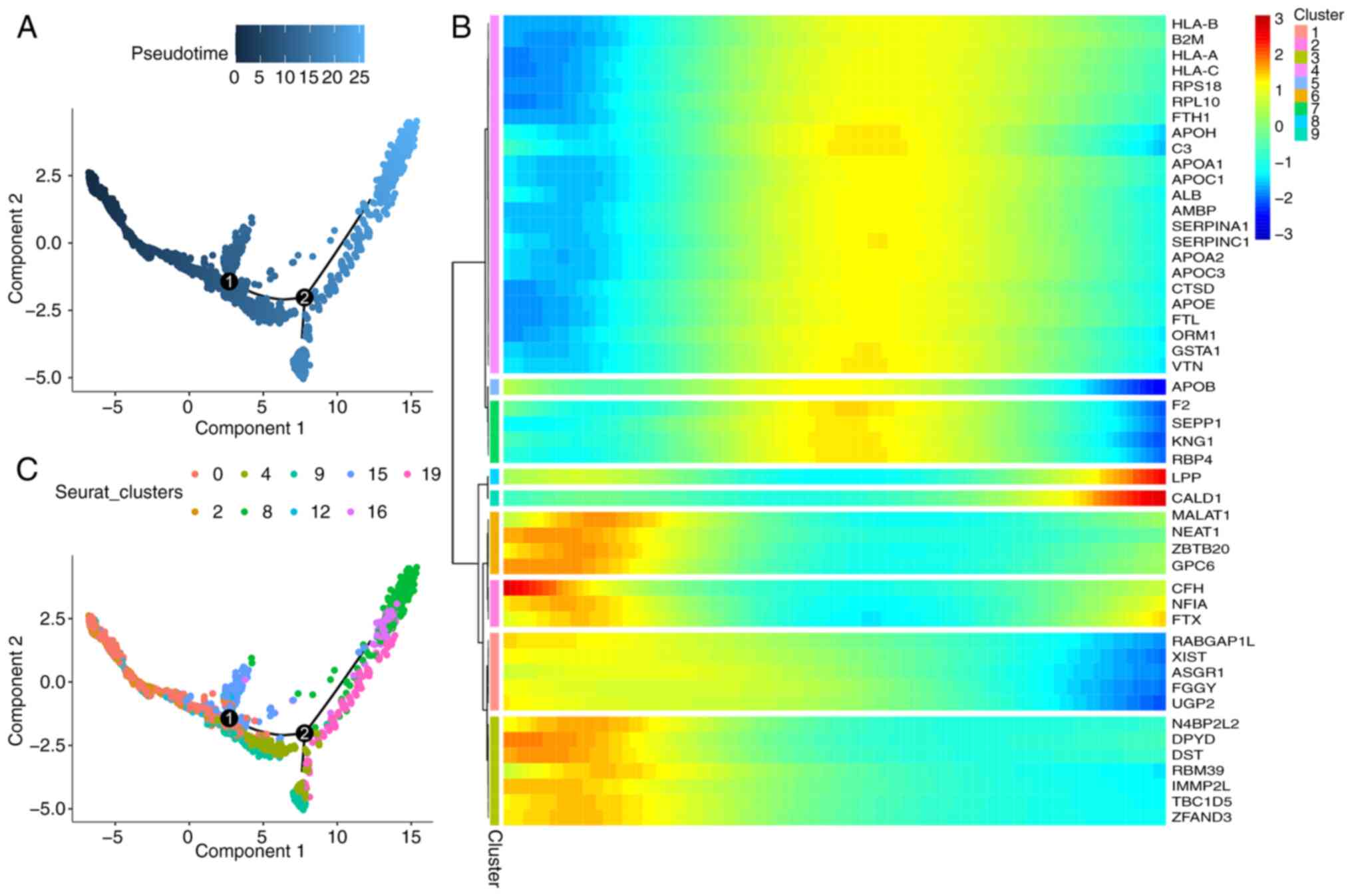
Shen Z, Ye Y, Jiang K and Wang S: Landscape of cell heterogeneity
and evolutionary trajectory in ulcerative colitis-associated colon
cancer revealed by single-cell RNA sequencing. Chin J Cancer Res.
33:271–288. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
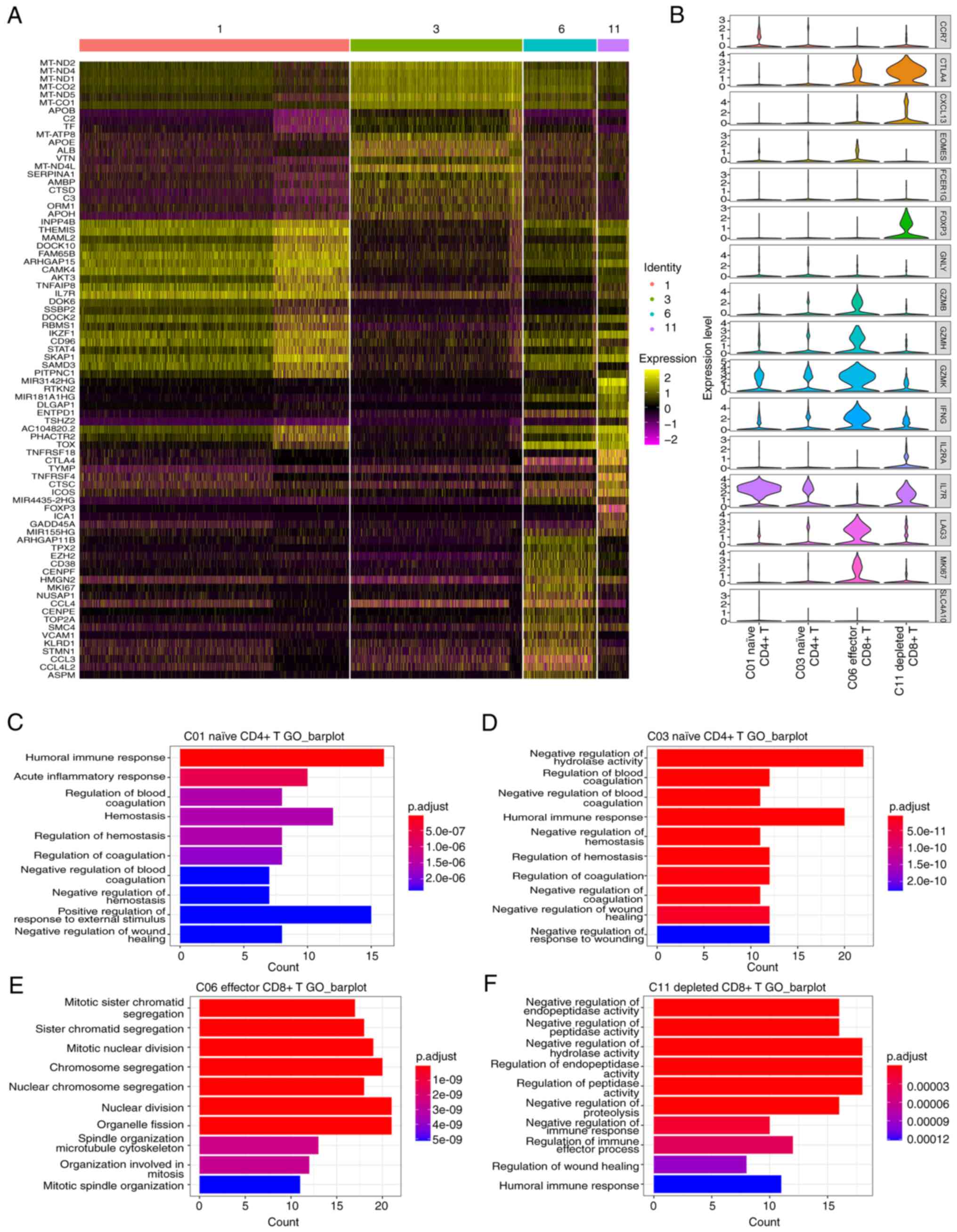
|
Habib N, Avraham-Davidi I, Basu A, Burks
T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K,
et al: Massively parallel single-nucleus RNA-seq with DroNc-seq.
Nat Methods. 14:955–958. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Hauwaert EL, Gammelmark E, Sarvari AK,
Larsen L, Nielsen R, Madsen JGS and Mandrup S: Isolation of nuclei
from mouse white adipose tissues for single-nucleus genomics. STAR
Protoc. 2:1006122021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Basile G, Kahraman S, Dirice E, Pan H,
Dreyfuss JM and Kulkarni RN: Using single-nucleus RNA-sequencing to
interrogate transcriptomic profiles of archived human pancreatic
islets. Genome Med. 13:1282021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fullard JF, Lee HC, Voloudakis G, Suo S,
Javidfar B, Shao Z, Peter C, Zhang W, Jiang S, Corvelo A, et al:
Single-nucleus transcriptome analysis of human brain immune
response in patients with severe COVID-19. Genome Med. 13:1182021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maitra M, Nagy C, Chawla A, Wang YC,
Nascimento C, Suderman M, Théroux JF, Mechawar N, Ragoussis J and
Turecki G: Extraction of nuclei from archived postmortem tissues
for single-nucleus sequencing applications. Nat Protoc.
16:2788–2801. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grindberg RV, Yee-Greenbaum JL, McConnell
MJ, Novotny M, O'Shaughnessy AL, Lambert GM, Araúzo-Bravo MJ, Lee
J, Fishman M, Robbins GE, et al: RNA-sequencing from single nuclei.
Proc Natl Acad Sci USA. 110:19802–19807. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu H, Kirita Y, Donnelly EL and Humphreys
BD: Advantages of single-nucleus over single-cell RNA sequencing of
adult kidney: Rare cell types and novel cell states revealed in
fibrosis. J Am Soc Nephrol. 30:23–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lake BB, Codeluppi S, Yung YC, Gao D, Chun
J, Kharchenko PV, Linnarsson S and Zhang K: A comparative strategy
for single-nucleus and single-cell transcriptomes confirms accuracy
in predicted cell-type expression from nuclear RNA. Sci Rep.
7:60312017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding J, Adiconis X, Simmons SK, Kowalczyk
MS, Hession CC, Marjanovic ND, Hughes TK, Wadsworth MH, Burks T,
Nguyen LT, et al: Systematic comparison of single-cell and
single-nucleus RNA-sequencing methods. Nat Biotechnol. 38:737–746.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zeng W, Jiang S, Kong X, El-Ali N, Ball AR
Jr, Ma CI, Hashimoto N, Yokomori K and Mortazavi A: Single-nucleus
RNA-seq of differentiating human myoblasts reveals the extent of
fate heterogeneity. Nucleic Acids Res. 44:e1582016.PubMed/NCBI
|
|
14
|
Thrupp N, Sala Frigerio C, Wolfs L, Skene
NG, Fattorelli N, Poovathingal S, Fourne Y, Matthews PM, Theys T,
Mancuso R, et al: Single-nucleus RNA-Seq is not suitable for
detection of microglial activation genes in humans. Cell Rep.
32:1081892020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morsey B, Niu M, Dyavar SR, Fletcher CV,
Lamberty BG, Emanuel K, Fangmeier A and Fox HS: Cryopreservation of
microglia enables single-cell RNA sequencing with minimal effects
on disease-related gene expression patterns. iScience.
24:1023572021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and national cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heinrich B, Gertz EM, Schäffer AA, Craig
A, Ruf B, Subramanyam V, McVey JC, Diggs LP, Heinrich S, Rosato U,
et al: The tumour microenvironment shapes innate lymphoid cells in
patients with hepatocellular carcinoma. Gut. 71:1161–1175. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Payen VL, Lavergne A, Alevra Sarika N,
Colonval M, Karim L, Deckers M, Najimi M, Coppieters W, Charloteaux
B, Sokal EM and El Taghdouini A: Single-cell RNA sequencing of
human liver reveals hepatic stellate cell heterogeneity. JHEP Rep.
3:1002782021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saviano A, Henderson NC and Baumert TF:
Single-cell genomics and spatial transcriptomics: Discovery of
novel cell states and cellular interactions in liver physiology and
disease biology. J Hepatol. 73:1219–1230. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qiu X, Mao Q, Tang Y, Wang L, Chawla R,
Pliner HA and Trapnell C: Reversed graph embedding resolves complex
single-cell trajectories. Nat Methods. 14:979–982. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
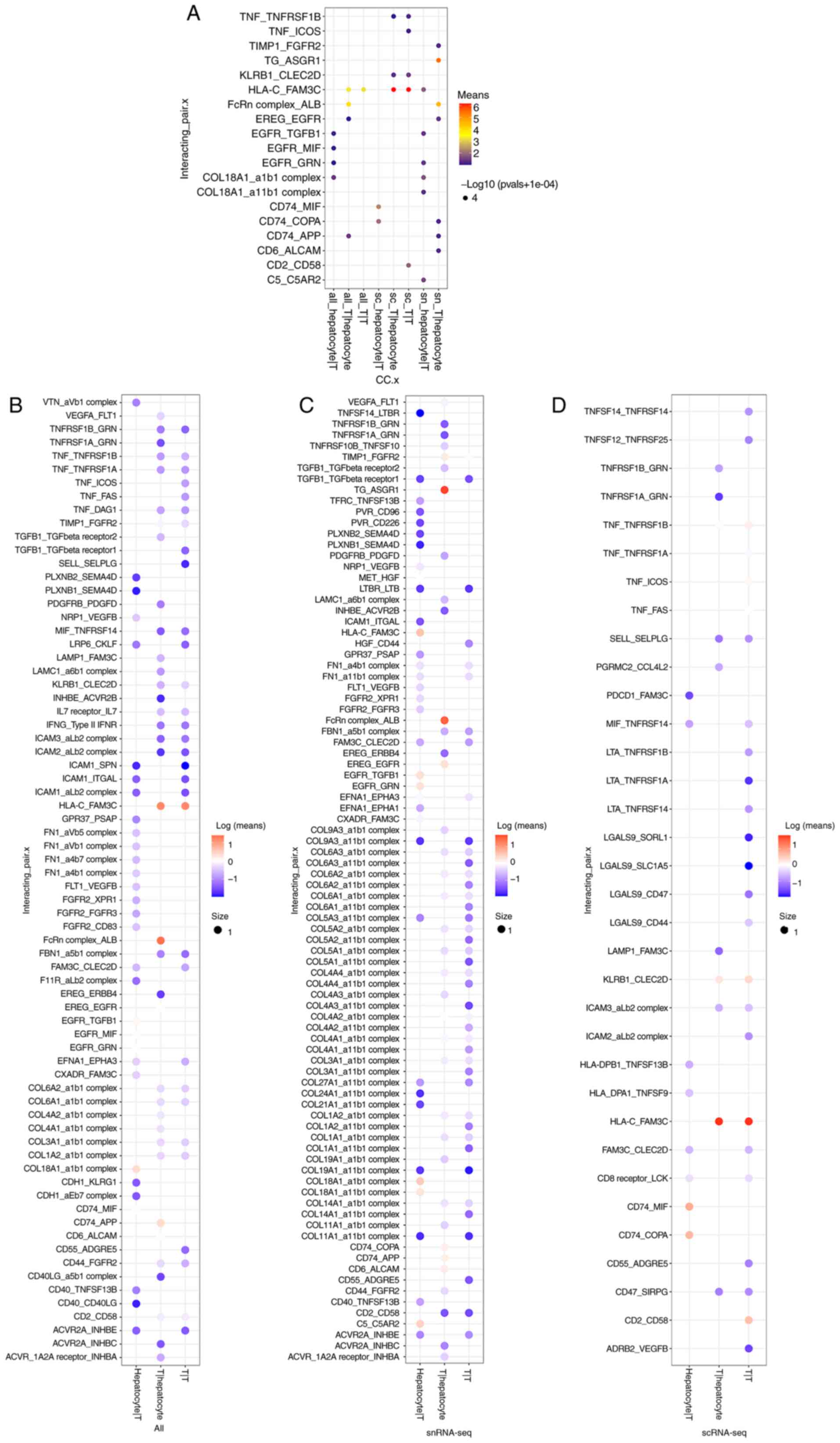
Efremova M, Vento-Tormo M, Teichmann SA
and Vento-Tormo R: CellPhoneDB: Inferring cell-cell communication
from combined expression of multi-subunit ligand-receptor
complexes. Nat Protoc. 15:1484–1506. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Uniken Venema WTC, Ramirez-Sanchez AD,
Bigaeva E, Withoff S, Jonkers I, McIntyre RE, Ghouraba M, Raine T,
Weersma RK, Franke L, et al: Gut mucosa dissociation protocols
influence cell type proportions and single-cell gene expression
levels. Sci Rep. 12:98972022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burja B, Paul D, Tastanova A, Edalat SG,
Gerber R, Houtman M, Elhai M, Bürki K, Staeger R, Restivo G, et al:
An optimized tissue dissociation protocol for single-cell RNA
sequencing analysis of fresh and cultured human skin biopsies.
Front Cell Dev Biol. 10:8726882022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aizarani N, Saviano A, Sagar Mailly L,
Durand S, Herman JS, Pessaux P, Baumert TF and Grün D: A human
liver cell atlas reveals heterogeneity and epithelial progenitors.
Nature. 572:199–204. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
MacParland SA, Liu JC, Ma XZ, Innes BT,
Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, et
al: Single cell RNA sequencing of human liver reveals distinct
intrahepatic macrophage populations. Nat Commun. 9:43832018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ng CKY, Dazert E, Boldanova T,
Coto-Llerena M, Nuciforo S, Ercan C, Suslov A, Meier MA, Bock T,
Schmidt A, et al: Integrative proteogenomic characterization of
hepatocellular carcinoma across etiologies and stages. Nat Commun.
13:24362022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Heinrich S, Craig AJ, Ma L, Heinrich B,
Greten TF and Wang XW: Understanding tumour cell heterogeneity and
its implication for immunotherapy in liver cancer using single-cell
analysis. J Hepatol. 74:700–715. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Deng R, Liu S, Shen S, Guo H and Sun J:
Circulating hepatitis B virus RNA: From biology to clinical
applications. Hepatology. Mar 28–2022.(Epub ahead of print).
View Article : Google Scholar
|
|
30
|
Shah PA, Patil R and Harrison SA:
NAFLD-related hepatocellular carcinoma: The growing challenge.
Hepatology. Apr 28–2022.(Epub ahead of print). View Article : Google Scholar
|