Introduction
Rheumatoid arthritis (RA) is a long-term, complex
inflammatory relapsing autoimmune disorder in which the immune
system mistakenly attacks the joints. RA has a prevalence of ~1% of
the global population (1). RA is
characterized by inflammation of the synovial joints, which
eventually leads to cartilage damage and bone destruction (2). The pathogenesis of RA is not fully
known due to the lack of knowledge regarding the etiology of this
disease (3). The preliminary event
in RA pathogenesis is believed to be the presence of immune
complexes in the bloodstream, which is considered the pre-articular
phase, during which the generation of autoantibodies against host
tissue occurs (4). Notably,
certain serological markers can be detected to diagnose disease
initiation during this phase, such as citrulline antibodies
(5). Subsequently, the transition
phase occurs, during which a number of autoantibodies are produced
and autoantigens are present in the articular joints. Autoantigens
bind to the Fc receptor γ of IgG antibodies to activate the innate
immune reaction via sentinel cells (6). Usually, dendritic cells (DCs) are
activated as the first line of defense, which bind to autoantigens
to induce the proliferation and differentiation of antigen-specific
T cells (7,8). Activated DCs increase major
histocompatibility complex II co-stimulatory surface molecules
CD80/86 to activate the production of cytokines from naive T cells
(9). Activated T-helper (Th) cells
can activate B cells to produce autoantibodies, such as rheumatoid
factor and anti-citrullinated protein antibodies, through plasma
cells and can be carried over through different pathways for
hyperplastic synovium, cartilage degradation and bone destruction
(10–12). Certain molecules, such as microRNA
(miR)-24, miR-125A-5p and miR-146a, have been identified as
biomarkers for RA that could increase diagnostic accuracy (6,13).
Unfortunately, there is currently no effective treatment that can
be used to cure RA in the clinic. Medications that are currently
used for the treatment of RA also induce prominent side effects
alongside their clinical efficacy (14). For example, gastrointestinal
adverse reactions such as abdominal pain, nausea, vomiting and
diarrhea are common. Furthermore, drug resistance is another
serious problem affecting RA treatment (15). Therefore, it is necessary to
develop novel drugs or therapeutic strategies for RA.
Ubiquitin D (UBD), also known as FAT10, is a
ubiquitin-like protein modifier that is mainly expressed in the
tissues and organs of the immune system, including the thymus and
lymph nodes (16). UBD expression
has been shown to be positively regulated by interferon-γ and TNF-α
(17,18). UBD may serve a significant role in
immunomodulation, including antigen presentation, immune response
and antiviral infection (19).
Emerging evidence has confirmed that UBD is involved in a number of
regulatory functions, including the cell cycle, apoptosis,
autophagy, DNA repair and tumorigenesis (20). High UBD expression has been
identified in a variety of tumor tissues, such as liver cancer
(21), colorectal cancer (22) and breast cancer (23). Growing evidence has indicated that
UBD has a pro-malignant role, due to its overexpression in a broad
spectrum of tumor tissues. Notably, forced UBD expression has been
reported to be associated with epirubicin resistance and the poor
prognosis of triple-negative breast cancer (23). Furthermore, patients with
UBD-positive colon cancer have a significantly higher recurrence
rate and poorer disease-free survival than those with low UBD
expression after radical surgery (24).
Notably, the role of UBD in RA remains to be
elucidated. Therefore, the present study aimed to investigate the
expression of UBD in RA samples from the Gene Expression Omnibus
(GEO) database, and determine the enriched Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathways related to the aberrantly
expressed UBD. Furthermore, the present study aimed to explore the
effects of UBD on the proliferation, apoptosis and inflammatory
cytokine production of RA-fibroblast-like synoviocytes (FLS), which
have been revealed to play a pathogenic role in RA (25,26),
as well as to confirm the underlying mechanism of UBD in RA.
Materials and methods
Identification of DEGs from GEO
datasets of RA
To identify differentially expressed genes (DEGs) in
RA, the GEO was used to assess RA data. The GSE55457 gene
expression profiles were downloaded from the GEO database
[https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55457;
GPL96 platform, Affymetrix Human Genome U133A Array] (27). The GSE55457 dataset contains data
from 79 samples, including 20 healthy control individuals, 33
patients with RA and 26 patients with osteoarthritis. The LIMMA
Bioconductor package (http://www.bioconductor.org/) was used to identify
DEGs by comparing the expression values between RA and normal
tissue samples. A classical unpaired Student's t-test was used to
identify DEGs that were statistically significant (P <0 .05 and
|log2FC|≥2). Subsequently, to present significant DEGs, volcano
plots were plotted using R software (version 3.4.0; http://www.r-project.org/).
KEGG pathway and gene ontology (GO)
analysis of DEGs
In the present study, the clusterProfiler
Bioconductor package (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
was used to perform the KEGG pathway analysis of the DEGs. The GO
enrichment analysis (https://david.ncifcrf.gov/) was conducted via R
software using the package ‘GO plot’ to explore the functions of
the DEGs. P<0.05 was considered to indicate a statistically
significant difference.
Cell culture
Normal human FLSs (cat. no. 408K-05a) were obtained
from Cell Applications, Inc. and the MH7A human RA-FLS cell line
(cat. no. C0878) was purchased from Shanghai Guandao Biological
Engineering Co., Ltd. Normal FLSs and RA-FLSs were incubated in
DMEM (cat. no. 12430054) supplemented with 10% fetal bovine serum
(FBS; cat. no. 10100147) and 1% penicillin/streptomycin (cat. no.
15070063) (all from Gibco; Thermo Fisher Scientific, Inc.). Normal
FLSs and RA-FLSs were grown in 75-cm2 flasks at 37°C in
an incubator containing 5% CO2. MAPK inhibitor SB202190
(10 µM; cat. no. HY-10295; MedChemExpress) was used to treat
RA-FLSs for 1 h at 37°C based on previous studies (28,29).
Vector construction and lentiviral
infection
The UBD overexpression lentiviral vector (GV492) was
purchased from Shanghai GeneChem Co., Ltd., and constructed based
on the full-length coding protein sequences of human UBD (GenBank
accession number NC_000006.12). A 3rd generation vector system was
used. Lentiviral vector (5 µg) was transfected into 293T cells
(cat. No. CRL-3216; American Type Culture Collection) cultured on
six-well plates with Lipofectamine® 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.). A total of 30 µg of plasmids were
used for lentivirus packaging, and the ratio of lentiviral plasmid:
GV492: Lipofectamine® 3000 was 2:1:1. The UBD
overexpression lentivirus (Lv-UBD) and blank GV492 plasmid vector
(NC) lentivirus was obtained after 293T cells were cultured at 37°C
for 4 days. RA-FLSs were then infected with the lentiviral vectors
at an MOI of 20 for 6 h followed by replacement with fresh medium.
RA-FLSs were grown for 48 h and subsequently treated with puromycin
(PURO; 1 µg/ml; InvivoGen) for 72 h to select transfected clones.
The infected cells were then collected 96 h after infection to
determine the infection efficiency.
Small interfering RNA (siRNA)
transfection
UBD siRNA and control siRNA (a non-targeting
siRNA-scrambled sequence) were constructed by Guangzhou Anernor
Biotechnology Co., Ltd. The siRNA sequences were as follows: UBD
siRNA 1#, 5′-ACCCATATGACAGCGTGAAAA-3′; UBD siRNA 2#,
5′-CCCATATGACAGCGTGAAAAA-3′; UBD siRNA 3#,
5′-CAGCGTGAAAAAAATCAAAGA-3′ and control siRNA,
5′-UUCUCCGAACGUGUCACGUTT-3′. SiRNAs (10 µg) were transfected into
5×106 RA-FLSs using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) for 24 h at 37°C according to the
manufacturer's protocols. The Mrna and protein expression levels of
UBD were measured at 48 h post-transfection by reverse
transcription-quantitative PCR (RT-qPCR) and western blotting,
respectively.
RT-qPCR
RA-FLSs were collected, centrifuged at 1,000 × g for
5 min at 4°C, the supernatant was removed and 1 ml SuPerfecTRI™
Total RNA Isolation Reagent (cat. no. 3101-100; Shanghai Pufei
Biotechnology Co., Ltd.) was added to the cell pellet to extract
the total RNA. The concentration and quality of RNA were determined
using an ND-2000 Spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Inc.). The extracted RNA was reverse transcribed into
cDNA using the Promega M-MLV kit (cat. No. M1705; Promega
Corporation). The samples were incubated for 60 min at 37°C. After
heat inactivation of reverse transcriptase (95°C, 2 min), the
first-strand cDNA was stored until use at −20°C. Subsequently, qPCR
was performed with the KAPA SYBR FAST qPCR kit (Kapa Biosystems;
Roche Diagnostics) using a SimpliAmp™ PCR System (Thermo Fisher
Scientific, Inc.). The relative Mrna expression levels of each
sample were calculated using the 2−ΔΔCq method (30). The primer sequences were as
follows: UBD, forward 5′-ATGCTTCCTGCCTCTGTGTG-3′, reverse
5′-TGCCCTTTCTGATGCCGTAA-3′; and GAPDH, forward
5′-CTGACTTCAACAGCGACACC-3′ and reverse
5′-GTGGTCCAGGGGTCTTACTC-3′.
Western blotting
Proteins were extracted from RA-FLSs using ice-cold
RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of
Biotechnology) and protein concentration was assessed using the BCA
method (cat. no. 23225; Thermo Fisher Scientific, Inc.). Proteins
(5 µg) were mixed with 2X SDS sample buffer, separated by SDS-PAGE
on 12% gels and transferred onto PVDF membranes, which were
incubated with 5% non-fat milk for 2 h at room temperature. The
membranes were then incubated overnight at 4°C with mouse anti-UBD
(1:1,000; cat. no. ab168680; Abcam), rabbit anti-phosphorylated
(p)-p38 (1:1,000; cat. no. ab178867; Abcam), rabbit anti-p38
(1:1,000; cat. no. ab170099; Abcam) and mouse anti-GAPDH (1:5,000;
cat. no. 60004-1-Ig; ProteinTech Group, Inc.). Horseradish
peroxidase-conjugated goat anti-mouse IgG (1:5,000; cat. no.
BA1051; Boster Biological Technology) and horseradish
peroxidase-conjugated mouse anti-rabbit IgG (1:5,000; cat. no.
BM2006; Boster Biological Technology) were used as secondary
antibodies to incubate the membranes for 1.5 h at room temperature.
Immunoreactive protein bands were detected using the ECL
hypersensitive chemiluminescence kit (cat. no. P0018M; Beyotime
Institute of Biotechnology) with the Odyssey Scanning System
(version 3.0; LI-COR Biosciences). ImageJ (version 1.8.0; National
Institutes of Health) was used for semi-quantification.
Cell counting kit 8 (CCK-8) assay
RA-FLSs were seeded at a density of 2,000 cells/well
in 96-well plates and were cultured in an incubator at 37°C with 5%
CO2 for 5 consecutive days. Subsequently, 10 µl CCK-8
solution (cat. no. 96992; MilliporeSigma) was added to each well
and incubated for 3 h at 37°C. The optical density (OD) value at
450 nm was measured using a Spectrafluor microreader plate
(Molecular Devices, LLC). These experiments were repeated three
times.
ELISA
RA-FLSs were cultured in DMEM containing 10% FBS for
24 h. ELISA was performed in accordance with the instructions of
the ELISA kits. In brief, the supernatant was collected after
centrifugation at 1,500 g for 20 min at 4°C to detect the levels of
IL-2, IL-6, IL-10 and TNF-α. IL-2 (cat. no. EH2IL22), IL-6 (cat.
no. EH2IL6), IL-10 (cat. no. EHIL10) and TNF-α (cat. no. BMS223HS)
ELISA kits were purchased from Thermo Fisher Scientific, Inc. The
calibration curves were plotted and the OD values of samples were
calculated from the standard curve for three assays.
EdU incorporation assay
RA-FLS proliferation was evaluated by assessing DNA
synthesis using an EdU incorporation assay (Click-iT™ EdU Cell
Proliferation Kit for Imaging, Alexa Fluor™ 488 dye; cat. no.
C10337; Invitrogen; Thermo Fisher Scientific, Inc.). UBD siRNA- and
control siRNA-treated RA-FLSs were incubated with 10 nM EdU for 6 h
at 37°C. Subsequently, RA-FLSs were harvested and fixed with
fixation buffer for 15 min at room temperature. After washing twice
with 2 ml permeabilization/washing buffer, the cells were incubated
with Click-iT EdU reaction cocktail for 30 min at room temperature.
After washing, the EdU-positive RA-FLSs were detected using a flow
cytometer (BD LSR II; BD Biosciences) and the data acquired with BD
FACSDiva 8.0.1 software (BD Biosciences).
Apoptosis analysis
The apoptotic RA-FLSs were measured using Annexin V
and PI staining (cat. no. V13242; Thermo Fisher Scientific).
RA-FLSs were washed twice with ice-cold PBS and centrifuged at 300
× g for 5 min at 4°C. Subsequently, RA-FLSs were resuspended in 195
µl Annexin V-FITC/PI binding buffer. Annexin V-FITC (5 µl) and PI
(10 µl) were supplemented following incubation in the dark for 30
min at 4°C. Apoptosis was analyzed using a flow cytometer (BD LSR
II). A total of 10,000 events were collected per sample, and data
were acquired and processed using CXP analysis software (version
2.0; Beckman Coulter, Inc.). Total apoptosis was considered the sum
of early- and late-stage apoptosis.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (8.0; GraphPad Software, Inc.). Each experiment was
repeated three times. Data are presented as the mean ± standard
deviation. Unpaired Student's t-test was used for two-group
comparisons and one-way ANOVA followed by Tukey's post hoc test was
used for multiple comparisons. P<0.05 was considered to indicate
a statistically significant difference.
Results
UBD is significantly increased in
RA
In the present study, microarray data for RA were
retrieved from the GEO. The DEGs between patients with RA and
healthy controls were identified from the GSE55457 dataset. A
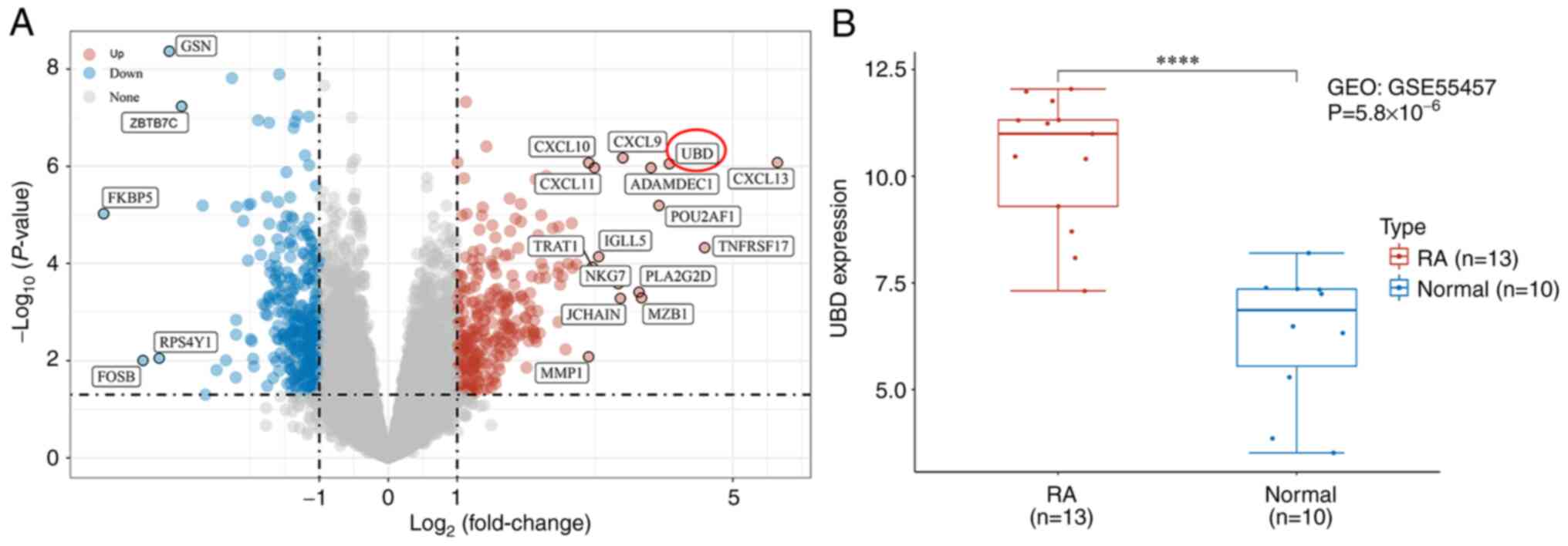
heatmap of the DEGs is shown in Fig.
1A. The results demonstrated that GSN, ZBTB7C, FKBP5, RPS4Y1
and FOSB were significantly decreased, whereas CXCL13, CXCL9,
CXCL10 and UBD were significantly increased in patients with RA.
UBD mRNA level was further confirmed to be markedly upregulated in
patients with RA compared with in the healthy controls (Fig. 1B).
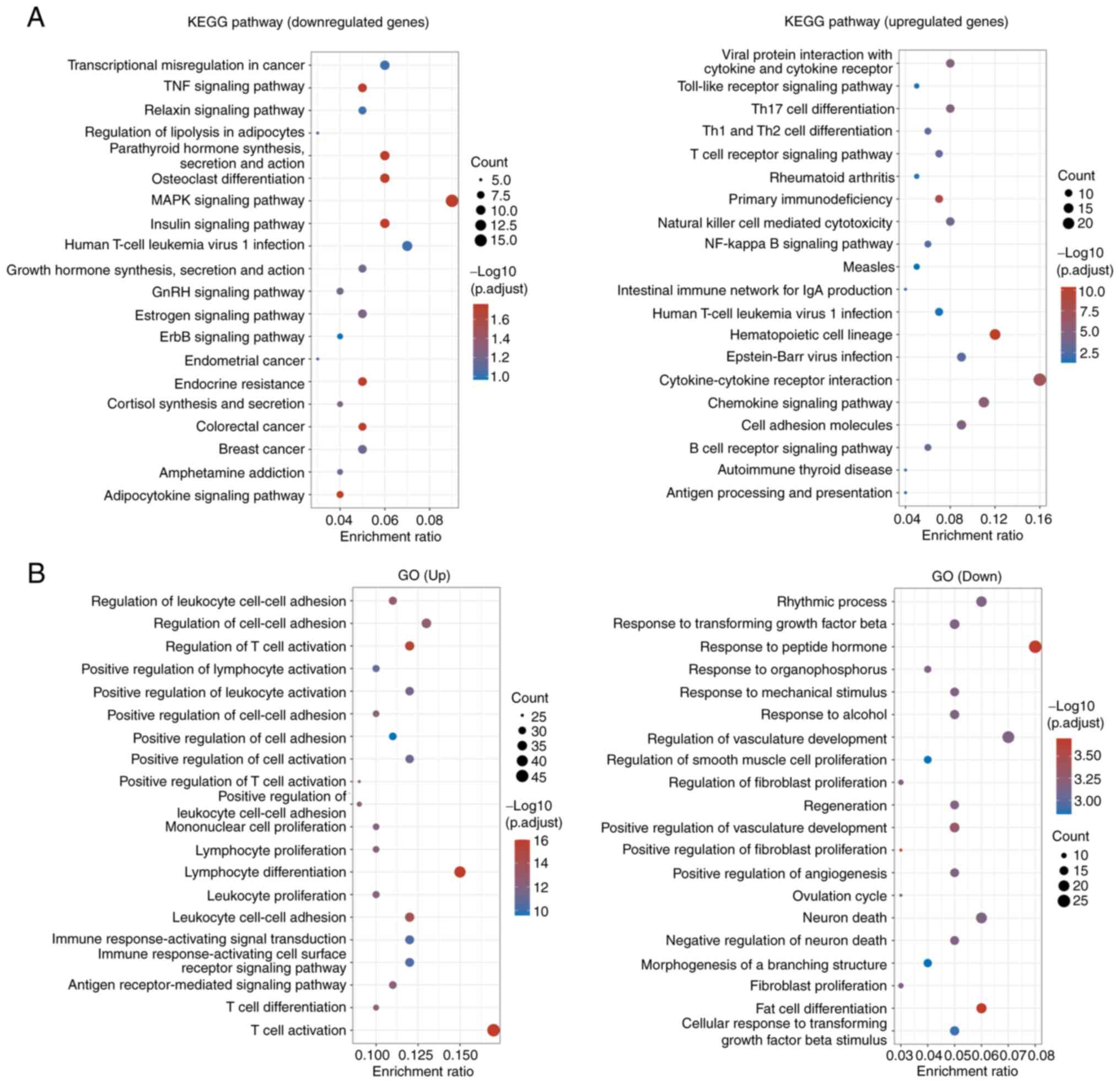
The DEGs were then subjected to KEGG pathway
enrichment and GO analysis. The results revealed that the
downregulated DEGs were enriched in pathways including
‘transcriptional misregulation in cancer’, ‘TNF signaling pathway’,
‘relaxin signaling pathway’, ‘regulation of lipolysis in
adipocytes’, ‘osteoclast differentiation’ and ‘MAPK signaling
pathway’, whereas the upregulated DEGs were enriched in pathways
including ‘viral protein interaction with cytokine and cytokine
receptor’, ‘Toll-like receptor signaling pathway’ and ‘Th17 cell
differentiation’ (Fig. 2A). The
aforementioned results demonstrated that UBD was overexpressed in
RA tissues and might be associated with MAPK pathway. GO enrichment
analysis revealed that the upregulated genes were enriched in terms
including ‘regulation of leukocyte cell-cell adhesion’, ‘regulation
of cell-cell adhesion’ and ‘regulation of T cell activation’,
whereas the downregulated genes were enriched in terms including
‘rhythmic process’, ‘response to transforming growth factor beta’
and ‘response to peptide hormone’ (Fig. 2B).
Elevated UBD increases the expression
of p-p38
The mRNA expression levels of UBD in normal FLSs and
RA-FLSs were measured by qPCR. UBD expression levels were
significantly increased in RA-FLSs compared with those in normal
FLSs (Fig. 3A). RA-FLSs were
subsequently transduced with NC lentivirus or UBD-expressing
lentivirus for 96 h, and the overexpression of UBD was confirmed by
qPCR and western blot. A significant increase was identified in the
mRNA and protein expression levels of UBD in the Lv-UBD group
compared with those in the uninfected cells or NC group (Fig. 3B and C). By contrast, the mRNA and
protein expression levels of UBD were significantly decreased in
the specific siRNA-transfected RA-FLSs compared with in those
transfected with the scramble siRNA (Fig. 3D and E). Furthermore, UBD
overexpression significantly promoted the protein expression levels
of p-p38 (Fig. 3F), whereas
siRNA-mediated UBD silencing significantly downregulated p-p38
expression (Fig. 3G). These
results suggested that upregulated UBD may activate the p38 MAPK
pathway in the progression of RA.
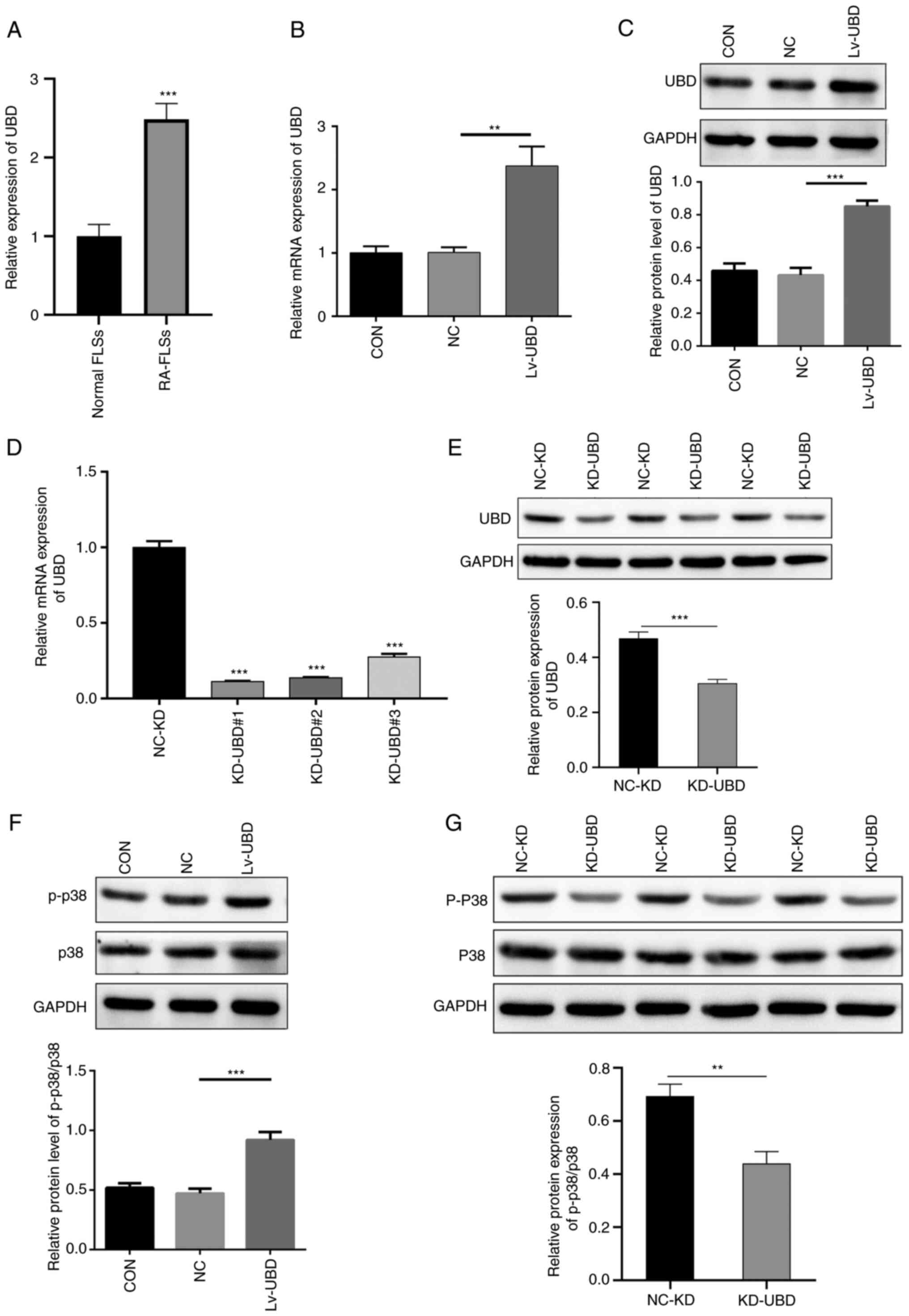 | Figure 3.UBD activates the p38 MAPK pathway.
RA-FLSs were transduced with a UBD overexpression lentiviral vector
or were transfected with UBD siRNAs. (A) mRNA expression levels of
UBD in normal-FLSs and RA-FLSs was detected using qPCR. (B) mRNA
and (C) protein expression levels of UBD in RA-FLSs transduced with
a UBD overexpression lentiviral vector were examined by qPCR and
western blotting, respectively. (D) mRNA and (E) protein expression
levels of UBD in RA-FLSs transfected with UBD siRNAs were examined
by qPCR and western blotting, respectively. Protein expression
levels of p-p38 and p38 in RA-FLSs with UBC (F) overexpression and
(G) KD, as determined by western blotting. Quantification of gene
and protein expression was normalized to GAPDH. Data are presented
as the mean ± SD (n=3). **P<0.01, ***P<0.001, as indicated or
vs. Normal FLSs or NC-KD. FLSs, fibroblast-like synoviocytes; KD,
knockdown; NC, negative control; p-, phosphorylated; qPCR,
quantitative PCR; RA, rheumatoid arthritis; siRNA, small
interfering RNA; UBD, ubiquitin D. |
UBD regulates RA-FLS cellular
processes via the p38 MAPK pathway
To further assess the relationship between UBD and
MAPK pathways, the p38 MAPK inhibitor SB202190 was used to treat
RA-FLSs. Overexpression of UBD significantly increased RA-FLSs
viability, whereas SB202190 administration suppressed the promoting
effect of UBD on RA-FLSs activity (Fig. 4A). In addition, UBD overexpression
resulted in a significant increase in the secretion of IL-2, IL-6,
IL-10 and TNF-α, whereas the administration of SB202190 blocked the
elevated secretion of IL-2, IL-6, IL-10 and TNF-α caused by UBD
overexpression (Fig. 4B). In
addition, overexpression of UBD enhanced the proliferation of
RA-FLSs, whereas SB202190 treatment suppressed the promoting effect
of UBD on RA-FLSs proliferation, as confirmed by flow cytometry
(Fig. 4C). By contrast, UBD
silencing significantly suppressed RA-FLSs proliferation (Fig. 4D). Notably, cell apoptosis was
significantly suppressed by UBD overexpression, whereas SB202190
administration alleviated the inhibitory effect of UBD
overexpression on cell apoptosis (Fig.
4E). By contrast, UBD silencing significantly promoted RA-FLSs
apoptosis (Fig. 4F). These results
revealed that UBD increased the viability, the release of
proinflammatory factors and the proliferation of RA-FLSs, whereas
treatment with the p38 MAPK inhibitor SB202190 exerted the opposite
effect on RA-FLSs, thus indicating that UBD regulated the cellular
processes of RA-FLSs via the p38 MAPK pathway.
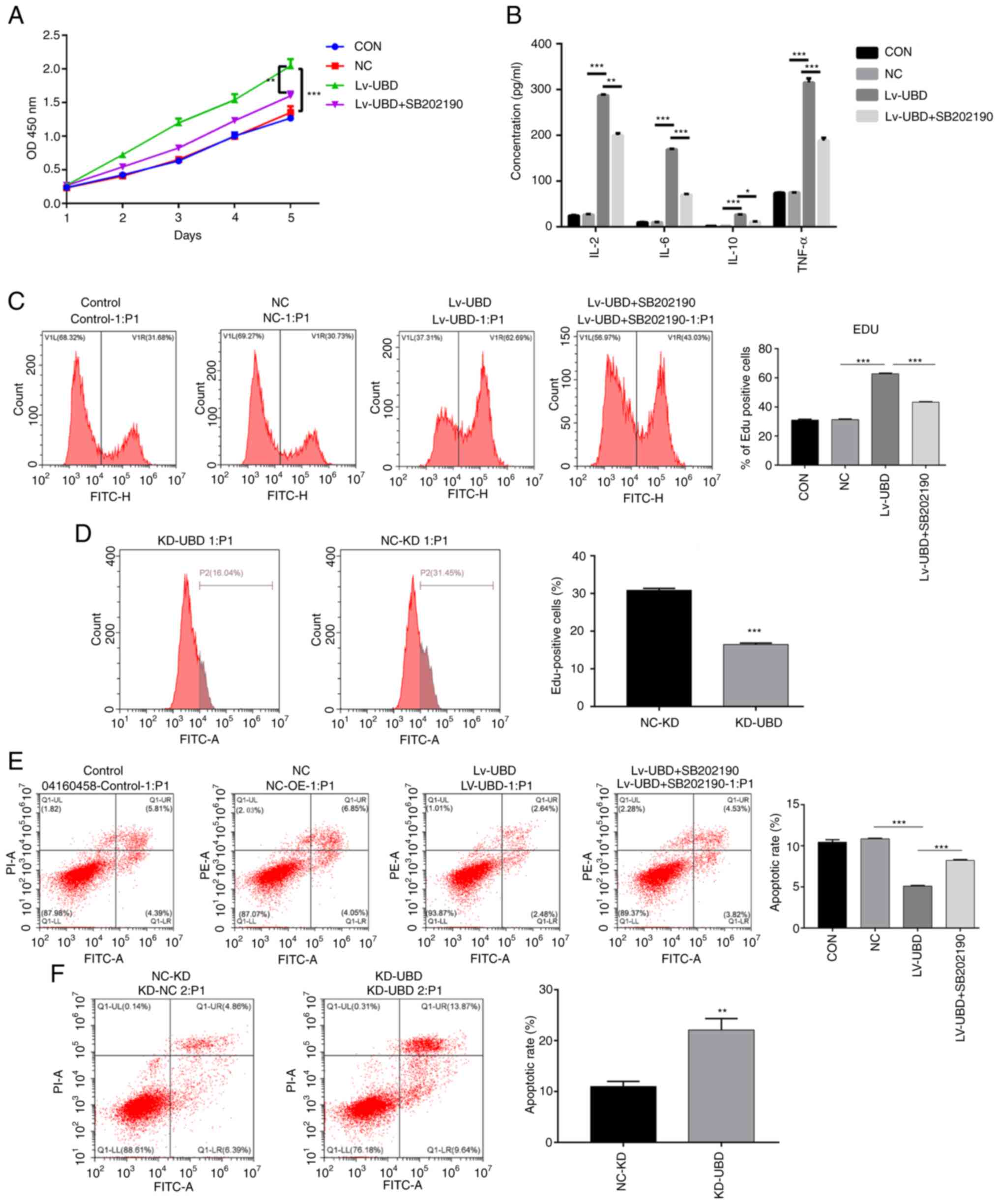 | Figure 4.UBD regulates the biological
processes of RA-FLSs by targeting the p38 MAPK pathway. RA-FLSs
transduced with a UBD overexpression lentiviral vector or UBD siRNA
were treated with or without the p38 MAPK inhibitor SB202190. (A)
Viability of RA-FLSs was detected by Cell Counting Kit-8. (B)
Levels of IL-2, IL-6, IL-10 and TNF-α were examined by ELISA. (C
and D) Proliferation of RA-FLSs was evaluated by assessing DNA
synthesis using the EdU incorporation assay. (E and F) Apoptosis of
RA-FLSs was detected by flow cytometric analysis. Data are
presented as the mean ± SD (n=3). *P<0.05, **P<0.01,
***P<0.001, as indicated or vs. NC-KD. FLSs, fibroblast-like
synoviocytes; KD, knockdown; NC, negative control; OD, optical
density; RA, rheumatoid arthritis; siRNA, small interfering RNA;
UBD, ubiquitin D. |
Discussion
Bioinformatics is an interdisciplinary field
combining molecular biology and information technology, which is
widely used to explore and reveal the molecular mechanism of
diseases (31). In the present
study, data from patients with RA, which is a type of inflammatory
arthritis of unknown etiology, were obtained from the GEO
(GSE55457) and were analyzed using bioinformatics tools. A series
of DEGs were identified between RA and healthy control samples,
among which chemokines related to Th1 cells (CXCL9, CXCL10), and T
follicular helper and B cells (CXCL13), were significantly
increased in RA tissues, indicating the important roles of these
chemokines in the progression of RA, which is in accordance with
previous reports (32–34). However, the role of another
significantly upregulated gene, UBD, which may have a role in the
development of RA remains to be elucidated.
UBD, an 18 kDa protein comprising 165 amino-acid
residues, is an immune system protein that is strongly induced by
proinflammatory cytokines (17).
UBD is the only modifier of all ubiquitin-like modifiers that acts
as an autonomous transferable signal for degradation by the 26S
proteasome, which can occur independently of ubiquitin (35). While UBD is primarily stimulated by
proinflammatory cytokines within the tumor microenvironment, a
growing number of studies has confirmed that the pro-malignant
ability of UBD itself largely underlies its upregulation in tumor
tissues (16,36). Upregulation of UBD has been
confirmed in various types of cancer where it promotes cell
migration, invasion and metastasis formation (37–39).
In addition, the expression of UBD has been reported to be markedly
increased during the maturation of DCs and epithelial cells within
the medulla of the thymus where it regulates T-cell selection
(40). In the present study, UBD
was confirmed as one of the most upregulated genes in RA samples
from a GEO dataset, indicating that UBD may have an important role
in the progression of RA. However, the expression of UBD in RA
clinical samples was not pursued further due to the lack of
clinical samples.
UBD has been reported to accelerate cell viability
and proliferation, and to suppress cell apoptosis (39,41–43).
In the present study, the biological function of UBD in RA was
further investigated. UBD overexpression significantly increased
the viability and proliferation of RA-FLSs, and inhibited their
apoptosis. A causal link between inflammation and the development
of RA is generally accepted (44).
IL-2, IL-6, IL-10 and TNF-α serve important roles in RA
pathogenesis, participating in Th1-mediated processes, and causing
cartilage and bone destruction (45–48).
The present study confirmed that overexpression of UBD
significantly induced the secretion of IL-2, IL-6, IL-10 and TNF-α
in RA-FLSs. These results suggested that UBD may be related to the
progression of RA by regulating proliferation, apoptosis and the
secretion of inflammatory factors. Autoantibodies serve essential
biological roles in the progression of RA (49). However, the present study mainly
demonstrated the function of UBD in RA-FLSs at the cellular level,
which is often poorly reflective of the in vivo situation;
thus, autoantibody testing was not performed in the current
study.
UBD modulates various signaling pathways involved in
tumor development, such as NF-κB and Wnt/β-catenin signaling
pathways (16,50). Notably, UBD accelerates the
progression of oral squamous cell carcinoma via NF-κB signaling
(50). Elevated UBD expression has
been shown to drive the invasion and metastasis of hepatocellular
carcinoma cells by binding to β-catenin to prevent its
ubiquitylation and degradation (51). In addition, UBD can directly bind
to target genes, including MAD and p53, to promote cell
proliferation, metastasis and migration via their regulation
(38,39). In the present study, KEGG pathway
analysis identified that the DEGs in RA samples were enriched in
pathways including ‘TNF signaling pathway’, ‘relaxin signaling
pathway’, ‘osteoclast differentiation’ and ‘MAPK signaling
pathway’. Furthermore, UBD overexpression significantly promoted
the protein expression levels of p-p38, whereas silencing of UBD
markedly inhibited p-p38 expression, indicating that UBD may
activate the p38 MAPK pathway in the progression of RA. To the best
of our knowledge, the relationship between UBD and p38 MAPK has not
been previously reported. To further investigate the targeting
regulatory relationship between UBD and p38 MAPK, the p38 MAPK
inhibitor SB202190 was used to treat RA-FLSs. Notably, the
application of SB202190 partially relieved the UBD-dependent
enhancing effects on cell viability and proliferation, as well as
the inhibitory effect on cell apoptosis. In addition, treatment
with SB202190 significantly blocked the enhancing effects of UBD
overexpression on the secretion of inflammatory factors. Taken
together, these results suggested that UBD may be a crucial
pathogenic factor for RA by activating the p38 MAPK pathway, which
provides additional opportunities for the intervention of RA.
In conclusion, the expression levels of UBD were
significantly increased in RA. Notably, the results revealed the
important role of UBD in RA, and also identified the novel
mechanism that UBD may regulate the biological and inflammatory
processes in RA by targeting p38 MAPK. Collectively, the present
study provided novel insights into the pathogenesis of RA and the
potential of UBD as a therapeutic target against RA.
Acknowledgements
Not applicable.
Funding
This work was supported by the Scientific Research Project of
Guangxi Zhuang Autonomous Region Administration of Traditional
Chinese Medicine (grant no. gzzc2019145), and the Scientific
Research Project of Guangxi Zhuang Autonomous Region Health
Committee (grant no. z20200163).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HC and HW designed the study. HC and LT performed
all of the experiments, interpreted the data and prepared the
manuscript. JL and CP analyzed the data. All authors read and
approved the final manuscript. HC and HW confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Approved by the Medical Ethics committee of the
Affiliated Hospital of Youjiang Medical University for
Nationalities, Baise, Guangxi, approval number YYFY-LL-2022-96.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
O'Neil LJ, Barrera-Vargas A,
Sandoval-Heglund D, Merayo-Chalico J, Aguirre-Aguilar E, Aponte AM,
Ruiz-Perdomo Y, Gucek M, El-Gabalawy H, Fox DA, et al:
Neutrophil-mediated carbamylation promotes articular damage in
rheumatoid arthritis. Sci Adv. 6:eabd26882020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith MH and Berman JR: What is rheumatoid
arthritis? JAMA. 327:11942022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee SH, Kwon JY, Kim SY, Jung K and Cho
ML: Interferon-gamma regulates inflammatory cell death by targeting
necroptosis in experimental autoimmune arthritis. Sci Rep.
7:101332017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shah N, Jadidi S and Bajaj P: Correction:
A review of non-surgical pain management in osteoarthritis. Cureus.
13:c452021.PubMed/NCBI
|
|
5
|
Mekic M and Hadzigrahic E: Anti-cyclic
citrullinated peptide antibody as a predictor of rheumathoid
arthritis complications. Med Arch. 74:183–186. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou Y, Abraham S, Andre P, Edelstein LC,
Shaw CA, Dangelmaier CA, Tsygankov AY, Kunapuli SP, Bray PF and
McKenzie SE: Anti-miR-148a regulates platelet FcγRIIA signaling and
decreases thrombosis in vivo in mice. Blood. 126:2871–2881. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Patin EC, Thompson A and Orr SJ: Pattern
recognition receptors in fungal immunity. Semin Cell Dev Biol.
89:24–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmid E, Bhandaru M, Nurbaeva MK, Yang W,
Szteyn K, Russo A, Leibrock C, Tyan L, Pearce D, Shumilina E and
Lang F: SGK3 regulates Ca(2+) entry and migration of dendritic
cells. Cell Physiol Biochem. 30:1423–1435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeon JH, Lee BC, Kim D, Cho D and Kim TS:
Hydrophilic astragalin galactoside induces T helper type 1-mediated
immune responses via dendritic cells. Int J Mol Sci. 19:31202018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuwabara T, Ishikawa F, Kondo M and
Kakiuchi T: The role of IL-17 and related cytokines in inflammatory
autoimmune diseases. Mediators Inflamm. 2017:39080612017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu S, Wang Y, Zhang J, Han B, Wang B, Gao
W, Zhang N, Zhang C, Yan F and Li Z: Efficacy and safety of
rituximab for systemic lupus erythematosus treatment: A
meta-analysis. Afr Health Sci. 20:871–884. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang F, Shi WX, Chen J, He K and Fang W:
Clinical therapeutic effects of combined diacerein and glucosamine
in the treatment of osteoarthritis: A protocol for systematic
review and meta-analysis. Medicine (Baltimore). 100:e275832021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Murata K, Furu M, Yoshitomi H, Ishikawa M,
Shibuya H, Hashimoto M, Imura Y, Fujii T, Ito H, Mimori T and
Matsuda S: Comprehensive microRNA analysis identifies miR-24 and
miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS
One. 8:e691182013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Donlin LT: Inching closer to precision
treatment for rheumatoid arthritis. Nat Med. 28:1129–1131. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lourenzi FM, Jones A, Pereira DF, Santos
JHCAD, Furtado RNV and Natour J: Effectiveness of an overall
progressive resistance strength program for improving the
functional capacity of patients with rheumatoid arthritis: A
randomized controlled trial. Clin Rehabil. 31:1482–1491. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aichem A and Groettrup M: The
ubiquitin-like modifier FAT10 in cancer development. Int J Biochem
Cell Biol. 79:451–461. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Raasi S, Schmidtke G, de Giuli R and
Groettrup M: A ubiquitin-like protein which is synergistically
inducible by interferon-gamma and tumor necrosis factor-alpha. Eur
J Immunol. 29:4030–4036. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu YC, Pan J, Zhang C, Fan W, Collinge M,
Bender JR and Weissman SM: A MHC-encoded ubiquitin-like protein
(FAT10) binds noncovalently to the spindle assembly checkpoint
protein MAD2. Proc Natl Acad Sci USA. 96:4313–4318. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Battaglia A, Fossati M, Buzzonetti A,
Scambia G and Fattorossi A: A robust immune system conditions the
response to abagovomab (anti-idiotypic monoclonal antibody
mimicking the CA125 protein) vaccination in ovarian cancer
patients. Immunol Lett. 191:35–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tanyi JL and George E: Personalized
vaccination against ovarian cancer: What are the possibilities?
Expert Rev Vaccines. 17:955–958. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Paijens ST, Leffers N, Daemen T, Helfrich
W, Boezen HM, Cohlen BJ, Melief CJ, de Bruyn M and Nijman HW:
Antigen-specific active immunotherapy for ovarian cancer. Cochrane
Database Syst Rev. 9:CD0072872018.PubMed/NCBI
|
|
22
|
Kalli KR, Block MS, Kasi PM, Erskine CL,
Hobday TJ, Dietz A, Padley D, Gustafson MP, Shreeder B,
Puglisi-Knutson D, et al: Folate receptor alpha peptide vaccine
generates immunity in breast and ovarian cancer patients. Clin
Cancer Res. 24:3014–3025. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han T, Liu Z, Li H, Xie W, Zhang R, Zhu L,
Guo F, Han Y, Sheng Y and Xie X: High expression of UBD correlates
with epirubicin resistance and indicates poor prognosis in
triple-negative breast cancer. Onco Targets Ther. 8:1643–1649.
2015.PubMed/NCBI
|
|
24
|
Yan DW, Li DW, Yang YX, Xia J, Wang XL,
Zhou CZ, Fan JW, Wen YG, Sun HC, Wang Q, et al: Ubiquitin D is
correlated with colon cancer progression and predicts recurrence
for stage II–III disease after curative surgery. Br J Cancer.
103:961–969. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bottini N and Firestein GS: Duality of
fibroblast-like synoviocytes in RA: Passive responders and
imprinted aggressors. Nat Rev Rheumatol. 9:24–33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bartok B and Firestein GS: Fibroblast-like
synoviocytes: Key effector cells in rheumatoid arthritis. Immunol
Rev. 233:233–255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
You R, Liu S and Tan J: Screening and
identification of osteoarthritis related differential genes and
construction of a risk prognosis model based on bioinformatics
analysis. Ann Transl Med. 10:4442022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang WC, Liou CJ, Shen SC, Hu S, Hsiao CY
and Wu SJ: Luteolin attenuates IL-1β-induced THP-1 adhesion to
ARPE-19 cells via suppression of NF-κB and MAPK pathways. Mediators
Inflamm. 2020:94213402020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang L, Yang H, Chu Y, Song Y, Ding L, Zhu
B, Zhai W, Wang X, Kuang Y, Ren F, et al: CREPT is required for
murine stem cell maintenance during intestinal regeneration. Nat
Commun. 12:2702021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liang X, Zhu W, Lv Z and Zou Q: Molecular
computing and bioinformatics. Molecules. 24:23582019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pandya JM, Lundell AC, Andersson K,
Nordström I, Theander E and Rudin A: Blood chemokine profile in
untreated early rheumatoid arthritis: CXCL10 as a disease activity
marker. Arthritis Res Ther. 19:202017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsai CH, Chen CJ, Gong CL, Liu SC, Chen
PC, Huang CC, Hu SL, Wang SW and Tang CH: CXCL13/CXCR5 axis
facilitates endothelial progenitor cell homing and angiogenesis
during rheumatoid arthritis progression. Cell Death Dis.
12:8462021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhong H, Xu LL, Bai MX and Su Y: Effect of
chemokines CXCL9 and CXCL10 on bone erosion in patients with
rheumatoid arthritis. Beijing Da Xue Xue Bao Yi Xue Ban.
53:1026–1031. 2021.(In Chinese). PubMed/NCBI
|
|
35
|
Schmidtke G, Kalveram B and Groettrup M:
Degradation of FAT10 by the 26S proteasome is independent of
ubiquitylation but relies on NUB1L. FEBS Lett. 583:591–594. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiang S, Shao X, Cao J, Yang B, He Q and
Ying M: FAT10: Function and relationship with cancer. Curr Mol
Pharmacol. 13:182–191. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee CGL, Ren J, Cheong ISY, Ban KHK, Ooi
LLPJ, Yong Tan SY, Kan A, Nuchprayoon I, Jin R, Lee KH, et al:
Expression of the FAT10 gene is highly upregulated in
hepatocellular carcinoma and other gastrointestinal and
gynecological cancers. Oncogene. 22:2592–2603. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Theng SS, Wang W, Mah WC, Chan C, Zhuo J,
Gao Y, Qin H, Lim L, Chong SS, Song J and Lee CG: Disruption of
FAT10-MAD2 binding inhibits tumor progression. Proc Natl Acad Sci
USA. 111:E5282–E5291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su H, Qin M, Liu Q, Jin B, Shi X and Xiang
Z: Ubiquitin-like protein UBD promotes cell proliferation in
colorectal cancer by facilitating p53 degradation. Front Oncol.
11:6913472021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Buerger S, Herrmann VL, Mundt S, Trautwein
N, Groettrup M and Basler M: The ubiquitin-like modifier FAT10 is
selectively expressed in medullary thymic epithelial cells and
modifies T cell selection. J Immunol. 195:4106–4116. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xiao Y, Diao Q, Liang Y, Peng Y and Zeng
K: MicroRNA-24-1-5p promotes malignant melanoma cell autophagy and
apoptosis via regulating ubiquitin D. Mol Med Rep. 16:8448–8454.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Taniai E, Yafune A, Hayashi H, Itahashi M,
Hara-Kudo Y, Suzuki K, Mitsumori K and Shibutani M: Aberrant
activation of ubiquitin D at G2 phase and apoptosis by carcinogens
that evoke cell proliferation after 28-day administration in rats.
J Toxicol Sci. 37:1093–1111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Brozzi F, Gerlo S, Grieco FA, Juusola M,
Balhuizen A, Lievens S, Gysemans C, Bugliani M, Mathieu C,
Marchetti P, et al: Ubiquitin D regulates IRE1α/c-Jun N-terminal
kinase (JNK) protein-dependent apoptosis in pancreatic beta cells.
J Biol Chem. 291:12040–12056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang H, Tu S, Yang S, Shen P, Huang Y, Ba
X, Lin W, Huang Y, Wang Y, Qin K and Chen Z: Berberine modulates
LPA function to inhibit the proliferation and inflammation of
FLS-RA via p38/ERK MAPK pathway mediated by LPA1. Evid
Based Complement Alternat Med. 2019:25802072019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Graßhoff H, Comdühr S, Monne LR, Müller A,
Lamprecht P, Riemekasten G and Humrich JY: Low-dose IL-2 therapy in
autoimmune and rheumatic diseases. Front Immunol. 12:6484082021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lauper K, Ludici M, Mongin D, Bergstra SA,
Choquette D, Codreanu C, Cordtz R, De Cock D, Dreyer L, Elkayam O,
et al: Effectiveness of TNF-inhibitors, abatacept, IL6-inhibitors
and JAK-inhibitors in 31 846 patients with rheumatoid arthritis in
19 registers from the ‘JAK-pot’ collaboration. Ann Rheum Dis.
81:1358–1366. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang J, Liu J, Wen JT and Wang X:
Correlation between circRNA0003353 in peripheral blood mononuclear
cells and immune inflammation in rheumatoid arthritis patients with
damp heat obstruction syndrome. Sichuan Da Xue Xue Bao Yi Xue Ban.
53:437–443. 2022.(In Chinese). PubMed/NCBI
|
|
48
|
Alturaiki W, Alhamad A, Alturaiqy M, Mir
SA, Iqbal D, Bin Dukhyil AA, Alaidarous M, Alshehri B, Alsagaby SA,
Almalki SG, et al: Assessment of IL-1β, IL-6, TNF-α, IL-8, and CCL
5 levels in newly diagnosed Saudi patients with rheumatoid
arthritis. Int J Rheum Dis. 25:1013–1019. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
van Delft MAM and Huizinga TWJ: An
overview of autoantibodies in rheumatoid arthritis. J Autoimmun.
110:1023922020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Song A, Wang Y, Jiang F, Yan E, Zhou J, Ye
J, Zhang H, Ding X, Li G, Wu Y, et al: Ubiquitin D promotes
progression of oral squamous cell carcinoma via NF-kappa B
signaling. Mol Cells. 44:468–480. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yuan R, Wang K, Hu J, Yan C, Li M, Yu X,
Liu X, Lei J, Guo W, Wu L, et al: Ubiquitin-like protein FAT10
promotes the invasion and metastasis of hepatocellular carcinoma by
modifying β-catenin degradation. Cancer Res. 74:5287–5300. 2014.
View Article : Google Scholar : PubMed/NCBI
|