|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leal YA, Torres J, Gamboa R,
Mantilla-Morales A, Piña-Sanchez P, Arrieta O, Bonifaz L, Meneses
A, Duque C and Piñeros M: Cancer incidence in Merida, Mexico
2015–2018: First report from the population-based cancer registry.
Arch Med Res. 53:859–866. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Early Breast Cancer Trialists'
Collaborative Group (EBCTCG), . Effects of chemotherapy and
hormonal therapy for early breast cancer on recurrence and 15-year
survival: An overview of the randomised trials. Lancet.
365:1687–1717. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nicolini A and Ferrari P: Targeted
therapies and drug resistance in advanced breast cancer,
alternative strategies and the way beyond. Cancers (Basel).
16:4662024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Unger-Saldaña K, Bandala-Jacques A,
Huerta-Gutierrez R, Zamora-Muñoz S, Hernández-Ávila JE,
Cabrera-Galeana P, Mohar A and Lajous M: Breast cancer survival in
Mexico between 2007 and 2016 in women without social security: A
retrospective cohort study. Lancet Reg Health Am.
23:1005412023.PubMed/NCBI
|
|
7
|
Grajales-Alvarez R, Gutiérrez-Mata A,
Pichardo-Piña C, Gutiérrez-De la Barrera M and Dip-Borunda K:
Survival outcomes of patients with breast cancer in a Mexican
population. JCO Glob Oncol. 10:e23002332024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calhoun BC and Collins LC: Predictive
markers in breast cancer: An update on ER and HER2 testing and
reporting. Semin Diagn Pathol. 32:362–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vieira AF and Schmitt F: An update on
breast cancer multigene prognostic tests-emergent clinical
biomarkers. Front Med (Lausanne). 5:2482018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sørlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karsli-Ceppioglu S, Dagdemir A, Judes G,
Lebert A, Penault-Llorca F, Bignon YJ and Bernard-Gallon D: The
epigenetic landscape of promoter genome-wide analysis in breast
cancer. Sci Rep. 7:65972017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Manjunath M and Choudhary B:
Triple-negative breast cancer: A run-through of features,
classification and current therapies (Review). Oncol Lett.
22:5122021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mohammed AA: The clinical behavior of
different molecular subtypes of breast cancer. Cancer Treat Res
Commun. 29:1004692021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lehmann BD, Jovanović B, Chen X, Estrada
MV, Johnson KN, Shyr Y, Moses HL, Sanders ME and Pietenpol JA:
Refinement of triple-negative breast cancer molecular subtypes:
Implications for neoadjuvant chemotherapy selection. PLoS One.
11:e01573682016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ahmad M, Weiswald LB, Poulain L, Denoyelle
C and Meryet-Figuiere M: Involvement of lncRNAs in cancer cells
migration, invasion and metastasis: Cytoskeleton and ECM crosstalk.
J Exp Clin Cancer Res. 42:1732023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sahu A, Singhal U and Chinnaiyan AM: Long
noncoding RNAs in cancer: From function to translation. Trends
Cancer. 1:93–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mattick JS, Amaral PP, Carninci P,
Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME,
Fitzgerald KA, et al: Long non-coding RNAs: Definitions, functions,
challenges and recommendations. Nat Rev Mol Cell Biol. 24:430–447.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Singh D, Assaraf YG and Gacche RN: Long
non-coding RNA mediated drug resistance in breast cancer. Drug
Resist Updat. 63:1008512022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taghvimi S, Abbaszadeh S, Banan FB, Fard
ES, Jamali Z, Najafabadi MA, Savardashtaki A and Movahedpour A:
LncRNAs roles in chemoresistance of cancer cells. Curr Mol Med.
22:691–702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tano K and Akimitsu N: Long non-coding
RNAs in cancer progression. Front Genet. 3:2192012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guzel E, Okyay TM, Yalcinkaya B,
Karacaoglu S, Gocmen M and Akcakuyu MH: Tumor suppressor and
oncogenic role of long non-coding RNAs in cancer. North Clin
Istanb. 22:81–86. 2019.
|
|
29
|
Kaushik AC, Mehmood A, Wang X, Wei DQ and
Dai X: Globally ncRNAs expression profiling of TNBC and screening
of functional lncRNA. Front Bioeng Biotechnol. 8:5231272021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xia M, Zu X, Chen Z, Wen G and Zhong J:
Noncoding RNAs in triple negative breast cancer: Mechanisms for
chemoresistance. Cancer Lett. 523:100–110. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lara-Medina F, Pérez-Sánchez V,
Saavedra-Pérez D, Blake-Cerda M, Arce C, Motola-Kuba D,
Villarreal-Garza C, González-Angulo AM, Bargalló E, Aguilar JL, et
al: Triple-negative breast cancer in Hispanic patients: High
prevalence, poor prognosis, and association with menopausal status,
body mass index, and parity. Cancer. 117:3658–3669. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Macari A, Soberanis-Pina P, Varela-Santoyo
E, Valle-Sanchez MA, Leal-Hidalgo JL, Torres-Guillen VM,
Motola-Kuba D, Ruiz-Morales JM and Dorantes-Heredia R: Prevalence
and molecular profile of breast carcinoma using
immunohistochemistry markers in Mexican women. World J Oncol.
12:119–123. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tuominen VJ, Ruotoistenmäki S, Viitanen A,
Jumppanen M and Isola J: ImmunoRatio: A publicly available web
application for quantitative image analysis of estrogen receptor
(ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res.
12:R562010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hammond MEH, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS,
Hayes M, et al: American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer (Unabridged Version). Arch Pathol Lab Med.
134:e48–e72. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Allison KH, Hammond MEH, Dowsett M,
McKernin SE, Carey LA, Fitzgibbons PL, Hayes DF, Lakhani SR,
Chavez-MacGregor M, Perlmutter J, et al: Estrogen and progesterone
receptor testing in breast cancer: ASCO/CAP guideline update. J
Clin Oncol. 38:1346–1366. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McCall MN, Bolstad BM and Irizarry RA:
Frozen robust multiarray analysis (fRMA). Biostatistics.
11:242–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Metsalu T and Vilo J: ClustVis: A web tool
for visualizing clustering of multivariate data using principal
component analysis and heatmap. Nucleic Acids Res. 43:W566–W570.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tiessen A, Cubedo-Ruiz EA and Winkler R:
Improved representation of biological information by using
correlation as distance function for heatmap cluster analysis. Am J
Plant Sci. 8:502–516. 2017. View Article : Google Scholar
|
|
39
|
Cedro-Tanda A, Ríos-Romero M,
Romero-Córdoba S, Cisneros-Villanueva M, Rebollar-Vega RG,
Alfaro-Ruiz LA, Jiménez-Morales S, Domínguez-Reyes C,
Villegas-Carlos F, Tenorio-Torres A, et al: A lncRNA landscape in
breast cancer reveals a potential role for AC009283.1 in
proliferation and apoptosis in HER2-enriched subtype. Sci Rep.
10:131462020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Han L, Roebuck P, Diao L, Liu L,
Yuan Y, Weinstein JN and Liang H: TANRIC: An interactive open
platform to explore the function of lncRNAs in cancer. Cancer Res.
75:3728–3737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meyer JS, Alvarez C, Milikowski C, Olson
N, Russo I, Russo J, Glass A, Zehnbauer BA, Lister K and Parwaresch
R; Cooperative Breast Cancer Tissue Resource, : Breast carcinoma
malignancy grading by Bloom-Richardson system vs proliferation
index: Reproducibility of grade and advantages of proliferation
index. Mod Pathol. 18:1067–1078. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bloom HJG and Richardson WW: Histological
grading and prognosis in breast cancer; a study of 1,409 cases of
which 359 have been followed for 15 years. Br J Cancer. 11:359–377.
1957. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Afifi N and Barrero CA: Understanding
breast cancer aggressiveness and its implications in diagnosis and
treatment. J Clin Med. 12:13752023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Holowatyj AN, Ruterbusch JJ, Ratnam M,
Gorski DH and Cote ML: HER2 status and disparities in luminal
breast cancers. Cancer Med. 5:2109–2116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lino MA, Palacios-Rodríguez Y,
Rodríguez-Cuevas S, Bautista-Piña V, Marchat LA, Ruíz-García E,
Astudillo-de la Vega H, González-Santiago AE, Flores-Pérez A,
Díaz-Chávez J, et al: Comparative proteomic profiling of
triple-negative breast cancer reveals that up-regulation of
RhoGDI-2 is associated to the inhibition of caspase 3 and caspase
9. J Proteomics. 111:198–211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hu X, Zhang Q, Xing W and Wang W: Role of
microRNA/lncRNA intertwined with the Wnt/β-Catenin axis in
regulating the pathogenesis of triple-negative breast cancer. Front
Pharmacol. 13:8149712022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang G, Liu P, Li J, Jin K, Zheng X and
Xie L: Novel prognosis and therapeutic response model of
immune-related lncRNA pairs in clear cell renal cell carcinoma.
Vaccines (Basel). 10:11612022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang W, Wu Y, Hou B, Wang Y, Deng D, Fu Z
and Xu Z: A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives
tumor growth and metastasis in hepatocellular carcinoma through the
Wnt/β-catenin pathway. Mol Oncol. 13:2194–2210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
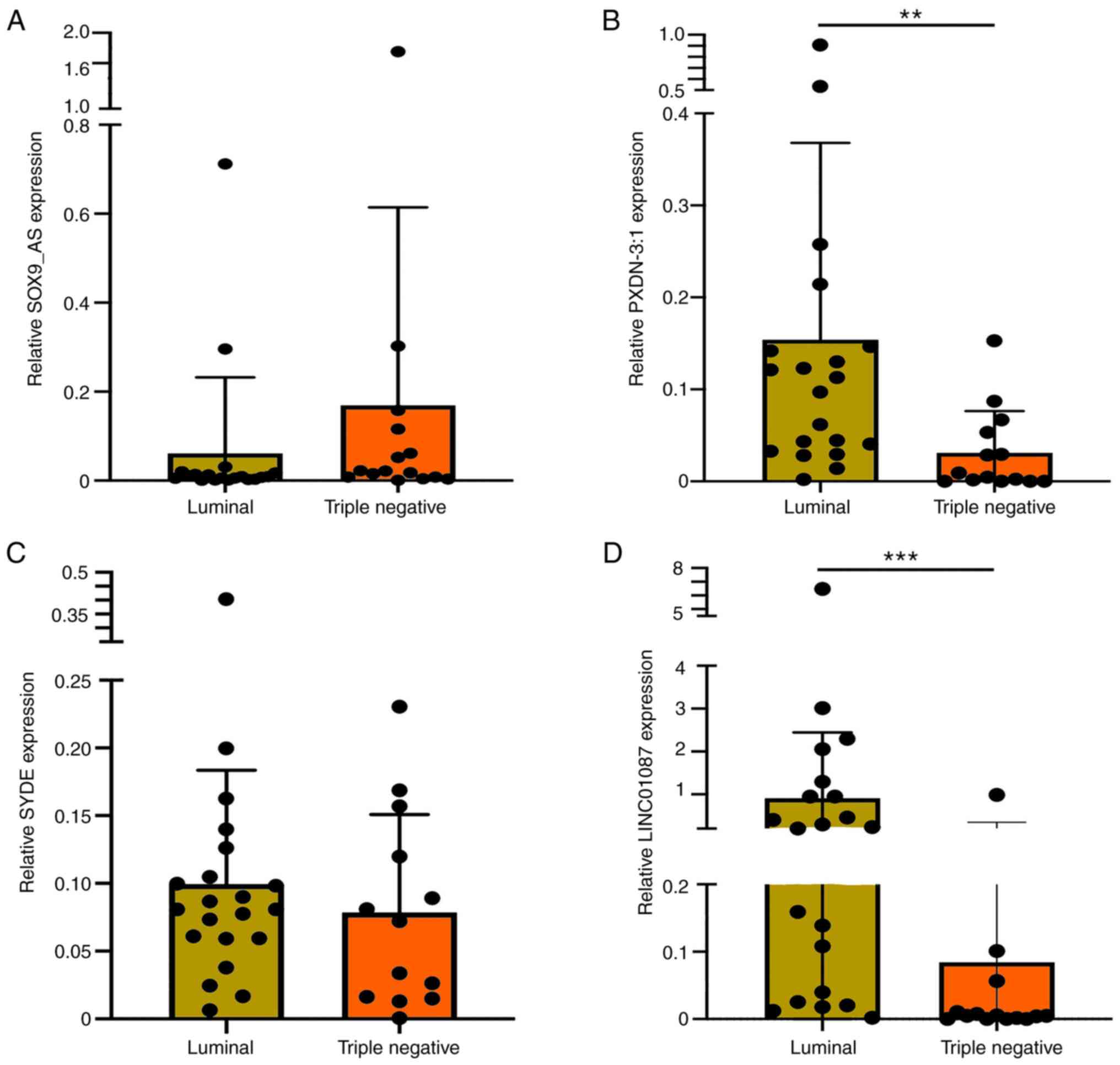
De Palma FDE, Del Monaco V, Pol JG, Kremer
M, D'Argenio V, Stoll G, Montanaro D, Uszczyńska-Ratajczak B, Klein
CC, Vlasova A, et al: The abundance of the long intergenic
non-coding RNA 01087 differentiates between luminal and
triple-negative breast cancers and predicts patient outcome.
Pharmacol Res. 161:1052492020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yuan S, Liu Q, Hu Z, Zhou Z, Wang G, Li C,
Xie W, Meng G, Xiang Y, Wu N, et al: Long non-coding RNA MUC5B-AS1
promotes metastasis through mutually regulating MUC5B expression in
lung adenocarcinoma. Cell Death Dis. 9:4502018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Garrido-Castro AC, Lin NU and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xie Y, Han J, Xie K and Gou Q: LncRNAs as
biomarkers for predicting radioresistance and survival in cancer: A
meta-analysis. Sci Rep. 12:184942022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kang J, Tang Q, He J, Li L, Yang N, Yu S,
Wang M, Zhang Y, Lin J, Cui T, et al: RNAInter v4.0: RNA
interactome repository with redefined confidence scoring system and
improved accessibility. Nucleic Acids Res. 50:D326–D332. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang KX, Ding C, Liu QH and Zhu DM:
Knockdown of LINC01087 inhibits gastric cancer malignant behavior
by regulating the miR-135a-5p/CAAP1 axis. Funct Integr Genomics.
23:2482023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yin Y, Huang J, Shi H, Huang Y, Huang Z,
Song M and Yin L: LINC01087 promotes the proliferation, migration,
and invasion of thyroid cancer cells by upregulating PPM1E. J
Oncol. 2022:1–12. 2022. View Article : Google Scholar
|
|
58
|
She JK, Fu DN, Zhen D, Gong GH and Zhang
B: LINC01087 is highly expressed in breast cancer and regulates the
malignant behavior of cancer cells through miR-335-5p/Rock1. Onco
Targets Ther. 13:9771–9783. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
De Palma FDE, Carbonnier V, Salvatore F,
Kroemer G, Pol JG and Maiuri MC: Systematic investigation of the
diagnostic and prognostic impact of LINC01087 in human cancers.
Cancers (Basel). 14:59802022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Polyak K: Heterogeneity in breast cancer.
J Clin Invest. 121:3786–3788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Mandor M, Atef MM, El-Sayed FM and
Abdel-Mohsen SE: Comparison of survival rate of triple negative
versus luminal B HER2 neu-positive breast cancer patients in
oncology medicine center in Suez Canal University Hospital. Suez
Canal University Medical J. 26:20–31. 2023. View Article : Google Scholar
|
|
62
|
Baeg S, Park I, Kim J, Park C, Cho H, Yang
K, Kim J, Shin Y, Park K and Gwak G: Comparative study for clinical
outcomes of triple-positive and triple-negative breast cancer:
Long-term results in 161 patients followed in a single center. J
Breast Dis. 8:78–84. 2020. View Article : Google Scholar
|
|
63
|
Bjørklund SS, Aure MR, Häkkinen J,
Vallon-Christersson J, Kumar S, Evensen KB, Fleischer T, Tost J;
OSBREAC; Sahlberg KK, ; et al: Subtype and cell type specific
expression of lncRNAs provide insight into breast cancer. Commun
Biol. 5:8342022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Merikhian P, Eisavand MR and Farahmand L:
Triple-negative breast cancer: Understanding wnt signaling in drug
resistance. Cancer Cell Int. 21:4192021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Menck K, Heinrichs S, Wlochowitz D, Sitte
M, Noeding H, Janshoff A, Treiber H, Ruhwedel T, Schatlo B, von der
Brelie C, et al: WNT11/ROR2 signaling is associated with tumor
invasion and poor survival in breast cancer. J Exp Clin Cancer Res.
40:3952021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang W, Wang X, Zhang C, Xue L and Yang
L: Expression and clinical significance of MAPK and EGFR in
triple-negative breast cancer. Oncol Lett. 19:1842–184.
2020.PubMed/NCBI
|
|
67
|
Crosas-Molist E, Samain R, Kohlhammer L,
Orgaz JL, George SL, Maiques O, Barcelo J and Sanz-Moreno V: Rho
GTPase signaling in cancer progression and dissemination. Physiol
Rev. 102:455–510. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kvokačková B, Remšík J, Jolly MK and
Souček K: Phenotypic heterogeneity of triple-negative breast cancer
mediated by epithelial-mesenchymal plasticity. Cancers (Basel).
13:21882021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hui R, Pearson A, Cortes J, Campbell C,
Poirot C, Azim HA Jr, Fumagalli D, Lambertini M, Daly F, Arahmani
A, et al: Lucitanib for the treatment of HR+/HER2-metastatic breast
cancer: Results from the multicohort phase II FINESSE study. Clin
Cancer Res. 26:354–363. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Goldstein LJ, Perez RP, Yardley D, Han LK,
Reuben JM, Gao H, McCanna S, Butler B, Ruffini PA, Liu Y, et al: A
window-of-opportunity trial of the CXCR1/2 inhibitor reparixin in
operable HER-2-negative breast cancer. Breast Cancer Research.
22:42020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xu Y, Ren W, Li Q, Duan C, Lin X, Bi Z,
You K, Hu Q, Xie N, Yu Y, et al: LncRNA Uc003×sl.1-mediated
activation of the NFκB/IL8 axis promotes progression of
triple-negative breast cancer. Cancer Res. 82:556–570. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Thapa R, Afzal O, Gupta G, Bhat AA,
Almalki WH, Alzarea SI, Kazmi I, Altamimi ASA, Subramaniyan V,
Thangavelu L, et al: Unveiling the connection: Long-chain
non-coding RNAs and critical signaling pathways in breast cancer.
Pathol Res Pract. 249:1547362023. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zheng S, Li M, Miao K and Xu H: lncRNA
GAS5-promoted apoptosis in triple-negative breast cancer by
targeting miR-378a-5p/SUFU signaling. J Cell Biochem.
121:2225–2235. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang Y, Bu N, Luan X, Song QQ, Ma BF, Hao
W, Yan JJ, Wang L, Zheng XL and Maimaitiyiming Y: Harnessing the
potential of long non-coding RNAs in breast cancer: From etiology
to treatment resistance and clinical applications. Front Oncol.
14:13375792024. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Pommier RM, Sanlaville A, Tonon L,
Kielbassa J, Thomas E, Ferrari A, Sertier AS, Hollande F, Martinez
P, Tissier A, et al: Comprehensive characterization of claudin-low
breast tumors reflects the impact of the cell-of-origin on cancer
evolution. Nat Commun. 11:34312020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Pan C, Xu A, Ma X, Yao Y, Zhao Y, Wang C
and Chen C: Research progress of Claudin-low breast cancer. Front
Oncol. 13:1226112023. View Article : Google Scholar
|
|
77
|
Cisneros-Villanueva M, Fonseca-Montaño MA,
Ríos-Romero M, López-Camarillo C, Jiménez-Morales S, Langley E,
Rosette-Rueda AS, Cedro-Tanda A, Hernández-Sotelo D and
Hidalgo-Miranda A: LncRNA SOX9-AS1 triggers a transcriptional
program involved in lipid metabolic reprogramming, cell migration
and invasion in triple-negative breast cancer. Sci Rep.
14:14832024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ye X, Cen Y, Li Q, Zhang Y, Li Q and Li J:
Immunosuppressive
<scp>SOX9</scp>-<scp>AS1</scp> resists
triple-negative breast cancer senescence via regulating wnt
signalling pathway. J Cell Mol Med. 28:e702082024. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Naorem LD, Prakash VS, Muthaiyan M and
Venkatesan A: Comprehensive analysis of dysregulated lncRNAs and
their competing endogenous RNA network in triple-negative breast
cancer. Int J Biol Macromol. 145:429–436. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tripathi R, Aier I, Chakraborty P and
Varadwaj PK: Unravelling the role of long non-coding RNA-LINC01087
in breast cancer. Noncoding RNA Res. 5:1–10. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li Y, Sundquist K, Zhang N, Wang X,
Sundquist J and Memon AA: Mitochondrial related genome-wide
Mendelian randomization identifies putatively causal genes for
multiple cancer types. EBioMedicine. 88:1044322023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Huang Y, Yao Z, Li L, Mao R, Huang W, Hu
Z, Hu Y, Wang Y, Guo R, Tang X, et al: Deep learning radiopathomics
based on preoperative US images and biopsy whole slide images can
distinguish between luminal and non-luminal tumors in early-stage
breast cancers. EBioMedicine. 94:1047062023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Atallah NM, Haque M, Quinn C, Toss MS,
Makhlouf S, Ibrahim A, Green AR, Alsaleem M, Rutland CS, Allegrucci
C, et al: Characterisation of luminal and triple-negative breast
cancer with HER2 low protein expression. Eur J Cancer.
195:1133712023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
González-Woge M, Contreras-Espinosa L,
García-Gordillo JA, Aguilar-Villanueva S, Bargallo-Rocha E,
Cabrera-Galeana P, Vasquez-Mata T, Cervantes-López X, Vargas-Lías
DS, Montiel-Manríquez R, et al: The expression profiles of lncRNAs
are associated with neoadjuvant chemotherapy resistance in locally
advanced, luminal B-type breast cancer. Int J Mol Sci. 25:80772024.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cocco S, Piezzo M, Calabrese A, Cianniello
D, Caputo R, Lauro VD, Fusco G, Gioia GD, Licenziato M and De
Laurentiis M: Biomarkers in triple-negative breast cancer:
State-of-the-art and future perspectives. Int J Mol Sci.
21:45792020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Price TJ, Hardingham JE, Lee CK, Townsend
AR, Wrin JW, Wilson K, Weickhardt A, Simes RJ, Murone C and Tebbutt
NC: Prognostic impact and the relevance of PTEN copy number
alterations in patients with advanced colorectal cancer (CRC)
receiving bevacizumab. Cancer Med. 2:277–285. 2013. View Article : Google Scholar : PubMed/NCBI
|