Introduction
Prostate cancer (PCa) is the most frequently
diagnosed neoplasm in men worldwide, affecting 1,276,106 of new
cases in 2018 (1). PCa incidence
has been rising in the last decades due to the increase in
population aging, obesity caused by dietary habits and lifestyle,
among other causes (2,3). The development and progression of PCa
is initially dependent on the stimulatory action of androgens,
which validate the use of therapies reducing the biosynthesis of
androgens and/or antagonizing the action of androgens through the
androgen receptor (AR) (4).
Bicalutamide (second generation) and enzalutamide and apalutamide
(third generation) are anti-androgens that antagonize the AR
activity, inhibiting gene expression associated with PCa
progression and consequently having therapeutic benefits in PCa
(5). However, several patients
become resistant to androgen-deprivation therapy (ADT), giving rise
to the so-called castrate-resistant PCa (6). Clinical trials have investigated the
efficacy of ADT in combination with other drugs, as strategies for
improved management of PCa and slowing the progression to
castrate-resistant stage (7). The
identification of new therapeutic targets, in combination with the
ADT, remains a fundamental aspect to improve PCa treatment and to
identify novel predictive biomarkers for response to ADT.
The human Six-Transmembrane Epithelial Antigen of
the Prostate 1 (STEAP1) is highly expressed in several types of
cancer, with special emphasis on PCa, where the STEAP1 protein
expression levels are 5–10 fold higher compared with other cancer
types (8). The mechanisms
underlying the overexpression of STEAP1 in cancer remain poorly
explored, but epigenetic changes associated with increased
expression levels in PCa have been identified in the STEAP1
promoter region (9). Among
non-tumoral tissues, STEAP1 is almost restricted to the epithelial
cells of the prostate gland, which makes it a promising biomarker
and/or therapeutic target for PCa (8,10).
The potential role of STEAP1 in cancer progression has been studied
extensively and its oncogenic role emphasized (11–13).
However, the physiological role of STEAP1 and the specific actions
driven the carcinogenic process remain to be elucidated.
Nevertheless, the STEAP1 protein seems to act as a channel for
small molecules, being involved in intercellular communication
(14). In addition, there is
evidence for formation of STEAP1-STEAP2 heterotrimers, which seem
to be associated with the activity of metal reductase and
superoxide synthase enzymes (15).
This is in accordance with a previous study demonstrating that
STEAP1 actions, favoring cancer progression, are associated with
oxidative stress (16).
Several research groups have demonstrated the
contribution of STEAP1 to tumor progression, namely, by its
involvement in promoting cell proliferation, migration and invasion
(17–21). Regarding PCa, it was shown that
silencing the STEAP1 gene reduces androgen-dependent PCa cell
viability and proliferation and increases the apoptosis rate. In
addition, the anti-proliferative and pro-apoptotic effects
triggered by STEAP1 knockdown are not abrogated by the treatment
with dihydrotestosterone (DHT), suggesting that STEAP1 inhibition
might be a good option of treatment to prevent the effects of DHT
in PCa (17). The effect of
androgens in regulating STEAP1 expression has also been studied,
with contradictory results. Some studies found STEAP1 as
androgen-stimulated, as androgen-inhibited, or as
androgen-independent in cell lines of PCa (8,22–24).
Concerning LNCaP cells, DHT downregulates STEAP1 expression, but
this effect should not directly involve the AR because no androgen
response elements are found in promoter region of the STEAP1
gene (23,25). This indirect action of AR in
downregulating STEAP1 means that de novo protein synthesis
should be required (23). In human
PCa biopsies, an association between STEAP1 overexpression and
metastasis was found, with the presence of more aggressive tumors
and the majority of patients becoming resistant to treatment with
anti-androgens (26). In addition,
a marginally positive significant association between STEAP1
overexpression and presurgical prostate specific antigen (PSA)
levels was detected, indicating a potential crosstalk between STEAP
and AR (26). Considering the use
of anti-androgens in PCa treatment and the potential of STEAP1 as
therapeutic target, associated with the putative relationship
between STEAP1 and AR, it was hypothesized that STEAP1 expression
may be regulated by anti-androgens. Also, it was hypothesized that
blocking the action of STEAP1 may sensitizes PCa cells to treatment
with anti-androgens drugs. Therefore, the main goal of the present
study was to investigate the effect of several types of
anti-androgens, bicalutamide, enzalutamide and apalutamide, in
LNCaP wild-type (LNCaP-WT) and LNCaP-STEAP1 knockdown cells.
Materials and methods
Cell culture
Human prostate adenocarcinoma cell line (LNCaP) was
purchased from the European Collection of Authenticated Cell
Cultures and maintained in Roswell Park Memorial Institute medium
(RPMI)-1640 (MilliporeSigma) supplemented with 10% fetal bovine
serum (FBS; Biochrom AG) and 1% penicillin/streptomycin (Gibco;
Thermo Fisher Scientific, Inc.), in a humidified chamber at 37°C
and a 5% CO2 atmosphere.
STEAP1 knockdown and experimental
design
LNCaP cells at 50% confluence in T-flasks or
multiwell plates were transfected with 20 nM of a small interfering
RNA (siRNA) targeting the STEAP1 (cat. no. s226093; Ambion; Thermo
Fisher Scientific, Inc.) using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at 37°C in
Opti-Minimum Essential Medium (MEM) (Invitrogen; Thermo Fisher
Scientific, Inc.) as recommended by the manufacturer. These cells
are referred to as LNCaP-STEAP1 knockdown. As a control for
STEAP1-knockdown, a scrambled siRNA sequence (cat. no. 4390846,
Ambion; Thermo Fisher Scientific, Inc.) was used and these control
cells are designated as LNCaP-wild type (WT) cells. The sequences
of STEAP1 and scramble siRNA were not provided by the manufacturer.
Then, 24 h following transfection, the cells were stimulated with
anti-androgens, 100 µM bicalutamide (MilliporeSigma), 10 µM
enzalutamide (MilliporeSigma) and 10 µM apalutamide (Alfa Aesar)
for 24 h at 37°C. All drugs stock solutions were prepared in
dimethyl sulfoxide (DMSO). Cells were harvested at 24 h after drugs
treatment and the efficiency of STEAP1 knockdown expression was
analyzed by reverse transcription-quantitative (RT-q) PCR and
western blotting.
MTT assay
In order to determine the viability of LNCaP cells
silenced for STEAP1 and exposed to the three anti-androgenic drugs,
3-(4,5-dimethylthiazol-2-thiazolyl)-2,5-diphenyltetrazolium bromide
(MTT) assay (MilliporeSigma) was used according to the
manufacturer's instructions. Briefly, 100 µl of MTT solution at 0.5
mg/ml concentration was added to cells. After 1 h of incubation at
37°C, the MTT solution was removed and 100 µl DMSO was added for
solubilization of the formazan crystals. Next, the optical density
was measured at 570 nm using the xMark Microplate Absorbance
Spectrophotometer (Bio-Rad Laboratories, Inc.).
Ki-67 fluorescence
immunocytochemistry
Fluorescent immunocytochemistry of the proliferation
marker Ki67 was used to estimate the proliferation index between
LNCaP cells knocked down for STEAP1 and LNCaP-WT, both treated with
the three drugs. LNCaP cells were fixed with 4% paraformaldehyde
(PFA) for 10 min at room temperature and permeabilized with 0.1%
Triton X-100 for 5 min also at RT. A blocking step was performed by
incubating cells with 20% FBS in phosphate buffer saline (PBS)
containing 0.1% Tween-20 (PBST) for 1 h at RT and then cells were
incubated with rabbit anti-Ki-67 (1:50; cat. no. 16667; Abcam) for
90 min at RT. The Alexa Fluor 546 goat anti-rabbit IgG (1:1,000;
Invitrogen; Thermo Fisher Scientific, Inc.) was used as a secondary
antibody. This incubation was performed at RT for 60 min. The
specificity of the staining was assessed by omission of the primary
antibody. Cell nuclei were stained with Hoechst 33342 (5 µg/ml;
Invitrogen; Thermo Fisher Scientific, Inc.) for 5 min at RT.
Coverslips were washed and mounted onto microscope slides with Dako
fluorescent mounting medium (Dako; Agilent Technologies, Inc.).
Images were acquired using a AxioImager Z2 optical microscope (Carl
Zeiss AG). Proliferation was determined by the percentage of
Ki-67-positive cells out of the total number of Hoechst-stained
nuclei in eight randomly selected fields per microscope cover
glass.
Terminal deoxynucleotidyl transferase
dUTP Nick End Labeling (TUNEL) assay
Cells were fixed with 4% PFA for 10 min at RT, and
then, permeabilized in 1% Triton X-100 with PBST for 5 min also at
RT. TUNEL reaction mixture (40 µl; Roche Applied Science) was added
to each sample for 1 h at room temperature in the dark. Cells were
washed in PBS and incubated for 5 min at RT with Hoechst-33342 (5
µg/ml, Invitrogen, Thermo Fisher Scientific, Inc.). Coverslips were
then mounted using Dako fluorescent mounting medium (Dako; Agilent
Technologies, Inc.) and analyzed by fluorescence microscopy using
the AxioImager Z2 Optical Microscopy (Carl Zeiss AG). The
percentage of apoptotic cells was estimated by counting the number
of TUNEL-positive cells and Hoechst-stained nuclei in eight
randomly selected ×40 magnification fields in each coverslip. The
ratio between the number of TUNEL-positive cells and total number
was calculated.
RNA extraction and RT-qPCR
Total RNA from 3×106 LNCaP cells was
obtained using TRI reagent (Grisp, Lda.) according to the
manufacturer's instructions. The RNA pellet was dried, resuspended
in 20 µl of diethylpyrocarbonate treated-water and maintained at
−80°C. In order to assess the quantity of total RNA, its optical
density was determined by measuring absorbance at 260 and 280 nm on
a nanospectrometer (Ultrospec 3000; GE Healthcare). Total RNA
integrity was verified by agarose gel electrophoresis. RT-qPCR was
used to determine expression levels of STEAP1, p21 and
kallikrein-related peptidase 3 (KLK3; encodes the PSA
protein) genes, using Power SYBR Green RNA-to-CT, 1-Step kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) on the CFX
connect real-time system (Bio-Rad Laboratories, Inc.) according to
the manufacturer's instructions. RT-qPCR was performed with 0.2 µg
of RNA in 10 µl of total reaction with specific primers for STEAP1
(sense: 5′GGCGATCCTACAGATACAAGTTGC3′ and anti-sense:
5′CCAATCCCACAATTCCCAGAGAC3′), p21 (sense: 5′TCCAGCGACCTTCCTCATC3′
and anti-sense: 5′AGCCTCTACTGCCACCATC3′), KLK3 (sense:
5′ACCAGAGGAGTTCTTGACCCC3′ and anti-sense 5′ CCCCAGAATCACCCGAGCAG3′)
and β-2-microglobulin housekeeping (β2M, sense:
5′ATGAGTATGCCTGCCGTGTG3′ and anti-sense: 5′CAAACCTCCATGCTGCTTAC3′).
All primers were synthesized by the services of STAB VIDA and were
previously characterized with qPCRs optimized by our research group
in previous papers (17,23). After cDNA synthesis at 48°C for 30
min, qPCR was performed with the following steps: Initial
denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C
for 30 sec, annealing temperature at 60°C for 30 sec and extension
at 72°C for 20 sec. The amplified PCR fragments were analyzed by
melting curves. β2M housekeeping was used as internal control to
normalize gene expression. Fold differences were calculated
following the mathematical model proposed by Pfaffl (27).
Protein extraction and western
blotting
LNCaP cells were lysed in an appropriate volume of
Radioimmunoprecipitation assay (150 mM NaCl, 1% Nonidet-P40
substitute, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate
and 50 mM Tris) supplemented with 10% phenylmethylsulfonyl fluoride
and 1% protease cocktail (A7779, PanReac AppliChem, ITW Reagents,
Ottoweg 2, Darmstadt, Germany). The total protein extract was
obtained after centrifugation of the cell lysate for 20 min at 18
620 g at 4°C. Quantification of the total protein was measured
using the Pierce 660 nm Protein assay reagent (Thermo Fisher
Scientific, Inc.). Then, ~20 µg of total protein were resolved on
10% TGX Stain-Free polyacrylamide gels (Bio-Rad Laboratories,
Inc.), scanned in the ChemiDoc MP Imaging System (Bio-Rad
Laboratories, Inc.) with one minute of exposure time and then,
transferred into a polyvinylidene difluoride membrane (Bio-Rad
Laboratories, Inc.). After blocking with 5% milk solution for 1 h
at room temperature, membranes were incubated overnight at 4°C with
following antibodies: Rabbit anti-STEAP1 (1:1,000; cat. no. D8B2V;
Lot 1; Cell Signaling Technology, Inc.), rabbit anti-p-AKT (1:500;
cat. no. 9271S; Lot 14; Cell Signaling Technology, Inc.), rabbit
anti-AKT (1:500; cat. no. 9272S; Lot 27; Cell Signaling Technology,
Inc.), rabbit anti-p-ERK (1:500; cat. no. 9101S; Lot 12; Cell
Signaling Technology, Inc.), rabbit anti-ERK (1:500; cat. no.
9102S; Lot 27; Cell Signaling Technology, Inc.), rabbit
anti-p-c-myc (1:500; cat. no. 13748S; Lot 4; Cell Signaling
Technology, Inc.), rabbit anti-c-myc (1:500; A-14; cat. no. sc-789;
Santa Cruz Biotechnology, Inc.), rabbit anti-Bcl-2 (1:1,000; cat.
no. 2876S; Lot 6; Cell Signaling Technology, Inc.), rabbit anti-Bax
(1:1,000; cat. no. 2772S; Lot 11; Cell Signaling Technology, Inc.)
and rabbit anti-p53 (1:1,000; FL-393; cat. no. sc-6243; Lot L2713,
Santa Cruz Biotechnology, Inc.). Membranes were incubated for 1 h
at room temperature with secondary anti-rabbit IgG-horseradish
peroxidase (1:15,000; cat. no. 9003-99-0; MilliporeSigma). After
this, immunoreactivity was visualized using the ChemiDoc MP Imaging
System (Bio-Rad Laboratories, Inc.) after the incubation with
enhanced chemiluminescence substrate (Bio-Rad Laboratories, Inc.).
Total protein normalization was carried out using the Image Lab 5.1
software (Bio-Rad Laboratories, Inc.), by opening a multichannel
image, configure two channels: channel 1, target protein blot and
channel 2, stain-free gel image. Normalization icon from the
Analysis Toolbox was used to detect bands and lanes and after were
adjusted if needed. Stain-free image was selected as normalization
channel and the normalized volumes are indicated in the Analysis
Table on the tool bar. The target protein band intensity value is
adjusted for variation in the protein total load on the gel,
following other studies (28,29).
Caspase-3-like activity assay
The caspase-3-like activity was determined after the
cleavage of the labeled substrate by the detection of the
chromophore p-nitroaniline (pNA), measured spectrophotometrically
at 405 nm. Total protein extract (25 µg) was incubated with a
reaction buffer [25 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 0.1%
3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, 10%
sucrose and supplemented with 10 mM dithiothreitol (pH 7.5)] and 2
mM of caspase-3 substrate (N-Acetyl-Asp-Glu-Val-Asp pNA and
Ac-DEVD-pNA, Sigma-Aldrich) for 2 h at 37°C. The amount of
generated pNA was calculated by extrapolation with a standard curve
of free pNA.
Statistical analysis
All experimental data are shown as mean ± standard
error of the mean. Statistical significance of differences among
experimental groups were evaluated by two-way analysis of variance
followed by Tukey's multiple comparisons test, using GraphPad Prism
v7.01 (GraphPad Software, Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
Effect of bicalutamide, enzalutamide
and apalutamide on STEAP1 expression in LNCaP-WT and LNCaP-STEAP1
knockdown cells
The effect of anti-androgens in regulating STEAP1
gene expression was evaluated in LNCaP-WT and LNCaP-STEAP1
knockdown cells. After stimulation with bicalutamide (100 µM),
enzalutamide (10 µM) or apalutamide (10 µM) for 24 h, the STEAP1
mRNA and protein expression was evaluated by qPCR and western
blotting, respectively. As can be seen in Fig. 1, using a siRNA targeting STEAP1
silenced STEAP1 gene was confirmed at mRNA (87±0.008% compared with
scramble siRNA; Fig. 1A) and
protein (80±0.053% compared with scramble siRNA; Fig. 1B) levels. The efficiency and
validation of the anti-androgen treatment was shown by analysing
the expression levels of an AR target gene, the KLK3 gene
that encodes the PSA protein. Fig.
1C showed that suppressing androgen actions with bicalutamide,
enzalutamide or apalutamide significantly reduced the KLK3
gene expression (0.15±0.01-, 0.16±0.03- and 0.16±0.01-fold
variation, respectively). It was also verified that STEAP1
knockdown decreased the KLK3 gene levels (0.573±0.04-
compared with 0.964±0.06-fold variation; Fig. 1C).
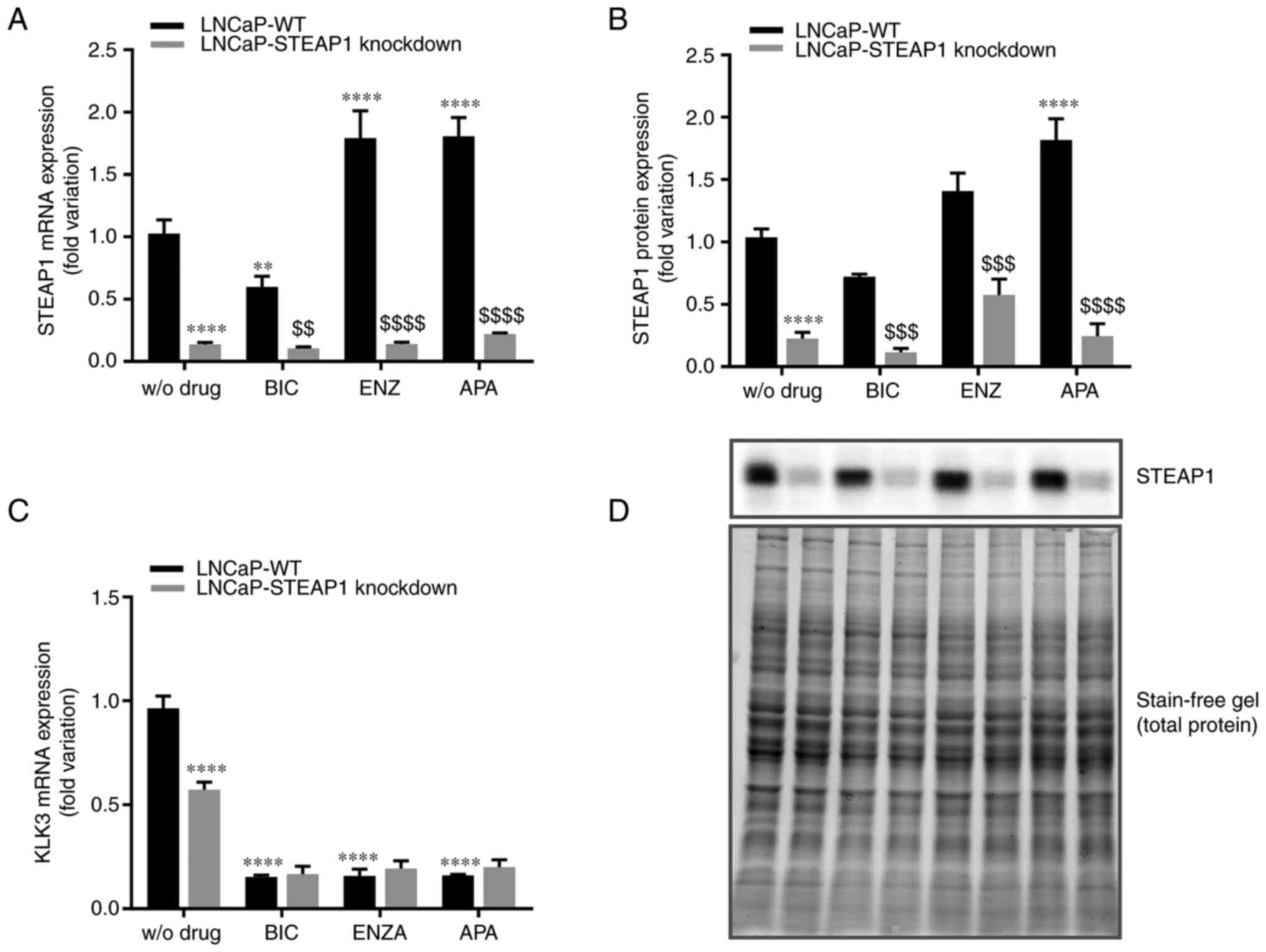 | Figure 1.Effect of bicalutamide, enzalutamide
and apalutamide on the expression of STEAP1 and KLK3 in LNCaP-WT
and LNCaP-STEAP1 knockdown prostate cancer cells. STEAP1 and KLK3
expression levels in LNCaP cells following transfection with siRNA
for 24 h following treatment with 100 µM of BIC, or 10 µM of ENZA
or 10 µM of APA for 24 h. Relative (A) STEAP1 mRNA, (B) STEAP1
protein and (C) KLK3 mRNA expression were determined by reverse
transcription-quantitative PCR following normalization with the β2M
housekeeping gene and western blot after normalization with total
protein as represented in (D). Representative immunoblots are also
showed in (D). Results are expressed as fold-variation relative to
LNCaP-WT (control group). Error bars indicate mean ± standard error
of the mean (n≥3). **P<0.01 and ****P<0.0001 vs. the LNCaP-WT
condition; $$P<0.01, $$$P<0.001 and
$$$$P<0.0001 vs. LNCaP-WT plus respective drug.
STEAP1, six transmembrane epithelial antigen of the prostate 1;
KLK3, kallikrein related peptidase 3; WT, wild type; siRNA, small
interfering RNA; BIC, bicalutamide; ENZA, enzalutamide; APA,
apalutamide; β2M, β-2-microglobulin. |
When LNCaP-WT cells were treated with bicalutamide,
there was a significant decrease in STEAP1 mRNA expression
(0.59±0.23-fold variation; Fig.
1A), but no significant effect was observed at STEAP1 protein
level (0.73±0.020-fold variation; P=0.108; Fig. 1B). On the other hand, enzalutamide
and apalutamide increased the expression of STEAP1 mRNA (1.79±0.018
and 1.77±0.275-fold variation, respectively; Fig. 1A). Concerning the STEAP1 protein
levels, the increase of STEAP1 in LNCaP-WT cells was observed with
the treatment of apalutamide (1.89±0.177-fold variation; Fig. 1B), but not with enzalutamide
(1.47±0.167-fold variation; P=0.108; Fig. 1B).
Regarding the effect of anti-androgens in
LNCaP-STEAP1 knockdown, no significant differences in STEAP1 mRNA
and protein expression were verified in comparison with LNCaP
STEAP1-knockdown without treatment with anti-androgens.
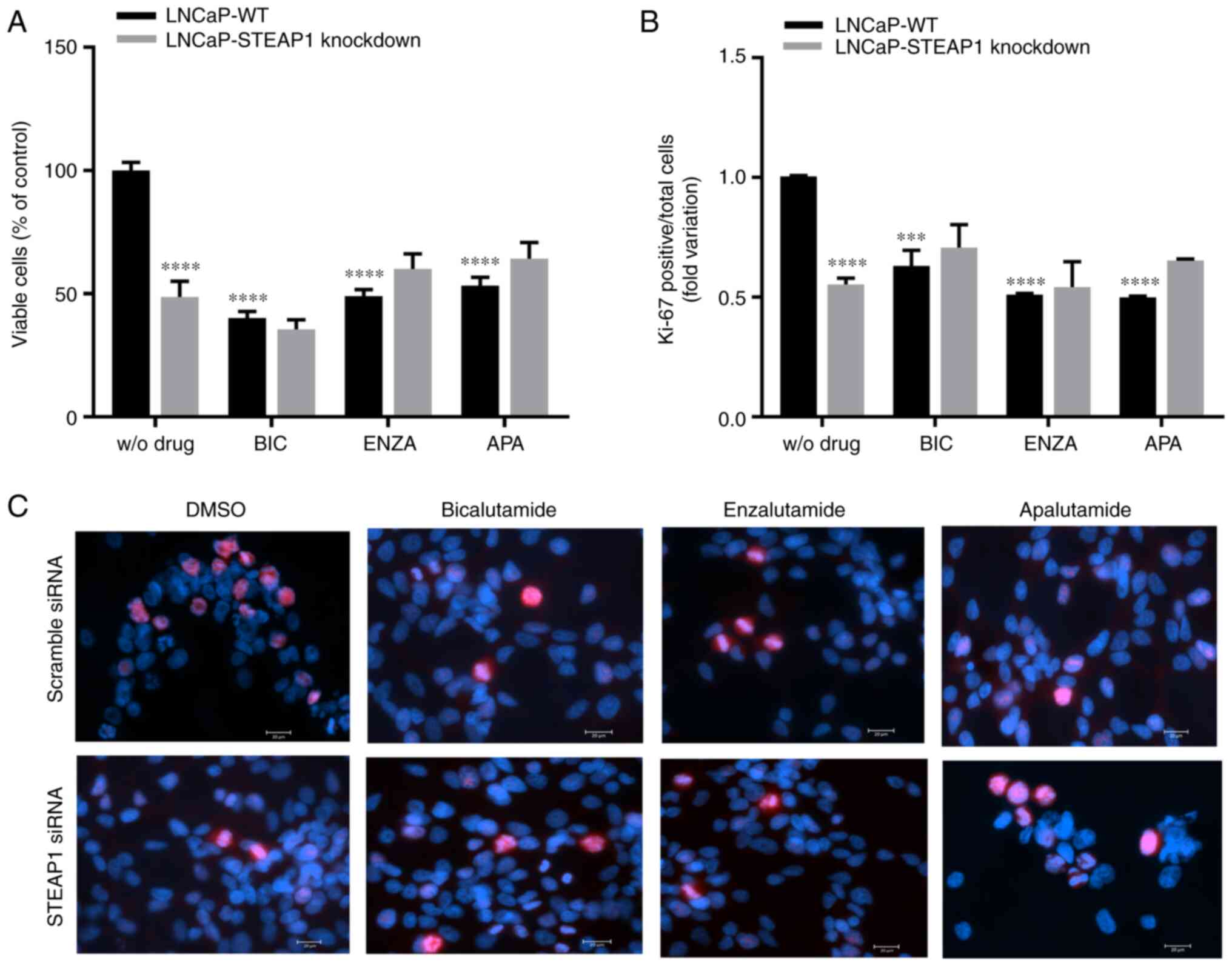
Cell viability and proliferation of
LNCaP-STEAP1 knockdown cells in response to anti-androgenic
drugs
The effect of bicalutamide, enzalutamide and
apalutamide anti-androgenic drugs on viability and proliferation of
LNCaP-STEAP1 knockdown cells were determined by the MTT assay and
immunofluorescent labelling of Ki-67, respectively. At 24 h after
knockdown of STEAP1, LNCaP cells were exposed to bicalutamide,
enzalutamide and apalutamide drugs. Viability of LNCaP cells
markedly decreased upon STEAP1 silencing (47.2±11.8% reduction
compared with scramble siRNA; Fig.
2A). Bicalutamide (100 µM), enzalutamide (10 µM) and
apalutamide (10 µM) significantly decreased the viability of
LNCaP-WT cells (59.55±5.5%, 50.68±3.7% and 46.1±6.7% of reduction,
respectively; Fig. 2A). The
silencing of STEAP1 in LNCaP cells did not significantly change
cell viability when treated with anti-androgenic drugs (Fig. 2A). The results of Ki-67 fluorescent
immunocytochemistry were similar to the results observed for the
MTT assay, also showing that the number of Ki-67-positive cells
relative to total cells (Hoechst-positive) was significantly
decreased in the LNCaP-STEAP1 knockdown cells when compared with
LNCaP-WT cells (0.553±0.03-compared with 1.01±0.003-fold variation;
Fig. 2B). Administration of
bicalutamide, enzalutamide and apalutamide drugs in LNCaP-WT cells
significantly decreased the number of Ki-67 positive cells compared
with LNCaP-WT without drug (0.630±0.06-, 0.511±0.01- and
0.500±0.01-fold variation, respectively; Fig. 2B). However, the STEAP1 gene
silencing in LNCaP cells did not significantly change the
Ki-67-positive cells number obtained when treated with
anti-androgenic drugs (Fig. 2B).
Representative fluorescent immunocytochemistry images of
Ki-67-labelled LNCaP cells in all experimental conditions were
represented in Fig. 2C.
Analysis of survival pathways in
LNCaP-STEAP1 knockdown cells in response to anti-androgenic
drugs
To understand the decreased viability and
proliferative activity of LNCaP cells in response STEAP1 knockdown
associated with anti-androgenic action, the expression of proteins
related with cell survival pathways was evaluated. Fig. 3 shows the western blot analysis for
the expression of the active phosphorylated (p-)AKT, ERK and c-myc
isoforms, respectively to the expression of total proteins. The
results showed that p-AKT/AKT and p-ERK/ERK ratio decreased in
LNCaP-STEAP1 knockdown cells when compared with LNCaP-WT
(0.487±0.04-compared with 0.989±0.01-fold variation and
0.701±0.02-compared with 1.02±0.024-fold variation, respectively;
Fig. 3A and B). Treatment of
LNCaP-WT cells with 100 µM of bicalutamide, 10 µM of enzalutamide
and 10 µM of apalutamide also decreased the p-AKT/AKT ratio
relatively to LNCaP-WT group without drug (0.748±0.003-compared
with 0.989±0.01-, 0.402±0.058-compared with 0.989±0.01- and
0.676±0.05-compared with 0.989±0.01-fold variation, respectively;
Fig. 3A). The silencing of STEAP1
did not alter the p-AKT/AKT ratio of LNCaP cells treated with
anti-androgens (Fig. 3A). No
statistically significant differences were found in the p-ERK/ERK
ratio in LNCaP-WT or LNCaP-STEAP1 knockdown cells, both treated
with bicalutamide, enzalutamide or apalutamide (Fig. 3B).
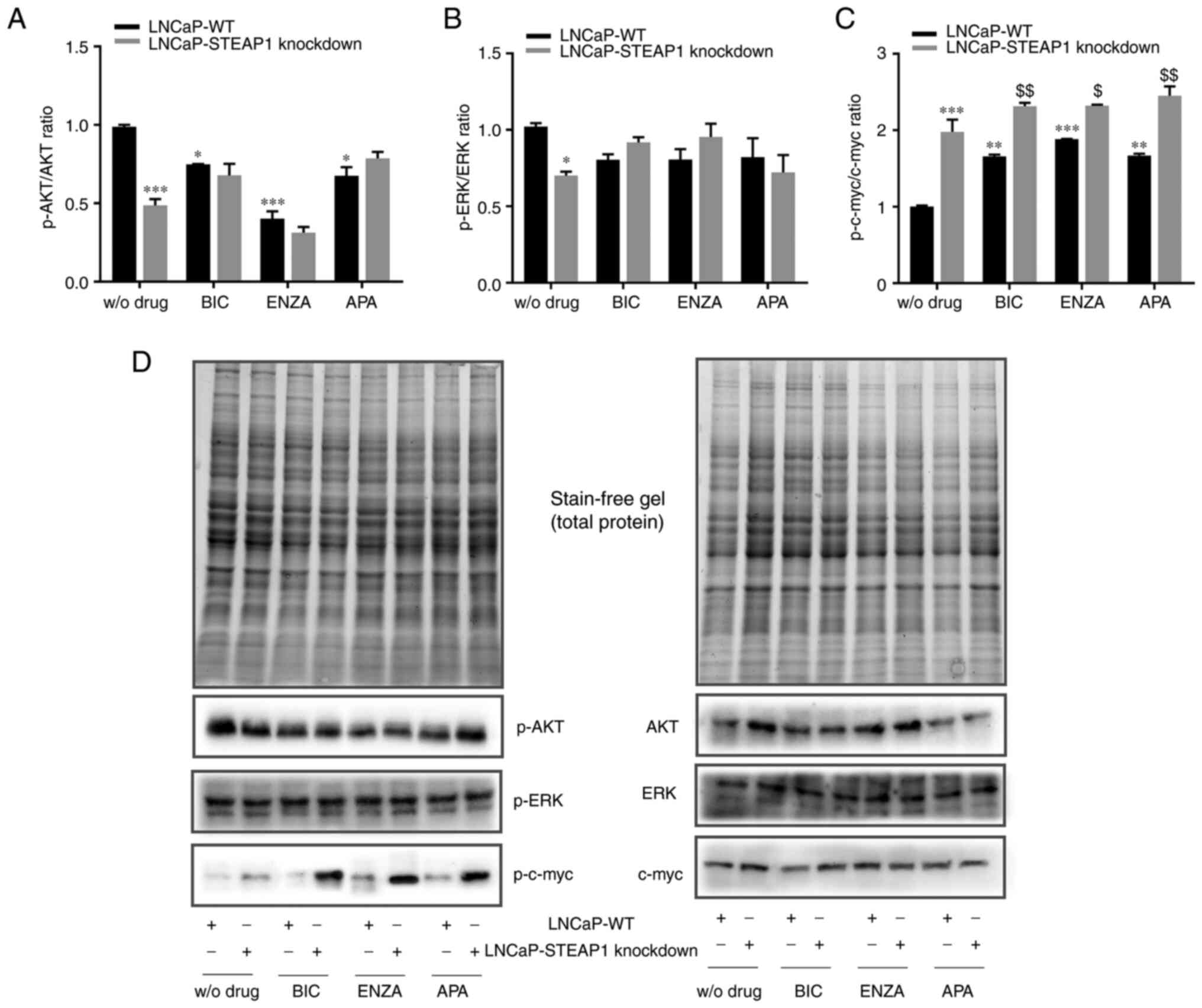 | Figure 3.Effect of BIC, ENZA and APA on the
expression of p-AKT, AKT, p-ERK, ERK, p-c-myc and c-myc in LNCaP-WT
and LNCaP-STEAP1 knockdown cells. LNCaP cells transfected with
siRNA targeting STEAP1 or scramble siRNA. 24 h after tranfection,
LNCaP cells were treated with 100 µM of BIC, or 10 µM of ENZA or 10
µM of APA for 24 h. Ratio of phospohorylated forms and total
protein of (A) AKT, (B) ERK and (C) c-myc were determined by
western blotting after independent normalization with total protein
load on gel as represented in (D) together with representative
immunoblots. Results are expressed as fold-variation relative to
LNCaP-WT (control group). Error bars indicate mean ± standard error
of the mean (n≥2). *P<0.05, **P<0.01 and ***P<0.001 vs.
the LNCaP-WT condition; $P<0.05 and
$$P<0.01 vs. LNCaP-WT plus respective drug. BIC,
bicalutamide; ENZA, enzalutamide; APA, apalutamide; p-,
phosphorylated; WT, wild type; STEAP1, six transmembrane epithelial
antigen of the prostate 1; siRNA, small interfering RNA. |
Regarding the levels of p-c-myc and c-myc, an
increased of p-c-myc/c-myc ratio was observed in LNCaP-STEAP1
knockdown when compared with the LNCaP-WT cells
(1.978±0.16-compared with 1.002±0.01-fold variation; Fig. 3C). Bicalutamide-, enzalutamide- and
apalutamide-treated group in LNCaP-WT cells, high levels of
p-c-myc/c-myc ratio was found in comparison with scramble siRNA
group (1.659±0.02-compared with 1.002±0.01-, 1.883±0.003-compared
with 1.002±0.01- and 1.668±0.03-compared with 1.002±0.01-fold
variation, respectively; Fig. 3C).
The p-c-myc/c-myc ratio in response to bicalutamide, enzalutamide
and apalutamide drugs was higher in LNCaP-STEAP1 knockdown cells
than in LNCaP-WT cells (2.315±0.04-compared with 1.659±0.02-fold
variation to bicalutamide, 2.320±0.01-compared with
1.883±0.003-fold variation to enzalutamide, 2.451±0.12-compared
with 1.668±0.03-fold variation to apalutamide; Fig. 3C).
In brief, anti-androgen treatment significantly
decreased p-AKT/AKT and increased p-c-myc/c-myc ratio expression
levels in LNCaP-WT cells. Moreover, a slight additive effect was
observed in p-c-myc/c-myc ratio in LNCaP cells knocked-down for
STEAP1 treated with bicalutamide and apalutamide.
Analysis of apoptotic pathways in
LNCaP-WT and LNCaP-STEAP1 knockdown cells treated with
anti-androgenic drugs
To determine whether the diminished
viability/proliferation of LNCaP-STEAP1 knockdown cells in response
to anti-androgens treatment was a consequence of increased
apoptosis, the expression levels and activity of several apoptotic
markers were evaluated. The knockdown of STEAP1 significantly
increased the expression of several regulators involved in the
apoptosis pathway, Bax/Bcl-2 ratio was increased 1.7-fold (Fig. 4A) and the expression of p53 protein
and p21 mRNA were also increased (2.004±0.08- and 2.161±0.16-fold
variation, respectively; Fig. 4B and
C). A notable end-point of apoptosis is the activation of
caspase-3 and the results showed an increased activity of caspase-3
in LNCaP-STEAP1 knockdown (1.944±0.27-fold variation; Fig. 4D) when compared with LNCaP-WT
cells. Also, the number of TUNEL-positive cells relative to total
cells was significantly increased in the LNCaP-STEAP1 knockdown
cells when compared with the scramble siRNA group (1.951±0.12-
compared with 1.002±0.004-fold variation; Fig. 4E). Treatment with 100 µM of
bicalutamide, 10 µM of enzalutamide and 10 µM of apalutamide
triggered an increased expression of apoptotic regulators (Fig. 4). The expression levels of pro- and
anti-apoptotic members (Bax and Bcl-2 respectively) led to enhanced
Bax/Bcl-2 ratio in response to anti-androgen drugs in LNCaP-WT
cells (1.6±0.08- to bicalutamide, 1.3±0.07- to enzalutamide and
1.6±0.08-fold variation to apalutamide; Fig. 4A). However, in LNCaP-STEAP1
knockdown cells, there appeared to be a potentiating effect of
Bax/Bcl-2 ratio with apalutamide treatment (1.9±0.04-compared with
1.6±0.08-fold variation; Fig. 4A),
but not with bicalutamide and enzalutamide.
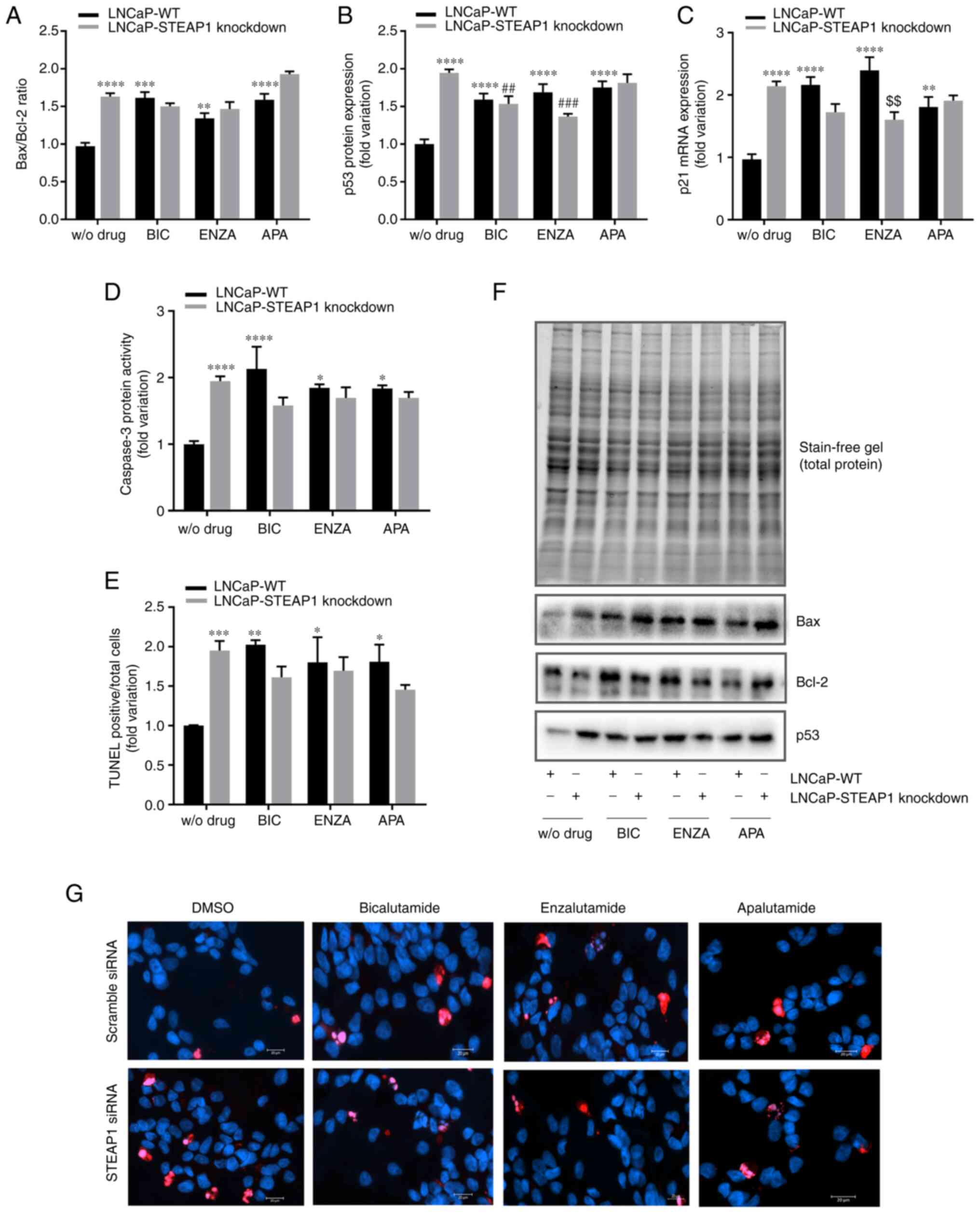 | Figure 4.Effect of BIC, ENZA and APA on the
expression of several apoptotic regulators in LNCaP-WT and
LNCaP-STEAP1 knockdown cells. LNCaP cells were transfected with
siRNA targeting STEAP1 or scramble siRNA. 24 h following
transfection, LNCaP cells were treated with 100 µM of BIC, or 10 µM
of ENZA or 10 µM of APA for 24 h. (A) Bax/Bcl-2 protein ratio, (B)
p53 protein expression and (C) p21 mRNA expression were determined
by western blotting and reverse transcription-quantitative PCR,
respectively. (D) Caspase-3 activity was measured
spectrophotometrically by the release of the product pNA and (E)
immunofluorescence analysis of TUNEL-positive cells was determined
by the TUNEL assay being the results expressed as the mean of
TUNEL-positive cells (red staining) relatively to the total cell
number [Hoechst 33342 (blue) staining]. (F) Relative protein
expression was normalized with total protein load on gel as
represented in and relative mRNA expression was normalized with the
β2M housekeeping gene. Representative immunoblots are also shown.
(G) Representative microscopy images showing TUNEL and Hoechst
staining in the different groups were obtained in the AxioImager Z2
fluorescence microscope (magnification, ×400). Results are
expressed as fold-variation relative to LNCaP-WT (control group).
Error bars indicate mean ± standard error of the mean (n≥2).
*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. the
LNCaP-WT condition; ##P<0.01 and
###P<0.001 vs. the LNCaP-STEAP1 knockdown condition;
$$P<0.01 vs. LNCaP-WT plus respective drug. BIC,
bicalutamide; ENZA, enzalutamide; APA, apalutamide; WT, wild type;
siRNA, small interfering RNA; STEAP1, six transmembrane epithelial
antigen of the prostate 1; β2M, β-2-microglobulin; WT, wild
type. |
The treatment of LNCaP-WT cells with anti-androgenic
drugs significantly increased the expression of p53 protein
(1.615±0.05- to bicalutamide, 1.689±0.16- to enzalutamide and
1.720±0.12-fold variation to apalutamide) and p21 mRNA (2.119±0.12-
to bicalutamide, 2.396±0.001- to enzalutamide and 1.810±0.09-fold
variation to apalutamide), as observed in Fig. 4B and C, respectively. The knockdown
of STEAP1 did not alter the effect of bicalutamide and enzalutamide
in expression of p53 protein (1.559±0.14-compared with 1.615±0.05-
and 1.813±0.16 compared with 1.720±0.12-fold variation,
respectively) and p21 mRNA (1.738±0.23-compared with 2.119±0.12-
and 1.902±0.05 compared with 1.810±0.09-fold variation,
respectively). However, the enzalutamide treatment seems to have
less effect in LNCaP-STEAP1 knockdown compared with LNCaP-WT cells
(1.366±0.02-compared with 1.689±0.16-fold variation to p53 levels
and 1.605±0.23 compared with 2.396±0.001-fold variation to p21
levels; Fig. 4B and C).
The activity of caspase-3 significantly increased in
LNCaP-STEAP1 knockdown cells treated with bicalutamide,
enzalutamide and apalutamide (2.11±0.27-, 1.849±0.24- and
1.836±0.07-fold variation, respectively, compared with
0.999±0.002-fold variation; Fig.
4D); this effect did not significantly alter with silencing of
STEAP1 (Fig. 4D).
The results of TUNEL fluorescent immunocytochemistry
assay showed that the number of TUNEL-positive LNCaP-STEAP1
knockdown cells were significantly increased with the treatment of
bicalutamide, enzalutamide and apalutamide when compared with
LNCaP-WT cells (2.154±0.13-, 1.801±0.32- and 1.809±0.22-fold
variation, respectively, compared with1.002±0.004-fold variation;
Fig. 4E). No significant effect
was observed in response to anti-androgen drugs in LNCaP-STEAP1
knockdown cells in comparison with LNCaP-WT cells (Fig. 4E).
In summary, anti-androgen treatment in LNCaP-WT
cells increased the expression and activity of several apoptosis
regulators. Silencing of STEAP1 did not significantly change the
effect observed by bicalutamide, enzalutamide and apalutamide
treatment.
Discussion
In the recent decades, the use of ADT to treat PCa
patients has notably increased (30,31).
However, ADT treatment alone becomes insufficient for the
management of PCa, since most patients with this pathology progress
to the castration-resistant disease within a few years (32,33).
A way of improving PCa treatment is to evaluate combined action
with other putative therapeutic targets. There are several proteins
that are dysregulated in PCa, including STEAP1 (8). This transmembrane protein has been
implicated in several forms of cancer due to its overexpression in
malignant tissue compared with their non-malignant counterparts
(11,13,34).
Considering the oncogenic role of STEAP1 in PCa, associated with a
lack of studies focusing on impact of ADT treatment in PCa cells
overexpressing STEAP1, the main goals of the present study were to
evaluate the effect of anti-androgens on expression of STEAP1 and
to investigate if the sensitivity of PCa cells to anti-androgen
drugs can be improved in response to STEAP1 knockdown. Thus, the
effect of bicalutamide, enzalutamide and apalutamide was evaluated
in LNCaP-WT and LNCaP-STEAP1 knockdown cells.
Deregulated cell proliferation and apoptosis are a
well-established cancer hallmarks and of the first deregulated
mechanisms underlying cancer progression (35). The silencing of STEAP1 was
confirmed 24 h following transfection (Fig. 1), decreasing the viability and
proliferation of LNCaP cells (Fig.
2 and 3) and accompanied by an
increasing of apoptosis (Fig. 4).
These results are in accordance with those previously described by
our research group (17).
Treatment of PCa cells with an antibody against the STEAP1 protein
was associated with inhibition of cell growth (36), supporting the present results
herein described and the role of STEAP1 as an oncoprotein. The high
activity of caspase-3, an effector caspase activated by intrinsic
and extrinsic pathway (37) and
the high number of TUNEL-positive cells, an established marker of
apoptosis by detection of free 3′-OH termini in single-stranded
breaks in high-molecular-weight nuclear DNA fragments (38), highlighted the enhanced apoptosis
of LNCaP cells in response to STEAP1 knockdown. The intrinsic
apoptotic pathway should be involved considering the up- and
downregulation of pro- and anti-apoptotic Bax and Bcl-2 proteins,
respectively (Fig. 4).
Furthermore, the inhibition of cell proliferation and the apoptotic
effect triggered by the knockdown of STEAP1 was also supported by
the upregulation of p53 and p21 levels (Fig. 4), which are involved in cell cycle
arrest at G1 and S phase (39); p53 is also an important inducer of
the apoptosis intrinsic pathway (40). The diminished viability and
proliferation of LNCaP cells in response to STEAP1 knockdown was
corroborated by the downregulation of p-AKT/total AKT and
p-ERK/total ERK ratios, two oncogenic survival pathways associated
with cancer progression (41,42).
These results also supported the hypothesis that reduced activity
of AKT may induce the expression of p53. In fact, AKT interacts
with the ubiquitin E3 ligase Mdm2, which controls the expression
levels and activity of p53 (43,44).
AKT enhances Mdm2-mediated p53 ubiquitination and degradation,
leading to cell survival (45).
Therefore, it is likely to assume that silencing of STEAP1 in LNCaP
cells decreased AKT activity, with increased levels of p53, which
is associated with the suppression of cell proliferation and
activation of apoptosis (40). The
precise mechanism underlying the role of STEAP1 in the activation
of AKT and ERK pathways remains to be elucidated, though it is
possible that AKT and ERK activation may be mediated by increased
levels of oxidative stress induced by STEAP1 overexpression
(16). This hypothesis is
supported by reports demonstrating AKT and ERK activation in
response to the increased levels of reactive oxygen species in
LNCaP cells (46). However, and in
order to overcome the limitation of this study, additional studies
should be carried out to clarify the relationship between STEAP1
and AKT/ERK using specific inhibitors, as well as its association
with oxidative stress.
The transcription factor c-myc is a master regulator
of the transcriptional program that controls cell survival and
proliferation (47). Increasing
evidence demonstrates that c-myc signaling has a tumor-promoting
role and is able to significantly increase proliferation and
metastasis of tumors (47–49). Iijima et al (21) showed that the knockdown of STEAP1
leads to cell-growth inhibition in liver cancer by targeting the
suppression of c-myc. Unexpectedly, it was observed that silencing
of STEAP1 increased c-myc expression in LNCaP PCa cells. Although
c-myc is associated with PCa progression, there are several studies
showing that it is among the most robust inducers of apoptosis in
hematologic diseases, as well as in solid tumors such as breast and
lung cancer (50–54). Murphy et al (55) showed that activation of the
p53-mediated apoptotic intrinsic pathway requires high levels of
c-myc. Thus, the knockdown of STEAP1 in LNCaP cells increased the
p-c-myc/total c-myc ratio, which may activate mechanisms of
surveillance, such as p53 induction. These changes ultimately
culminate in apoptosis, as a way to eliminate cancer cells. On the
other hand, the increased levels of c-myc may also be a strategy of
cancer cells to overcome the inhibitory effect triggered by STEAP1
knockdown. This possibility cannot be ignored, and more studies
should be addressed in the future to clarify the relationship
between STEAP1 and c-myc.
It is well documented that the treatment of PCa with
anti-androgens, such as bicalutamide, enzalutamide and apalutamide,
result in blockage of PCa cell growth due to antagonistic effects
on AR transactivation (30,56).
At present, it is being evaluated the use of anti-androgens in
combination with other therapeutic targets. To date, no studies
have focused on effect of anti-androgens on expression of STEAP1 or
the effect of combined action between anti-androgens and STEAP1
inhibition in PCa treatment. Therefore, the present study intended
to determine the effect of anti-androgens in STEAP1 expression, as
well as to evaluate the hypothesis that silencing STEAP1 may
improve the effectiveness of anti-androgen therapy.
The present study observed that AR inhibition in
LNCaP-WT cells affected STEAP1 expression (Fig. 1). Overall, bicalutamide decreased
STEAP1 expression, but enzalutamide and apalutamide increased the
levels of STEAP1. Using microarray analysis, Carter et al
(57) also showed that
STEAP1 is downregulated in LNCaP cells treated with
bicalutamide. In contrast to the findings of the present study,
Doran et al (25) showed
that treatment of CWR22 PCa cells with enzalutamide and apalutamide
represses STEAP1 mRNA and protein expression. Beyond the slightly
different concentrations used in that Doran study and the present
study, the effect may differ between cell lines. Although the
precise explanation for different effects between bicalutamide and
enzalutamide/apalutamide in STEAP1 expression is unclear, the
differences observed might be due to different affinities of these
drugs to AR. In fact, it the conformational dynamics of AR with
bicalutamide, enzalutamide and apalutamide was evaluated and it was
shown that enzalutamide and apalutamide induce different
conformational changes in AR compared with bicalutamide (58). In addition, point mutations in AR,
namely F877L and T878A, are associated with resistance to
enzalutamide and apalutamide, but not to bicalutamide (59–61).
Considering that T878A mutation in AR is found in LNCaP cells, one
could hypothesize that upregulation of STEAP1 in response to
enzalutamide and apalutamide may occur as a mechanism of resistance
(62). However, further studies
should be performed to clarify the role of STEAP1 in resistance to
these drugs.
As expected, anti-androgen drugs efficiently reduced
the expression of the KLK3 gene, which is an AR target gene.
It suggested that the effect of anti-androgen on STEAP1 expression
was dependent on other factors besides the expression of AR.
Nevertheless, it should be highlighted that in LNCaP cells, the
knockdown of STEAP1 inhibited the expression of the KLK3
gene. This result is in accordance with the results previously
described by our research group, demonstrating that the
proliferative effect of DHT is abrogated in LNCaP cells knocked
down for STEAP1 (17). Moreover,
these findings are also supported by Ihlaseh-Catalano et al
(26), who describe a positive
strong trend between STEAP1 and PSA levels. The present study
explored the cellular pathways of proliferation and apoptosis of
anti-androgen treatment in LNCaP cells. Data obtained from LNCaP-WT
cells treated with bicalutamide, enzalutamide and apalutamide
showed a reduction on cell viability, determined by MTT assay, and
cell proliferation, as indicated by the estimated cell
proliferation index assessed by the Ki-67 fluorescent
immunocytochemistry (Fig. 2).
Anti-androgen effects modulating PCa cells behavior has been
underpinned by alterations on key protein targets associated with
cell proliferation, survival and oncogenic pathways, namely the AKT
and ERK signaling pathway (41,42,63).
The results of the present study showed a significant decreased of
p-AKT/AKT ratio in response to bicalutamide, enzalutamide and
apalutamide treatment in LNCaP-WT cells (Fig. 3), but no significant differences in
p-ERK/ERK ratio were observed. Altogether, these results suggested
that treatment of LNCaP cells with anti-androgen drugs inhibited
the signaling pathways associated with cell proliferation and
survival in an independent manner of their effect in expression of
STEAP1, at least in early treatment phase.
Considering the coordinated action of c-myc and AR
in PCa development (64,65), the effect of anti-androgens was
evaluated. The results showed an increased expression of p-c-myc
with bicalutamide, enzalutamide and apalutamide exposure of
LNCaP-WT cells (Fig. 3). These
findings are in line with a recent study showing that androgen
deprivation in vitro and castration in vivo leads to
rapid and persistent increases in c-myc expression (66). This observation suggests that
decreased AR activity can be compensated by increased levels of
c-myc, contributing to progression to castrate-resistant PCa
following ADT.
Anti-androgen treatment in LNCaP-WT cells was also
characterized by the increased expression and activity of several
apoptosis regulators (Fig. 4).
These results are in agreement with other studies describing the
apoptotic effect of anti-androgens in PCa cells (67–69).
Furthermore, a few reports have shown that administration of
anti-androgens in combination with other anti-cancer drugs trigger
cytotoxic effects in PCa (70–72).
The co-administration of enzalutamide and abiraterone (an inhibitor
of the steroidal enzyme CYP17A1) inhibits the proliferation and
promotes the apoptosis of LNCaP cells (70) and enzalutamide combined with
AS602801 (an inhibitor of c-Jun N-terminal kinase) synergistically
kills PCa cells, decreasing their migration and invasion capacity
(71). In addition, apalutamide,
in combination with autophagy inhibitors, provides a significantly
elevated anti-tumor effect in LNCaP cells (72). However, the present study is the
first, to the best of the authors' knowledge, to evaluate the
combined effect of STEAP1 knockdown with anti-androgen therapy on
PCa cells. LNCaP-STEAP1 knockdown cells treated with bicalutamide,
enzalutamide and apalutamide exhibited a decrease on cell viability
and proliferation, but no significant differences were observed in
comparison with the effect of these anti-androgens in LNCaP-WT
cells (Fig. 2). Similar data was
observed regarding the effect of bicalutamide and enzalutamide in
p-AKT/AKT and p-ERK/ERK ratios (Fig.
3). These observations are in accordance with increased
expression and activity of regulators/effectors of apoptosis in
response to anti-androgen treatment and no significant differences
between LNCaP-WT and LNCaP-STEAP1 knockdown cells were detected
(Fig. 4). Thus, the results
suggested that there is no synergistic effect between the silencing
of STEAP1 and anti-androgens treatment, at least in LNCaP cells
treated for 24 h with anti-androgen drugs.
Unexpectedly, an additive effect of c-myc expression
levels in LNCaP cells knocked-down for STEAP1 and treated with
bicalutamide, enzalutamide and apalutamide was observed (Fig. 3). To the best of the authors'
knowledge, there are no studies corroborating these discoveries and
further studies are needed to improve understanding of the role of
c-myc in response to combined action between anti-androgen and
STEAP1 knockdown in PCa cells.
In conclusion, the present findings showed that
anti-androgen drugs affected the regulation of STEAP1 expression,
but inhibition of STEAP1 did not alter the response of LNCaP cells
to anti-androgen treatment. Although the levels of STEAP1 in LNCaP
cells did not seem to change the effect of anti-androgens in cell
proliferation and apoptosis, the synergic effect in p-c-myc levels
deserves attention in future studies. Despite the limitations
concerning the use of only one PCa cell line and the unique
concentration and time of exposure tested for the anti-androgen
drugs, the present study strengthened the potential use of STEAP1
knockdown in PCa therapy, as well as opening new avenues of
research aimed at exploring the mechanisms underlying the role of
STEAP1 in human PCa. Further studies deepening the role of STEAP1
in response to anti-androgen drugs and investigating its actions in
the development of PCa resistance to treatments would be
fundamental for improved management of the disease.
Acknowledgments
Not applicable.
Funding
The authors acknowledge the Sandra M Rocha's individual PhD
Fellowship (grant nos. SFRH/BD/115693/2016 and
COVID/BD/151732/2021) from FCT-Fundação para a Ciência e
Tecnologia. The present study was funded by FEDER funds through the
POCI-COMPETE 2020-Operational Program Competitiveness and
Internationalization in Axis I-Strengthening research,
technological development and innovation (Project No. 029114) and
developed within the scope of the CICS-UBI projects UIDB/00709/2020
and UIDP/00709/2020, financed by national funds through the
Portuguese Foundation for Science and Technology/MCTES. This work
was also supported by the European Regional Development Fund
through the Programa Operacional Regional do Centro (Centro
2020)-Sistema de Apoio à Investigação Científica e
Tecnológica-Programas Integrados de IC&DT (Project
Centro-01-0145-FEDER-000019-C4-Centro de Competências em Cloud
Computing). The microscopy facility used in the development of this
work is part of the PPBI-Portuguese Platform of BioImaging and is
partially supported by the Project POCI-01-0145-FEDER-022122.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SMR and CJM conceived and designed the study and
wrote the manuscript. SMR, DN and AMC performed experiments. LP
analyzed and interpreted data. SS participated in the study design,
data analysis and revision of the manuscript. SMR and CJM confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rawla P: Epidemiology of prostate cancer.
World J Oncol. 10:63–89. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shafi AA, Yen AE and Weigel NL: Androgen
receptors in hormone-dependent and castration-resistant prostate
cancer. Pharmacol Ther. 140:223–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Crawford ED, Schellhammer PF, McLeod DG,
Moul JW, Higano CS, Shore N, Denis L, Iversen P, Eisenberger MA and
Labrie F: Androgen receptor targeted treatments of prostate cancer:
35 years of progress with antiandrogens. J Urol. 200:956–966. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murray TBJ: The pathogenesis of prostate
cancer. Prostate Cancer [Internet]. Bott SRJ and Ng KL: Exon
Publications; Brisbane, AU: pp. 29–42. 2021, View Article : Google Scholar
|
|
7
|
Teo MY, Rathkopf DE and Kantoff P:
Treatment of advanced prostate cancer. Annu Rev Med. 70:479–499.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hubert RS, Vivanco I, Chen E, Rastegar S,
Leong K, Mitchell SC, Madraswala R, Zhou Y, Kuo J, Raitano AB, et
al: STEAP: A prostate-specific cell-surface antigen highly
expressed in human prostate tumors. Proc Natl Acad Sci USA.
96:14523–14528. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rocha SM, Sousa I, Gomes IM, Arinto P,
Costa-Pinheiro P, Coutinho E, Santos CR, Jerónimo C, Lemos MC,
Passarinha LA, et al: Promoter demethylation upregulates STEAP1
gene expression in human prostate cancer: In vitro and in silico
analysis. Life (Basel). 11:12512021.PubMed/NCBI
|
|
10
|
Maitland NJ, Frame FM, Polson ES, Lewis JL
and Collins AT: Prostate cancer stem cells: Do they have a basal or
luminal phenotype? Horm Cancer. 2:47–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen WJ, Wu HT, Li CL, Lin YK, Fang ZX,
Lin WT and Liu J: Regulatory roles of six-transmembrane epithelial
antigen of the prostate family members in the occurrence and
development of malignant tumors. Front Cell Dev Biol. 9:7524262021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barroca-Ferreira J, Pais JP, Santos MM,
Goncalves AM, Gomes IM, Sousa I, Rocha SM, Passarinha LA and Maia
CJ: Targeting STEAP1 protein in human cancer: Current trends and
future challenges. Curr Cancer Drug Targets. 18:222–230. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rocha SM, Socorro S, Passarinha LA and
Maia CJ: Comprehensive landscape of STEAP family members expression
in human cancers: Unraveling the potential usefulness in clinical
practice using integrated bioinformatics analysis. Data. 7:642022.
View Article : Google Scholar
|
|
14
|
Challita-Eid PM, Morrison K, Etessami S,
An Z, Morrison KJ, Perez-Villar JJ, Raitano AB, Jia XC, Gudas JM,
Kanner SB and Jakobovits A: Monoclonal antibodies to
six-transmembrane epithelial antigen of the prostate-1 inhibit
intercellular communication in vitro and growth of human tumor
xenografts in vivo. Cancer Res. 67:5798–5805. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim K, Mitra S, Wu G, Berka V, Song J, Yu
Y, Poget S, Wang DN, Tsai AL and Zhou M: Six-transmembrane
epithelial antigen of prostate 1 (STEAP1) has a single b heme and
is capable of reducing metal ion complexes and oxygen.
Biochemistry. 55:6673–6684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grunewald TGP, Diebold I, Esposito I,
Plehm S, Hauer K, Thiel U, da Silva-Buttkus P, Neff F, Unland R,
Müller-Tidow C, et al: STEAP1 is associated with the invasive and
oxidative stress phenotype of Ewing tumors. Mol Cancer Res.
10:52–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gomes IM, Rocha SM, Gaspar C, Alvelos MI,
Santos CR, Socorro S and Maia CJ: Knockdown of STEAP1 inhibits cell
growth and induces apoptosis in LNCaP prostate cancer cells
counteracting the effect of androgens. Med Oncol. 35:402018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huo SF, Shang WL, Yu M, Ren XP, Wen HX,
Chai CY, Sun L, Hui K, Liu LH, Wei SH, et al: STEAP1 facilitates
metastasis and epithelial-mesenchymal transition of lung
adenocarcinoma via the JAK2/STAT3 signaling pathway. Biosci Rep.
40:BSR201931692020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiao Z, Huang L, Sun J, Xie J, Wang T, Yin
X, Zhang H and Chen J: Six-transmembrane epithelial antigen of the
prostate 1 expression promotes ovarian cancer metastasis by aiding
progression of epithelial-to-mesenchymal transition. Histochem Cell
Biol. 154:215–230. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Z, Hou WB, Zhang C, Tan YE, Zhang
DD, An W, Pan SW, Wu WD, Chen QC and Xu HM: A research of STEAP1
regulated gastric cancer cell proliferation, migration and invasion
in vitro and in vivos. J Cell Mol Med. 24:14217–14230. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iijima K, Nakamura H, Takada K, Hayasaka
N, Kubo T, Umeyama Y, Iyama S, Miyanishi K, Kobune M and Kato J:
Six-transmembrane epithelial antigen of the prostate 1 accelerates
cell proliferation by targeting c-Myc in liver cancer cells. Oncol
Lett. 22:5462021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marques RB, Dits NF, Erkens-Schulze S, van
Weerden WM and Jenster G: Bypass mechanisms of the androgen
receptor pathway in therapy-resistant prostate cancer cell models.
PLoS One. 5:e135002010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gomes IM, Santos CR, Socorro S and Maia
CJ: Six transmembrane epithelial antigen of the prostate 1 is
down-regulated by sex hormones in prostate cells. Prostate.
73:605–613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marques RB, Dits NF, Erkens-Schulze S, van
IJcken WFJ, van Weerden WM and Jenster G: Modulation of androgen
receptor signaling in hormonal therapy-resistant prostate cancer
cell lines. PLoS One. 6:e231442011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Doran MG, Watson PA, Cheal SM, Spratt DE,
Wongvipat J, Steckler JM, Carrasquillo JA, Evans MJ and Lewis JS:
Annotating STEAP1 regulation in prostate cancer with 89Zr
Immuno-PET. J Nucl Med. 55:2045–2049. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ihlaseh-Catalano SM, Drigo SA, de Jesus
CMN, Domingues MAC, Aparecida C, Filho JCS, de Camargo JLV and
Rogatto SR: STEAP1 protein overexpression is an independent marker
for biochemical recurrence in prostate carcinoma. Histopathology.
63:678–685. 2013.PubMed/NCBI
|
|
27
|
Pfaffl MW: Quantification strategies in
real-time PCR. A-Z of Quantitative PCR. Bustin SA: International
University Line (IUL); La Jolla: pp. 89–113. 2004
|
|
28
|
Neris RLS, Dobles AMC and Gomes AV:
Western blotting using in-gel protein labeling as a normalization
control: Advantages of stain-free technology. Methods Mol Biol.
2261:443–456. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Posch A, Kohn J, Oh K, Hammond M and Liu
N: V3 stain-free workflow for a practical, convenient, and reliable
total protein loading control in western blotting. J Vis Exp.
30:509482013.PubMed/NCBI
|
|
30
|
Nguyen PL, Alibhai SM, Basaria S, D'Amico
AV, Kantoff PW, Keating NL, Penson DF, Rosario DJ, Tombal B and
Smith MR: Adverse effects of androgen deprivation therapy and
strategies to mitigate them. Eur Urol. 67:825–836. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lanz C, Bennamoun M, Macek P, Cathelineau
X and Sanchez-Salas R: The importance of antiandrogen in prostate
cancer treatment. Ann Transl Med. 7:S362. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gillessen S, Attard G, Beer TM, Beltran H,
Bossi A, Bristow R, Carver B, Castellano D, Chung BH, Clarke N, et
al: Management of patients with advanced prostate cancer: The
report of the advanced prostate cancer consensus conference APCCC
2017. Eur Urol. 73:178–211. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Morgans AK and Beltran H: Isn't androgen
deprivation enough? Optimal treatment for newly diagnosed
metastatic prostate cancer. J Clin Oncol. 40:818–824. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gomes IM, Arinto P, Lopes C, Santos CR and
Maia CJ: STEAP1 is overexpressed in prostate cancer and prostatic
intraepithelial neoplasia lesions, and it is positively associated
with Gleason score. Urol Oncol Semin Orig Investig.
32:53.e23–53.e29. 2014.PubMed/NCBI
|
|
35
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamamoto T, Tamura Y, Kobayashi JI,
Kamiguchi K, Hirohashi Y, Miyazaki A, Torigoe T, Asanuma H,
Hiratsuka H and Sato N: Six-transmembrane epithelial antigen of the
prostate-1 plays a role for in vivo tumor growth via intercellular
communication. Exp Cell Res. 319:2617–2626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Green DR: Caspases and their substrates.
Cold Spring Harb Perspect Biol. 14:a0410122022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kyrylkova K, Kyryachenko S, Leid M and
Kioussi C: Detection of apoptosis by TUNEL assay. Methods Mol Biol.
887:41–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He G, Siddik ZH, Huang Z, Wang R, Koomen
J, Kobayashi R, Khokhar AR and Kuang J: Induction of p21 by p53
following DNA damage inhibits both Cdk4 and Cdk2 activities.
Oncogene. 24:2929–2943. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Demir Ö, Barros EP, Offutt TL, Rosenfeld M
and Amaro RE: An integrated view of p53 dynamics, function, and
reactivation. Curr Opin Struct Biol. 67:187–194. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao Z, Liao Q, Su M, Huang K, Jin J and
Cao D: AKT and ERK dual inhibitors: The way forward? Cancer Lett.
459:30–40. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gottlieb TM, Leal JFM, Seger R, Taya Y and
Oren M: Cross-talk between Akt, p53 and Mdm2: Possible implications
for the regulation of apoptosis. Oncogene. 21:1299–303. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Momand J, Wu HH and Dasgupta G:
MDM2-master regulator of the p53 tumor suppressor protein. Gene.
242:15–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ogawara Y, Kishishita S, Obata T, Isazawa
Y, Suzuki T, Tanaka K, Masuyama N and Gotoh Y: Akt enhances
Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem.
277:21843–21850. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kumar B, Koul S, Khandrika L, Meacham RB
and Koul HK: Oxidative stress is inherent in prostate cancer cells
and is required for aggressive phenotype. Cancer Res. 68:1777–1785.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hsieh AL, Walton ZE, Altman BJ, Stine ZE
and Dang CV: MYC and metabolism on the path to cancer. Semin Cell
Dev Biol. 43:11–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Labbé DP and Brown M: Transcriptional
regulation in prostate cancer. Cold Spring Harb Perspect Med.
8:a0304372018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Faskhoudi MA, Molaei P, Sadrkhanloo M,
Orouei S, Hashemi M, Bokaie S, Rashidi M, Entezari M, Zarrabi A,
Hushmandi K, et al: Molecular landscape of c-Myc signaling in
prostate cancer: A roadmap to clinical translation. Pathol Res
Pract. 233:1538512022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Uribesalgo I, Benitah SA and Croce LD:
From oncogene to tumor suppressor: The dual role of Myc in
leukemia. Cell Cycle. 11:1757–1764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
McMahon SB: MYC and the control of
apoptosis. Cold Spring Harb Perspect Med. 4:a0144072014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Adams CM and Eischen CM: Histone
deacetylase inhibition reveals a tumor-suppressive function of
MYC-regulated miRNA in breast and lung carcinoma. Cell Death
Differ. 23:1312–1321. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Muthalagu N, Junttila MR, Wiese KE, Wolf
E, Morton J, Bauer B, Evan GI, Eilers M and Murphy DJ: BIM is the
primary mediator of MYC-induced apoptosis in multiple solid
tissues. Cell Rep. 8:1347–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Prendergast GC: Mechanisms of apoptosis by
c-Myc. Oncogene. 18:2967–2987. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Murphy DJ, Junttila MR, Pouyet L, Karnezis
A, Shchors K, Bui DA, Brown-Swigart L, Johnson L and Evan GI:
Distinct thresholds govern Myc's biological output in vivo. Cancer
Cell. 14:447–457. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Student S, Hejmo T, Poterała-Hejmo A,
Leśniak A and Bułdak R: Anti-androgen hormonal therapy for cancer
and other diseases. Eur J Pharmacol. 866:1727832020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Carter SL, Centenera MM, Tilley WD, Selth
LA and Butler LM: IκBα mediates prostate cancer cell death induced
by combinatorial targeting of the androgen receptor. BMC Cancer.
16:1412016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gim HJ, Park J, Jung ME and Houk KN:
Conformational dynamics of androgen receptors bound to agonists and
antagonists. Sci Rep. 11:158872021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rathkopf DE, Smith MR, Ryan CJ, Berry WR,
Shore ND, Liu G, Higano CS, Alumkal JJ, Hauke R, Tutrone RF, et al:
Androgen receptor mutations in patients with castration-resistant
prostate cancer treated with apalutamide. Ann Oncol. 28:2264–2271.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Balbas MD, Evans MJ, Hosfield DJ,
Wongvipat J, Arora VK, Watson PA, Chen Y, Greene GL, Shen Y and
Sawyers CL: Overcoming mutation-based resistance to antiandrogens
with rational drug design. Elife. 2:e004992013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Joseph JD, Lu N, Qian J, Sensintaffar J,
Shao G, Brigham D, Moon M, Maneval EC, Chen I, Darimont B and Hager
JH: A clinically relevant androgen receptor mutation confers
resistance to second-generation antiandrogens enzalutamide and
ARN-509. Cancer Discov. 3:1020–1029. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun C, Shi Y, Xu LL, Nageswararao C, Davis
LD, Segawa T, Dobi A, McLeod DG and Srivastava S: Androgen receptor
mutation (T877A) promotes prostate cancer cell growth and cell
survival. Oncogene. 25:3905–3913. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shorning BY, Dass MS, Smalley MJ and
Pearson HB: The PI3K-AKT-mTOR pathway and prostate cancer: At the
crossroads of AR, MAPK, and WNT signaling. Int J Mol Sci.
21:45072020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Qiu X, Boufaied N, Hallal T, Feit A, de
Polo A, Luoma AM, Alahmadi W, Larocque J, Zadra G, Xie Y, et al:
MYC drives aggressive prostate cancer by disrupting transcriptional
pause release at androgen receptor targets. Nat Commun.
13:25592022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Barfeld SJ, Urbanucci A, Itkonen HM, Fazli
L, Hicks JL, Thiede B, Rennie PS, Yegnasubramanian S, DeMarzo AM
and Mills IG: c-myc antagonises the transcriptional activity of the
androgen receptor in prostate cancer affecting key gene networks.
EBioMedicine. 18:83–93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Guo H, Wu Y, Nouri M, Spisak S, Russo JW,
Sowalsky AG, Pomerantz MM, Wei Z, Korthauer K, Seo JH, et al:
Androgen receptor and MYC equilibration centralizes on
developmental super-enhancer. Nat Commun. 12:73082021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lee ECY, Zhan P, Schallhom R, Packman K
and Tenniswood M: Antiandrogen-induced cell death in LNCaP human
prostate cancer cells. Cell Death Differ. 10:761–771. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Guerrero J, Alfaro IE, Gómez F, Protter AA
and Bernales S: Enzalutamide, an androgen receptor signaling
inhibitor, induces tumor regression in a mouse model of
castration-resistant prostate cancer. Prostate. 73:1291–1305. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Koukourakis MI, Kakouratos C, Kalamida D,
Mitrakas A, Pouliliou S, Xanthopoulou E, Papadopoulou E, Fasoulaki
V and Giatromanolaki A: Comparison of the effect of the
antiandrogen apalutamide (ARN-509) versus bicalutamide on the
androgen receptor pathway in prostate cancer cell lines. Anticancer
Drugs. 29:323–333. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Han J, Zhang J, Zhang W, Zhang D, Li Y,
Zhang J, Zhang Y, Diao T, Cui L, Li W, et al: Abiraterone and
MDV3100 inhibits the proliferation and promotes the apoptosis of
prostate cancer cells through mitophagy. Cancer Cell Int.
19:3322019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li Z, Sun C, Tao S, Osunkoya AO, Arnold
RS, Petros JA, Zu X and Moreno CS: The JNK inhibitor AS602801
synergizes with enzalutamide to kill prostate cancer cells in vitro
and in vivo and inhibit androgen receptor expression. Transl Oncol.
13:1007512020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Eberli D, Kranzbühler B, Mortezavi A,
Sulser T and Salemi S: Apalutamide in combination with autophagy
inhibitors improves treatment effects in prostate cancer cells.
Urol Oncol. 38:683.e19–683.e26. 2020. View Article : Google Scholar : PubMed/NCBI
|