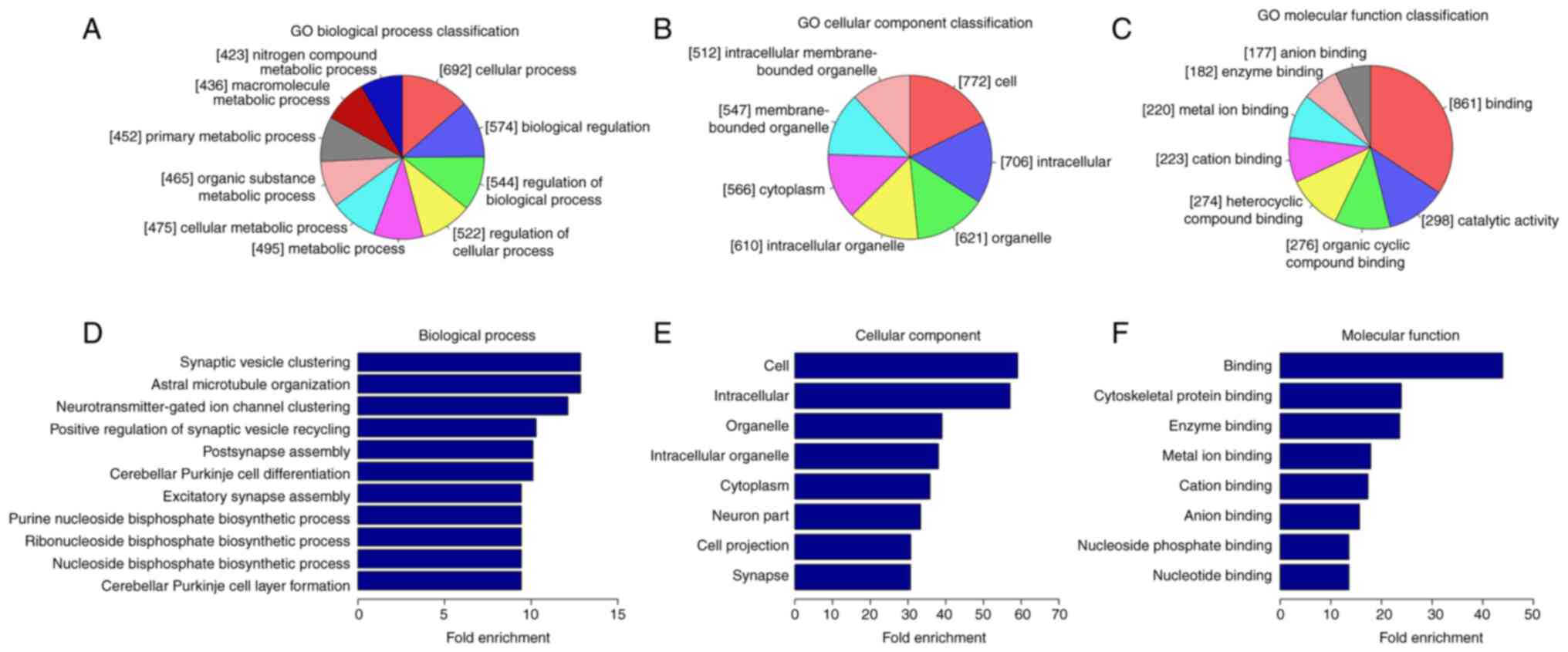
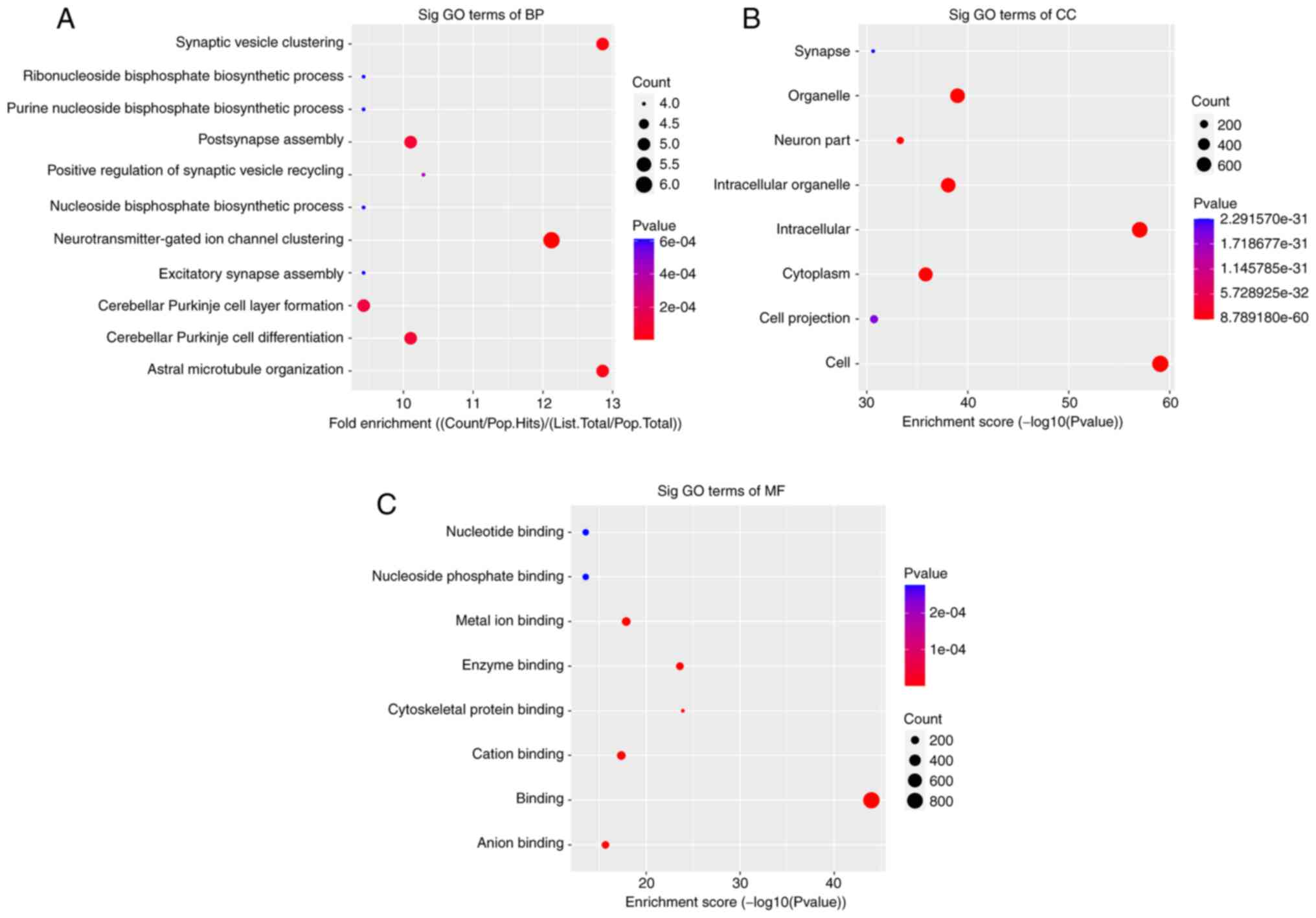
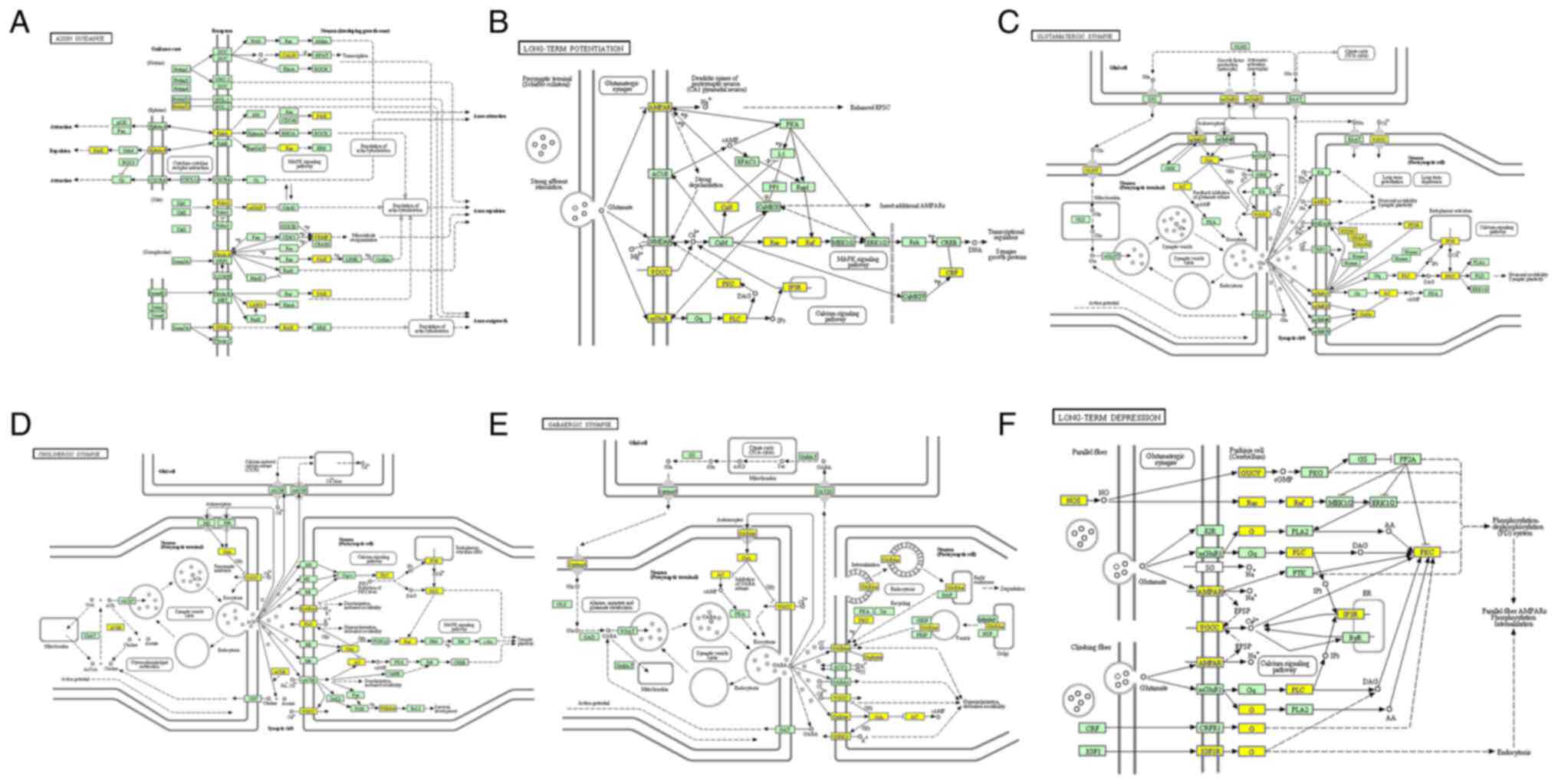
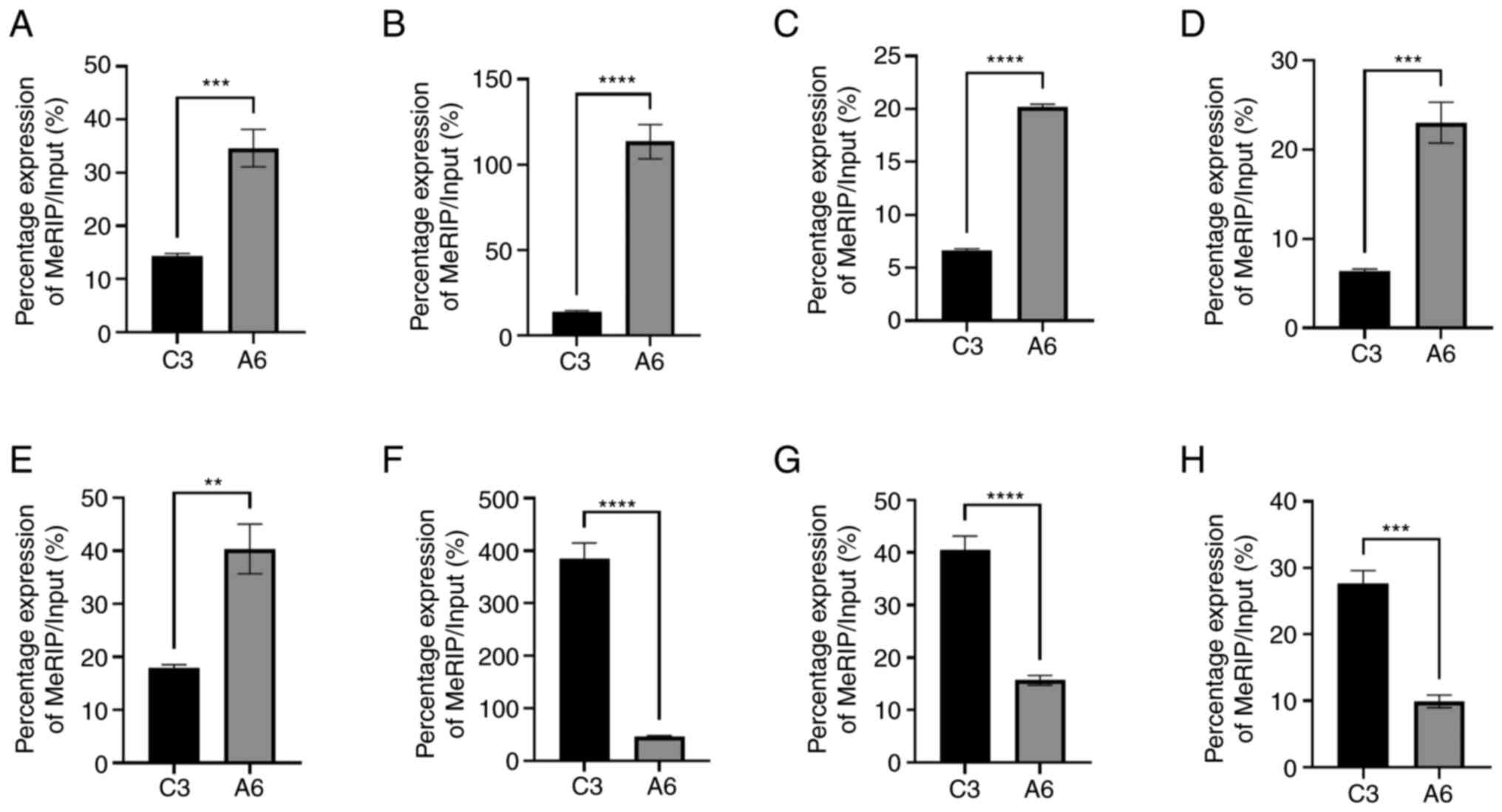
|
1
|
Lane CA, Hardy J and Schott JM:
Alzheimer's disease. Eur J Neurol. 25:59–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yiannopoulou KG and Papageorgiou SG:
Current and future treatments in Alzheimer disease: An update. J
Cent Nerv Syst Dis. 12:11795735209073972020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang JZ, Wang ZH and Tian Q: Tau
hyperphosphorylation induces apoptotic escape and triggers
neurodegeneration in Alzheimer's disease. Neurosci Bull.
30:359–366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kwok JB: Role of epigenetics in
Alzheimer's and Parkinson's disease. Epigenomics. 2:671–682. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma X, Liu L and Meng J: MicroRNA-125b
promotes neurons cell apoptosis and tau phosphorylation in
Alzheimer's disease. Neurosci Lett. 661:57–62. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patrick E, Rajagopal S, Wong HA, McCabe C,
Xu J, Tang A, Imboywa SH, Schneider JA, Pochet N, Krichevsky AM, et
al: Dissecting the role of non-coding RNAs in the accumulation of
amyloid and tau neuropathologies in Alzheimer's disease. Mol
Neurodegener. 12:512017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Irier HA and Jin P: Dynamics of DNA
methylation in aging and Alzheimer's disease. DNA Cell Biol. 31
(Suppl 1):S42–S48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Unnikrishnan A, Freeman WM, Jackson J,
Wren JD, Porter H and Richardson A: The role of DNA methylation in
epigenetics of aging. Pharmacol Ther. 195:172–185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Y, Chen YS, Sun BF and Yang YG: RNA
methylation: Regulations and mechanisms. Yi Chuan. 40:964–976.
2018.(In Chinese). PubMed/NCBI
|
|
10
|
Shafik AM, Zhang F, Guo Z, Dai Q, Pajdzik
K, Li Y, Kang Y, Yao B, Wu H, He C, et al: N6-methyladenosine
dynamics in neurodevelopment and aging, and its potential role in
Alzheimer's disease. Genome Biol. 22:172021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haussmann IU, Bodi Z, Sanchez-Moran E,
Mongan NP, Archer N, Fray RG and Soller M: m6A
potentiates Sxl alternative pre-mRNA splicing for robust Drosophila
sex determination. Nature. 540:301–304. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoon KJ, Ringeling FR, Vissers C, Jacob F,
Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al:
Temporal control of mammalian cortical neurogenesis by
m6A methylation. Cell. 171:877–889.e17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Widagdo J and Anggono V: The
m6A-epitranscriptomic signature in neurobiology: From
neurodevelopment to brain plasticity. J Neurochem. 147:137–152.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Flamand MN and Meyer KD: The
epitranscriptome and synaptic plasticity. Curr Opin Neurobiol.
59:41–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Du K, Wang J, Nie Y, Lee T and
Sun T: Unique and specific m6A RNA methylation in mouse
embryonic and postnatal cerebral cortices. Genes (Basel).
11:11392020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang H, Camats-Perna J, Medeiros R,
Anggono V and Widagdo J: Altered expression of the m6A
methyltransferase METTL3 in Alzheimer's disease. eNeuro.
7:ENEURO.0125–20.2020. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao F, Xu Y, Gao S, Qin L, Austria Q,
Siedlak SL, Pajdzik K, Dai Q, He C, Wang W, et al: METTL3-dependent
RNA m6A dysregulation contributes to neurodegeneration
in Alzheimer's disease through aberrant cell cycle events. Mol
Neurodegener. 16:702021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han M, Liu Z, Xu Y, Liu X, Wang D, Li F,
Wang Y and Bi J: Abnormality of m6A mRNA methylation is involved in
Alzheimer's disease. Front Neurosci. 14:982020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matsuzaki M, Honkura N, Ellis-Davies GC
and Kasai H: Structural basis of long-term potentiation in single
dendritic spines. Nature. 429:761–766. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Noureddini M and Bagheri-Mohammadi S:
Adult hippocampal neurogenesis and Alzheimer's disease: Novel
application of mesenchymal stem cells and their role in hippocampal
neurogenesis. Int J Mol Cell Med. 10:1–10. 2021.PubMed/NCBI
|
|
22
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang JL, Su M and Wu DP: Functional roles
of circular RNAs in Alzheimer's disease. Ageing Res Rev.
60:1010582020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang JL, Qin MC, Zhou Y, Xu ZH, Yang SM,
Zhang F, Zhong J, Liang MK, Chen B, Zhang WY, et al: Comprehensive
analysis of differentially expressed profiles of Alzheimer's
disease associated circular RNAs in an Alzheimer's disease mouse
model. Aging (Albany NY). 10:253–265. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Xu P, Chen B, Zhang Z, Zhang C,
Zhan Q, Huang S, Xia ZA and Peng W: Identifying
circRNA-associated-ceRNA networks in the hippocampus of
Aβ1-42-induced Alzheimer's disease-like rats using microarray
analysis. Aging (Albany NY). 10:775–788. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin
D, Liu J and Sun Z: The role of N6-methyladenosine
(m6A) modification in the regulation of circRNAs. Mol
Cancer. 19:1052020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li W and Jin G: The circRNA and role in
Alzheimer's disease: From regulation to therapeutic and diagnostic
targets. Hippocampus. 2022.PubMed/NCBI
|
|
28
|
Akhter R: Circular RNA and Alzheimer's
disease. Adv Exp Med Biol. 1087:239–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lok K, Zhao H, Shen H, Wang Z, Gao X, Zhao
W and Yin M: Characterization of the APP/PS1 mouse model of
Alzheimer's disease in senescence accelerated background. Neurosci
Lett. 557(Pt B): 84–89. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiong H, Callaghan D, Wodzinska J, Xu J,
Premyslova M, Liu QY, Connelly J and Zhang W: Biochemical and
behavioral characterization of the double transgenic mouse model
(APPswe/PS1dE9) of Alzheimer's disease. Neurosci Bull. 27:221–232.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Heneka MT, Kummer MP, Stutz A, Delekate A,
Schwartz S, Vieira-Saecker A, Griep A, Axt D, Remus A, Tzeng TC, et
al: NLRP3 is activated in Alzheimer's disease and contributes to
pathology in APP/PS1 mice. Nature. 493:674–678. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Izco M, Martínez P, Corrales A, Fandos N,
García S, Insua D, Montañes M, Pérez-Grijalba V, Rueda N, Vidal V,
et al: Changes in the brain and plasma Aβ peptide levels with age
and its relationship with cognitive impairment in the APPswe/PS1dE9
mouse model of Alzheimer's disease. Neuroscience. 263:269–279.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Denver P, English A and McClean PL:
Inflammation, insulin signaling and cognitive function in aged
APP/PS1 mice. Brain Behav Immun. 70:423–434. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Götz J, Bodea LG and Goedert M: Rodent
models for Alzheimer disease. Nat Rev Neurosci. 19:583–598. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Webster SJ, Bachstetter AD, Nelson PT,
Schmitt FA and Van Eldik LJ: Using mice to model Alzheimer's
dementia: An overview of the clinical disease and the preclinical
behavioral changes in 10 mouse models. Front Genet. 5:882014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Li Y, Yue M, Wang J, Kumar S,
Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G and Zhao
JC: N6-methyladenosine RNA modification regulates
embryonic neural stem cell self-renewal through histone
modifications. Nat Neurosci. 21:195–206. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu
Z, Li J, Hao P, Zhang Y, Zhang F, et al: m6A facilitates
hippocampus-dependent learning and memory through YTHDF1. Nature.
563:249–253. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chang M, Lv H, Zhang W, Ma C, He X, Zhao
S, Zhang ZW, Zeng YX, Song S, Niu Y and Tong WM: Region-specific
RNA m6A methylation represents a new layer of control in
the gene regulatory network in the mouse brain. Open Biol.
7:1701662017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He
X, Wu G, Zhao S, Zhang Y, Wang D, et al: RNA m6A
methylation participates in regulation of postnatal development of
the mouse cerebellum. Genome Biol. 19:682018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weng YL, Wang X, An R, Cassin J, Vissers
C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, et al: Epitranscriptomic
m6A regulation of axon regeneration in the adult
mammalian nervous system. Neuron. 97:313–325.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dube U, Del-Aguila JL, Li Z, Budde JP,
Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, et al:
An atlas of cortical circular RNA expression in Alzheimer disease
brains demonstrates clinical and pathological associations. Nat
Neurosci. 22:1903–1912. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li Y, Fan H, Sun J, Ni M, Zhang L, Chen C,
Hong X, Fang F, Zhang W and Ma P: Circular RNA expression profile
of Alzheimer's disease and its clinical significance as biomarkers
for the disease risk and progression. Int J Biochem Cell Biol.
123:1057472020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kashyap G, Bapat D, Das D, Gowaikar R,
Amritkar RE, Rangarajan G, Ravindranath V and Ambika G: Synapse
loss and progress of Alzheimer's disease-a network model. Sci Rep.
9:65552019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
John A and Reddy PH: Synaptic basis of
Alzheimer's disease: Focus on synaptic amyloid beta, P-tau and
mitochondria. Ageing Res Rev. 65:1012082021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chokkalla AK, Mehta SL and Vemuganti R:
Epitranscriptomic regulation by m6A RNA methylation in
brain development and diseases. J Cereb Blood Flow Metab.
40:2331–2349. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Madugalle SU, Meyer K, Wang DO and Bredy
TW: RNA N6-methyladenosine and the regulation of RNA
localization and function in the brain. Trends Neurosci.
43:1011–1023. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zimmerman AJ, Hafez AK, Amoah SK,
Rodriguez BA, Dell'Orco M, Lozano E, Hartley BJ, Alural B, Lalonde
J, Chander P, et al: A psychiatric disease-related circular RNA
controls synaptic gene expression and cognition. Mol Psychiatry.
25:2712–2727. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang X, Zhou X, Li G, Zhang Y, Wu Y and
Song W: Modifications and trafficking of APP in the pathogenesis of
Alzheimer's disease. Front Mol Neurosci. 10:2942017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Herskowitz JH, Feng Y, Mattheyses AL,
Hales CM, Higginbotham LA, Duong DM, Montine TJ, Troncoso JC,
Thambisetty M, Seyfried NT, et al: Pharmacologic inhibition of
ROCK2 suppresses amyloid-β production in an Alzheimer's disease
mouse model. J Neurosci. 33:19086–19098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang XW, Feng N, Liu YC, Guo Q, Wang JK,
Bai YZ, Ye XM, Yang Z, Yang H, Liu Y, et al: Neuroinflammation
inhibition by small-molecule targeting USP7 noncatalytic domain for
neurodegenerative disease therapy. Sci Adv. 8:eabo07892022.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jia L, Piña-Crespo J and Li Y: Restoring
Wnt/β-catenin signaling is a promising therapeutic strategy for
Alzheimer's disease. Mol Brain. 12:1042019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
McPhie DL, Coopersmith R, Hines-Peralta A,
Chen Y, Ivins KJ, Manly SP, Kozlowski MR, Neve KA and Neve RL: DNA
synthesis and neuronal apoptosis caused by familial Alzheimer
disease mutants of the amyloid precursor protein are mediated by
the p21 activated kinase PAK3. J Neurosci. 23:6914–6927. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Park D, Choi EK, Cho TH, Joo SS and Kim
YB: Human neural stem cells encoding ChAT gene restore cognitive
function via acetylcholine synthesis, Aβ elimination, and
neuroregeneration in APPswe/PS1dE9 mice. Int J Mol Sci.
21:39582020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang H, Zhang L, Zhou D, Li H and Xu Y:
ErbB4 mediates amyloid β-induced neurotoxicity through JNK/tau
pathway activation: Implications for Alzheimer's disease. J Comp
Neurol. 529:3497–3512. 2021. View Article : Google Scholar : PubMed/NCBI
|