Introduction
Stem cell therapy has been reported to be effective
for injury healing. Growth, wound healing and cell replacement are
considered to be a function of stem cells. The mesenchymal stem
cells (MSCs) are the most well studied (1) and can differentiate into epidermal
cells of skin appendages, including the sebaceous and sweat glands
(2). The potential of stem cells
to heal skin wounds is evident. However, basic stem cell
transplantation does not guarantee successful skin wound healing
(3). Therefore, it is necessary
to investigate different therapeutic approaches using stem cells in
skin wound healing.
Epidermal stem cells (EpSCs) are located at the
epidermal base layer and their potential for stem cell production
for tissue repair is significant (4,5).
Clinical approaches in wound treatment closely rely on EpSCs to
maintain skin homeostasis and facilitate wound healing (6). Moreover, enrichment using EpSCs
within cultured epidermal autografts helps treat severe wounds
(7,8). The biological activity of stem cells
can also further improve wound healing (9). For example, Nanog controls the fate
of pluripotent inner cell masses during embryonic development via
maintaining pluripotent epiblasts and preventing differentiation
(10). Furthermore, Nanog, which
regulates induced pluripotent stem cell pluripotency and
reprogramming, binds to the OCT4 promoter and enhances embryonic
stem cell (ESC) self-renewal via mutation (11). Numerous studies have reported that
Nanog is a key transcription factor of stem cells (12,13). Our previous study demonstrated
that Nanog is inversely related to the differentiation of EpSCs
(14). Furthermore, it was also
reported that Nanog and the β-catenin/wnt signaling pathway
function in EpSCs self-renewal and differentiation (15). Therefore, the aim of the present
study was to upregulate Nanog expression and subsequently assess
the proliferation of EpSCs. These cells were then used to explore
healing in a scalded rat.
Materials and methods
Experimental animals
In total, 15 specific-pathogen-free male
Sprague-Dawley (SD) rats (weight, 140–260 g, 6 weeks old) were
purchased from Sanxia University (Hubei, China). The rearing
environment was relative humidity of 50–60%, and artificial light
and dark for 12 h each. The rats were fed in SPF condition for 7
days. All rats were housed at 22–26°C for 21 days. All animal care
and experimental procedures were approved by the Ethics Committee
of The Third Affiliated Hospital of Guangzhou Medical University
(No. 2020122) (Guangzhou, China). All animal experiments were
performed in accordance with the United States National Institutes
of Health Guide for the Care and Use of Laboratory Animals (NIH
Publication No. 85–23, revised 2011) (16). All efforts were made to minimize
pain and distress of the experimental animals, and all operations
comply with the requirements of Guangzhou Medical University on the
welfare of laboratory animals, such as operation under anesthesia,
euthanasia.
Isolation of mouse EpSCs
Skin tissue was obtained from the back of neonatal
SD rats via plastic surgical procedures, washed in PBS and the
connective tissue and subcutaneous fat were removed. The skin
sample was sterilized with 70% ethanol, rinsed in PBS and minced
into 5-mm wide strips using a sharp scalpel. The strips were
treated with 0.25% dispase II (Sigma-Aldrich; Merck KGaA) solution
at 4°C overnight. The epidermis was mechanically separated from the
dermis and incubated in a solution of 0.25% trypsin (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C for 30 min to dissociate the
cells. Enzyme activity was subsequently blocked using DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and the cells were suspended with a
pipette. The cell suspension was filtered through a stainless-steel
mesh attached to a 60-mm cell culture plate to remove any remaining
tissue pieces. Subsequently, the cells were transferred to a 15-ml
centrifuge tube and collected using centrifugation for 5 min at 800
g and room temperature (RT). To select stem cells, 1×106
dissociated epidermal cells were plated onto collagen type IV (100
µg/ml; Sigma-Aldrich; Merck KGaA)-coated dishes at room temperature
for 10 min. The unattached cells were removed and the rapidly
adherent epidermal cells were cultured in keratinocyte serum-free
medium supplemented with epidermal growth factor, bovine pituitary
extract (all from Gibco; Thermo Fisher Scientific, Inc.) and 0.05
mM CaCl2 (Sigma-Aldrich; Merck KGaA). These cells were
cultured at 37°C with 5% CO2 in a humidified incubator
for two days before replacing the medium. The medium was changed
every other day. The EpSC isolation procedure was regarded as a
standard protocol, which was first reported by Liu et al
(17) and Jensen et al
(18). Rat EpSCs were
successfully isolated and identified using CK15, CK19 and
β1-integrin with immunofluorescence as previously described
(14).
Lentivirus vector construction and
EpSCs transduction
The coding sequence of rat Nanog were amplified
using PCR from the GV208: Rat Nanog primers (Chengdu Jingming
Haorui Biotechnology Co. Ltd.) with AgeI/AgeI overhangs, were used
and the fragment was cloned into pTZ58 (Fermentas; Thermo Fisher
Scientific, Inc.). The AgeI/AgeI fragment was sub-cloned into pUbi
and pEGFP-C1 (Takara Bio USA, Inc.) to generate
ubiquitin-Nanog-EGFP encoding plasmids. To produce transduced
lentivirus, pBABE-puro plasmids were co-transfected along with
helper plasmids into 293T cells (ATCC, CRL-3216) and the medium was
harvested at 36 and 72 h. The lentiviral was used 3rd generation
system. Quantity of lentiviral plasmid used 5 µg for transfection,
and the ratio is 3:2:1 ratio of the lentivirus, packaging and
envelope plasmids. The MOI is 10, the interval time is 72 h. The
lentivirus vector was construction by OriGene Wuxi Biotechnology
Co., Ltd. EpSC transduction was performed by incubating the cells
in virus-enriched medium for 12 h, which contained 4 µg/ml
polybrene. The transduced EpSCs were divided into a
lentivirus-Nanog overexpression (LV-Nanog) group and a control
group (LV), which was transduced with the control lentivirus vector
(OriGene Wuxi Biotechnology Co., Ltd.) which is empty vector. The
Nanog primer sequences are: (accession no. NM001100781) forward,
CCGTTGGGCTGACATGAGCGT and reverse, GGCAGGCATCGGCGAGGAAT.
Establishment of EpSCs with stable
knockdown of Nanog via transfection of small interfering
(si)RNA
The isolated EpSCs were cultured at 37°C in a
12-well plate (5×104 cells/well) for 24 h prior to
transfection and the 12-well plate was coated with collagen type IV
(100 µg/ml). Transfections were performed using
Lipofectamine® RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions for
30 mins at RT. Following transfection, incubated cells for 1–3 days
at 37°C. Then, analyze transfected cells and do the subsequent
experiments. The following siRNAs were purchased from Qiagen
(QIAGEN Shenzhen Company Limited): AllStars Negative Control siRNA
(10 µm, cat. no. 1027281); AllStars Hs Cell Death siRNA (10 µm,
cat. no. 1027298); Rn_Nanog_2 FlexiTube siRNA (10 µm, cat. no.
NM_001100781; GeneGlobe ID, SI02949135). All the siRNA work
concentration were kept as 5–10 nM.
Cell proliferation assay
The EpSCs were seeded into 96-well plates, which
were coated with collagen type IV, at a density of 2×103
cells/100 µl media/well. The cells were subsequently divided into
the following five groups: i) Cells without any treatment; ii)
cells transfected with negative control siRNA cells; iii) cells
transfected with Nanog-siRNA; iv) cells transduced with LV; and v)
cells transduced with LV-Nanog. In brief, 20 µl CellTiter
96® AQueous One Solution Reagent was added to each well
(100 µl media/20 µl MTS reagent; cat. no. G3582; Promega
Corporation). Subsequently, 20 µl of MTS was added to each well
after seeding on day 1–5 of incubation and cultured in the
incubator at 37°C. After 1 h of incubation, a microplate reader
(Thermo Fisher Scientific, Inc.) was used to assess the absorbance
at 490 nm. All experiments were repeated independently three times
and measurements from three duplicate wells were used to average
each sample.
Wound healing assay
The EpSCs were seeded into six-well plates, which
were coated with collagen type IV, at a density of 2×105
cells/1,000 µl media/well. The cells were incubated at 37°C for 24
h to let them adhere to the plate without serum. Scratch wounds
were created in confluent cell monolayers using a sterile 200-µl
pipette tip and 80% Confluence on either side of the wound at the
start of the assay. After 48 h of incubation suspended cells were
removed by washing with PBS. Images of the cells were captured
immediately using the EVOS Cell Imaging System f1 (Thermo Fisher
Scientific, Inc. cat. no: AMF5000). Digitized images of the wounds
were analyzed using ImageJ software 3.0 (National Institutes of
Health). Wound closure rates were determined as the difference
between the wound width at 0 and 48 h.
Western blotting
The control and treated cells were lysed in ice-cold
RIPA lysis buffer (Thermo Fisher Scientific, 89900) with protease
inhibitors (Thermo Fisher Scientific, 78440) and the protein
concentration was determined using the Bradford protein assay
(Bio-Rad Laboratories, Inc.) according to the manufacturer's
protocol. A total of 30 µg protein/lane was separated by 4–12%
NuPage Bis-Tirs gels (Thermo Fisher Scientific, NP0336) and then
transferred to a PVDF membranes (Bio-Rad, 1620264) on ice (300 mA
for 2 h). The membrane was subsequently blocked with 5% non-fat
milk dissolved in TBST (10% TBS, 1% tween) buffer with for 1 h at
room temperature. Subsequently, the membranes were incubated with
the following primary antibodies against: Nanog (1:1,000; cat. no.
3580); cellular (c)-Myc (1:1,000; cat. no. 9402); p53 (1:1,000;
cat. no. 9282); and β-actin (1:10,000; cat. no. 5125) (all
purchased from Cell Signaling Technology, Inc.) overnight at 4°C.
After vigorous washing with TBST (6×5 min), the membranes were
subjected to incubation with anti-mouse-IgG-HRP (purchased from
Cell Signaling Technology, Inc. 1:1,000; cat. no. 7076) for 1 h at
room temperature. The membranes were then washed again with TBST
(6×5 min), incubated with Pierce ECL Western Blotting Substrate
(Thermo Fisher Scientific, Inc.cat. no. 32209) and imaged using
Bio-Rad equipment. Further analysis, as well as image processing
and quantification of the bands, was performed using the program
Image Lab3.0 (Bio-rad Laboratories, Inc.).
Establishment of the rat scald model
and EpSC treatment
The rats were randomly divided into three groups:
the normal EpSCs group, the LV-Nanog EpSCs group, the Nanog-siRNA
EpSCs group. The rats were intraperitoneally anesthetized using 3%
sodium pentobarbital (60 mg/kg) before scalding. All animal
procedures conformed to the Ethics Committee of The Third
Affiliated Hospital of Guangzhou Medical University (No. 2020122;
Guangzhou, China) A self-made constant temperature and constant
pressure scald instrument was used to create the model. Immediately
following the scald, 5 ml sodium lactate Ringer's solution was
injected intraperitoneally for anti-shock. The burn model was
produced by the specialized machine under the following conditions:
Temperature, 85°C; injury pressure of the scald stick, 0.5 kg;
scalding head area, 5 cm2; and scalding time, 8 sec. A
stable second-degree scald model was obtained. The area of the
scald was 2.25 cm2 (diameter, 1.5 cm). Each rat had one
wound on the middle back and bleeding was stopped by appropriate
pressure. On the second day of modeling, the test group was
injected with 1×105 EpSCs with different levels of Nanog
expression using wet compresses, and the control group was injected
with PBS using wet compresses. All wounds received pressure
dressings. All the group was injected with 200 µl PBS with cells or
without cells.
Sample collection and H&E
staining
Images of the scalded areas of five rats in each
group were captured on days 0, 5, 10, 14 and 21 following injury.
The area of each scald was imaged immediately (data not shown). On
day 21 following injury, three rats were randomly selected in each
group and anesthetized with 3% sodium pentobarbital. 100 mg/kg
sodium pentobarbital was used to sacrifice the animal to make all
the rats were dead at the end of the experiment. No rats died
unexpectedly during our experiments. Death was verified by the
absence of a heartbeat and no breathing. For rodents, overdose of
sodium pentobarbital is the most preferable euthanizing agent
(19,20). Samples from the backs of the rats
were received and fixed at room temperature (RT) using 4%
polyphosphate formaldehyde (PFA) for 24 h. The samples were
embedded in paraffin and sliced into 3-µm thick sections. Slices
were dewaxed with xylene for two times (10 min each), hydrated and
stained at RT with hematoxylin for 3–6 min, washed with distilled
water for 2 min, treated with a mixture of 1% hydrochloric acid in
70% ethanol for 1–3 sec, stained at RT with eosin for 2–3 min,
washed with distilled water for 1–2 sec, hydrated with 80% ethanol
and 95% ethanol 15–30 sec each and absolute ethanol 1–2 sec,
treated with xylene for 2–3 sec twice. The edges of the tissue
slices were cleaned using neutral resin and covered with
coverslips. Pathological changes were observed using a Leica
microscope system with light mode and Leica Application Suite X
(Leica Microsystems, Inc.).
Statistical analysis
All the experimental repeats three times and the
data was analyzed using PRISM 5.0 software (GraphPad Software,
Inc.). Data are presented as the mean ± SEM. Since there was no
statistical difference in all data analysis between male and female
groups, the data from the male and female groups was pooled. The
unpaired Student's t-test was used to determine the statistical
differences between the control and experimental groups. One-way
ANOVA or multifactorial ANOVA were used for comparisons between
multiple groups. A Bonferroni post hoc test was used for pairwise
comparisons where appropriate. P<0.05 was considered to indicate
a statistically significant difference.
Results
EpSCs proliferative ability is
influenced by Nanog expression
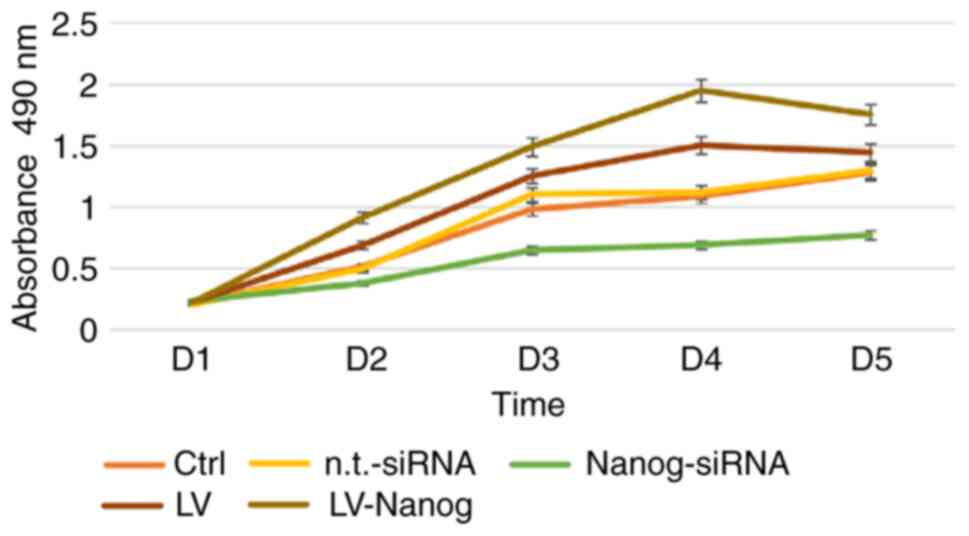
The proliferative capability of EpSCs was improved
when the cells were transduced with LV-Nanog (Fig. 1). Cell proliferation reached its
peak on day 4. Compared with the overexpression of Nanog, the cells
treated with Nanog-siRNA exhibited significantly reduced
proliferation capabilities (P<0.01). Moreover, both the
aforementioned increased proliferation and decreased proliferation
were significant compared with the vehicle control group
(P<0.05). However, the n.t.-siRNA group and LV control group did
not significantly affect cell proliferation.
Western blotting analysis of
transduced EpSCs
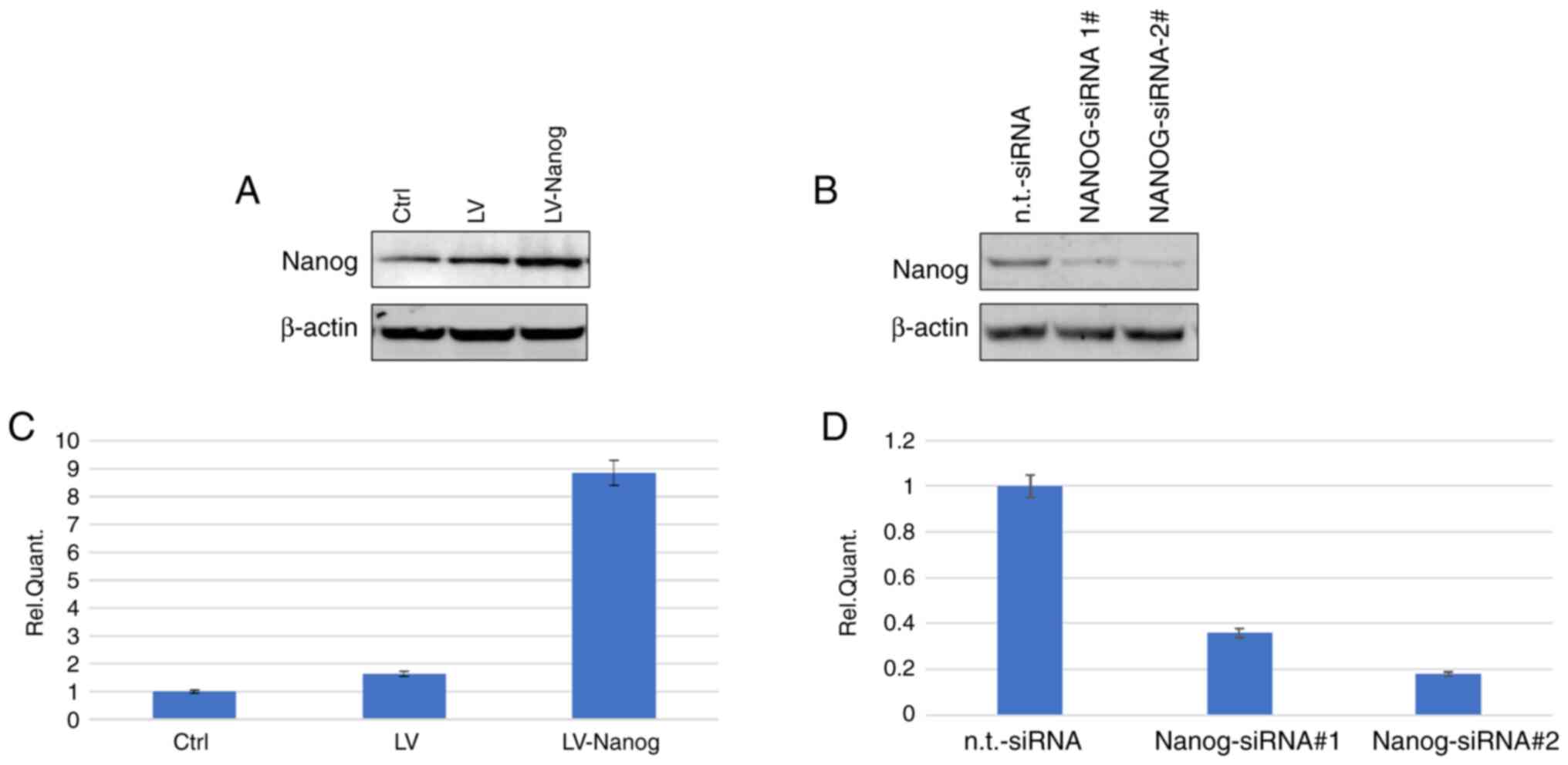
The successful transduction of LV-Nanog EpSCs was
confirmed via western blotting following transduction with LV-Nanog
for 4 days. The protein expression levels of Nanog were increased
in the LV-Nanog EpSCs compared with the LV-control group (Fig. 2A and C).
Western blotting analysis of
Nanog-knockdown EpSCs
EpSCs transfected with siRNAs were subsequently
analyzed via western blotting. The results demonstrated that the
n.t.-siRNA group did not significantly affect the Nanog protein
expression levels. However, in the Nanog-siRNA group Nanog protein
expression levels were significantly downregulated (Fig. 2B and D).
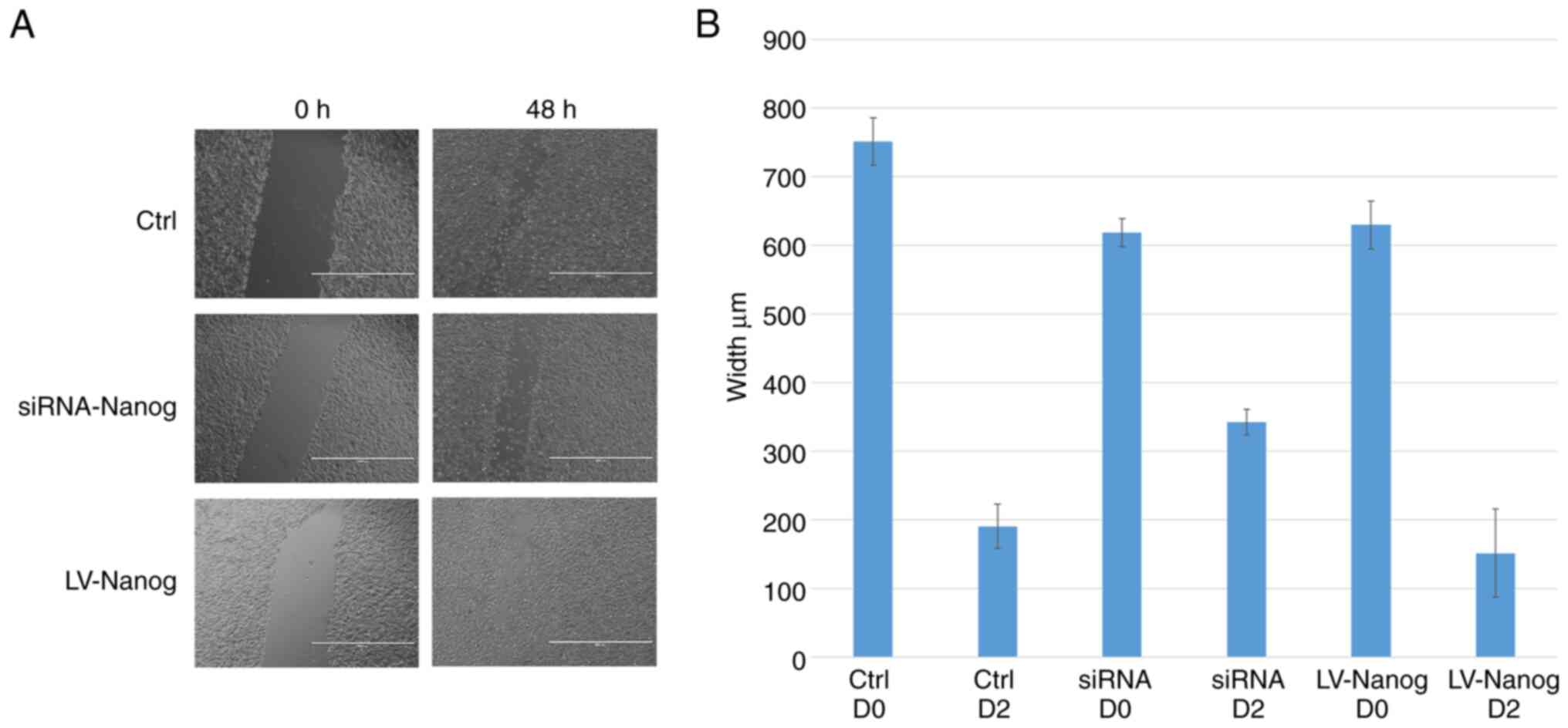
Nanog expression affects EpSCs wound
healing effectiveness
The wound healing assay was used to detect the
effect of Nanog on the healing ability of EpSCs in vitro.
The LV-Nanog EpSCs were demonstrated to heal faster compared with
the vehicle control, with the wound being almost completely healed
after 48 h (Fig. 3A). Moreover,
the EpSCs transfected with Nanog-siRNA were demonstrated to heal
slower compared with the vehicle control (Fig. 3B).
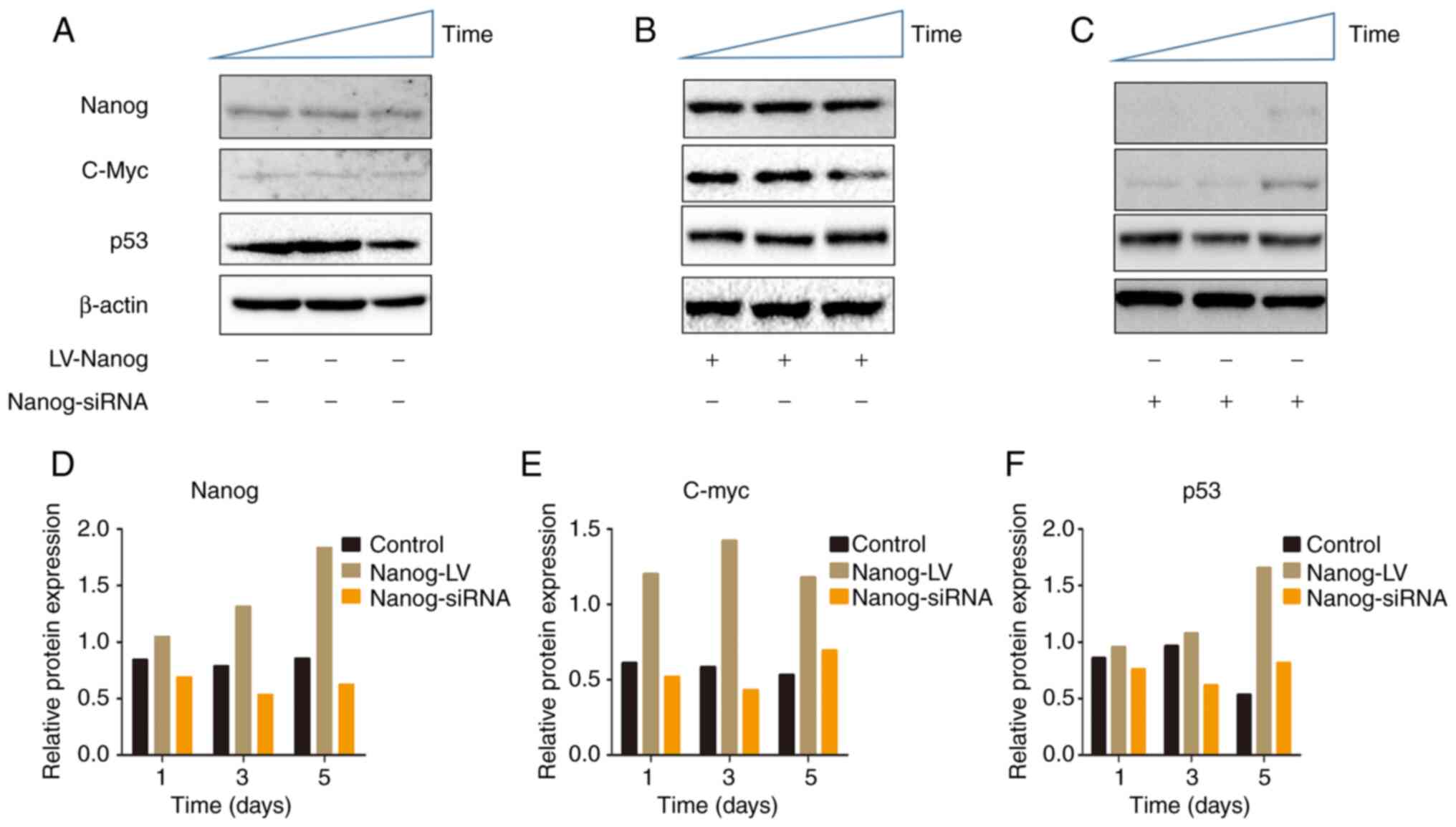
Nanog promotes EpSCs proliferation via
the activation of c-Myc expression
In the aforementioned results it was demonstrated
that LV-Nanog EpSCs exhibited increased proliferation.
Subsequently, it was demonstrated via western blotting that Nanog
protein expression levels were upregulated and the c-Myc protein
expression levels were also upregulated in LV-Nanog group (Fig. 4A and B). In the present study, it
was demonstrated that when EpSC proliferation is improved via the
upregulation of Nanog expression, c-Myc expression is also
upregulated. Furthermore, a similar trend was observed in
Nanog-siRNA EpSCs, whereby Nanog is downregulated as well as c-Myc
(Fig. 4C). The abnormal
upregulation of c-Myc expression is directly associated with up to
70% of all human cancers (21,22). The present study demonstrated that
c-Myc was upregulated alongside Nanog and potentially improved EpSC
proliferation.
The p53 or Arf checkpoints are activated by the
persistent expression of oncogenic Myc. Loss of checkpoint
regulation via mutations in p53 or Arf demonstrates the full
tumorigenic potential of Myc (21). The results of the present study
demonstrated that in the LV-Nanog EpSCs c-Myc expression levels
were upregulated, along with Nanog expression levels, on day 1 and
these peak on day 3. Subsequently, c-Myc expression levels
decrease; however, p53 expression levels increase. On day 5, the
p53 expression levels were even further upregulated and c-Myc
expression levels were decreased compared with those on day 3.
Moreover, EpSCs proliferation also reached its peak on day 4 and
then decreased. These results suggested that p53 expression was
affected by c-Myc expression levels, which potentially may help to
avoid c-Myc overexpression. Nanog expression levels in the
Nanog-siRNA EpSCs were only slightly upregulated on day 5, which
may be due to the efficacy of the siRNA decreasing over time.
Moreover, the c-myc expression levels were upregulated on D5
slightly. These results supported the hypothesis that an underlying
mechanism of Nanog may be to regulate c-Myc expression to improve
the proliferation of EpSCs, a process that may be further
supervised by p53.
Scald model of rats treated with EpSCs
expressing different levels of Nanog
Once the rat scald model was successfully
established, wound healing was investigated on days 0, 5, 10, 14
and 21 (data not shown). The LV-Nanog group healed faster,
particularly on day 14 (Fig. 5A).
Wound healing was observed in H&E-stained scalded tissue
specimen on day 21. The wounds in the control group were the most
re-epithelialized. Furthermore, the wounds in the LV-Nanog group
were almost completely re-epithelialized on day 21, whereas the
wounds in the Nanog-siRNA group were only partially
re-epithelialized (Fig. 5B).
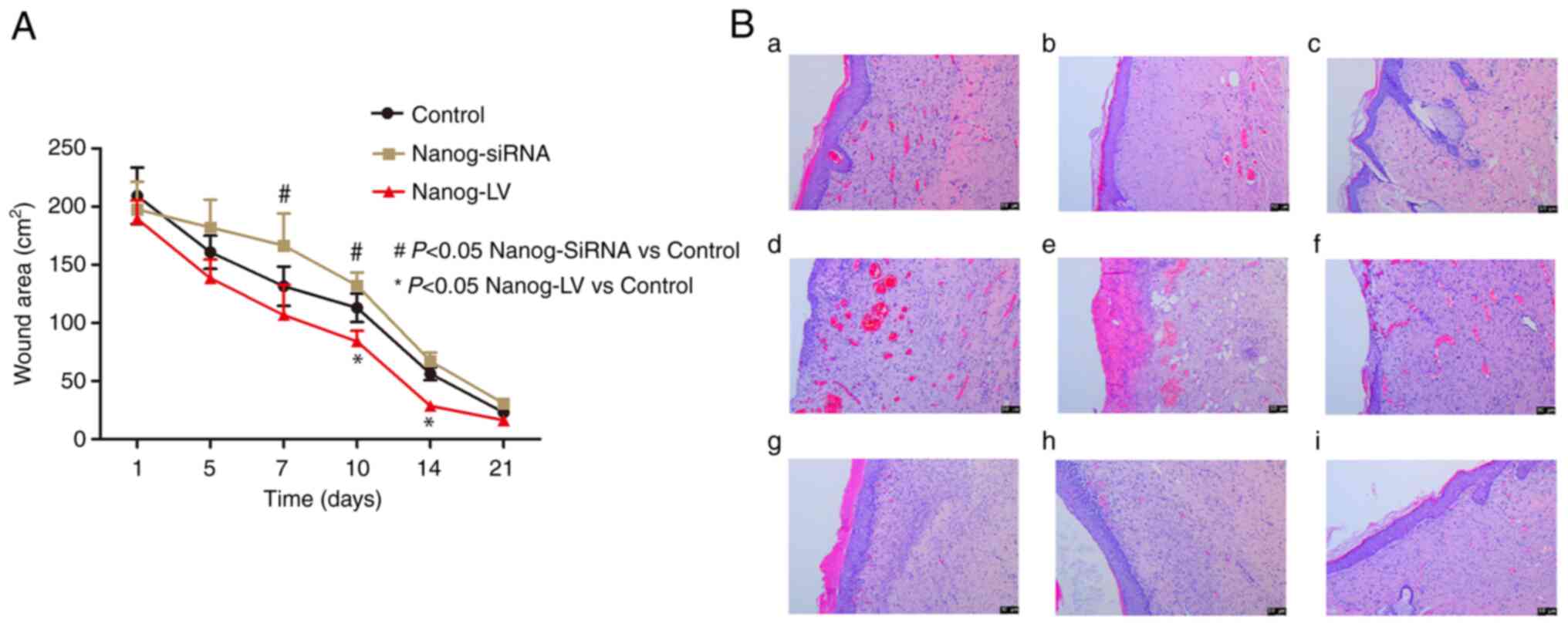 | Figure 5.Effect of Nanog overexpression and
knockdown in EpSCs on scald healing in vivo. (A) The control
group of the scald model on days 0, 5, 10, 14 and 21. The
Nanog-siRNA group, the LV-Nanog group's scald healing state,
Healing curves of different intervention groups. (B) (a-c) Repeated
samples within the control group, showed a small number of
inflammatory cells and scattered capillaries under the skin via HE
staining results of wound tissues on day 21. (d-f) Repeated samples
within the Nanog-siRNA group, showed that a large number of
inflammatory cells in the subcutaneous tissue, and more scattered
capillaries than in the control group via HE staining results of
wound tissues on day 21. (g-i) Repeated samples within the Nanog-LV
group, showed a dense epidermal layer under the skin, with no
obvious capillaries or peripheral inflammatory cells via HE
staining results of wound tissues on day 21. H&E staining was
observed using a Leica microscope system with light mode. Scale
bar=50 µm. Data are presented as the mean ± SEM (n=3). EpSC,
epidermal stem cell; siRNA, small interfering RNA; LV, lentivirus
overexpression vector. |
Furthermore, the epidermal structure was visible in
the normal EpSCs group with only a few inflammatory cells visible
on day 21. An intact epidermis was visible in the LV-Nanog group
and fresh hair follicle tissues were visible in the dermis.
Discussion
It has previously been demonstrated that EpSCs serve
a key role in the wound healing process (4,6,23,24). Furthermore, multipotent EpSCs
could enrich from keratinocyte isolates, with their specific
location in the hair follicle bulge (5). This result was inconsistent with
previous research (15), in this
study, the EpSCs proliferation reached its peak on day 4, which was
potentially a result of the cell density used and the different of
the Nanog expression. Nanog expression maintains EpSC proliferation
and the expression of pluripotency genes is maintained by Nanog
overexpression (25).
The self-renewal gene, Nanog, is highly expressed in
self-renewing embryonic stem cells and has been well studied
(12). Nanog is known as one of
three ‘core’ factors that allow for stem cell pluripotency
(26). Furthermore. Nanog has
been reported to also be expressed in several types of adult stem
cells, including EpSCs. However, Nanog is nearly not expressed in
differentiated cells. In our previous study, it was demonstrated
that Nanog served a key role in the regulation of EpSC
proliferation (14,15). Stem cell marker expression, such
as that of Nanog and Sox-2, is gradually lost as stem cells age
(27). Furthermore, our previous
study also reported that Nanog expression is reduced in
differentiated cells (14). In
the present study, it was demonstrated that high Nanog expression
levels improved EpSC proliferation and migration. Previous studies
have also suggested that in all Nanog target promoters, genes are
equally expressed or repressed, whereas in Nanog unique targets
genes were predominantly inactivated or suppressed in response to
the expression of a subset of target genes. This observation is
common to all factors except Myc (28). In an extended transcriptional
network for the pluripotency of ESCs it was demonstrated that Nanog
and Myc have a large overlap of target promoters; however, Myc also
has its own distinct cluster (28). Myc promotes cell proliferation, as
previously demonstrated (29–32). Moreover, dominant negative mutants
of Myc antagonizes self-renewal and promotes cell differentiation
(29). These results suggested
that other transcription factors may be involved in this
process.
Myc is a transcription factor that regulates cell
proliferation (33). Moreover,
Myc is widely expressed during embryogenesis, but also in highly
proliferative adult tissues, such as the epidermis and gut
(34). Myc is sufficient to
stimulate proliferation in quiescent cells via overexpression
(35). Furthermore, Myc is known
as a co-regulator of cell proliferation and metabolism in numerous
types of stem cells (28,36). Therefore, the expression of Myc is
finely regulated by transcriptional, post-transcriptional and
post-translational regulatory mechanisms in adult tissue
homeostasis (37,38). Myc is usually maintained at low
levels or restricted to regeneration and cell proliferation, such
as in the epidermis and the gut. In mouse ES cells, Myc is required
to prevent MAPK activation forcing differentiation of cells into
primitive endoderm and serves a role in cell proliferation
(30). Together, these
aforementioned studies demonstrated that c-Myc functionally
upregulates energy production and biosynthetic processes required
for successful cellular replication, thereby directly coordinating
proliferative metabolism and cell cycle progression. In the present
study, the results demonstrated that c-Myc protein expression
levels were upregulated when Nanog was overexpressed, which
potentially increased EpSC proliferation.
Nanog and Myc, along with Krüppel-like factor 4, are
naïve pluripotent markers that drive the transition of EpSCs to
ESCs (39). Moreover, Nanog and
Myc gene expression are independent of each other in the
transcriptional regulatory pathways and do not directly regulate
each other, but the Nanog and Myc both stimulate cell proliferation
(28). Our previous hypothesis
that Nanog overexpression, with high c-Myc expression levels,
increased EpSC proliferation, was confirmed by the results of the
present study. The result was inconsistent with previous research
(12). In this study, the EpSCs
reach proliferation peak earlier, which was potentially a result of
the cell density used.
Furthermore, in the present study it was
demonstrated that Nanog overexpression in EpSCs could potentially
improve cell proliferation via enhancing c-Myc expression. However,
there is also a protection mechanism to prevent excessive
proliferation and tumor development (40).
p53 is a well-known critical tumor suppressor and
transcription factor (40,41).
In the present study, the results demonstrated that c-Myc protein
expression was induced when Nanog was overexpressed in EpSCs, which
led to EpSC proliferation. However, overexpression of c-Myc protein
can induce hepatocellular carcinoma and other types of tumor
(42,43). Co-occupancy regions and
cis-overlapping motifs of p53 and c-Myc proteins suggest that these
two transcription factors interact and regulate gene expression via
competitive binding (44). A
previous study also demonstrated that intact p53 suppresses Nanog
function (45).
Wound healing is a complex process. Self-repair
following an injury occurs underneath the skin and tissue where
pluripotent adult stem cells have the ability to self-renew and
give rise to different cells types (46). Stem cells give rise to progenitor
cells, which cannot self-renew but can give rise to numerous cell
types. The extent to which stem cells are involved in skin wound
healing is complex and not fully understood (3,46).
If epithelium formation in the injured area is rapid, healing will
lead to regeneration, or scarring will develop over weeks or
months. Therefore, epithelization is the one of the most important
factors in wound healing and EpSCs are responsible for the process
of epidermalization. The rate of epidermalization also depends on
the cell proliferation (47). In
the present study, according to the rat scald model, it was
demonstrated that following treatment with EpSCs being injected
into the wound, the wounds in the LV-Nanog group were
re-epithelialized by day 21, which was faster compared with the
control group. This led to regeneration that did not result in a
scar.
Consequently, to better understand the
Nanog-stimulated wound healing, the wound healing assay was used
with transfected EpSCs. The results demonstrated that Nanog
overexpression in EpSCs increased the wound repair capabilities,
whereas in EpSCs with Nanog knockdown wound closure was delayed.
Different levels of Nanog in the EpSCs may therefore affect the
wound closure rate of the scratch. These results suggested that
Nanog potential provides a proliferative advantage to EpSCs and
that Nanog may be very active in inducing and supporting the wound
healing process.
In the present study, the effectiveness of Nanog on
EpSCs during the scald healing process was demonstrated. The data
suggested that Nanog overexpression in EpSCs could potentially
increase the cell proliferation and enhance the wound closure rate.
In vivo experiments also supported the hypothesis. When
EpSCs expressing LV-Nanog were injected into the rat scald model,
the skin re-epithelialized earlier compared with the control
group.
In conclusion, in the present study the role of
Nanog in EpSC proliferation and migration was investigated. The
results demonstrated that in EpSCs, Nanog potentially stimulates
cell proliferation and cells that overexpress Nanog may be able to
improve wound healing in the rat scald model. Therefore,
modifications to EpSC proliferation potential may be an effective
way of speeding up the healing of scalds. The present study also
demonstrated that the function of Nanog in EpSCs may also be
influenced by c-Myc upregulation and p53 control. These data have
therefore proposed that this approach could be an effective
treatment option to repair wounds to a high quality.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the Third Affiliated
Hospital of Guangzhou Medical University and the Guangdong Medical
Science and Technology Research Fund (grant no. B2016026).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding authors on reasonable
request.
Authors' contributions
DLY performed data curation, software and formal
analysis, validation, investigation, provided methodology, acquired
funding, wrote the original draft, wrote reviewed and edited the
manuscript. XHZ, QYJ, SL and YL performed data curation, software,
formal analysis, validation, investigation and provided
methodology. PC conducted software and formal analysis,
investigation, wrote, reviewed and edited the manuscript. GQJ
conceptualized and supervised the study, provided resources,
performed data curation, validation, project administration, wrote,
reviewed and edited the manuscript. CYL conceptualized and
supervised the study, provided resources, project administration,
wrote, reviewed and edited the manuscript. All authors read and
approved the final manuscript. DLY and GQJ confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
All animal care and experimental procedures in the
present study were approved by the Ethics Committee of The Third
Affiliated Hospital of Guangzhou Medical University (No. 2020122)
(Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wei X, Yang X, Han ZP, Qu FF, Shao L and
Shi YF: Mesenchymal stem cells: A new trend for cell therapy. Acta
Pharmacol Sin. 34:747–754. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jeschke MG, Rehou S, McCann MR and
Shahrokhi S: Allogeneic mesenchymal stem cells for treatment of
severe burn injury. Stem Cell Res Ther. 10:3372019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kosaric N, Kiwanuka H and Gurtner GC: Stem
cell therapies for wound healing. Expert Opin Biol Ther.
19:575–585. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang R, Liu F, Wang J, Chen X, Xie J and
Xiong K: Epidermal stem cells in wound healing and their clinical
applications. Stem Cell Res Ther. 10:2292019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morasso MI and Tomic-Canic M: Epidermal
stem cells: The cradle of epidermal determination, differentiation
and wound healing. Biol Cell. 97:173–183. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Zhang J, Yue J, Gou X and Wu X:
Epidermal stem cells in skin wound healing. Adv Wound Care (New
Rochelle). 6:297–307. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brockmann I, Ehrenpfordt J, Sturmheit T,
Brandenburger M, Kruse C, Zille M, Rose D and Boltze J:
Skin-derived stem cells for wound treatment using cultured
epidermal autografts: Clinical applications and challenges. Stem
Cells Int. 2018:46236152018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Teng M, Huang Y and Zhang H: Application
of stems cells in wound healing-an update. Wound Repair Regen.
22:151–160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dash BC, Xu Z, Lin L, Koo A, Ndon S,
Berthiaume F, Dardik A and Hsia H: Stem cells and engineered
scaffolds for regenerative wound healing. Bioengineering (Basel).
5:232018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silva J, Chambers I, Pollard S and Smith
A: Nanog promotes transfer of pluripotency after cell fusion.
Nature. 441:997–1001. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayashi Y, Caboni L, Das D, Yumoto F,
Clayton T, Deller MC, Nguyen P, Farr CL, Chiu HJ, Miller MD, et al:
Structure-based discovery of NANOG variant with enhanced properties
to promote self-renewal and reprogramming of pluripotent stem
cells. Proc Natl Acad Sci USA. 112:4666–4671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitsui K, Tokuzawa Y, Itoh H, Segawa K,
Murakami M, Takahashi K, Maruyama M, Maeda M and Yamanaka S: The
homeoprotein Nanog is required for maintenance of pluripotency in
mouse epiblast and ES cells. Cell. 113:631–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pashaiasl M, Khodadadi K, Kayvanjoo AH,
Pashaei-Asl R, Ebrahimie E and Ebrahimi M: Unravelling evolution of
Nanog, the key transcription factor involved in self-renewal of
undifferentiated embryonic stem cells, by pattern recognition in
nucleotide and tandem repeats characteristics. Gene. 578:194–204.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin D, Tian L, Ye Y, Li K, Wang J, Cheng
P, Chen A, Guo F and Huang H: Nanog and β-catenin: A new
convergence point in EpSC proliferation and differentiation. Int J
Mol Med. 29:587–592. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng P, Sun X, Yin D, Xu F, Yang K, Qin
L, Dong Y, Guo F, Chen A, Zhang W and Huang H: Nanog down-regulates
the Wnt signaling pathway via β-catenin phosphorylation during
epidermal stem cell proliferation and differentiation. Cell Biosci.
5:52015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
National Research Council, . Guide for the
care and use of laboratory animals. 8th edition. The National
Academies Press; Washington, DC: pp. 2462011
|
|
17
|
Liu Y, Zhou H and Gao F: Isolation and
identification of stem cells from adult cashmere goat skin. Int J
Dermatol. 47:551–556. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jensen KB, Driskell RR and Watt FM:
Assaying proliferation and differentiation capacity of stem cells
using disaggregated adult mouse epidermis. Nat Protoc. 5:898–911.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wong D, Makowska IJ and Weary DM: Rat
aversion to isoflurane versus carbon dioxide. Biol Lett.
9:201210002012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hickman DL and Johnson SW: Evaluation of
the aesthetics of physical methods of euthanasia of anesthetized
rats. J Am Assoc Lab Anim Sci. 50:695–701. 2011.PubMed/NCBI
|
|
21
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gabay M, Li Y and Felsher DW: MYC
activation is a hallmark of cancer initiation and maintenance. Cold
Spring Harb Perspect Med. 4:a0142412014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Gu W, Du J, Reid B, Deng X, Liu Z,
Zong Z, Wang H, Yao B, Yang C, et al: Electric fields guide
migration of epidermal stem cells and promote skin wound healing.
Wound Repair Regen. 20:840–851. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang R, Wang J, Chen X, Shi Y and Xie J:
Epidermal stem cells in wound healing and regeneration. Stem Cells
Int. 2020:91483102020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Finley LWS, Vardhana SA, Carey BW,
Alonso-Curbelo D, Koche R, Chen Y, Wen D, King B, Radler MR, Rafii
S, et al: Pluripotency transcription factors and Tet1/2 maintain
Brd4-independent stem cell identity. Nat Cell Biol. 20:565–574.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan G and Thomson JA: Nanog and
transcriptional networks in embryonic stem cell pluripotency. Cell
Res. 17:42–49. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bijonowski BM, Yuan X, Jeske R, Li Y and
Grant SC: Cyclical aggregation extends in vitro expansion potential
of human mesenchymal stem cells. Sci Rep. 10:204482020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim J, Chu J, Shen X, Wang J and Orkin SH:
An extended transcriptional network for pluripotency of embryonic
stem cells. Cell. 132:1049–1061. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cartwright P, McLean C, Sheppard A, Rivett
D, Jones K and Dalton S: LIF/STAT3 controls ES cell self-renewal
and pluripotency by a Myc-dependent mechanism. Development.
132:885–896. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wernig M, Meissner A, Cassady JP and
Jaenisch R: c-Myc is dispensable for direct reprogramming of mouse
fibroblasts. Cell Stem Cell. 2:10–12. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fagnocchi L, Cherubini A, Hatsuda H,
Fasciani A, Mazzoleni S, Poli V, Berno V, Rossi RL, Reinbold R,
Endele M, et al: A Myc-driven self-reinforcing regulatory network
maintains mouse embryonic stem cell identity. Nat Commun.
7:119032016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Carroll PA, Freie BW, Mathsyaraja H and
Eisenman RN: The MYC transcription factor network: Balancing
metabolism, proliferation and oncogenesis. Front Med. 12:412–425.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maoz M, Devir M, Inbar M, Inbar-Daniel Z,
Sherill-Rofe D, Bloch I, Meir K, Edelman D, Azzam S, Nechushtan H,
et al: Clinical implications of sub-grouping HER2 positive tumors
by amplicon structure and co-amplified genes. Sci Rep. 9:187952019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Spencer CA and Groudine M: Control of
c-myc regulation in normal and neoplastic cells. Adv Cancer Res.
56:1–48. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Eilers M, Schirm S and Bishop JM: The MYC
protein activates transcription of the alpha-prothymosin gene. EMBO
J. 10:133–141. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yoon K, Lim YS, Yu SB, Kim DS, Ryu SJ, Kim
KH, Jang TH and Kim SH: The expression of survivin and its related
genes in adipocyte-derived stem cell by demethylation. Korean J
Anesthesiol. 58:383–390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Levens D: You don't muck with MYC. Genes
Cancer. 1:547–554. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Farrell AS and Sears RC: MYC degradation.
Cold Spring Harb Perspect Med. 4:a0143652014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Swaidan NT, Salloum-Asfar S, Palangi F,
Errafii K, Soliman NH, Aboughalia AT, Wali AHS, Abdulla SA and
Emara MM: Identification of potential transcription factors that
enhance human iPSC generation. Sci Rep. 10:219502020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pitolli C, Wang Y, Candi E, Shi Y, Melino
G and Amelio I: p53-mediated tumor suppression: DNA-damage response
and alternative mechanisms. Cancers (Basel). 11:19832019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Parfenyev S, Singh A, Fedorova O, Daks A,
Kulshreshtha R and Barlev NA: Interplay between p53 and non-coding
RNAs in the regulation of EMT in breast cancer. Cell Death Dis.
12:172021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang P, Jiang Y, Pan Y, Ding X, Rhea P,
Ding J, Hawke DH, Felsher D, Narla G, Lu Z and Lee RT: Mistletoe
extract Fraxini inhibits the proliferation of liver cancer by
down-regulating c-Myc expression. Sci Rep. 9:64282019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nakagawa R, Toboso-Navasa A, Schips M,
Young G, Bhaw-Rosun L, Llorian-Sopena M, Chakravarty P, Sesay AK,
Kassiotis G, Meyer-Hermann M and Calado DP: Permissive selection
followed by affinity-based proliferation of GC light zone B cells
dictates cell fate and ensures clonal breadth. Proc Natl Acad Sci
USA. 118:e20164251182021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kin K, Chen X, Gonzalez-Garay M and
Fakhouri WD: The effect of non-coding DNA variations on P53 and
cMYC competitive inhibition at cis-overlapping motifs. Hum Mol
Genet. 25:1517–1527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Brandner S: Nanog, Gli, and p53: A new
network of stemness in development and cancer. EMBO J.
29:2475–2476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nourian Dehkordi A, Mirahmadi Babaheydari
F, Chehelgerdi M and Raeisi Dehkordi S: Skin tissue engineering:
Wound healing based on stem-cell-based therapeutic strategies. Stem
Cell Res Ther. 10:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pastar I, Stojadinovic O, Yin NC, Ramirez
H, Nusbaum AG, Sawaya A, Patel SB, Khalid L, Isseroff RR and
Tomic-Canic M: Epithelialization in wound healing: A comprehensive
review. Adv Wound Care (New Rochelle). 3:445–464. 2014. View Article : Google Scholar : PubMed/NCBI
|